Abstract
Dihydrochalcomycin (1) and chalcomycin (2), two known chalcomycins, and chalcomycin E (3), a new compound, were isolated from marine-derived Streptomyces sp. HK-2006-1. Their structures were elucidated by detailed spectroscopic and X-ray crystallographic analysis. The antimicrobial activities against Staphylococcus aureus, Escherichia coli, Candida albicans, and Aspergillus niger of 1–3 were evaluated. Compounds 1–2 exhibited activities against S. aureus with minimal inhibitory concentrations (MICs) of 32 µg/mL and 4 µg/mL, respectively. The fact that 1–2 showed stronger activity against S. aureus 209P than 3 indicated that the epoxy unit was important for antimicrobial activity. This structure–activity tendency of chalcomycins against S. aureus is different from that of aldgamycins reported in our previous research, which provide a valuable example for the phenomenon that 16-membered macrolides with different sugars do not have parallel structure–activity relationships.
Keywords: marine-derived Streptomyces, secondary metabolite, 16-membered macrolide, chalcomycin E, antimicrobial activity
1. Introduction
Infectious diseases seriously imperil human health. Antibiotics are important medicines against infectious diseases [1]. However, the prolonged, extensive, and indiscriminate use of antibiotics has triggered widespread resistance [2]. The global epidemic of continually rising resistance has become a critical threat to human health and therefore the discovery of new antibiotics is urgently needed [2]. Macrolide antibiotics such as erythromycins, tylosins, avermectins, and milbemycins have significant activity against a broad spectrum of Gram-positive bacteria [3,4,5], playing an important role in the chemotherapy of infectious diseases [6,7]. Macrolides are usually characterized by a 12-, 14-, 16-, 18-, 20-, 22-, or 24-membered lactone ring with one or more sugar moieties [3,8]. Different types of macrolides have different structure-antimicrobial activity relationships. For example, 16-membered macrolides with different sugars have no parallel structure–activity tendencies. Omura reported that the structure–activity relationships of some 16-membered macrolides (rosamicins, angolamycins, and neutramycins) differed from the evidence found in other 16-membered macrolides (leucomycins) [9]. The 16-membered macrolides with different sugar moiety for instance spiramycins, neospiramycins, and forocidins have different structure–activity relationships [10].
Many interesting strains were obtained in our continuing investigations on active components from microorganisms. Among our recent discoveries [11,12,13,14,15,16,17,18,19,20,21], we recently reported that a strain of Streptomyces sp. HK-2006-1 produced both aldgamycins and chalcomycins, which are 16-membered macrolides [11,21]. Chalcomycin and seven aldgamycins were isolated from this strain, and chalcomycin showed more potent antibacteria activity against Staphylococcus aureus than aldgamycins [11]. Chalcomycin, the first member of chalcomycins, was reported with its activity against bacteria as early as 1962 [22]. However, there have only been seven chalcomycins (chalcomycin, chalcomycins B-D, dihydrochalcomycin, 8-deoxy-chalcomycin, 250-144C) reported until now [23,24,25,26,27], and there is no discussion on the structure-antimicrobial activity relationship of chalcomycins against S. aureus. Thus, in this study, the fermentation volume of this strain Streptomyces sp. HK-2006-1 was scaled up in search of more chalcomycins. The crude extract of the culture of the strain was subjected to column chromatography (CC) over silica gel, Sephadex LH-20, octadecylsilane (ODS), and high performance liquid chromatography (HPLC), yielding three chalcomycins, dihydrochalcomycin (1), chalcomycin (2), and a new compound, chalcomycin E (3) (Figure 1). In addition, their antimicrobial activities against two bacteria, Gram-positive S. aureus 209P and Gram-negative Escherichia coli ATCC0111, as well as two fungi, Candida albicans FIM709 and Aspergillus niger R330, were evaluated. Details of the isolation, structural elucidation, and antimicrobial activities of compounds 1–3 are presented herein.
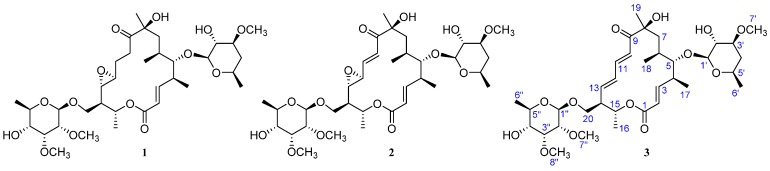
Figure 1.
The structures of compounds 1–3.
