Abstract
Structural maintenance of chromosomes (SMC) complexes play critical roles in chromosome dynamics in virtually all organisms but how they function remains poorly understood. In Bacillus subtilis, SMC condensin complexes are topologically loaded at centromeric sites adjacent to the replication origin. Here we provide evidence that these ring-shaped assemblies tether the left and right chromosome arms together while traveling from the origin to the terminus (>2 Mb) at rates >50kb/min. Condensin movement scales linearly with time arguing for an active transport mechanism. These data support a model in which SMC complexes function by processively enlarging DNA loops. Loop formation followed by processive enlargement provides a mechanism for how condensin complexes compact and resolve sister chromatids in mitosis and how cohesin generates topologically associating domains (TADs) during interphase.
Keywords: SMC, ParB, condensin, cohesion, loop extrusion, TAD
Recent chromosome conformation capture (Hi-C) studies (1-4), and polymer simulations (2, 5-7) have reinvigorated a model proposed over a decade ago (8) in which SMC complexes generate DNA loops via processive loop enlargement (also referred to as loop extrusion) (5, 6). In this model, these ring-shaped assemblies encircle the DNA flanking their loading site, tethering the DNA duplexes together. As these tethers move away from their loading site they generate loops. Moreover, if rings are continuously loaded at the same site then the DNA duplexes within the loop segment become juxtaposed (Fig. 1E). De novo loop generation along chromosome arms provides a simple solution to how condensin complexes could compact and resolve replicated chromosomes into rod-shaped structures during mitosis and a mechanism for the formation of TADs by the SMC cohesin complex during interphase (3, 6, 9). Although compelling in its simplicity, this model remains largely untested.
Fig. 1. DNA juxtaposition propagates from condensin's loading site.
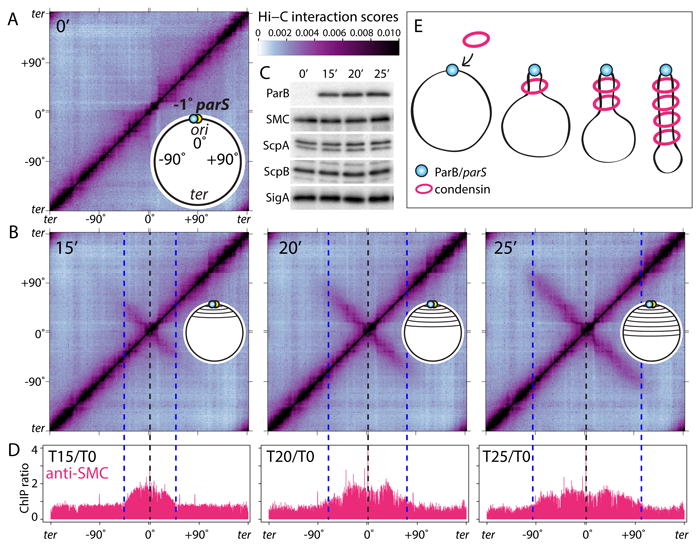
(A-B) Normalized Hi-C interaction maps displaying contact frequencies for pairs of 10 kb bins. The strain (BWX3352) contains a single parS site at -1° and an IPTG-inducible gfp-parB fusion. Maps show interaction frequencies before (A) and after induction (B) with IPTG. Axes present genome positions in degrees and are oriented with the replication origin at the center. Schematics of the juxtaposed regions are shown. The scale bar depicts Hi-C interaction scores for all contact maps presented in this study. The dotted lines indicate the -1° parS site (black) and the leading edge of the juxtaposed DNA (blue). (C) Immunoblot analysis of the same samples from (A) showing GFP-ParB accumulation, the levels of the condensin complex (SMC, ScpA, ScpB) and SigA to control for loading. (D) Anti-SMC ChIP-seq performed under the same condition as in (A-B). Sequencing reads from ChIP and input samples were normalized to the total number of reads. The ratio of ChIP enrichment (ChIP/Input) at each time point relative to time 0 is shown in 1 kb bins. (E) Schematic interpretation of the Hi-C and ChIP-seq data. Progressive juxtaposition enlarges a DNA loop centered on parS.
