ABSTRACT
The use of culture-independent techniques has allowed us to appreciate that the upper and lower respiratory tract contain a diverse community of microbes in health and disease. Research has only recently explored the effects of the microbiome on the host immune response. The exposure of the human body to the bacterial environment is an important factor for immunological development; thus, the interaction between the microbiome and its host is critical to understanding the pathogenesis of disease. In this article, we discuss the mechanisms that determine the composition of the airway microbiome and its effects on the host immune response. With the use of ecological principles, we have learned how the lower airways constitute a unique niche subjected to frequent microbial migration (e.g., through aspiration) and constant immunological pressure. The discussion will focus on the possible inflammatory pathways that are up- and downregulated when the immune system is challenged by dysbiosis. Identification of potential markers and microbial targets to address the modulation of inflammation in early disease, when changes may have the most effect, will be critical for future therapies.
INTRODUCTION
In 2007, the Human Microbiome Project (HMP) was added to the National Institutes of Health (NIH) Roadmap for Medical Research, and since then, over $200 million has been invested in the exploration of the human microbiome. Several sites on the human body, including the nares, oral cavity, skin, gastrointestinal tract, and urogenital tract, have been studied by the HMP (1). The gastrointestinal tract remains the most thoroughly investigated organ-microbiome interaction studied thus far, and its role in shaping the host immune response is rapidly becoming defined within the context of inflammatory response (2–4). Observations have noted associations of specific microbes and the gut microbiome in obesity (5–8), coronary artery disease (9–12), Clostridium difficile colitis (13, 14), type 2 diabetes (15, 16), and inflammatory bowel disease (4, 17, 18). While the nares and oral cavity were included as locations to be studied for the HMP, the lower airway respiratory system was not included as a location of interest. The microbial community of the oropharynx had been well described even before the advances of next-generation sequencing and multiplexed data (19, 20). Microbiome approaches to the upper and lower respiratory systems created a deluge of associations between host and microbes in health and disease (21–23). Over the past few years, our understanding of the airway microbiome has shifted, upending the old adage of the lungs being a sterile field (24) to a new paradigm of a continuous organ system with a rich and vibrant mucosal surface that embodies complex interactions between the microbiome and its host that can propagate or resist disease (21, 24). Moreover, little is known regarding how the respiratory microbiome interacts with the host immune response. While multiple research projects have focused on the effects of the gut microbiota on the immune response of the gastrointestinal mucosa (25–27), few papers have focused on the effects of the lower airway microbiota on the respiratory mucosa that it inhabits (23, 28–30).
Some of the unique challenges in studying the lower airway microbiome include the technical difficulty of sampling the lower respiratory tract, the extremely low bacterial burden in healthy lung leading to a ratio of low signal to high noise, and the lack of animal models to study the lung microbiome (31, 32). Shifts in gastrointestinal microorganism composition in organ systems that contain high bacterial biomass, such as the gastrointestinal tract, can lead to broad observations resulting in disease states (33). However, the lung microbiome is a low-bacterial-burden organ in which transient perturbations of its microbiome can have dramatic effects on the diversity of its composition and the host inflammatory response to dysbiosis (23). Thus, the upper and lower respiratory tract microbiome is unique, as it represents an organ with a significant biomass gradient, moving from a high-bacterial-burden reservoir in the upper respiratory tract to a very-low-bacterial-burden area in the lung microbiome (32, 34). We will discuss first the microbiomes of the upper and lower respiratory tracts and then the possible mechanisms for microbial migration and colonization, followed by the recent finding of the host immune response to the microbiome and the inflammatory pathways that the microbiome may impact, and finally we will address possible microbial targets or markers for specific diseases.
The airway microbiome can only be understood as the sum of its individual parts. In the upper airways, the nasal and oral cavities contain a very distinct microbiome. In the nasal cavity, the microbiota is characterized by enrichment with Streptococcus, Acinetobacter, Lactococcus, Staphylococcus, and Corynebacterium. In the oral cavity, the microbiota is characterized by enrichment with Prevotella, Streptococcus, Fusobacterium, Neisseria, Leptotrichia, and Veillonella (35). Further defining the qualities of each location, the upper airways are characterized by a constant exposure to airborne microbes and microbes ingested as part of each individual’s diet. Although the respiratory system and its mucosa are a continuum, the lower airway microbiota has very distinctive features that set it apart from that of the upper respiratory tract. The boundary between what we define as the upper and the lower airways is classically defined by anatomical structures, with the vocal cords being analogous to a natural “dam” against aspiration (36, 37). Similar to the water held by a dam, the upper airways hold a high bacterial burden, while the lower-airway microbiota has about 100 to 10,000 times less of a bacterial burden than the upper microbiota based on 16S rRNA copies (32, 38). Microaspiration occurs in healthy individuals (39), and its prevalence is higher in several lung diseases, including chronic obstructive pulmonary disease (COPD), asthma, obstructive sleep apnea, cystic fibrosis, and lung infections due to atypical (such as nontuberculous mycobacteria) and typical microorganisms (40–45).
Charlson and colleagues, who in initial studies examined bronchoscopy results from six healthy patients, concluded that the microbiome of the lower respiratory tract resembled that of the upper respiratory tract (32). Further microbiome work studying cell-free bronchoalveolar lavage (BAL) fluids has noted that the bacteria characteristic of the oral cavity are found in about 45% of relatively healthy subjects and that the remaining subjects had a lung microbiome whose members resembled the background taxa (23, 30). Other studies have found bacteria characteristic of the oral cavity at a higher prevalence, although it is unclear if this is related to the sample type utilized (acellular BAL fluid versus whole BAL fluid versus airway brush sample) or differences in cohorts (32, 46–48). Importantly, a dichotomized view of the lower airway microbiota (i.e., enriched with upper airway microbes or not) may be an oversimplification of a more likely physiological continuum of similarity between the lower and upper airway microbiotas. Of note, older studies using radiotracers have found that a similar proportion of healthy individuals microaspirate (39, 49). Using culture-independent techniques, various studies have shown that bacteria commonly found in the upper airways are also frequently found in multiple other disease states, such as asthma (29, 50–58), cystic fibrosis (59–62), HIV infection (47, 63, 64), and advanced COPD (21, 23, 28, 65, 66). Considering the continuity of the airway mucosa, it is no surprise that the upper airway microbiota with its high bacterial burden is the major source of microbes for the lower airways. However, even when the lower airways are enriched with microbes found in the oral cavity, differences can still be noted between the upper and lower airway microbiotas (30, 32). This suggests that there are other important factors that affect the dynamic homeostasis of the lower airway. Several questions remained unanswered regarding (i) what other microbiotas from different mucosae contribute to the “cross pollination” of microbes to the lower airways, (ii) what selection pressures microbes suffer as they enter the lower airways, and (iii) how this selection pressure changes in different compartments of the lower airways.
UNIQUE CHALLENGES IN EVALUATING THE LUNG MICROBIOME
One particular challenge that research in the lung microbiome has yet to resolve is the microbiome signal-to-background noise ratio during health and early disease (Fig. 1) (31). While this might also be important in advanced disease states, the signal-to-noise ratio is highly unfavorable in early stages of disease, when there is low microbial biomass in lower airway samples. Functionally, the lungs have a 100- to 10,000 times-lower bacterial burden than the upper airways (23, 38). Thus, the low-bacterial-burden nature of the pulmonary system poses unique challenges: signal-to-noise ratios that are characterized by high background noise signals (31), lack of standardized methods for background removal, and limited ability to detect signals from low-abundance taxa. These challenges confound researchers and make the lung microbiome difficult to study. One consistent finding in multiple studies is that background environmental samples, such as sterile saline and prebronchoscopic wash through the bronchoscope (prior to the procedure), contain microbial DNA that is characterized as belonging to a diverse environmental background microbiota (30, 31). This is understandable and predictable, since instruments are “clean” but not necessarily DNA free. Furthermore, reagents utilized for DNA isolation and library preparation may also contribute to background DNA (67). A large percentage of healthy subjects have a lower airway microbiota enriched with microbial DNA that is characteristic of background samples (21, 23). Thus, the background microbiome may confound observations in a low-biomass host microbiome in approximately 60% of healthy subjects that will have a microbiome enriched with background predominant taxa (also referred as pneumotypeBPT) (23, 30). Even in these subjects, bioinformatic approaches that identify the contribution of background taxa (e.g., SourceTracker) to a sample showed that close to one-half of the composition of a BAL sample was not present in background samples (e.g., bronchoscope) (30). This result suggests that there is still a significant amount of information to be gained from these low-biomass samples. Consider for comparison the microbiome of the gastrointestinal tract. This high-bacterial-burden and diverse microbial community is subjected to multiple microbial challenges (e.g., food intake, medications, etc.). In this setting, the background microbiota is rarely seen as an important source of contamination. However, the background microbiota may influence the recovery of rare or less abundant microorganisms or, in the setting of gross contamination, may play a role in obscuring the bacterial composition. Microbial changes due to different microbial challenges or active disease processes are more likely to be detected in the gastrointestinal tract (Fig. 1A and C). Conversely, the low bacterial burden found in the respiratory tract makes the microbial assessment using culture-independent techniques commonly influenced by the background microbiome. Thus, when microbial challenges that disturb the underlying lung microbiome occur, they may not be apparent due to the low signal that needs to overcome a high background noise (Fig. 1B and D). This model also suggests that the signal can be transient and subjected to the dynamics that dictate the microbial host interaction in the lower airways. The low microbial load of the respiratory system may lead to large shifts in the microbiome when faced with upper airway tract microbial challenges. The microbial challenges caused by microaspiration may then be seen as the enrichment of the lower airway microbiome with upper airway microbes (also referred as pneumotypeSPT, where SPT stands for supraglottic predominant taxa) (23, 30). The lower microbial biomass and low signal-to-noise ratio present a distinct challenge when studying the lung microbiome. It is therefore important to include the study of the background microbiome in microbiome research involving low-biomass samples.
FIGURE 1.
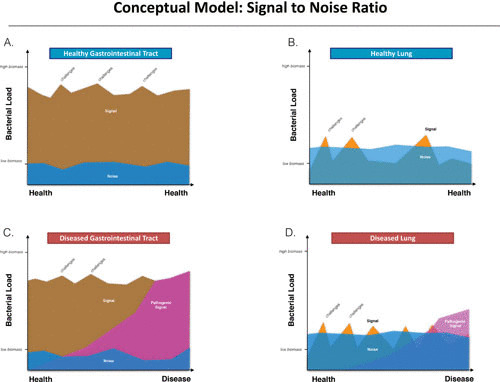
Conceptual model: signal-to-noise ratio. (A) Healthy gastrointestinal microbiome, where there is an organ of healthy biomass and background “noise” or signal amplified by background (e.g., background microbiota present in the colonoscope) that does not represent the gut microbiome. This background microbiome is overwhelmed by the large biomass present the sample. (B) Healthy lung microbiome, where there is relatively low biomass and background signals tend to overwhelm the lung microbiome signal. (C) Diseased gastrointestinal microbiome, where the pathogenic signal (dysbiosis) will eventually overcome the high underlying biomass. The pathogenic signal will overpower the background microbiome and be apparent given the high amount of biomass present in the gut. (D) Diseased lung microbiome. Unlike the diseased gut microbiome, the pathogenic signal may be confounded by the background noise and may not be apparent until sufficient progression of disease supports the altered dysbiosis.