2. Results and Discussion
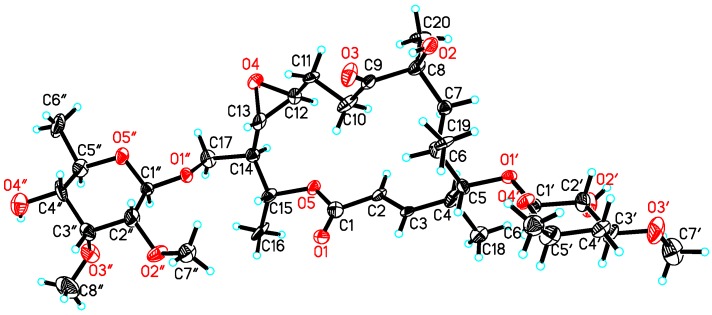
Compounds 1 and 2 were established as dihydrochalcomycin and chalcomycin respectively by precisely comparing the nuclear magnetic resonance (NMR) data with literature values [11,24,28]. The single-crystal X-ray crystallographic analysis of dihydrochalcomycin (1) was reported for the first time (Figure 2). Chalcomycin (2) was also obtained and identified in our previous study on the strain of Streptomyces sp. HK-2006-1 [11].
Figure 2.
X-ray structure of 1.
Compound 3 was obtained as a white amorphous powder. The quasi-molecular ion at m/z 707.3616 [M + Na]+ by high resolution electrospray ionization mass spectroscopy (HRESIMS) indicated that the molecular formula of 3 was C35H56O13 (eight degrees of unsaturation), which was 16 atomic mass unit (O) less than 2. Analysis of its 1H and 13C NMR spectroscopic data (Table 1) revealed nearly identical structure features to 2, except that two mono-oxygenated methine carbons at δC 59.0 and 58.7 disappeared, and two olefinic carbons at δC 143.3 and 133.0 appeared. Analysis of 1H−1H COSY and the coupling values of the protons revealed the presence of the spin system C-10−C-11−C-12−C-13−C-14(C-20)−C-15−C-16. Therefore, 3 was the reduction product of 2 at C-12/C-13. The geometrical configuration of the double bond moiety (C-12/C-13) was deduced as E on the basis of the coupling constant of the olefinic protons (J12,13 = 14.1 Hz). Thus, compound 3 can be recognized as a new member of the chalcomycin family, consisting of the 16-membered lactone ring, mycinose, and chalcose, and its structure was further confirmed by two-dimensional NMR (2D NMR) data (Table 1 and Table S1). The observed rotating frame overhauser effect spectroscopy (ROESY) correlations (Figure 3) were consistent with the stereochemistry of the 16-membered lactone ring. All the reported mycinose and chalcose units in natural products have D configurations. The mycinose and chalcose units in the isolated macrolides from the strain of Streptomycetes sp. HK-2006-1 also had D configurations [11]. Therefore, the absolute configurations of the mycinose and chalcose units in 3 were assumed to be D. The relative configurations of the two units were established as β from the coupling constants of the anomeric protons (H-1′ and H-1′′). Thus, the structure of 3 was elucidated as (3E,5S,6S,7S,9S,11E,13E,15R,16R)-9-hydroxy-15-(((2R,3R,4R,5R,6R)-5-hydroxy-3,4-dimethoxy-6-methyltetrahydro-2H-pyran-2-yloxy)methyl)-6-((2S,3R,4S,6R)-3-hydroxy-4-methoxy-6-methyltetrahydro-2H-pyran-2-yl)oxy)-5,7,9,16-tetramethyloxacyclohexadeca-3,11,13-triene-2,10-dione, and named as chalcomycin E.
Table 1.
NMR (600 MHz, CDCl3) data for 3.