In the bacterium Bacillus subtilis, the SMC condensin complex is required for the resolution and segregation of newly replicated sister origins (10, 11). Condensin is recruited to the origin by the broadly conserved partitioning protein ParB bound to centromeric parS sites located adjacent to the origin (12-14). Like its eukaryotic counterparts, B. subtilis condensin encircles chromosomal DNA in vivo and topological entrapment is strongly reduced in the absence of ParB, suggesting that most condensin is loaded onto the chromosome at parS sites (15). Recent Hi-C studies in B. subtilis revealed that recruitment of condensin to origin-proximal parS sites is required for the alignment of the left and right chromosome arms (1, 4). However, the mechanism by which condensin promotes the juxtaposition of DNA flanking its loading site remains unknown.
To investigate whether condensin-dependent DNA juxtaposition (or “zip-up”) initiates at parS and progresses down the flanking DNA, we used a strain with a single origin-proximal parS site (at -1°) that harbors an IPTG-inducible allele of parB as the sole source of the loader, enabling us to follow zip-up dynamics. Inter-arm interactions were monitored by Hi-C at 5-minute intervals after ParB induction (Fig. 1A-C). Prior to the addition of IPTG, interactions between the left and right chromosome arms were undetectable (Fig. 1A) as observed previously in the absence of ParB (1, 4). Fifteen minutes after ParB induction, ∼500 kb of DNA on either side of parS were juxtaposed and 5 minutes later the juxtaposition had progressed ∼290 kb further down the left and right arms. Analysis of all seven Hi-C time points indicated that the two arms zipped-up at a nearly constant rate of 52±5 kb/min (Fig. S1, S2). ChIP-seq using anti-SMC antibodies before and after ParB induction revealed modest SMC enrichment that correlated with the extent of inter-arm interaction observed by Hi-C (Fig. 1D, S1). These results and previous studies (1, 4, 15, 16) suggest that condensin is loaded at parS and then progressively accumulates along the flanking DNA (Fig. 1E, S3A-C).
Condensin promotes the juxtaposition of large tracks of DNA flanking ectopic parS sites (4). To determine whether SMC was specifically enriched along these juxtaposed regions, we performed Hi-C and ChIP-seq on four strains each with an ectopic parS site at a different position along the left chromosome arm (Fig. 2, S4). The Hi-C contact maps indicated that DNA flanking the ectopic parS sites interacted giving characteristic zip-up patterns (Fig. 2A, S4A). As observed previously, the zip-ups were asymmetric containing more terminus-proximal than origin-proximal DNA (discussed below). Importantly, the ChIP-seq profiles revealed strong SMC enrichment along the DNA flanking the parS sites that correlated with the extent of juxtaposition (Fig. 2B, S4B, S5, S6AC). ChIP-seq using antibodies against the other two subunits of the condensin complex (ScpA and ScpB) showed similar enrichment profiles (Fig. S4D) while ParB enrichment was limited to small chromosomal regions (12-23kb) centered on parS (12) (Fig. S4C).
Figure 2. Condensin is specifically enriched along juxtaposed DNA.
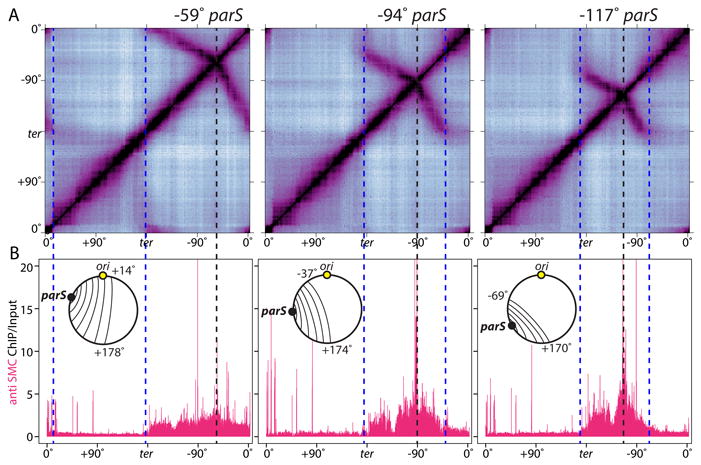
(A) Hi-C contact maps of strains harboring single parS sites at -59° (BWX3377), -94° (BWX3270), and -117° (BWX3381). To visualize interactions in the terminus region, the genome was oriented with the terminus (ter) at the center of the maps. The position of the parS sites and the extent of DNA juxtaposition are indicated by black and blue dotted lines, respectively. (B) Anti-SMC ChIP-seq was performed on the same samples as in (A). ChIP enrichment (ChIP/Input) was plotted in 1 kb bins. Schematics of the juxtaposed regions are shown. SMC enrichment at the highly transcribed genes outside the juxtaposed regions is probably nonspecific (see Figure S3).