Questions remain regarding what to do with background taxa when studying the lower airway microbiome. Curation of the sequencing data commonly includes removal of technical contaminating taxa that can be done as part of the upstream processing of the data. Neutral modeling is a technique that can be utilized to identify taxa preferentially enriched in one sample type (68). Taxa that are found but deviate from this model are the most likely to be true inhabitants of the area that was sampled. For example, this technique can then be used to remove taxa found in the bronchoscope to identify microbes that are resident in the lung. SourceTracker is another technique utilized to estimate the degree of microbial contribution that sources of background microbiota (e.g., bronchoscope) may have on a biological sample (69). While these methods have been utilized in lung microbiome research, there is a lack of consensus on how to standardize the analysis. One concern is that in samples with a predominance of background taxa (e.g., pneumotypeBPT), the removal of background taxa may yield low reads and, given the relative-abundance nature of the microbiome data, will inflate the relative proportion of the taxa that were not removed. Also, taxon removal approaches have the potential of excluding taxa existing in both the lung and the possible contamination sources (e.g., bronchoscope) that might be meaningful to comprehend the lower airway microbiota. Finally, bacteria may have a significant pathogenic role in a disease state despite its low abundance in a microbial community, and removal of background taxa may in fact change our understanding of the microbe’s role in the lung (70). For example, sequence analysis of experimental diarrhea induced by enterotoxigenic Escherichia coli has shown that even low abundance of this organism can cause significant disease (71). Thus, in the lung, the high abundance of background noise represents a major challenge to detect a signal that occurs in microorganisms from low-abundance taxa that may be biologically important to health and disease.
SOURCES OF MICROBIAL CHALLENGE TO THE LOWER AIRWAYS
Predictably, the most straightforward explanation for the source of microbes to the lower airways is the delivery of microbes from the upper respiratory tract through microaspiration. Although it is generally accepted that aspiration of oral cavity secretions likely affects the lung microbiota, other possible sources must be considered. The contribution of the airborne microbiome present in ambient air may be a significant contributor to the lower airways. The human body ventilates approximately 7,000 to 20,000 liters of ambient air per day (24, 72), potentially exposing the lower airways to microbes present in the air (airborne microbiota) in conjunction with other well-recognized pollutants, chemicals, dust, allergens, and other particulate matter from the atmosphere. While there is significant knowledge on pathogens that are aerosolized and can transmit infections, such as mycobacteria (73, 74), much less is known about the presence and relevance of other microorganisms present in the air. It is possible that different airborne microbiota members may also significantly contribute to the lower airway microbiome, a subject that needs further investigation.
There is an increasing understanding of the complex airborne microbiome and its contributions to the lower airway microbiome. Adams and colleagues found differences between indoor and outdoor environments that were dictated by human-associated microorganisms (75). In indoor environments, they found an enrichment of Corynebacterium, Streptococcus, Staphylococcus, Propionibacterium, Lactococcus, and Enterobacteriaceae. In outdoor environments, there was enrichment with Pseudomonas, Acinetobacter, and Sphingomonas (75). Also, Meadow and colleagues identified a distinct “microbial cloud” that supports the notion that in an enclosed space, ambient air can be impacted by its occupants (76). However, the effects of exposure to different airborne microbiotas on the lung microbiome or the effects of the exhaled air microbiota on the airborne ambient microbiome have not been studied. The nasal microbiota is distinct from the oral cavity microbiota (35) and likely represents an important microbial reservoir to the lower airways that is enriched by inhalation of ambient air.
Culture-independent methods in chronic rhinosinusitis have shown enrichment with Haemophilus influenzae (77, 78) and other pathogens that are frequently associated with development of pneumonia. Postnasal drip is highly associated with cough and bronchial inflammatory diseases, but its effect on the lower airway microbiota has not been elucidated. The gastrointestinal tract represents another potential source of microbial challenge to the lung microbiome. Gastroesophageal reflux disease (GERD) is frequently present in many disease states. The presence of a distinct microbiome in the gut dominated by Helicobacter pylori has been associated with decreased risk for asthma (79, 80). The use of proton pump inhibitors affects the composition of not only the gastric microbiota but also the lung microbiome (81). Further, the lower gastrointestinal tract should be considered a potential source of microbial challenge to the lung. Animal models of sepsis have shown that germ-free animals are protected from lung injury (82) while selective gastrointestinal tract decontamination reduces the development of multiple-organ dysfunction syndrome in 50% of the subjects studied (83).
However, the most frequently observed microorganisms in the lower airways are those characteristically present in the oral cavity. Multiple culture-independent investigations have shown that different subjects have various degrees of enrichment of the lower airway microbiota with oral microbes such as Prevotella, Veillonella, Rothia, Streptococcus, and Porphyromonas (23, 30, 65, 84, 85). Thus, multiple potential sources may contribute to the lower airway microbiome through different routes of microbial challenge, e.g., aspiration, regurgitation, hematogenous, and aerosolized routes. These microbial challenges will then be subjects of unique selection pressures present in the lower airway environment. Although there is limited understanding of what these selection pressures are at individual levels, it is likely that they have a very significant effect on the structure of the microbial communities that may reside in the lower airways.
UNIQUE SELECTION PRESSURES OF THE LOWER AIRWAY MICROBIOME
Several different theories have been adapted from biology, ecology, and environmental studies to explain the composition of the lung microbiome. One of the most purported has been the “adapted island model” (86–88). Similar to islands, the lungs are influenced by a balance between immigration of microbes from the “mainland” and their extinction (86, 87). Based on this theory, the lungs can be viewed as conceptual “islands” that receive immigration challenges, most commonly from the upper respiratory tract. Thus, the diversity in the lungs is driven mainly by the diversity from the sources of microbial challenges and the selection pressure on a microorganism’s viability.
The proximal airways, including the trachea and the main stem bronchi, may be viewed as large swaths of area that due to their location in relation to the upper airway more closely resemble the upper respiratory tract (32). As one progresses distally in the lower respiratory tract, the conducting airways may represent isolated islands (87, 89), potentially forming a unique microbial niche. Thus, diversity is influenced by three main factors: the rate of immigration, the rate of elimination, and the rate of reproduction of the community members (86–88, 90). In health, the diversity of these islands is dictated mainly by immigration from the upper respiratory tract and elimination by various mechanisms including the mucociliary ladder, bacteriophages, and alveolar macrophages. Frequent episodes of microaspiration observed in healthy individuals are likely to be the most dominant source of microbial airway challenge. In the event of disease, different factors may influence the lung microbiome. For example, in active pneumonia, chronic airway colonization in COPD, or cystic fibrosis, the reproduction rate of the organism in the lower airway will offset the mechanisms of immigration and elimination.
Furthermore, the airway mucosa has several unique features: it is a hollow system of mucosa-lined tubes in constant exposure to ambient air with a role to perform gas exchange; it contains high levels of phospholipids in the form of surfactant (91); and as a result of its gas exchange properties, it contains gradients of oxygen and carbon dioxide tension (92). Thus, microbes, as they enter the lower airways, have to overcome or adapt to the changing environment of the lung. Recent investigations have demonstrated that by evaluating microbial genetic variations within a host, multiple lineages can coexist, providing evidence of selective pressures in the lower airways (93). Several environmental factors present in the lower airways potentially shape the lower airway microbiota. The phospholipid-rich environment not only is important to maintain surface tension and aerated alveoli but also has significant effects on microbial metabolism (94). Multiple antimicrobial peptides such as surfactant protein A, lactoferrin, and defensins are present in the airways (94). Experimental evidence shows that secretory immunoglobulin A (SIgA) has an important role in the airway microbiota and its deficiency leads to increased inflammation in response to the resident lung microbiota and progressive airway remodeling and emphysema (95). Thus, the antimicrobial properties of the pulmonary mucosa represent a significant hurdle for microbial colonization. Microbes have developed and evolved specific mechanisms in order to evade or ameliorate the effects of the host defense. As an example, biofilm constitutes its own microbial niche in which anaerobes can find an optimal anaerobic niche (96). For example, Pseudomonas aeruginosa advantageously uses biofilm to create its own microenvironment (97). Biofilms, close proximity to aerobes and anaerobes, and development of oxygen-depleted microenvironments may allow for microbial colonization under anaerobic conditions (98). Thus, although the lung is a highly aerated organ, anaerobes are frequently described in the lower airway microbiome, suggesting that biofilms may have an important role in the co-occurrence of aerobes and anaerobes in the lower airways (96, 98). Microbes with different adhesion properties are also affected differently by mechanical selection pressure such as mucociliary clearance (99, 100). Different bacteria express adhesins, pilin, and flagellin in order to overcome mucociliary removal from the lower airways (101, 102). These are important virulence factors for bacterial colonization and pathogenesis, and not all bacteria express these advantages. Thus, the microorganisms found in the respiratory system likely reflect highly specialized strains or species that are in constant balance with a highly immune-active mucosa (103, 104). Experimental approaches are needed to study how these factors affect the selection pressures that shape the lower airway microbiome.
The lung environment ecosystem can also be thought of as a series of forward and feedback loops that suppresses or allows the underlying lung bacteria to grow (88). A major risk factor for the development of pneumonia in the hospital setting is the use of antibiotics (105). Evidence suggests that it is likely that the pathogen responsible for the development of pneumonia exists in the lung prior to the onset of disease. In a murine model of this theory, Poroyko and colleagues conducted a study of lipopolysaccharide (LPS)-induced acute lung injury and studied the changes in the murine lung microbiome (106). The mice notably had an increase in bacterial burden without an increase in the number of species. There was also an increase in abundance of the family of Proteobacteria and a decrease in Firmicutes. These findings suggest that challenge with LPS may overwhelm the feedback mechanisms responsible for equilibrium in the lungs, allowing the resident microorganisms in the lung to proliferate unchecked during times of stress and inflammation. Nutrition availability may also affect the alterations in the lung microbiome (107). Issues such as iron availability and nutritional immunity may select for microorganisms that are able to adapt to the lack of or abundance of certain nutrients. Moreover, coinfection with viruses may affect the equilibrium in the lung as well. Molyneaux and colleagues demonstrated that there are intrinsic changes in the lower respiratory tract microbiome when it is challenged with rhinovirus infection. In this study, healthy subjects and those with a history of COPD were challenged with rhinovirus and monitored longitudinally over a period of 42 days (108). Using induced sputum, the authors showed that rhinovirus challenge led to an increase in Proteobacteria, driven mainly by an increase in Haemophilus influenzae (108). This may explain why lower airway pneumonia occurs after viral infection.
Changes in the host immune system will affect the selection pressure on the lower airway microbiota. An example of this selection is the enrichment with Tropheryma whipplei, the etiological agent of Whipple’s disease, in the lungs of treatment-naive HIV subjects and the subsequent decline of the relative abundance of T. whipplei following antiretroviral therapy (64). Importantly, since this microbe was not found in the upper airway samples or background controls, it seems to reflect a true inhabitant of the lower airways, where growth is favored or promoted by the host’s immunodeficient state. Recently, broader microbial differences were noted in advanced HIV and partially corrected with immune reconstitution (109). Twigg and colleagues identified higher relative abundance of Streptococcus, Prevotella, and Veillonella taxa in the lungs of the HIV-infected patients after 1 year of treatment with antiretroviral therapy as compared to the same patients prior to the start of treatment. Moreover, there is increasing evidence of the effects of immune deficiency on nonbacterial microbes in the lung, such as in the mycobiome, the collection of fungi found in a community (110). The response of the host’s respiratory system to this environmental and microorganism challenge is an integral facet to understanding the roles of the upper and lower respiratory microbiomes in health and disease (21, 23, 30).
Specific deficiencies in the host immune response play a significant role in the microbiome. Richmond and colleagues explored changes in mucosal immunity by studying the impact of specific SIgA immunodeficiency (95). The researchers developed mouse models that were deficient in the polymeric immunoglobulin receptor (pIgR) and were not able to produce SIgA on mucosal surfaces. The pIgR deficiency results in the persistent activation of inflammation signaling due to lung microbiota invasion (95). Mice with a pIgR deficiency contained an increase in taxa (400 versus 194) in the lung. Discriminating taxa found in higher relative abundance in pIgR-deficient mice than in wild-type mice included Veillonella, Prevotella, Neisseriaceae, Bacillus, Actinomycetaceae, Tissierellaceae, and Ruminococcus (95). Furthermore, germ-free pIgR-deficient mice did not develop the COPD-like phenotype, in contrast to pIgR-deficient mice raised in a conventional environment. Once the germ-free pIgR mice were removed from their germ-free environment and their microbiotas were conventionalized, similar levels of airway wall remodeling, emphysema, and inflammation were measured and found to be comparable to those of pIgR mice raised under conventional (non-germ-free) conditions for the same duration of exposure (95). Upregulation of NF-κB was found in the lungs of pIgR-deficient mice when compared to wild-type age-matched controls (95). This is important, since structural abnormalities in COPD correlate with decreased expression of pIgR and a disruption of the protective mucosal barrier (111, 112).