| Position | δC, Mult. | δH (J in Hz) § | 1H, 1H-COSY | HMBC | ROESY |
|---|---|---|---|---|---|
| aglycone | |||||
| 1 | 165.6, C | – | – | – | – |
| 2 | 121.4, CH | 5.75 d (15.4) | 3 | 4 | 4, 17 |
| 3 | 151.7, CH | 6.62 dd (15.4, 9.5) | 2, 4 | 1 | 5, 6, 17 |
| 4 | 41.0, CH | 2.66 | 3, 5, 17 | 2, 3 | 2, 6, 7a, 7b |
| 5 | 88.1, CH | 3.19 | 4, 6 | 3, 4, 6, 7, 17, 18, 1′ | 3, 6, 17, 18, 1′ |
| 6 | 34.0, CH | 1.30 | 5, 7a, 7b, 18 | – | 3, 4, 5, 7a, 10 |
| 7 | 37.4, CH2 | 1.89, Ha | 6, 7b | 6, 8, 9, 18 | 4, 6, 10, 19 |
| 1.83, Hb | 6, 7a | 6, 18 | 4, 18, 19 | ||
| 8 | 78.3, C | – | – | – | – |
| 9 | 202.0, C | – | – | – | – |
| 10 | 122.0, CH | 6.18 d (15.1) | 11 | 8, 9, 11, 12 | 6, 7a, 19 |
| 11 | 144.1, CH | 7.30 dd (15.1, 10.1) | 10, 12 | 9, 12, 13 | – |
| 12 | 133.0, CH | 6.15 dd (14.1, 10.1) | 11, 13 | 10, 11, 13, 14 | – |
| 13 | 143.3, CH | 6.14 dd (14.1, 9.2) | 12, 14 | 11, 12, 14, 20 | 15, 20b |
| 14 | 51.2, CH | 2.47 | 13, 15, 20a, 20b | 12, 13, 15 | 16, 20a |
| 15 | 69.2, CH | 5.06 dq (10.2, 6.2) | 14, 16 | 1, 13, 14 | 13, 20a, 20b |
| 16 | 18.6, CH3 | 1.36 d (6.3) | 15 | 14, 15 | 14, 20a, 20b |
| 17 | 19.2, CH3 | 1.18 d (6.9) | 4 | 3, 4, 5 | 2, ,3, 5, 1′ |
| 18 | 19.3, CH3 | 1.00 d (6.9) | 6 | 5, 6, 7 | 5, 7b |
| 19 | 27.9, CH3 | 1.38 s | – | 7, 8, 9 | 7a, 7b, 10 |
| 20 | 68.4, CH2 | 4.04 dd (9.6, 3.7), Ha | 14, 20b | 13, 14, 15, 1″ | 14, 15, 16, 20b, 1″ |
| 3.57 dd (9.6, 6.1), Hb | 14, 20a | 13, 14, 15, 1″ | 13, 15, 16, 20a, 1″ | ||
| β-d-chalcose unit | |||||
| 1′ | 103.0, CH | 4.19 d (7.6) | 2′ | 5, 5′ | 5, 17, 3′, 5′ |
| 2′ | 75.1, CH | 3.32 dd (8.8, 7.6) | 1′, 3′ | 1′, 3′, 4′ | 4′b |
| 3′ | 80.4, CH | 3.22 | 2′, 4′a, 4′b | 1′, 2′, 4′, 7′ | 1′, 4′a, 5′ |
| 4′ | 36.8, CH2 | 2.04 ddd (12.7, 4.9, 1.9), Ha | 3′, 4′b, 5′ | 2′, 3′ | 3′, 5′, 6′ |
| 1.25, Hb | 3′, 4′a, 5′ | 2′, 3′, 5′ | 2′, 6′ | ||
| 5′ | 67.8, CH | 3.48 | 4′a, 4′b, 6′ | 1′ | 1′, 3′, 4′a |
| 6′ | 20.9, CH3 | 1.23 d (6.2) | 5′ | 4′, 5′ | 4′a, 4′b |
| 7′ | 56.7, CH3 | 3.41 s | – | 3′ | – |
| β-d-mycinose unit | |||||
| 1″ | 101.1, CH | 4.58 d (7.8) | 2″ | 20, 3″, 5″ | 20a, 20b, 5″, 8″ |
| 2″ | 81.9, CH | 3.04 dd (7.8, 3.1) | 1″, 3″ | 1″, 7″ | 3″, 4″, 7″ |
| 3″ | 79.8, CH | 3.76 t (3.1) | 2″, 4″ | 1″, 2″, 4″, 5″, 8″ | 2″, 4″, 8″ |
| 4″ | 72.7, CH | 3.18 | 3″, 5″ | 2″ | 2″, 3″, 6″ |
| 5″ | 70.6, CH | 3.52 | 4″, 6″ | 3″, 4″ | 1″ |
| 6″ | 17.8, CH3 | 1.27 d (6.2) | 5″ | 4″, 5″ | 4″ |
| 7″ | 59.8, CH3 | 3.52 s | – | 2″ | 2″ |
| 8″ | 61.8, CH3 | 3.62 s | – | 3″ | 1″, 3″ |
§ Indiscernible signals owing to overlapping or having complex multiplicity are reported without designating multiplicity. NMR: nuclear magnetic resonance; 1H, 1H COSY: 1H, 1H chemical shift correlated spectroscopy; HMBC: heteronuclear multiple-bond correlation; ROESY: rotating frame overhauser effect spectroscopy.