For unknown reasons, regardless of where the ectopic parS site was inserted along the left or right arm, the zip-up did not extend beyond a ∼170 kb region surrounding the replication terminus (Fig. 2A, S4A, S7). Examination of SMC enrichment in the ChIP-seq profiles indicated that condensin did not appreciably accumulate in this region (Fig. 2B, S4B, S6A), suggesting the complexes were actively dissociated. Similarly, SMC did not accumulate where enrichment ended on the origin-proximal side of parS (Fig. 2B, S4B, S6A). The mechanism by which condensin is released from the DNA is currently unknown. A comparison of the Hi-C contact maps from the four strains revealed that the greater the distance between the ectopic parS site and the terminus region, the greater the extent of juxtaposition from parS toward the origin (Fig. 2, S4AB, S5AC). Similarly, the distance between the ectopic parS and the terminus region correlated with the extent of SMC enrichment from the parS toward the origin. These data indicate that condensin on one side of parS is influenced by condensin on the other. The simplest explanation for this “communication” between complexes separated by 100s of kilobases is that condensin functions as a tether holding the juxtaposed DNA duplexes together. In the context of this model, removal of condensin at the terminus also dissociates condensin at its juxtaposed position on the origin-proximal side of parS (Fig. S6D).
How condensin accumulates along the DNA flanking parS is not known. In one scenario condensin rings topologically loaded at parS travel the length of the chromosome arms. Alternatively, after loading, the condensin tether might not travel far away from parS (17), but, by aligning the two arms, generates a template for loading new condensin complexes further down the arms (5) (Fig. S8B). To distinguish between these models we used a strain with a wild-type copy of smc and an IPTG-inducible gfp-smc fusion and monitored where newly synthesized GFP-SMC accumulated along the DNA (Fig. 3). Because SMC enrichment along the chromosome arms was difficult to detect when loaded at origin-proximal parS sites (Fig. 1D, S1), we used a strain harboring an ectopic parS at -94°. We performed ChIP-seq using anti-GFP antibodies on an asynchronously growing culture prior to induction and at 5 min intervals after the addition of IPTG. Ten minutes after induction, GFP-SMC was specifically enriched along a ∼310 kb region surrounding parS (Fig. 3B, S8AD). Five minutes later, GFP-SMC occupied a larger expanse of DNA (∼650 kb) flanking parS. Enrichment along this region was greater than at the previous time point (Fig. 3B, S8AD), which correlated with the increase in GFP-SMC protein levels (Fig. 3A, S8C). However, enrichment was virtually undetectable at sites outside this 650 kb region. Ten minutes later, the enrichment profile largely overlapped the profile of untagged SMC prior to induction (Fig. S8A). The rate of GFP-SMC accumulation from parS toward the terminus was 46±5 kb/min (Fig. 3C), similar to the rate at which the left and right arms became juxtaposed (Fig. S2B). Interestingly, the rate of GFP-SMC accumulation from parS towards the origin was slower (26±2 kb/min), consistent with the asymmetric juxtaposition observed by Hi-C (Fig. 2A). Altogether, these results are consistent with a model in which condensin rings are loaded at parS and then travel down the flanking DNA tethering the duplexes together.
Figure 3. Condensin complexes loaded at parS travel down the flanking DNA.