The use of medications, especially corticosteroids, may also affect the host response to microorganisms. The host immune response determines host susceptibility to microbes, thereby affecting the distinction between pathogens and commensals. In cases where the immune response is inadequate or altered, microorganisms that may regularly colonize or be cleared by the host may have the potential to cause disease. Mucosa-associated invariant T (MAIT) cells are a prevalent and unique T-cell population in humans with the capacity to detect intracellular infections with bacteria, including Mycobacterium tuberculosis (113). MAIT cells recognize early intermediates in bacterial riboflavin synthesis that can be potent antigens for these cells (113). Although they are able to recognize a wide variety of bacterial microorganisms, bacteria that do not express riboflavin synthesis pathways do not stimulate MAIT cells (114). Hinks and colleagues demonstrated that in 11 patients who were exposed to inhaled corticosteroids, peripheral serum and bronchial biopsy specimens contained fewer MAIT cells than did healthy controls (115). In in vitro studies, nontypeable Haemophilus influenzae-infected macrophages obtained from healthy subjects were exposed to fluticasone or budesonide. In the presence of these corticosteroids, significant impairment of the upregulation of MR1, a stimulatory receptor on macrophages for MAIT cells, as well as a decrease in nontypeable H. influenzae-induced gamma interferon expression from MAIT cells was found in these experiments (115). Therefore, steroid-induced suppression of MAIT cells may be responsible for the increased risk of pneumonia in subjects with airway diseases (115).
Unique selection pressures of the lung may have a profound impact on the lung microbiome. There is increasing evidence that the adapted island model may help model the microbial challenges to the lower airways from microaspiration. Once in the lower airways, the oral microorganisms are likely exposed to multiple selection pressures that dictate their persistence or transient nature in the lungs. Microbes that evolutionarily develop advantages to mitigate and avoid the host immune response are more likely to persist in the lower airway microbiome. Their ability to overcome the innate and adaptive immune systems may serve to guide future studies to address new pathways that are regulated by microbes and particular molecular patterns that may dictate these interactions. Most importantly, host responses and medications deserve particular attention, as these mechanisms remain largely unexplored in the current literature in relation to their effects on the lung microbiome.
THE LUNG MICROBIOME IN DISEASE STATES
Morris and colleagues studied the microbiomes of healthy subjects and healthy smokers in a study of the lung microbiome (85). They found that the most common genera in both oral wash and BAL fluid were microorganisms commonly found in the oral cavity (Prevotella, Streptococcus, and Veillonella). Although the study did detect organisms found to be more abundant in the lung than in the oral cavity, including Haemophilus, Enterobacteriaceae, and Tropheryma whipplei, differences in the lung microbiota between smokers and nonsmokers were not found (85). However, despite no differences found in the lung microbiome, microbiota differences were found in the oropharynx of smokers compared with that of nonsmokers. Neisseria, Porphyromonas, and Gemella were depleted in the oral washes of the former set of subjects (85). Several other studies have shown that smoking alone is sufficient to change the upper airway microbiota (20, 35) but not the lower airway microbiota (23, 85).
The exposure to tobacco smoke may also greatly increase an individual’s risk of developing COPD. In contrast to what is seen in individuals with exposure to tobacco smoke, subjects with advanced stages of COPD have significant alterations within the underlying lung microbiome. Erb-Downward and colleagues described these differences in lung microbiome in the lungs of end-stage COPD patients (84). Importantly, different areas of the lung have different microbiotas. Presumably, changes in the underlying lung architecture and decreased mucociliary clearance lead to enrichment with Pseudomonas and Streptococcus (84). Other studies have also described differences in the lung microbiome of COPD (28, 65, 116). However, most COPD microbiome studies have focused on samples from GOLD stage IV COPD patients and samples collected from explanted lungs, where other confounders, such as use of inhaled steroids and frequent courses of antibiotics, are commonly present and may present confusing results regarding the lung microbiome. Hilty and colleagues also described a disordered lung microbiome in patients with asthma and moderate COPD (58). Notably, 60% of these patients were on inhaled corticosteroid therapy.
Dysbiosis may also play a role in COPD exacerbations. Huang and colleagues studied a group of patients (n = 8) who were admitted to their hospital for COPD exacerbations. All of the patients studied were mechanically ventilated and intubated and received antibiotics (117). The subjects studied showed variations in their interpersonal bacterial richness, with subjects with decreased richness having more Pseudomonaceae and Enterobacteriaceae and subjects with increased richness possessing higher relative abundance of Clostridiaceae, Lachnospiraceae, Bacillaceae, and Peptostreptococcaceae (117). One challenging issue with interpreting some of these results may be that the subjects received different lengths and types of antibiotic treatment at the time of the study. Thus, changes in the lung microbiome that may be related to a subject having a COPD exacerbation may be lost early in the disease and the changes seen may be due to an antibiotic effect on the lung microbiome (117).
Pragman and colleagues examined the lung microbiomes of moderate and severe COPD subjects with relatively stable disease and compared them to healthy subjects. They found that while the moderate and severe COPD groups had more operational taxonomic units in common, very few operational taxonomic units (approximately 13%) were shared among all three groups (116). Notably, the study found several anaerobes within the lung, consisting of Bacteroidetes, Fusobacteria, and the genus Clostridium, extending observations that anaerobic bacteria are found in an aerobic environment; however, it is not known if these anaerobic bacteria are metabolically active (116). In a subanalysis of the study, the authors found that the most discriminant factor for a distinct lung microbiome was the use of inhaled steroids. Emphasizing the effects of medications on the microbiome, a recent investigation showed that in subjects with early emphysema, the use of azithromycin affected the diversity of the lower airway microbiome and increased stress-related microbial metabolites that might have anti-inflammatory properties (118). These data further support the role of the lower airway microbiome on the immune phenotype of the lower airway mucosa and illustrate the selective pressures of medications.
Using core biopsy specimens from explanted lungs, Sze and colleagues also found differences in advanced COPD, with an increase in the relative abundance of Firmicutes, including Lactobacillus and Proteobacteria (65). However, in a follow-up study, the same researchers found that there was a relative expansion of the phylum Proteobacteria and a reduction of the phylum Firmicutes when they compared the lung specimens from COPD subjects to lungs from transplant donors (28), reinforcing that it remains difficult to define a consistent “core” COPD microbiota. One important finding of the last study was that there were several taxa that were positively and negatively associated with the host immune response, including neutrophil infiltration, eosinophil infiltration, and B-cell infiltration (28).
Despite studies that have examined the lung microbiome in GOLD stage IV COPD patients, early and moderate COPD patients and active smokers remain difficult to study and represent a knowledge deficit in lung microbiome research. Attention should be given to these populations due to several challenging issues, including a need to study early events in the disease process, a way to control and limit possible confounders such as inhaled corticosteroids, and a focus on longitudinal changes in the lung microbiome.
In asthma, there is evidence of enrichment with Proteobacteria, including the Haemophilus genus (29, 119). Importantly, distinct immunological phenotypes have been associated with changes in the lower airway microbiome. The respiratory tract of asthmatic patients tends to have a higher bacterial burden than that of subjects without asthma (119). Exposure to diverse allergens and microbes (from animal or environmental exposure) in youth proves to be protective against developing asthma, thus supporting the “hygiene hypothesis” (120, 121). In contrast, several taxa in the lung microbiome, including Streptococcus pneumoniae and Haemophilus influenzae, have been identified as increasing the susceptibility of developing asthma once colonization is established in a neonatal host (122). Huang and colleagues studied asthmatic patients with a disordered lung microbiome. They identified that a greater airway diversity was strongly associated with increased bronchial hyperresponsiveness (119). Moreover, they found that the taxa that were strongly associated with bronchial hyperresponsiveness belonged primarily to the Proteobacteria phylum. The treatment of these patients with clarithromycin showed that those showing improvement in their bronchial reactivity following clarithromycin treatment had an increase in their Shannon diversity index (α diversity) (119).
In severe asthmatics, Huang and colleagues also sought to characterize the microbiome associated with subjects with different phenotypes of asthma. They observed that enrichment with Bacteroidetes and the Firmicutes phylum (including families Prevotellaceae, Mycoplasmataceae, Lachnospiraceae, and Spirochaetaceae) was associated with obese subjects who were also asthmatics. They also identified certain taxa associated with worsening asthma symptoms such as the Proteobacteria phylum (including families Pasteurellaceae, Enterobacteriaceae, Neisseriaceae, Burkholderiaceae, and Pseudomonadaceae) (123). The researchers demonstrated that different asthma phenotypes might harbor distinct microbiomes, supporting an important aspect of the lung microbiome on the host immune phenotype (29).
In cystic fibrosis, disordered airway clearance is a result of dysfunction of the sodium chloride channel, leading to thickened secretions and progressive microbial airway colonization and infection. Culture-based data have identified clinically significant bacteria, including Pseudomonas aeruginosa (124), Burkholderia cepacia complex (124), methicillin-resistant Staphylococcus aureus (125), and Mycobacterium avium complex (126), in the airways of cystic fibrosis patients. Early microbiome studies in cystic fibrosis patients identified a possible core microbiome represented by 15 taxa from seven genera including Catonella, Neisseria, Porphyromonas, Prevotella, Pseudomonas, Streptococcus, and Veillonella (127). The lungs of cystic fibrosis subjects may contain high spatial heterogeneity (128). In a study involving 269 cystic fibrosis patients conducted by Coburn and colleagues, the researchers studied a broad range of ages and disease stages (128, 129). The study reaffirmed particular species as belonging to a possible core microbiome including Streptococcus, Prevotella, Rothia, Veillonella, and Actinomyces (129). More typical cystic fibrosis-related microorganisms (e.g., Pseudomonas, Burkholderia, Stenotrophomonas, and Achromobacter) were less prevalent but, when present in sputum specimens, tended to dominate the relative abundance within the samples (129). Furthermore, Coburn and colleagues showed that community diversity and lung function were greatest in patients less than 10 years of age, reaching a plateau at age 25, with a subsequent decreasing community diversity in following years correlated with worsening lung function (129). Bacteria that once were thought to be oral contaminants may be important oral microorganisms in lung ecology (90, 130) and may also represent a potential source of inflammatory response by the host in cystic fibrosis (131).
THE LUNG-GUT AXIS AND ITS ROLE IN DISEASE
The gut is a major reservoir for the microbiome within the human body. Evidence suggests that the gastrointestinal tract plays an important role for the immunological priming of the host. This immunological function is not limited to the gut mucosa or systemic circulation, but there is growing evidence of its relevance to the lung through immunological cross talk between the gut and the lung. Thus, the gastrointestinal tract not only may have direct interaction (e.g., aspiration) with the lower airways but also may serve as an immune-modulating organ with ability to cross talk with other organs, contributing cells related to the innate and adaptive immune systems (dendritic cells and macrophages) and possibly inflammatory cytokines and chemokines in times of disease to the respiratory tract (60, 132, 133).
Microbiome studies of asthma patients have also identified possible lung and gut interactions. The exposure to livestock or canines during youth significantly decreases the risk of developing asthma (134, 135). Until recently, the mechanism that protects the subject from the development of asthma remained elusive. Fujimura and colleagues explored this association with experiments using dust collected from households with canines (25). Mice exposed to dust from canine-inhabited houses exhibited protective responses to allergens. In particular, the mice that received canine household dust had elevated relative abundance of Lactobacillus johnsonii in their gastrointestinal tract (25). Oral supplementation of this single species of Lactobacillus decreased bronchoresponsiveness to allergens and respiratory syncytial virus challenge. Furthermore, the authors purport that while they were able to detect L. johnsonii in cecal samples, they did not detect any in the lung samples, providing evidence that the gastrointestinal microbiome affects the local and systemic inflammatory responses (25).
Cystic fibrosis murine models show that there is an appreciable modulation of the lung microbiome due to changes from the intestinal microbiome in response to antibiotic treatment (136, 137). Bazett and colleagues observed that treatment with streptomycin, an antibiotic that is not systemically absorbed from the gut in a murine cystic fibrosis model, was associated with a decrease in pulmonary interleukin-17 (IL-17) and γδ T cells, in contrast to the increase seen when wild-type mice were treated with streptomycin (136). Thus, the gut microbiome also plays an important immune-modulating role in cystic fibrosis (138, 139).