Figure 3.
The observed rotating frame overhauser effect spectroscopy (ROESY) correlations (dashed double arrow in blue) of C-4−C-5−C-6−C-7−C-8 and C-14−C-15 in 3.
Until now, only seven chalcomycins had been reported. The discovery of chalcomycin E (3) adds a new member to chalcomycins. The single-crystal X-ray crystallographic analysis of dihydrochalcomycin (1) was firstly reported. Compounds 1–3 were tested for antimicrobial activities against two bacteria, Gram-positive S. aureus 209P and Gram-negative E. coli ATCC0111, as well as two fungi, C. albicans FIM709 and A. niger R330 (Table 2). Compounds 1–2 showed activities against S. aureus, but no activity against the other test strains. The fact that 1 and 2 exhibited stronger activity against S. aureus 209P than 3 suggested that the epoxy unit was important for antimicrobial activity. However, the replacement of the double bond in C-10 to C-13 by the epoxy unit in aldgamycins is not beneficial for antimicrobial activity. The difference in structure between aldgamycins and chalcomycins is just the sugar type at C-5, but the two types of macrolides have different structure–activity tendencies. Our findings provide a valuable example for the phenomenon that 16-membered macrolide antibiotics with different sugars do not have parallel structure–activity relationships [9,10].
Table 2.
Antimicrobial activities of 1–3 (minimal inhibitory concentrations (MICs): µg/mL).
| Compound | Bacteria | Fungi | ||
|---|---|---|---|---|
| S. aureus | E. coli | C. albicans | A. niger | |
| 1 | 32 | >512 | >512 | >512 |
| 2 | 4 | >512 | >512 | >512 |
| 3 | >512 | >512 | >512 | >512 |
| Tobramycin | 0.4 | 2 | NT | NT |
| Actidione | NT | NT | 64 | 32 |
NT: not tested.
3. Conclusions
Two known chalcomycins, dihydrochalcomycin (1) and chalcomycin (2), together with a new one, chalcomycin E (3) were isolated from marine-derived Streptomyces sp. HK-2006-1. Their structures were determined by detailed spectroscopic and X-ray crystallographic analysis. The discovery of chalcomycin E (3) adds a new member to chalcomycins. The antimicrobial activities of 1–3 were tested against S. aureus, E. coli, C. albicans, and A. niger. Compounds 1–2 showed activities against S. aureus with minimal inhibitory concentrations (MICs) of 32 µg/mL and 4 µg/mL, respectively. Compounds 1–2 showed stronger activity against S. aureus 209P than 3, which suggested a different structure–activity tendency against S. aureus from that of aldgamycins. This case indicated that 16-membered macrolide antibiotics with different sugars do not have parallel structure–activity relationships.
Acknowledgments
This work was financially supported by grants from the National Natural Science Foundation of China (81373306, 81422054), Fok Ying Tung Education Foundation (121039) from the Ministry of Education of China, the Guangdong Natural Science Funds for Distinguished Young Scholar (S2013050014287), Guangdong Special Support Program (2016TX03R280), Guangdong Province Universities and Colleges Pearl River Scholar Funded Scheme (Hao Gao, 2014), Wong Kwan Cheng Education Foundation (Hao Gao, 2016), and “Challenge Cup” National Undergraduate Curricular Academic Science and Technology Works of Jinan University (16112001).
Supplementary Materials
The following are available online at www.mdpi.com/1660-3397/15/6/153/s1, materials and methods, one-dimensional NMR (1D NMR) data and spectra for 1 and 2, and 1D/2D NMR, ultraviolet (UV), and HRESIMS spectra for 3.