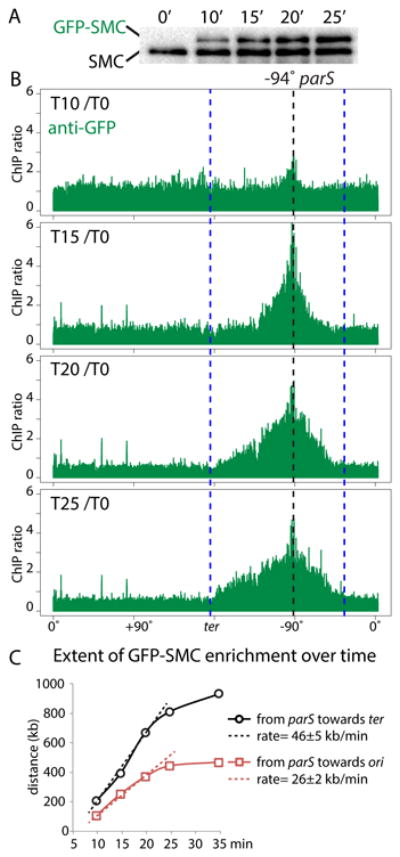
Cells (BWX3690) harboring a single parS site at -94° with wild-type SMC and an IPTG-inducible gfp-smc fusion we analyzed before an after induction. (A) Immunoblot analysis of GFP-SMC levels (using anti-SMC antibodies) during the induction time course. (B) ChIP-seq enrichment using anti-GFP antibodies. The ratio of ChIP enrichment at the indicated time point relative to time 0 (before induction) is plotted in 1 kb bins. Dotted lines indicate the position of the parS site (black) and the extent of ChIP enrichment of untagged SMC at time 0 (blue). (C) The accumulation of GFP-SMC from the parS towards the origin (red squares) and terminus (black circles) are plotted. The rates of GFP-SMC accumulation were calculated using the first three time points. See Figure S8 for unprocessed data and additional analysis.
In the context of this model, condensin movement is likely to be influenced by DNA transactions encountered along its path. We investigated the impact of convergent transcription on ring movement by analyzing the zip-up generated by a parS site at +26°. The ∼300 kb region between +26° and the origin contains many highly transcribed genes that are co-oriented with replication, including an 80 kb region between +6° and +15° that contains 5 ribosomal RNA operons and a ∼35 kb operon that encodes abundant ribosomal proteins and translation factors (18) (Fig. S9). For comparison, we analyzed the zip-up generated from a parS site at -27°. The region between -27° and the origin has few highly transcribed genes (18) (Fig. S9). The zip-up generated by the +26° parS was significantly more asymmetric than the one generated by the -27° parS (Fig. 4A). Specifically, the 80 kb region between +6° and +15° interacted with >500 kb of DNA on the terminus-proximal side of the +26° parS. This asymmetry suggests that movement of condensin rings through the convergently transcribed genes is impaired while movement along the DNA on the terminus-proximal side of parS is not. Condensin complexes were ultimately able to traverse this region and continue juxtaposing origin- and terminus-proximal DNA. Once condensin crossed the origin its movement along the DNA was co-oriented with the transcription of most genes (18) and the resulting zip-up was symmetric.
Figure 4. Highly transcribed genes influence DNA juxtaposition.
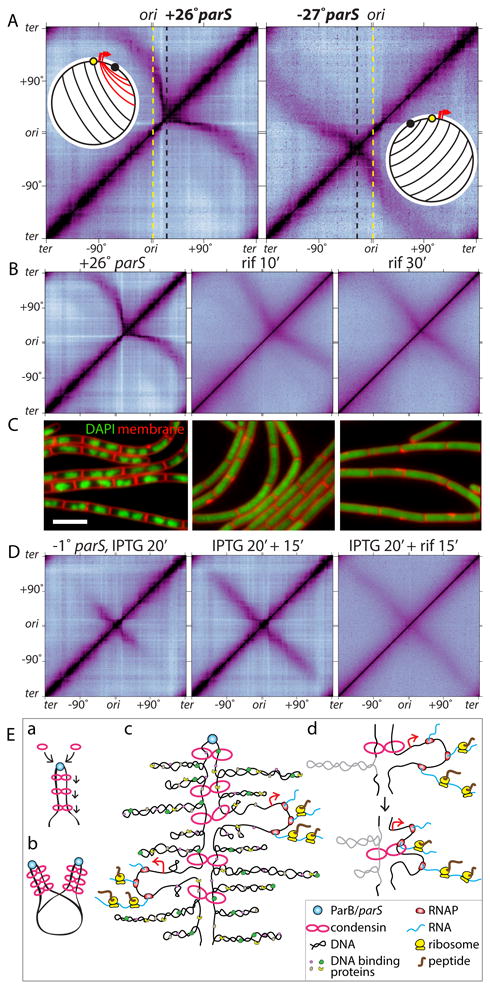
(A) Hi-C contact maps of strains harboring a single parS site at +26° (BWX3403) or -27° (BWX3268). The maps are oriented with the origin at the center of the axes. Dotted lines indicate the positions of the origin (yellow) and the parS sites (black). Schematics show DNA juxtaposition. Red lines highlight the asymmetric juxtaposition adjacent to the highly transcribed genes (red arrows). (B) Hi-C contact maps of the strain harboring the +26° parS site (BWX3403) in the absence or presence of 25 μg/ml rifampicin (rif) for 10 and 30 min. (C) Representative images of cells from (B). DAPI-stained DNA (green) and membranes (red) are shown. Bar, 4 μm. (D) Hi-C contact maps of a strain (BWX3352) harboring a single parS site at -1° and gfp-parB under IPTG control. Cells were induced for 20 min with IPTG, then treated with or without 25 μg/ml rifampicin for 15 min. (E) Schematic model for condensin function. (a) Condensin rings topologically loaded by ParB bound to parS travel down the flanking DNA as handcuffs, (b) resolving newly replicated sister origins by processive loop enlargement. Alternatively, a single ring composed of one or more condensin complexes could encircle the DNA on both sides of parS (not shown). (c) Condensin tethers encounter supercoiled plectonemes, DNA binding proteins, RNA polymerase, and ribosomes translating nascent transcripts. (d) Schematic model of encounters between tethered rings and either a plectoneme (gray) or a transcription complex.