Changes in the gut microbiome have also been associated with pulmonary complications in the setting of allogeneic hematopoietic cell transplantation in humans (140). Domination of the fecal microbiota by Gammaproteobacteria, of which Klebsiella pneumoniae and Klebsiella oxytoca are members, was associated with pulmonary complications and doubling of the risk for mortality (140), suggesting that the gut contributes to the translocation of gut bacteria early in transplantation or is able to contribute to indirect lung injury from microbiota-induced local or systemic inflammatory pathways. The gut microbiome may also attenuate and protect against sepsis (141–143). Eradication of the gastrointestinal microbiota with antibiotics is associated with a dramatic reduction in survival in mice intranasally challenged with a pathogenic strain of Streptococcus pneumoniae, as shown in a study in which cytokine production of alveolar macrophages, such as IL-1β, IL-6, and CXCL1, was upregulated while IL-10 and tumor necrosis factor alpha were downregulated (144).
The full extent of gut-lung interactions still requires further clarification. There is clear cross talk between the two organs, and both organs represent large areas of mucosal surfaces that likely share similar immune pathways. Thus, the gut-and-lung interactions expose shortcomings that occur when the immunological effects of the lung or gut microbiome are described in isolation. New evidence is beginning to suggest that the gut-lung axis represents an area of communication and sharing of information between these two organs. The future direction of research is to define and refine these interactions and to take advantage of them to benefit the host.
MICROBIOTA EFFECTS ON THE ACTIVATION OF INFLAMMATORY PATHWAYS OF THE LOWER AIRWAY ENVIRONMENT
The interaction of the host’s respiratory system with the environmental and microbiome challenges is an integral component of understanding the role of the upper and lower respiratory microbiomes in health and disease (21, 23, 30). Although examples are currently scant in the pulmonary microbiome literature, several articles have demonstrated the role of the gastrointestinal microbiome in the maturation of the Th17 response in the mucosal immune system (2, 145–147). Within the gastrointestinal literature, there is evidence that the gastrointestinal microbiome plays a significant immune-modulating role in health and disease (2, 146, 147). In particular, the Th17 response on mucosal surfaces is of interest. Ivanov and colleagues demonstrated that segmented filamentous bacilli appear to play a role in the differentiation of Th17 cells in the gastrointestinal tract (2, 147). Furthermore, Suzuki and colleagues demonstrated that in the IgA-deficient mouse model, aberrant and uncontrolled immune responses were associated with the proliferation of segmented filamentous bacilli in these mice (148). In all, the gut microbiome was found to play a necessary role in the maturation of the immune response. However, the lung microbiome may also play a significant role in a mucosa-based immune response.
In the lung mucosa, changes in microbiome in healthy patients are associated with subclinical inflammation (23). In a dichotomized grouping of the lung microbiome, two distinct lung microbiomes (pneumotypes) could be identified: one characterized by enrichment with supraglottic predominant taxa (pneumotypeSPT) (Fig. 2) and another characterized by enrichment with background predominant taxa (pneumotypeBPT) (Fig. 2) (23). The degree of similarity with the upper airway microbiome can also be used to characterize the lung microbiome, where lower UniFrac distances (a measurement of dissimilarity based on relative abundance and a phylogenetic tree) occur in BAL samples showing enrichment with supraglottic predominant taxa (pneumotypeSPT); however, the metabolic activity of these taxa is currently unknown (93, 149). Moreover, the organization of the cohorts into these two pneumotypes (Fig. 2) was supported by contributed phage data (30). The fact that phage data were recovered implies that different phages selected for different taxa in the lungs of our subjects, suggesting that the ecological gradients of the lung may affect the virome as well as the microbiome. The phage data from the cohort also support the existence of an actively replicating phage population; therefore, having an active phage population implies that there also is an actively replicating microbial community (30). Importantly, the lower airway microbiome enriched with oral taxa is associated with a distinct genomic potential and associated with several metabolites of microbial origin. This provides evidence of active microbial metabolism in the lower airways. The host is not merely a bystander in its interaction with the lower respiratory tract microbiome; it can also be affected or benefited by the by-products of bacterial metabolism. A recent investigation has shown that the use of azithromycin in early emphysema promotes the bacterial production of anti-inflammatory metabolites (118), suggesting that some of the anti-inflammatory effects of macrolides might be mediated by their effects on the microbial metabolism in the lower airways.
FIGURE 2.
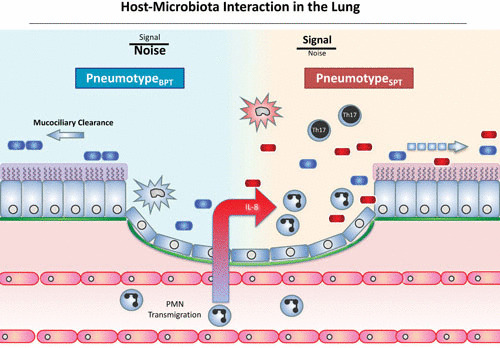
Host-microbiota interaction in the lung. This schema represents the normal lung microbiome and its dysbiosis. In this model, enrichment with background taxa (represented as blue bacteria) in pneumotypeBPT occurs in a lung with preserved mucociliary clearance of microorganisms and minimal inflammatory signals within the lung. In the presence of enrichment of the lower airway microbiome with oral taxa (represented as red bacteria) in pneumotypeSPT, there will be upregulation of the Th17 inflammatory phenotype and recruitment of neutrophils and lymphocytes. PMN, polymorphonuclear leukocyte.
Further, differences in lung microbiome are associated with a distinct immunological phenotype, demonstrating that the lung microbiome is not a bystander commensal but rather interacts with the host immune system. Specifically, enrichment with supraglottic predominant taxa (pneumotypeSPT) is associated with the Th17 inflammatory phenotype (Fig. 2). The lower airway microbiome is most likely an active participant in pneumotypeSPT, and samples that resembled those of the upper respiratory tract were also associated with Th17-chemoattractant cytokines: IL-1α, IL-1β, fractalkine, and IL-7 (30). Furthermore, transcriptome analysis of airway brushes showed that factors important for a Th17 differentiation, such as STAT3 and thymic stromal lymphopoietin, were upregulated in association with pneumotypeSPT. Given that these microbiome signals were found in both healthy volunteers and those who were exposed to cigarette smoke, enrichment of the lower airways must occur with or without the context of disease (Fig. 2).
Other investigations have also found microbial signatures associated with a Th17 phenotype in severe asthma (29). The mechanism by which this phenotype occurs in the lung is still not clear. Huang and colleagues’ study of severe asthmatic patients found that the presence of Proteobacteria correlates with Th17-associated gene expression. Members of the Proteobacteria taxa include Pasteurellaceae, Enterobacteriaceae, and Bacillaceae. Although both FKBP5 (a gene associated with steroid responsiveness in asthma) and genes associated with Th17 inflammation correlated with taxa from the phylum Proteobacteria, particular families did not significantly overlap, suggesting that different microorganisms, despite sharing a common phylum, are responsible for different effects on the lower airway (29). Thus, distinct airway microbiotas may promote Th17-associated inflammation but may not be associated with FKBP5-related steroid-induced effects (29). The identification of microorganisms that are associated with Th17 inflammation may represent another pathway of inflammation that is independent of a Th2 and eosinophilic response. The Th17 inflammatory phenotype may represent an important immunomodulatory target in some asthmatics, especially those with a neutrophilic phenotype (29). The Th2 phenotype, however, was not significantly correlated with the microbiome data (29).
Moreover, research by Yadava and colleagues involving an experimental mouse model found that exposure to LPS and elastase leads to a dysbiotic lung microbiome, with an increase of IL-17A expression due to an increase in the γδ+ T cell phenotype (150). This murine inflammatory phenotype is associated with airway abnormalities and emphysematous changes that resembled human COPD. BAL fluids obtained from mice challenged with LPS and elastase demonstrated a decreased α diversity and an increase of relative abundances of Pseudomonas, Lactobacillus, and Chryseobacterium (150). The researchers found that the microbiota enhanced the production of IL-17A by γδ+ T cells by using microbiota-depleted mice. Importantly, researchers recapitulated the upregulated IL-17A immunological phenotype by transfer of enriched microbiotas from LPS- and elastase-treated mice and concurrent challenge with LPS and elastase into antibiotic-treated mice (150). This study demonstrated experimentally that the lung microbiome has a functional role on an immunological phenotype; however, the individual components of this role still need to be clearly elucidated. The use of LPS and elastase in this study raises the question, as does Poroyko and colleagues’ study, of whether it is living complex microbial communities or pathogen-associated molecular patterns that are required to induce a Th17 phenotype. Furthermore, LPS may have variable effects depending on the source organism, commonly being E. coli, and thus may not be derived from microbes frequently found in the lower airways (106, 150). Given the complexity of the lower airway microbiome, it is likely that other microbial products also play an important role on the host immune phenotype in the lower airways (106, 150).
The presence of the Th17 phenotype and the upregulation of IL-17 have important implications for microbial communities in the lower airway. The lower respiratory tract microbiome, instead of being a nonresponsive passenger, stimulates and attenuates the development of the innate and adaptive immune response. Dysbiosis, here defined by a microbiome that resembles the upper respiratory tract microbiome, is associated with a subclinical proinflammatory phenotype (30). Complex metabolic interactions are present in relationship with this dysbiotic microbiota. Further, there is also evidence of activation of counterregulatory mechanisms being triggered by a distinct lung microbiome. Pulmonary alveolar macrophages collected from subjects who had a lower airway microbiome characterized as pneumotypeSPT were associated with a blunted Toll-like receptor 4 response with decreased production of tumor necrosis factor alpha, macrophage-derived chemokine, IL-6, and granulocyte-macrophage colony-stimulating factor in response to LPS stimulation (30). This attenuated innate immune response in this distinct lung microbiome may therefore be an important counterregulatory mechanism present in alveolar macrophages, needed to attenuate the inflammatory immune response in the lung mucosa (3, 30, 145, 147). This is in line with multiple other counterregulatory mechanisms found in other organ systems, including the gastrointestinal tract (3, 145–147).
The Th17 inflammatory pathway and IL-17 upregulation are not the only pathways that may be upregulated in a host with lung dysbiosis. Richmond and colleagues’ study also demonstrated in pIgR-deficient mice that the loss of mucosal immunity resulted in microorganism invasion into the epithelium (95). In contrast, the percentage of bacteria present in the lumen of the airways did not differ between the wild-type mice and pIgR-deficient mice, suggesting that the effects were due to the invasion of microorganisms alone. Mice with pIgR deficiency also demonstrated activation of the NF-κB pathway with associated NF-κB-dependent chemokine keratinocyte chemoattractant in BAL fluid (95). All these results indicate that this mouse model of mucosal immunodeficiency is an example of another pathway that is activated by the interaction between innate microorganisms in the lower airway and represents a target for study.
Metabolites and products of bacterial metabolism will also have immunomodulating properties on the lung microbiome. Chronic inflammation typifies the effects of cystic fibrosis on the pulmonary system. Mechanisms of inflammation revolve around the colonization of typical cystic fibrosis microorganisms but also around the anaerobic metabolism from the facultative anaerobes that make up a smaller portion of the microbiome composition (90). The microorganisms capable of facultative anaerobic metabolism are frequently found in the mouth and have been identified in the lower airway of cystic fibrosis patients. Short-chain fatty acids, a by-product of anaerobic metabolism, have been shown to alter and influence the inflammation in cystic fibrosis (131).
The lung mucosal surface is an important interface with the exterior world, given its roles in the innate and adaptive immune responses that scavenge and respond to microbes and in providing alveolar macrophages that maintain an important function in presenting antigens from opsonized microbes, as well as its ability to regulate signaling cytokines and chemokines (151). The data described above show that there are multiple lines of evidence that support that the lower airway microbiome has an important role in maintaining the immunological tone of the lower airway mucosa. This response may be part of a “healthy” immunological tone required to prime the immune system or may play an important role in the inflammatory process of several inflammatory diseases of the airways.