Author Contributions
H.G. and X.Y. initiated and coordinated the project. H.G. and C.W. wrote the paper. S.J., L.Z., and C.W. performed the extraction, isolation, and structural identification of the compounds. K.H. supplied the strain. K.H. and D.H. performed the identification of the strain. G.C. performed the X-ray crystallographic analysis. X.P. and F.D. performed the fermentation of the strain.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Aminov R. History of Antimicrobial Drug Discovery: Major Classes and Health Impact. Biochem. Pharmacol. 2017;133:4–19. doi: 10.1016/j.bcp.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 2.Chaudhary A.S. A Review of Global Initiatives to Fight Antibiotic Resistance and Recent Antibiotics Discovery. Acta Pharm. Sin. B. 2016;6:552–556. doi: 10.1016/j.apsb.2016.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gamerdinger M., Deuerling E. Macrolides: The Plug Is Out. Cell. 2012;151:469–471. doi: 10.1016/j.cell.2012.10.015. [DOI] [PubMed] [Google Scholar]
- 4.Kwiatkowska B., Maslinska M. Macrolide Therapy in Chronic Inflammatory diseases. Mediat. Inflamm. 2012;2012:1–7. doi: 10.1155/2012/636157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hansen J.L., Ippolito J.A., Ban N., Nissen P., Moore P.B., Steitz T.A. The Structures of Four Macrolide Antibiotics Bound to the Large Ribosomal Subunit. Mol. Cell. 2002;10:117–128. doi: 10.1016/S1097-2765(02)00570-1. [DOI] [PubMed] [Google Scholar]
- 6.Blondeau J.M., DeCarolis E., Metzler K.L., Hansen G.T. The macrolides. Expert Opin. Investig. Drugs. 2002;11:189–215. doi: 10.1517/13543784.11.2.189. [DOI] [PubMed] [Google Scholar]
- 7.Zuckerman J.M., Qamar F., Bono B.R. Review of Macrolides (Azithromycin, Clarithromycin), Ketolids (Telithromycin) and Glycylcyclines (Tigecycline) Med. Clin. N. Am. 2011;95:761–791. doi: 10.1016/j.mcna.2011.03.012. [DOI] [PubMed] [Google Scholar]
- 8.Elshahawi S.I., Shaaban K.A., Kharel M.K., Thorson J.S. A comprehensive review of glycosylated bacterial natural products. Chem. Soc. Rev. 2015;44:7591–7697. doi: 10.1039/C4CS00426D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Omura S., Nakagawa A. Chemical and biological studies on 16-membered macrolide antibiotics. J. Antibiot. 1975;28:401–433. doi: 10.7164/antibiotics.28.401. [DOI] [PubMed] [Google Scholar]
- 10.Corcoran J.W., Hahn F.E. Antibiotics, Vol. 3: Mechanism of Action of Antimicrobial and Antitumor Agents. Springer; Berlin/Heidelberg, Germany: 1975. pp. 459–479. [Google Scholar]
- 11.Wang C.X., Ding R., Jiang S.T., Tang J.S., Hu D., Chen G.D., Lin F., Hong K., Yao X.S., Gao H. Aldgamycins J–O, 16-membered Macrolides with a Branched Octose Unit from Streptomycetes sp. and their Antibacterial Activities. J. Nat. Prod. 2016;79:2446–2454. doi: 10.1021/acs.jnatprod.6b00200. [DOI] [PubMed] [Google Scholar]
- 12.Zhao H., Chen G.D., Zou J., He R.R., Qin S.Y., Hu D., Li G.Q., Guo L.D., Yao X.S., Gao H. Dimericbiscognienyne A: A meroterpenoid dimer from Biscogniauxia sp. with new skeleton and its activity. Org. Lett. 2017;19:38–41. doi: 10.1021/acs.orglett.6b03264. [DOI] [PubMed] [Google Scholar]
- 13.Sun T.Y., Zou J., Chen G.D., Hu D., Wu B., Liu X.Z., Yao X.S., Gao H. A set of interesting sequoiatones stereoisomers from a wetland soil-derived fungus Talaromyces flavus. Acta Pharm. Sin. B. 2017 doi: 10.1016/j.apsb.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang C.X., Chen G.D., Feng C.C., He R.R., Qin S.Y., Hu D., Chen H.R., Liu X.Z., Yao X.Z., Gao H. Same Data, different structures: diastereoisomers with substantiallyidentical NMR data from nature. Chem. Commun. 2016;52:1250–1253. doi: 10.1039/C5CC07141K. [DOI] [PubMed] [Google Scholar]