To more directly test whether transcription influences DNA juxtaposition, we treated the cells containing the +26° parS site with the transcription inhibitor rifampicin and monitored the impact by Hi-C (Fig. 4B). As observed previously in Caulobacter crescentus (19), short-range interactions along the chromosome arms were rapidly lost upon inhibition of transcription, while the interactions between the DNA flanking the parS site became progressively more symmetric (Fig. 4B). To investigate whether condensin complexes loaded after transcription inhibition were responsible for the shift to a symmetric zip-up, we used the IPTG-inducible parB strain described in Figure 1 and added rifampicin 20 minutes after induction of ParB. As can be seen in Figure 4D, DNA juxtaposition continued unabated after transcription was inhibited. The rate of zip-up appeared to be slightly faster in the presence of drug than in its absence suggesting that transcription along the arms also affects the rate of condensin movement from the origin. As reported previously in E. coli (20), inhibition of transcription caused a rapid and pronounced decompaction of B. subtilis chromosomes (Fig. 4C). That the DNA flanking the +26° parS site remained juxtaposed and the zip-up continued unabated after decompaction provide additional evidence that condensin physically tethers the DNA duplexes. Altogether, these results indicate that transcription influences condensin-mediated DNA juxtaposition and provide additional support for the idea that condensin rings traverse the flanking DNA as they tether the duplexes together.
Finally, to assess the rate of juxtaposition away from the +26° parS as a proxy for condensin movement, we followed zip-up progression by Hi-C after induction of parB, as described in Figure 1 (Fig. S10, S11). The rate of DNA incorporated into the zip-up from parS heading toward the terminus was 50±5 kb/min (Fig. S10B, S11), similar to the rate observed for a parS site adjacent to the origin (52±5 kb/min). The rate of DNA incorporated into the zip-up heading toward the origin appeared biphasic (Fig. S10B, S11). The zip-up from parS through the highly transcribed genes was slow (∼8 kb/min) but once the juxtaposition traversed this region, the rate (50±4 kb/min) was similar to the zip-up of the terminus-proximal DNA. These data suggest that convergent transcription on one DNA duplex impairs condensin movement along this track of DNA but does not influence the rate of condensin movement on the partner DNA duplex. A similar explanation can account for the differential rates of GFP-SMC movement on either side of the -94° parS (Fig. 3C). These results and the asymmetric enrichment of SMC on either side of the ectopic parS sites (Fig. 2B, S6B) are consistent with a model in which two condensin rings each encircle a DNA duplex on either side of parS and tether them together by handcuffing (Fig. 4E, S6D).