Microbiome studies have uncovered a complex and diverse microbial community that inhabits within us. With the use of multiomics, we can evaluate the dynamics between the microbes and the host. This new vision of a complex microbial community interacting with the host immune system forces us to reconsider the classical Koch’s postulates for our understanding of disease. The great challenge is how to recognize that complex communities of microbes may have a mutualistic and active role in influencing their microenvironment, including the host immune response (152). The effects of communities of microorganisms causing disease represent a challenge to Koch’s postulates as they are commonly used, i.e., in positing that a single microbe is the cause of a disease. These postulates have been used to categorize microbes as pathogens (153). In the context of microbial communities interacting with the host and affecting its environment, it is likely that the “pathogenic” role of a microbe and its community will have to be redefined (154, 155). It is then possible that pathogenicity should be viewed in the context of the interaction between the microorganisms and the host, an interface affected by the microorganism’s own virulence factors, the associated microbiota, and the host immune response.
Our understanding of many lung diseases is currently limited to association studies, and functional studies have yet to be done in human and mouse models. These different interactions and mechanisms can be further elucidated with the study of the metagenome, metabolomics, metatranscriptomic, metaproteomic, viromics, and other omics studies (22, 23, 30, 155). The use of these approaches will be important to understand how the microbiome communities affect health and disease and to challenge the classical approach to Koch’s postulates by addressing the community’s role in health and disease as a whole (155).
CONCLUSION
The respiratory mucosa represents a broad and encompassing border between the human host and the environment. The differences between the gastrointestinal tract and the respiratory tract, such as the concentration of microbes, length of organ, ease of sampling, and physiological parameters, represent challenging nuances in the study of the lung microbiome. Attention has shifted from understanding the ecological composition of the respiratory tract microbiome to several key issues, including the colonization and persistence of particular communities of microbes, inflammatory pathways that are upregulated and downregulated in the presence of particular communities, and the functional and mechanistic causes of these inflammatory pathways (23, 30).
The lungs see multiple microbial challenges through different mechanisms such as aerodigestive reflux and microaspiration during the course of the day and night (38). These repetitive microbial challenges may play an important role in the host immune response (23, 30, 156). The effects on the host immunological tone may represent beneficial immunological priming or contribute to a pathogenic immunological process relevant for disease development.
Future directions in the study of the lower respiratory lung microbiome will require focus on dissecting the microbial community function, understanding microbe-microbe interactions, and developing experimental models to uncover causality. Ultimately, the goal is to find potential microbial targets for a more precise manipulation of the lower airway microbiota, such as refined use of antibiotics, phage therapy, or molecules that target specific microbial metabolic pathways, in order to alter the natural course of pulmonary disease processes.
ACKNOWLEDGMENTS
Research support funding was provided by K23 AI102970 (NIH/NIAID), the Flight Attendant Medical Research Institute (FAMRI), a Stony Wold-Herbert Fund Fellowship Award, and 1UL1TR001445 (NIH/NCATS).
REFERENCES
- 1.Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. 2007. The human microbiome project. Nature 449:804–810. 10.1038/nature06244 [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ivanov II, Frutos RL, Manel N, Yoshinaga K, Rifkin DB, Sartor RB, Finlay BB, Littman DR. 2008. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe 4:337–349. 10.1016/j.chom.2008.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, Taniguchi T, Takeda K, Hori S, Ivanov II, Umesaki Y, Itoh K, Honda K. 2011. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 331:337–341. 10.1126/science.1198469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wright EK, Kamm MA, Teo SM, Inouye M, Wagner J, Kirkwood CD. 2015. Recent advances in characterizing the gastrointestinal microbiome in Crohn’s disease: a systematic review. Inflamm Bowel Dis 21:1219–1228. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bäckhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI. 2004. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA 101:15718–15723. 10.1073/pnas.0407076101 [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. 2011. Human nutrition, the gut microbiome and the immune system. Nature 474:327–336. 10.1038/nature10213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turnbaugh PJ, Gordon JI. 2009. The core gut microbiome, energy balance and obesity. J Physiol 587:4153–4158. 10.1113/jphysiol.2009.174136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. 2009. A core gut microbiome in obese and lean twins. Nature 457:480–484. 10.1038/nature07540 [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, Britt EB, Fu X, Wu Y, Li L, Smith JD, DiDonato JA, Chen J, Li H, Wu GD, Lewis JD, Warrier M, Brown JM, Krauss RM, Tang WH, Bushman FD, Lusis AJ, Hazen SL. 2013. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med 19:576–585. 10.1038/nm.3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang WH, Wang Z, Levison BS, Koeth RA, Britt EB, Fu X, Wu Y, Hazen SL. 2013. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N Engl J Med 368:1575–1584. 10.1056/NEJMoa1109400 [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang WH, Wang Z, Shrestha K, Borowski AG, Wu Y, Troughton RW, Klein AL, Hazen SL. 2015. Intestinal microbiota-dependent phosphatidylcholine metabolites, diastolic dysfunction, and adverse clinical outcomes in chronic systolic heart failure. J Card Fail 21:91–96. 10.1016/j.cardfail.2014.11.006 [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, Feldstein AE, Britt EB, Fu X, Chung YM, Wu Y, Schauer P, Smith JD, Allayee H, Tang WH, DiDonato JA, Lusis AJ, Hazen SL. 2011. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 472:57–63. 10.1038/nature09922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang JY, Antonopoulos DA, Kalra A, Tonelli A, Khalife WT, Schmidt TM, Young VB. 2008. Decreased diversity of the fecal microbiome in recurrent Clostridium difficile-associated diarrhea. J Infect Dis 197:435–438. 10.1086/525047 [DOI] [PubMed] [Google Scholar]
- 14.Lawley TD, Clare S, Walker AW, Stares MD, Connor TR, Raisen C, Goulding D, Rad R, Schreiber F, Brandt C, Deakin LJ, Pickard DJ, Duncan SH, Flint HJ, Clark TG, Parkhill J, Dougan G. 2012. Targeted restoration of the intestinal microbiota with a simple, defined bacteriotherapy resolves relapsing Clostridium difficile disease in mice. PLoS Pathog 8:e1002995. 10.1371/journal.ppat.1002995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qin J, et al. 2012. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490:55–60. 10.1038/nature11450 [DOI] [PubMed] [Google Scholar]
- 16.Forslund K, Hildebrand F, Nielsen T, Falony G, Le Chatelier E, Sunagawa S, Prifti E, Vieira-Silva S, Gudmundsdottir V, Krogh Pedersen H, Arumugam M, Kristiansen K, Voigt AY, Vestergaard H, Hercog R, Igor Costea P, Kultima JR, Li J, Jørgensen T, Levenez F, Dore J, MetaHIT Consortium, Nielsen HB, Brunak S, Raes J, Hansen T, Wang J, Ehrlich SD, Bork P, Pedersen O. 2015. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature 528:262–266. 10.1038/nature15766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, Frangeul L, Nalin R, Jarrin C, Chardon P, Marteau P, Roca J, Dore J. 2006. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut 55:205–211. 10.1136/gut.2005.073817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morgan XC, Kabakchiev B, Waldron L, Tyler AD, Tickle TL, Milgrom R, Stempak JM, Gevers D, Xavier RJ, Silverberg MS, Huttenhower C. 2015. Associations between host gene expression, the mucosal microbiome, and clinical outcome in the pelvic pouch of patients with inflammatory bowel disease. Genome Biol 16:67. 10.1186/s13059-015-0637-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Metzker ML. 2010. Sequencing technologies—the next generation. Nat Rev Genet 11:31–46. 10.1038/nrg2626 [DOI] [PubMed] [Google Scholar]
- 20.Wu J, Peters BA, Dominianni C, Zhang Y, Pei Z, Yang L, Ma Y, Purdue MP, Jacobs EJ, Gapstur SM, Li H, Alekseyenko AV, Hayes RB, Ahn J. 2016. Cigarette smoking and the oral microbiome in a large study of American adults. ISME J 10:2435–2446. 10.1038/ismej.2016.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Segal LN, Rom WN, Weiden MD. 2014. Lung microbiome for clinicians. New discoveries about bugs in healthy and diseased lungs. Ann Am Thorac Soc 11:108–116. 10.1513/AnnalsATS.201310-339FR [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Segal LN, Blaser MJ. 2014. A brave new world: the lung microbiota in an era of change. Ann Am Thorac Soc 11(Suppl 1):S21–S27. 10.1513/AnnalsATS.201306-189MG [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Segal LN, Alekseyenko AV, Clemente JC, Kulkarni R, Wu B, Gao Z, Chen H, Berger KI, Goldring RM, Rom WN, Blaser MJ, Weiden MD. 2013. Enrichment of lung microbiome with supraglottic taxa is associated with increased pulmonary inflammation. Microbiome 1:19. 10.1186/2049-2618-1-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dickson RP, Erb-Downward JR, Martinez FJ, Huffnagle GB. 2016. The microbiome and the respiratory tract. Annu Rev Physiol 78:481–504. 10.1146/annurev-physiol-021115-105238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fujimura KE, Demoor T, Rauch M, Faruqi AA, Jang S, Johnson CC, Boushey HA, Zoratti E, Ownby D, Lukacs NW, Lynch SV. 2014. House dust exposure mediates gut microbiome Lactobacillus enrichment and airway immune defense against allergens and virus infection. Proc Natl Acad Sci USA 111:805–810. 10.1073/pnas.1310750111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Noverr MC, Falkowski NR, McDonald RA, McKenzie AN, Huffnagle GB. 2005. Development of allergic airway disease in mice following antibiotic therapy and fungal microbiota increase: role of host genetics, antigen, and interleukin-13. Infect Immun 73:30–38. 10.1128/IAI.73.1.30-38.2005 [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Noverr MC, Noggle RM, Toews GB, Huffnagle GB. 2004. Role of antibiotics and fungal microbiota in driving pulmonary allergic responses. Infect Immun 72:4996–5003. 10.1128/IAI.72.9.4996-5003.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sze MA, Dimitriu PA, Suzuki M, McDonough JE, Campbell JD, Brothers JF, Erb-Downward JR, Huffnagle GB, Hayashi S, Elliott WM, Cooper J, Sin DD, Lenburg ME, Spira A, Mohn WW, Hogg JC. 2015. Host response to the lung microbiome in chronic obstructive pulmonary disease. Am J Respir Crit Care Med 192:438–445. 10.1164/rccm.201502-0223OC [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang YJ, Nariya S, Harris JM, Lynch SV, Choy DF, Arron JR, Boushey H. 2015. The airway microbiome in patients with severe asthma: associations with disease features and severity. J Allergy Clin Immunol 136:874–884. 10.1016/j.jaci.2015.05.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Segal LN, Clemente JC, Tsay JCJ, Koralov SB, Keller BC, Wu BG, Li Y, Shen N, Ghedin E, Morris A, Diaz P, Huang L, Wikoff WR, Ubeda C, Artacho A, Rom WN, Sterman DH, Collman RG, Blaser MJ, Weiden MD. 2016. Enrichment of the lung microbiome with oral taxa is associated with lung inflammation of a Th17 phenotype. Nat Microbiol 1:16031. 10.1038/nmicrobiol.2016.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Twigg HL III, Morris A, Ghedin E, Curtis JL, Huffnagle GB, Crothers K, Campbell TB, Flores SC, Fontenot AP, Beck JM, Huang L, Lynch S, Knox KS, Weinstock G, Lung HIV Microbiome Project. 2013. Use of bronchoalveolar lavage to assess the respiratory microbiome: signal in the noise. Lancet Respir Med 1:354–356. 10.1016/S2213-2600(13)70117-6 [DOI] [PubMed] [Google Scholar]