- 15.Gao Y.M., Sun T.Y., Ma M., Chen G.D., Zhou Z.Q., Wang C.X., Hu D., Chen L.G., Yao X.S., Gao H. Adeninealkylresorcinol, the first alkylresorcinol tethered with nucleobase from Lasiodiplodia sp. Fitoterapia. 2016;112:254–259. doi: 10.1016/j.fitote.2016.06.011. [DOI] [PubMed] [Google Scholar]
- 16.Sun T.Y., Kuang R.Q., Chen G.D., Qin S.Y., Wang C.X., Hu D., Wu B., Liu X.Z., Yao X.S., Gao H. Three pairs of new isopentenyl dibenzo[b,e]oxepinone enantiomers from Talaromyces flavus, a wetland soil-derived fungus. Molecules. 2016;21:1184. doi: 10.3390/molecules21091184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He J.W., Wang C.X., Yang L., Chen G.D., Hu D., Guo L.D., Yao X.S., Gao H. A pair of new polyketide enantiomers from three endolichenic fungal strains Nigrospora sphaerica, Alternaria alternata, and Phialophora sp. Nat. Prod. Commun. 2016;11:829–831. [PubMed] [Google Scholar]
- 18.Zhao Q., Wang C.X., Yu Y., Wang G.Q., Zheng Q.C., Chen G.D., Lian Y.Y., Lin F., Guo L.D., Gao H. Nodulisporipyrones A-D, new bioactive α-pyrone derivatives from Nodulisporium sp. J. Asian Nat. Prod. Res. 2015;17:567–575. doi: 10.1080/10286020.2015.1040776. [DOI] [PubMed] [Google Scholar]
- 19.Zou J., Li J., Wu Z.Y., Zhao Q., Wang G.Q., Zhao H., Chen G.D., Sun X., Guo L.D., Gao H. New α-pyrone and phthalide from the Xylariaceae fungus. J. Asian Nat. Prod. Res. 2015;17:705–710. doi: 10.1080/10286020.2015.1054816. [DOI] [PubMed] [Google Scholar]
- 20.Wu Y.H., Chen G.D., Wang C.X., Hu D., Li X.X., Lian Y.Y., Lin F., Guo L.D., Gao H. Pericoterpenoid A, a new bioactive cadinane-type sesquiterpene from Periconia sp. J. Asian Nat. Prod. Res. 2015;17:671–675. doi: 10.1080/10286020.2015.1049162. [DOI] [PubMed] [Google Scholar]
- 21.Tang X.L., Dai P., Gao H., Wang C.X., Chen G.D., Hong K., Hu D., Yao X.S. A Single Gene Cluster for Chalcomycins and Aldgamycins: Genetic Basis for Bifurcation of Their Biosynthesis. ChemBioChem. 2016;17:1241–1249. doi: 10.1002/cbic.201600118. [DOI] [PubMed] [Google Scholar]
- 22.Frohardt R.P., Pittillo R.F., Ehrlich J. Chalcomycin and Its Fermentative Production. US 3065137. U.S. Patent. 1962 Nov 20;
- 23.Hauske J.R., Dibrino J., Guadliana M., Kostek G. Structure Elucidation of a New Neutral Macrolide Antibiotic. J. Org. Chem. 1986;51:2808–2814. doi: 10.1021/jo00364a037. [DOI] [Google Scholar]
- 24.Kim S.D., Ryoo I.J., Kim C.J., Kim W.G., Kim J.P., Kong J.Y., Koshino H., Uramoto M., Yoo I.D. GERI-155, a New Macrolide Antibiotic Related to Chalcomycin. J. Antibiot. 1996;49:955–957. doi: 10.7164/antibiotics.49.955. [DOI] [PubMed] [Google Scholar]
- 25.Goo Y.M., Lee Y.Y., Kim B.T. A New 16-membered Chalcomycin Type Macrolide Antibiotic, 250-144C. J. Antibiot. 1997;50:85–88. doi: 10.7164/antibiotics.50.85. [DOI] [PubMed] [Google Scholar]
- 26.Asolkar R.N., Maskey R.P., Helmke E., Laatsch H. Chalcomycin B, a New Macrolide Antibiotic from the Marine Isolate Streptomyces sp. B7064. J. Antibiot. 2002;55:893–898. doi: 10.7164/antibiotics.55.893. [DOI] [PubMed] [Google Scholar]
- 27.Ding L., Qin S., Li F., Zhang W., Lachi H. Method for Preparation of Chalcomycin-like Compounds from Marine Streptomyces and Its Application as Antitumor Agents. CN 101624413. C.N. Patent. 2010
- 28.Morisaki N., Hashimoto Y., Furihata K., Yazawa K., Tamura M., Mikami Y. Glycosylative Inactivation of Chalcomycin and Tylosin by a Clinically Isolated Nocardia asteroides Strain. J. Antibiot. 2001;54:157–165. doi: 10.7164/antibiotics.54.157. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.