In B. subtilis, SMC complexes are required to segregated newly replicated origins (10, 11). Our data provide support for the idea that these ring-shaped complexes resolve replicated origins by processive loop enlargement (Fig. 4E). Loop formation draws contiguous DNA in on itself and away from non-contiguous DNA and facilitates the removal of entanglements by Topoisomerase IV (21). It can also explain how a limited number of condensin complexes in B. subtilis (∼30 per replication origin) (15) can organize such large expanses of DNA. Our data suggest that processive loop enlargement underlies condensin's role in resolving replicated chromosomes in all organism and support the idea that eukaryotic cohesin complexes act similarly to generate TADs. How B. subtilis condensin rings move down the DNA is unknown. However, condensin's progression away from the parS site scaled linearly with time (Fig. 3C, S2, S10B), consistent with an active transport mechanism. While the prevailing view is that the ATPase cycle of SMC complexes is critical for topologically loading onto DNA, its role in providing energy for DNA translocation remains largely untested (5, 8, 16, 22, 23). Biophysical studies on B. subtilis condensin suggest that the ATPase cycle induces large structural transitions from rod- to ring-like conformations (24) and could represent a mechanochemical cycle that mediates DNA translocation (16). This structural transition could also help explain how condensin is able to thread large nucleoprotein complexes through its pore. Finally, our data suggesting that each condensin ring encircles a single DNA duplex and therefore each complex acts on a single helix provide a simple model in which tethered condensin motors extrude DNA loops.
Supplementary Material
Acknowledgments
Hi-C and ChIP-seq data were deposited in the Gene Expression Omnibus (accession no. GSE85612). We thank members of the Rudner and Bernhardt labs and J. Loparo for stimulating discussions, K. Marquis, C. Morlot, A. Grossman for antibodies, S. Dove and K. Ramsey for advice on ChIP-seq. Support for this work comes from the National Institute of Health Grants GM086466 and GM073831 (D.Z.R.), and GM082899 (M.T.L). H.B.B. was supported by a Natural Sciences and Engineering Research Council of Canada Fellowship. M.T.L. is an investigator of the Howard Hughes Medical Institute.
References
- 1.Marbouty M, et al. Condensin- and Replication-Mediated Bacterial Chromosome Folding and Origin Condensation Revealed by Hi-C and Super-resolution Imaging. Mol Cell. 2015;59:588–602. doi: 10.1016/j.molcel.2015.07.020. [DOI] [PubMed] [Google Scholar]
- 2.Naumova N, et al. Organization of the mitotic chromosome. Science. 2013;342:948–953. doi: 10.1126/science.1236083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sanborn AL, et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc Natl Acad Sci U S A. 2015;112:E6456–6465. doi: 10.1073/pnas.1518552112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang X, et al. Condensin promotes the juxtaposition of DNA flanking its loading site in Bacillus subtilis. Genes Dev. 2015;29:1661–1675. doi: 10.1101/gad.265876.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alipour E, Marko JF. Self-organization of domain structures by DNA-loop-extruding enzymes. Nucleic Acids Res. 2012;40:11202–11212. doi: 10.1093/nar/gks925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dekker J, Mirny L. The 3D Genome as Moderator of Chromosomal Communication. Cell. 2016;164:1110–1121. doi: 10.1016/j.cell.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goloborodko A, Imakaev MV, Marko JF, Mirny L. Compaction and segregation of sister chromatids via active loop extrusion. eLife. 2016;5 doi: 10.7554/eLife.14864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nasmyth K. Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu Rev Genet. 2001;35:673–745. doi: 10.1146/annurev.genet.35.102401.091334. [DOI] [PubMed] [Google Scholar]