- 32.Charlson ES, Bittinger K, Haas AR, Fitzgerald AS, Frank I, Yadav A, Bushman FD, Collman RG. 2011. Topographical continuity of bacterial populations in the healthy human respiratory tract. Am J Respir Crit Care Med 184:957–963. 10.1164/rccm.201104-0655OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, No D, Liu H, Kinnebrew M, Viale A, Littmann E, van den Brink MR, Jenq RR, Taur Y, Sander C, Cross JR, Toussaint NC, Xavier JB, Pamer EG. 2015. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517:205–208. 10.1038/nature13828 [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Charlson ES, Bittinger K, Chen J, Diamond JM, Li H, Collman RG, Bushman FD. 2012. Assessing bacterial populations in the lung by replicate analysis of samples from the upper and lower respiratory tracts. PLoS One 7:e42786. 10.1371/journal.pone.0042786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Charlson ES, Chen J, Custers-Allen R, Bittinger K, Li H, Sinha R, Hwang J, Bushman FD, Collman RG. 2010. Disordered microbial communities in the upper respiratory tract of cigarette smokers. PLoS One 5:e15216. 10.1371/journal.pone.0015216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shaker R, Hogan WJ. 2000. Reflex-mediated enhancement of airway protective mechanisms. Am J Med 108(Suppl 4a):8S–14S. 10.1016/S0002-9343(99)00289-2 [DOI] [PubMed] [Google Scholar]
- 37.Kronenberger MB, Meyers AD. 1994. Dysphagia following head and neck cancer surgery. Dysphagia 9:236–244. 10.1007/BF00301917 [DOI] [PubMed] [Google Scholar]
- 38.Bassis CM, Erb-Downward JR, Dickson RP, Freeman CM, Schmidt TM, Young VB, Beck JM, Curtis JL, Huffnagle GB. 2015. Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. MBio 6:e00037. 10.1128/mBio.00037-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gleeson K, Eggli DF, Maxwell SL. 1997. Quantitative aspiration during sleep in normal subjects. Chest 111:1266–1272. 10.1378/chest.111.5.1266 [PubMed] [DOI] [PubMed] [Google Scholar]
- 40.Cvejic L, Harding R, Churchward T, Turton A, Finlay P, Massey D, Bardin PG, Guy P. 2011. Laryngeal penetration and aspiration in individuals with stable COPD. Respirology 16:269–275. 10.1111/j.1440-1843.2010.01875.x [PubMed] [DOI] [PubMed] [Google Scholar]
- 41.Morse CA, Quan SF, Mays MZ, Green C, Stephen G, Fass R. 2004. Is there a relationship between obstructive sleep apnea and gastroesophageal reflux disease? Clin Gastroenterol Hepatol 2:761–768. 10.1016/S1542-3565(04)00347-7 [DOI] [PubMed] [Google Scholar]
- 42.Teramoto S, Ohga E, Matsui H, Ishii T, Matsuse T, Ouchi Y. 1999. Obstructive sleep apnea syndrome may be a significant cause of gastroesophageal reflux disease in older people. J Am Geriatr Soc 47:1273–1274. 10.1111/j.1532-5415.1999.tb05216.x [DOI] [PubMed] [Google Scholar]
- 43.Field SK, Underwood M, Brant R, Cowie RL. 1996. Prevalence of gastroesophageal reflux symptoms in asthma. Chest 109:316–322. 10.1378/chest.109.2.316 [DOI] [PubMed] [Google Scholar]
- 44.Scott RB, O’Loughlin EV, Gall DG. 1985. Gastroesophageal reflux in patients with cystic fibrosis. J Pediatr 106:223–227. 10.1016/S0022-3476(85)80291-2 [DOI] [PubMed] [Google Scholar]
- 45.Koh WJ, Lee JH, Kwon YS, Lee KS, Suh GY, Chung MP, Kim H, Kwon OJ. 2007. Prevalence of gastroesophageal reflux disease in patients with nontuberculous mycobacterial lung disease. Chest 131:1825–1830. 10.1378/chest.06-2280 [DOI] [PubMed] [Google Scholar]
- 46.Dickson RP, Erb-Downward JR, Prescott HC, Martinez FJ, Curtis JL, Lama VN, Huffnagle GB. 2014. Cell-associated bacteria in the human lung microbiome. Microbiome 2:28. 10.1186/2049-2618-2-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beck JM, Schloss PD, Venkataraman A, Twigg H III, Jablonski KA, Bushman FD, Campbell TB, Charlson ES, Collman RG, Crothers K, Curtis JL, Drews KL, Flores SC, Fontenot AP, Foulkes MA, Frank I, Ghedin E, Huang L, Lynch SV, Morris A, Palmer BE, Schmidt TM, Sodergren E, Weinstock GM, Young VB, Lung HIV Microbiome Project. 2015. Multicenter comparison of lung and oral microbiomes of HIV-infected and HIV-uninfected individuals. Am J Respir Crit Care Med 192:1335–1344. 10.1164/rccm.201501-0128OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Segal LN, Dickson RP. 2016. The lung microbiome in HIV. Getting to the HAART of the host-microbe interface. Am J Respir Crit Care Med 194:136–137. 10.1164/rccm.201602-0280ED [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huxley EJ, Viroslav J, Gray WR, Pierce AK. 1978. Pharyngeal aspiration in normal adults and patients with depressed consciousness. Am J Med 64:564–568. 10.1016/0002-9343(78)90574-0 [DOI] [PubMed] [Google Scholar]
- 50.Simpson JL, Daly J, Baines KJ, Yang IA, Upham JW, Reynolds PN, Hodge S, James AL, Hugenholtz P, Willner D, Gibson PG. 2016. Airway dysbiosis: Haemophilus influenzae and Tropheryma in poorly controlled asthma. Eur Respir J 47:792–800. 10.1183/13993003.00405-2015 [DOI] [PubMed] [Google Scholar]
- 51.Smits HH, Hiemstra PS, Prazeres da Costa C, Ege M, Edwards M, Garn H, Howarth PH, Jartti T, de Jong EC, Maizels RM, Marsland BJ, McSorley HJ, Müller A, Pfefferle PI, Savelkoul H, Schwarze J, Unger WW, von Mutius E, Yazdanbakhsh M, Taube C. 2016. Microbes and asthma: opportunities for intervention. J Allergy Clin Immunol 137:690–697. 10.1016/j.jaci.2016.01.004 [DOI] [PubMed] [Google Scholar]
- 52.Huang YJ. 2015. The respiratory microbiome and innate immunity in asthma. Curr Opin Pulm Med 21:27–32. 10.1097/MCP.0000000000000124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang YJ. 2013. Asthma microbiome studies and the potential for new therapeutic strategies. Curr Allergy Asthma Rep 13:453–461. 10.1007/s11882-013-0355-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang YJ, Boushey HA. 2015. The microbiome in asthma. J Allergy Clin Immunol 135:25–30. 10.1016/j.jaci.2014.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang YJ, Boushey HA. 2014. The microbiome and asthma. Ann Am Thorac Soc 11(Suppl 1):S48–S51. 10.1513/AnnalsATS.201306-187MG [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang YJ, Boushey HA. 2013. The bronchial microbiome and asthma phenotypes. Am J Respir Crit Care Med 188:1178–1180. 10.1164/rccm.201309-1702ED [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang YJ, Charlson ES, Collman RG, Colombini-Hatch S, Martinez FD, Senior RM. 2013. The role of the lung microbiome in health and disease. A National Heart, Lung, and Blood Institute workshop report. Am J Respir Crit Care Med 187:1382–1387. 10.1164/rccm.201303-0488WS [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hilty M, Burke C, Pedro H, Cardenas P, Bush A, Bossley C, Davies J, Ervine A, Poulter L, Pachter L, Moffatt MF, Cookson WO. 2010. Disordered microbial communities in asthmatic airways. PLoS One 5:e8578. 10.1371/journal.pone.0008578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lynch SV, Bruce KD. 2013. The cystic fibrosis airway microbiome. Cold Spring Harb Perspect Med 3:a009738. 10.1101/cshperspect.a009738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huang YJ, LiPuma JJ. 2016. The microbiome in cystic fibrosis. Clin Chest Med 37:59–67. 10.1016/j.ccm.2015.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Whelan FJ, Surette MG. 2015. Clinical insights into pulmonary exacerbations in cystic fibrosis from the microbiome. What are we missing? Ann Am Thorac Soc 12(Suppl 2):S207–S211. [PubMed] [DOI] [PubMed] [Google Scholar]
- 62.Caverly LJ, Zhao J, LiPuma JJ. 2015. Cystic fibrosis lung microbiome: opportunities to reconsider management of airway infection. Pediatr Pulmonol 50(Suppl 40):S31–S38. 10.1002/ppul.23243 [DOI] [PubMed] [Google Scholar]
- 63.Twigg HL III, Knox KS, Zhou J, Crothers KA, Nelson DE, Toh E, Day RB, Lin H, Gao X, Dong Q, Mi D, Katz BP, Sodergren E, Weinstock GM. 2016. Effect of advanced HIV infection on the respiratory microbiome. Am J Respir Crit Care Med 194:226–235. 10.1164/rccm.201509-1875OC [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lozupone C, Cota-Gomez A, Palmer BE, Linderman DJ, Charlson ES, Sodergren E, Mitreva M, Abubucker S, Martin J, Yao G, Campbell TB, Flores SC, Ackerman G, Stombaugh J, Ursell L, Beck JM, Curtis JL, Young VB, Lynch SV, Huang L, Weinstock GM, Knox KS, Twigg H, Morris A, Ghedin E, Bushman FD, Collman RG, Knight R, Fontenot AP, Lung HIV Microbiome Project. 2013. Widespread colonization of the lung by Tropheryma whipplei in HIV infection. Am J Respir Crit Care Med 187:1110–1117. 10.1164/rccm.201211-2145OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sze MA, Dimitriu PA, Hayashi S, Elliott WM, McDonough JE, Gosselink JV, Cooper J, Sin DD, Mohn WW, Hogg JC. 2012. The lung tissue microbiome in chronic obstructive pulmonary disease. Am J Respir Crit Care Med 185:1073–1080. 10.1164/rccm.201111-2075OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sze MA, Hogg JC, Sin DD. 2014. Bacterial microbiome of lungs in COPD. Int J Chron Obstruct Pulmon Dis 9:229–238. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, Moffatt MF, Turner P, Parkhill J, Loman NJ, Walker AW. 2014. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol 12:87. 10.1186/s12915-014-0087-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Venkataraman A, Bassis CM, Beck JM, Young VB, Curtis JL, Huffnagle GB, Schmidt TM. 2015. Application of a neutral community model to assess structuring of the human lung microbiome. mBio 6:e02284-14. 10.1128/mBio.02284-14 [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Knights D, Kuczynski J, Charlson ES, Zaneveld J, Mozer MC, Collman RG, Bushman FD, Knight R, Kelley ST. 2011. Bayesian community-wide culture-independent microbial source tracking. Nat Methods 8:761–763. 10.1038/nmeth.1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hajishengallis G, Liang S, Payne MA, Hashim A, Jotwani R, Eskan MA, McIntosh ML, Alsam A, Kirkwood KL, Lambris JD, Darveau RP, Curtis MA. 2011. Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe 10:497–506. 10.1016/j.chom.2011.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pop M, Paulson JN, Chakraborty S, Astrovskaya I, Lindsay BR, Li S, Bravo HC, Harro C, Parkhill J, Walker AW, Walker RI, Sack DA, Stine OC. 2016. Individual-specific changes in the human gut microbiota after challenge with enterotoxigenic Escherichia coli and subsequent ciprofloxacin treatment. BMC Genomics 17:440. 10.1186/s12864-016-2777-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schwartz DA, Quinn TJ, Thorne PS, Sayeed S, Yi AK, Krieg AM. 1997. CpG motifs in bacterial DNA cause inflammation in the lower respiratory tract. J Clin Invest 100:68–73. 10.1172/JCI119523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Riley RL. 1957. Aerial dissemination of pulmonary tuberculosis. Am Rev Tuberc 76:931–941. [PubMed] [DOI] [PubMed] [Google Scholar]
- 74.Riley RL, Mills CC, O’Grady F, Sultan LU, Wittstadt F, Shivpuri DN. 1962. Infectiousness of air from a tuberculosis ward. Ultraviolet irradiation of infected air: comparative infectiousness of different patients. Am Rev Respir Dis 85:511–525. [PubMed] [DOI] [PubMed] [Google Scholar]
- 75.Adams RI, Bateman AC, Bik HM, Meadow JF. 2015. Microbiota of the indoor environment: a meta-analysis. Microbiome 3:49. 10.1186/s40168-015-0108-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Meadow JF, Altrichter AE, Bateman AC, Stenson J, Brown GZ, Green JL, Bohannan BJ. 2015. Humans differ in their personal microbial cloud. PeerJ 3:e1258. 10.7717/peerj.1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stephenson MF, Mfuna L, Dowd SE, Wolcott RD, Barbeau J, Poisson M, James G, Desrosiers M. 2010. Molecular characterization of the polymicrobial flora in chronic rhinosinusitis. J Otolaryngol Head Neck Surg 39:182–187. [PubMed] [PubMed] [Google Scholar]
- 78.Boase S, Foreman A, Cleland E, Tan L, Melton-Kreft R, Pant H, Hu FZ, Ehrlich GD, Wormald PJ. 2013. The microbiome of chronic rhinosinusitis: culture, molecular diagnostics and biofilm detection. BMC Infect Dis 13:210. 10.1186/1471-2334-13-210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Blaser MJ, Chen Y, Reibman J. 2008. Does Helicobacter pylori protect against asthma and allergy? Gut 57:561–567. 10.1136/gut.2007.133462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Reibman J, Marmor M, Filner J, Fernandez-Beros ME, Rogers L, Perez-Perez GI, Blaser MJ. 2008. Asthma is inversely associated with Helicobacter pylori status in an urban population. PLoS One 3:e4060. 10.1371/journal.pone.0004060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rosen R, Hu L, Amirault J, Khatwa U, Ward DV, Onderdonk A. 2015. 16S community profiling identifies proton pump inhibitor related differences in gastric, lung, and oropharyngeal microflora. J Pediatr 166:917–923. 10.1016/j.jpeds.2014.12.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Souza DG, Vieira AT, Soares AC, Pinho V, Nicoli JR, Vieira LQ, Teixeira MM. 2004. The essential role of the intestinal microbiota in facilitating acute inflammatory responses. J Immunol 173:4137–4146. 10.4049/jimmunol.173.6.4137 [DOI] [PubMed] [Google Scholar]
- 83.Silvestri L, van Saene HK, Zandstra DF, Marshall JC, Gregori D, Gullo A. 2010. Impact of selective decontamination of the digestive tract on multiple organ dysfunction syndrome: systematic review of randomized controlled trials. Crit Care Med 38:1370–1376. 10.1097/CCM.0b013e3181d9db8c [PubMed] [DOI] [PubMed] [Google Scholar]
- 84.Erb-Downward JR, Thompson DL, Han MK, Freeman CM, McCloskey L, Schmidt LA, Young VB, Toews GB, Curtis JL, Sundaram B, Martinez FJ, Huffnagle GB. 2011. Analysis of the lung microbiome in the “healthy” smoker and in COPD. PLoS One 6:e16384. 10.1371/journal.pone.0016384 [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Morris A, Beck JM, Schloss PD, Campbell TB, Crothers K, Curtis JL, Flores SC, Fontenot AP, Ghedin E, Huang L, Jablonski K, Kleerup E, Lynch SV, Sodergren E, Twigg H, Young VB, Bassis CM, Venkataraman A, Schmidt TM, Weinstock GM, Lung HIV Microbiome Project. 2013. Comparison of the respiratory microbiome in healthy nonsmokers and smokers. Am J Respir Crit Care Med 187:1067–1075. 10.1164/rccm.201210-1913OC [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lomolino MV, Brown JH. 2009. The reticulating phylogeny of island biogeography theory. Q Rev Biol 84:357–390. 10.1086/648123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dickson RP, Erb-Downward JR, Freeman CM, McCloskey L, Beck JM, Huffnagle GB, Curtis JL. 2015. Spatial variation in the healthy human lung microbiome and the adapted island model of lung biogeography. Ann Am Thorac Soc 12:821–830. 10.1513/AnnalsATS.201501-029OC [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dickson RP, Erb-Downward JR, Huffnagle GB. 2014. Towards an ecology of the lung: new conceptual models of pulmonary microbiology and pneumonia pathogenesis. Lancet Respir Med 2:238–246. 10.1016/S2213-2600(14)70028-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dickson RP, Martinez FJ, Huffnagle GB. 2014. The role of the microbiome in exacerbations of chronic lung diseases. Lancet 384:691–702. 10.1016/S0140-6736(14)61136-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Whiteson KL, Bailey B, Bergkessel M, Conrad D, Delhaes L, Felts B, Harris JK, Hunter R, Lim YW, Maughan H, Quinn R, Salamon P, Sullivan J, Wagner BD, Rainey PB. 2014. The upper respiratory tract as a microbial source for pulmonary infections in cystic fibrosis. Parallels from island biogeography. Am J Respir Crit Care Med 189:1309–1315. 10.1164/rccm.201312-2129PP [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Veldhuizen R, Nag K, Orgeig S, Possmayer F. 1998. The role of lipids in pulmonary surfactant. Biochim Biophys Acta 1408:90–108. 10.1016/S0925-4439(98)00061-1 [DOI] [PubMed] [Google Scholar]
- 92.West JB. 2012. Respiratory Physiology: The Essentials, 9th ed. Wolters Kluwer Health/Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 93.Lieberman TD, Flett KB, Yelin I, Martin TR, McAdam AJ, Priebe GP, Kishony R. 2014. Genetic variation of a bacterial pathogen within individuals with cystic fibrosis provides a record of selective pressures. Nat Genet 46:82–87. 10.1038/ng.2848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rogan MP, Geraghty P, Greene CM, O’Neill SJ, Taggart CC, McElvaney NG. 2006. Antimicrobial proteins and polypeptides in pulmonary innate defence. Respir Res 7:29. 10.1186/1465-9921-7-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Richmond BW, Brucker RM, Han W, Du RH, Zhang Y, Cheng DS, Gleaves L, Abdolrasulnia R, Polosukhina D, Clark PE, Bordenstein SR, Blackwell TS, Polosukhin VV. 2016. Airway bacteria drive a progressive COPD-like phenotype in mice with polymeric immunoglobulin receptor deficiency. Nat Commun 7:11240. 10.1038/ncomms11240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Costerton JW, Lewandowski Z, DeBeer D, Caldwell D, Korber D, James G. 1994. Biofilms, the customized microniche. J Bacteriol 176:2137–2142. 10.1128/jb.176.8.2137-2142.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. 2002. Extracellular DNA required for bacterial biofilm formation. Science 295:1487. 10.1126/science.295.5559.1487 [DOI] [PubMed] [Google Scholar]
- 98.Costerton JW, Stewart PS, Greenberg EP. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322. 10.1126/science.284.5418.1318 [PubMed] [DOI] [PubMed] [Google Scholar]
- 99.Twigg HL III. 1998. Pulmonary host defenses. J Thorac Imaging 13:221–233. 10.1097/00005382-199810000-00003 [DOI] [PubMed] [Google Scholar]
- 100.Renshaw SA, Parmar JS, Singleton V, Rowe SJ, Dockrell DH, Dower SK, Bingle CD, Chilvers ER, Whyte MK. 2003. Acceleration of human neutrophil apoptosis by TRAIL. J Immunol 170:1027–1033. 10.4049/jimmunol.170.2.1027 [PubMed] [DOI] [PubMed] [Google Scholar]
- 101.Chmiel JF, Davis PB. 2003. State of the art: why do the lungs of patients with cystic fibrosis become infected and why can’t they clear the infection? Respir Res 4:8. 10.1186/1465-9921-4-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Klemm P, Schembri MA. 2000. Bacterial adhesins: function and structure. Int J Med Microbiol 290:27–35. 10.1016/S1438-4221(00)80102-2 [DOI] [PubMed] [Google Scholar]
- 103.Deslée G, Mal H, Dutau H, Bourdin A, Vergnon JM, Pison C, Kessler R, Jounieaux V, Thiberville L, Leroy S, Marceau A, Laroumagne S, Mallet JP, Dukic S, Barbe C, Bulsei J, Jolly D, Durand-Zaleski I, Marquette CH, REVOLENS Study Group. 2016. Lung volume reduction coil treatment vs usual care in patients with severe emphysema: the REVOLENS randomized clinical trial. JAMA 315:175–184. 10.1001/jama.2015.17821 [PubMed] [DOI] [PubMed] [Google Scholar]
- 104.Oliver A, Cantón R, Campo P, Baquero F, Blázquez J. 2000. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288:1251–1254. 10.1126/science.288.5469.1251 [DOI] [PubMed] [Google Scholar]
- 105.Rello J, Ausina V, Ricart M, Castella J, Prats G. 1993. Impact of previous antimicrobial therapy on the etiology and outcome of ventilator-associated pneumonia. Chest 104:1230–1235. 10.1378/chest.104.4.1230 [DOI] [PubMed] [Google Scholar]
- 106.Poroyko V, Meng F, Meliton A, Afonyushkin T, Ulanov A, Semenyuk E, Latif O, Tesic V, Birukova AA, Birukov KG. 2015. Alterations of lung microbiota in a mouse model of LPS-induced lung injury. Am J Physiol Lung Cell Mol Physiol 309:L76–L83. 10.1152/ajplung.00061.2014 [PubMed][CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dickson RP, Erb-Downward JR, Huffnagle GB. 2015. Homeostasis and its disruption in the lung microbiome. Am J Physiol Lung Cell Mol Physiol 309:L1047–L1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Molyneaux PL, Mallia P, Cox MJ, Footitt J, Willis-Owen SA, Homola D, Trujillo-Torralbo MB, Elkin S, Kon OM, Cookson WO, Moffatt MF, Johnston SL. 2013. Outgrowth of the bacterial airway microbiome after rhinovirus exacerbation of chronic obstructive pulmonary disease. Am J Respir Crit Care Med 188:1224–1231. 10.1164/rccm.201302-0341OC [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Twigg H, Knox KS, Zhou J, Crothers K, Nelson D, Toh E, Day RB, Lin H, Gao X, Dong Q, Mi D, Katz BP, Sodergren E, Weinstock G. 2016. Effect of advanced HIV infection on the respiratory microbiome. Am J Respir Crit Care Med 194:226–235. 10.1164/rccm.201509-1875OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cui L, Lucht L, Tipton L, Rogers MB, Fitch A, Kessinger C, Camp D, Kingsley L, Leo N, Greenblatt RM, Fong S, Stone S, Dermand JC, Kleerup EC, Huang L, Morris A, Ghedin E. 2015. Topographic diversity of the respiratory tract mycobiome and alteration in HIV and lung disease. Am J Respir Crit Care Med 191:932–942. 10.1164/rccm.201409-1583OC [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gohy ST, Detry BR, Lecocq M, Bouzin C, Weynand BA, Amatngalim GD, Sibille YM, Pilette C. 2014. Polymeric immunoglobulin receptor down-regulation in chronic obstructive pulmonary disease. Persistence in the cultured epithelium and role of transforming growth factor-β. Am J Respir Crit Care Med 190:509–521. 10.1164/rccm.201311-1971OC [PubMed] [DOI] [PubMed] [Google Scholar]
- 112.Polosukhin VV, Cates JM, Lawson WE, Zaynagetdinov R, Milstone AP, Massion PP, Ocak S, Ware LB, Lee JW, Bowler RP, Kononov AV, Randell SH, Blackwell TS. 2011. Bronchial secretory immunoglobulin a deficiency correlates with airway inflammation and progression of chronic obstructive pulmonary disease. Am J Respir Crit Care Med 184:317–327. 10.1164/rccm.201010-1629OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Corbett AJ, Eckle SB, Birkinshaw RW, Liu L, Patel O, Mahony J, Chen Z, Reantragoon R, Meehan B, Cao H, Williamson NA, Strugnell RA, Van Sinderen D, Mak JY, Fairlie DP, Kjer-Nielsen L, Rossjohn J, McCluskey J. 2014. T-cell activation by transitory neo-antigens derived from distinct microbial pathways. Nature 509:361–365. 10.1038/nature13160 [PubMed] [DOI] [PubMed] [Google Scholar]
- 114.Gapin L. 2014. Check MAIT. J Immunol 192:4475–4480. 10.4049/jimmunol.1400119 [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hinks TS, Wallington JC, Williams AP, Djukanovič R, Staples KJ, Wilkinson TM. 2016. Steroid-induced deficiency of mucosal-associated invariant T cells in the COPD lung: implications for NTHi infection. Am J Respir Crit Care Med 194:1208–1218. 10.1164/rccm.201601-0002OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Pragman AA, Kim HB, Reilly CS, Wendt C, Isaacson RE. 2012. The lung microbiome in moderate and severe chronic obstructive pulmonary disease. PLoS One 7:e47305. 10.1371/journal.pone.0047305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Huang YJ, Kim E, Cox MJ, Brodie EL, Brown R, Wiener-Kronish JP, Lynch SV. 2010. A persistent and diverse airway microbiota present during chronic obstructive pulmonary disease exacerbations. OMICS 14:9–59. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Segal LN, Clemente JC, Wu BG, Wikoff WR, Gao Z, Li Y, Ko JP, Rom WN, Blaser MJ, Weiden MD. 2016. Randomised, double-blind, placebo-controlled trial with azithromycin selects for anti-inflammatory microbial metabolites in the emphysematous lung. Thorax 72:13–22. 10.1136/thoraxjnl-2016-208599. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Huang YJ, Nelson CE, Brodie EL, Desantis TZ, Baek MS, Liu J, Woyke T, Allgaier M, Bristow J, Wiener-Kronish JP, Sutherland ER, King TS, Icitovic N, Martin RJ, Calhoun WJ, Castro M, Denlinger LC, Dimango E, Kraft M, Peters SP, Wasserman SI, Wechsler ME, Boushey HA, Lynch SV, National Heart, Lung, and Blood Institute’s Asthma Clinical Research Network. 2011. Airway microbiota and bronchial hyperresponsiveness in patients with suboptimally controlled asthma. J Allergy Clin Immunol 127:372–381.e1-3. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Fujimura KE, Johnson CC, Ownby DR, Cox MJ, Brodie EL, Havstad SL, Zoratti EM, Woodcroft KJ, Bobbitt KR, Wegienka G, Boushey HA, Lynch SV. 2010. Man’s best friend? The effect of pet ownership on house dust microbial communities. J Allergy Clin Immunol 126:410–412.e1-3. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ege MJ, Mayer M, Normand AC, Genuneit J, Cookson WO, Braun-Fahrländer C, Heederik D, Piarroux R, von Mutius E, GABRIELA Transregio 22 Study Group. 2011. Exposure to environmental microorganisms and childhood asthma. N Engl J Med 364:701–709. 10.1056/NEJMoa1007302 [PubMed] [DOI] [PubMed] [Google Scholar]
- 122.Bisgaard H, Hermansen MN, Buchvald F, Loland L, Halkjaer LB, Bønnelykke K, Brasholt M, Heltberg A, Vissing NH, Thorsen SV, Stage M, Pipper CB. 2007. Childhood asthma after bacterial colonization of the airway in neonates. N Engl J Med 357:1487–1495. 10.1056/NEJMoa052632 [PubMed] [DOI] [PubMed] [Google Scholar]
- 123.Huang EY, Inoue T, Leone VA, Dalal S, Touw K, Wang Y, Musch MW, Theriault B, Higuchi K, Donovan S, Gilbert J, Chang EB. 2015. Using corticosteroids to reshape the gut microbiome: implications for inflammatory bowel diseases. Inflamm Bowel Dis 21:963–972. 10.1097/MIB.0000000000000332 [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gilligan PH. 2014. Infections in patients with cystic fibrosis: diagnostic microbiology update. Clin Lab Med 34:197–217. 10.1016/j.cll.2014.02.001 [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Dasenbrook EC, Checkley W, Merlo CA, Konstan MW, Lechtzin N, Boyle MP. 2010. Association between respiratory tract methicillin-resistant Staphylococcus aureus and survival in cystic fibrosis. JAMA 303:2386–2392. 10.1001/jama.2010.791 [DOI] [PubMed] [Google Scholar]
- 126.Martiniano SL, Nick JA. 2015. Nontuberculous mycobacterial infections in cystic fibrosis. Clin Chest Med 36:101–115. 10.1016/j.ccm.2014.11.003 [PubMed] [DOI] [PubMed] [Google Scholar]
- 127.van der Gast CJ, Walker AW, Stressmann FA, Rogers GB, Scott P, Daniels TW, Carroll MP, Parkhill J, Bruce KD. 2011. Partitioning core and satellite taxa from within cystic fibrosis lung bacterial communities. ISME J 5:780–791. 10.1038/ismej.2010.175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Willner D, Haynes MR, Furlan M, Schmieder R, Lim YW, Rainey PB, Rohwer F, Conrad D. 2012. Spatial distribution of microbial communities in the cystic fibrosis lung. ISME J 6:471–474. 10.1038/ismej.2011.104 [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Coburn B, Wang PW, Diaz Caballero J, Clark ST, Brahma V, Donaldson S, Zhang Y, Surendra A, Gong Y, Elizabeth Tullis D, Yau YC, Waters VJ, Hwang DM, Guttman DS. 2015. Lung microbiota across age and disease stage in cystic fibrosis. Sci Rep 5:10241. 10.1038/srep10241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Goddard AF, Staudinger BJ, Dowd SE, Joshi-Datar A, Wolcott RD, Aitken ML, Fligner CL, Singh PK. 2012. Direct sampling of cystic fibrosis lungs indicates that DNA-based analyses of upper-airway specimens can misrepresent lung microbiota. Proc Natl Acad Sci USA 109:13769–13774. 10.1073/pnas.1107435109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ghorbani P, Santhakumar P, Hu Q, Djiadeu P, Wolever TM, Palaniyar N, Grasemann H. 2015. Short-chain fatty acids affect cystic fibrosis airway inflammation and bacterial growth. Eur Respir J 46:1033–1045. 10.1183/09031936.00143614 [DOI] [PubMed] [Google Scholar]
- 132.Clark JA, Coopersmith CM. 2007. Intestinal crosstalk: a new paradigm for understanding the gut as the “motor” of critical illness. Shock 28:384–393. 10.1097/shk.0b013e31805569df [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Klingensmith NJ, Coopersmith CM. 2016. The gut as the motor of multiple organ dysfunction in critical illness. Crit Care Clin 32:203–212. 10.1016/j.ccc.2015.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ownby DR, Johnson CC, Peterson EL. 2002. Exposure to dogs and cats in the first year of life and risk of allergic sensitization at 6 to 7 years of age. JAMA 288:963–972. 10.1001/jama.288.8.963 [DOI] [PubMed] [Google Scholar]
- 135.von Mutius E, Vercelli D. 2010. Farm living: effects on childhood asthma and allergy. Nat Rev Immunol 10:861–868. 10.1038/nri2871 [PubMed] [DOI] [PubMed] [Google Scholar]
- 136.Bazett M, Bergeron ME, Haston CK. 2016. Streptomycin treatment alters the intestinal microbiome, pulmonary T cell profile and airway hyperresponsiveness in a cystic fibrosis mouse model. Sci Rep 6:19189. 10.1038/srep19189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Bazett M, Honeyman L, Stefanov AN, Pope CE, Hoffman LR, Haston CK. 2015. Cystic fibrosis mouse model-dependent intestinal structure and gut microbiome. Mamm Genome 26:222–234. 10.1007/s00335-015-9560-4 [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Segal LN, Blaser MJ. 2015. Harnessing the early-life microbiota to protect children with cystic fibrosis. J Pediatr 167:16–18.e11. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hoen AG, Li J, Moulton LA, O’Toole GA, Housman ML, Koestler DC, Guill MF, Moore JH, Hibberd PL, Morrison HG, Sogin ML, Karagas MR, Madan JC. 2015. Associations between gut microbial colonization in early life and respiratory outcomes in cystic fibrosis. J Pediatr 167:138–147.e1-3. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Harris B, Morjaria SM, Littmann ER, Geyer AI, Stover DE, Barker JN, Giralt SA, Taur Y, Pamer EG. 2016. Gut microbiota predict pulmonary infiltrates after allogeneic hematopoietic cell transplantation. Am J Respir Crit Care Med 194:450–463. 10.1164/rccm.201507-1491OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Deshmukh HS, Liu Y, Menkiti OR, Mei J, Dai N, O’Leary CE, Oliver PM, Kolls JK, Weiser JN, Worthen GS. 2014. The microbiota regulates neutrophil homeostasis and host resistance to Escherichia coli K1 sepsis in neonatal mice. Nat Med 20:524–530. 10.1038/nm.3542 [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Caballero S, Pamer EG. 2015. Microbiota-mediated inflammation and antimicrobial defense in the intestine. Annu Rev Immunol 33:227–256. 10.1146/annurev-immunol-032713-120238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Schuijt TJ, van der Poll T, de Vos WM, Wiersinga WJ. 2013. The intestinal microbiota and host immune interactions in the critically ill. Trends Microbiol 21:221–229. 10.1016/j.tim.2013.02.001 [DOI] [PubMed] [Google Scholar]
- 144.Schuijt TJ, Lankelma JM, Scicluna BP, de Sousa e Melo F, Roelofs JJ, de Boer JD, Hoogendijk AJ, de Beer R, de Vos A, Belzer C, de Vos WM, van der Poll T, Wiersinga WJ. 2016. The gut microbiota plays a protective role in the host defence against pneumococcal pneumonia. Gut 65:575–583. 10.1136/gutjnl-2015-309728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kumar P, Monin L, Castillo P, Elsegeiny W, Horne W, Eddens T, Vikram A, Good M, Schoenborn AA, Bibby K, Montelaro RC, Metzger DW, Gulati AS, Kolls JK. 2016. Intestinal interleukin-17 receptor signaling mediates reciprocal control of the gut microbiota and autoimmune inflammation. Immunity 44:659–671. 10.1016/j.immuni.2016.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Tanabe S. 2013. The effect of probiotics and gut microbiota on Th17 cells. Int Rev Immunol 32:511–525. 10.3109/08830185.2013.839665 [DOI] [PubMed] [Google Scholar]
- 147.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, Tanoue T, Imaoka A, Itoh K, Takeda K, Umesaki Y, Honda K, Littman DR. 2009. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139:485–498. 10.1016/j.cell.2009.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Suzuki K, Meek B, Doi Y, Muramatsu M, Chiba T, Honjo T, Fagarasan S. 2004. Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. Proc Natl Acad Sci USA 101:1981–1986. 10.1073/pnas.0307317101 [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lozupone C, Lladser ME, Knights D, Stombaugh J, Knight R. 2011. UniFrac: an effective distance metric for microbial community comparison. ISME J 5:169–172. 10.1038/ismej.2010.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Yadava K, Pattaroni C, Sichelstiel AK, Trompette A, Gollwitzer ES, Salami O, von Garnier C, Nicod LP, Marsland BJ. 2016. Microbiota promotes chronic pulmonary inflammation by enhancing IL-17A and autoantibodies. Am J Respir Crit Care Med 193:975–987. 10.1164/rccm.201504-0779OC [PubMed] [DOI] [PubMed] [Google Scholar]
- 151.McDermott AJ, Huffnagle GB. 2014. The microbiome and regulation of mucosal immunity. Immunology 142:24–31. 10.1111/imm.12231 [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Byrd AL, Segre JA. 2016. Infectious disease. Adapting Koch’s postulates. Science 351:224–226. 10.1126/science.aad6753 [DOI] [PubMed] [Google Scholar]
- 153.Koch R. 1952. Tuberculosis etiology. Dtsch Gesundheitsw 7:457–465. (In German.) [PubMed] [PubMed] [Google Scholar]
- 154.Fredricks DN, Relman DA. 1996. Sequence-based identification of microbial pathogens: a reconsideration of Koch’s postulates. Clin Microbiol Rev 9:18–33. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wommack KE, Ravel J. 2013. Microbiome, demystifying the role of microbial communities in the biosphere. Microbiome 1:1. 10.1186/2049-2618-1-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Dickson RP. 2016. The microbiome and critical illness. Lancet Respir Med 4:59–72. 10.1016/S2213-2600(15)00427-0 [DOI] [PMC free article] [PubMed] [Google Scholar]