- 9.Fudenberg G, et al. Formation of Chromosomal Domains by Loop Extrusion. Cell reports. 2016;15:2038–2049. doi: 10.1016/j.celrep.2016.04.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gruber S, et al. Interlinked Sister Chromosomes Arise in the Absence of Condensin during Fast Replication in B. subtilis. Curr Biol. 2014;24:293–298. doi: 10.1016/j.cub.2013.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X, Tang OW, Riley EP, Rudner DZ. The SMC condensin complex is required for origin segregation in Bacillus subtilis. Curr Biol. 2014;24:287–292. doi: 10.1016/j.cub.2013.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Breier AM, Grossman AD. Whole-genome analysis of the chromosome partitioning and sporulation protein Spo0J (ParB) reveals spreading and origin-distal sites on the Bacillus subtilis chromosome. Mol Microbiol. 2007;64:703–718. doi: 10.1111/j.1365-2958.2007.05690.x. [DOI] [PubMed] [Google Scholar]
- 13.Gruber S, Errington J. Recruitment of condensin to replication origin regions by ParB/SpoOJ promotes chromosome segregation in B. subtilis. Cell. 2009;137:685–696. doi: 10.1016/j.cell.2009.02.035. [DOI] [PubMed] [Google Scholar]
- 14.Sullivan NL, Marquis KA, Rudner DZ. Recruitment of SMC by ParB-parS organizes the origin region and promotes efficient chromosome segregation. Cell. 2009;137:697–707. doi: 10.1016/j.cell.2009.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilhelm L, et al. SMC condensin entraps chromosomal DNA by an ATP hydrolysis dependent loading mechanism in Bacillus subtilis. eLife. 2015;10 doi: 10.7554/eLife.06659. 7554/eLife.06659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Minnen A, et al. Control of Smc Coiled Coil Architecture by the ATPase Heads Facilitates Targeting to Chromosomal ParB/parS and Release onto Flanking DNA. Cell reports. 2016;14:2003–2016. doi: 10.1016/j.celrep.2016.01.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stigler J, Camdere GO, Koshland DE, Greene EC. Single-Molecule Imaging Reveals a Collapsed Conformational State for DNA-Bound Cohesin. Cell reports. 2016;15:988–998. doi: 10.1016/j.celrep.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nicolas P, et al. Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis. Science. 2012;335:1103–1106. doi: 10.1126/science.1206848. [DOI] [PubMed] [Google Scholar]
- 19.Le TB, Imakaev MV, Mirny LA, Laub MT. High-resolution mapping of the spatial organization of a bacterial chromosome. Science. 2013;342:731–734. doi: 10.1126/science.1242059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cabrera JE, Jin DJ. The distribution of RNA polymerase in Escherichia coli is dynamic and sensitive to environmental cues. Mol Microbiol. 2003;50:1493–1505. doi: 10.1046/j.1365-2958.2003.03805.x. [DOI] [PubMed] [Google Scholar]
- 21.Marko JF. Micromechanical studies of mitotic chromosomes. Chromosome research : an international journal on the molecular, supramolecular and evolutionary aspects of chromosome biology. 2008;16:469–497. doi: 10.1007/s10577-008-1233-7. [DOI] [PubMed] [Google Scholar]
- 22.Strick TR, Kawaguchi T, Hirano T. Real-time detection of single-molecule DNA compaction by condensin I. Curr Biol. 2004;14:874–880. doi: 10.1016/j.cub.2004.04.038. [DOI] [PubMed] [Google Scholar]
- 23.Kim H, Loparo JJ. Multistep assembly of DNA condensation clusters by SMC. Nature communications. 2016;7:10200. doi: 10.1038/ncomms10200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soh YM, et al. Molecular basis for SMC rod formation and its dissolution upon DNA binding. Mol Cell. 2015;57:290–303. doi: 10.1016/j.molcel.2014.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Youngman PJ, Perkins JB, Losick R. Genetic transposition and insertional mutagenesis in Bacillus subtilis with Streptococcus faecalis transposon Tn917. Proc Natl Acad Sci U S A. 1983;80:2305–2309. doi: 10.1073/pnas.80.8.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harwood CR, Cutting SM. Molecular Biological Methods for Bacillus. Wiley; New York: 1990. [Google Scholar]
- 27.Imakaev M, et al. Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nat Methods. 2012;9:999–1003. doi: 10.1038/nmeth.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rousseeuw PJ, Croux C. Alternatives to the Median Absolute Deviation. Journal of the American Statistical Association. 1983;88:1273–1283. [Google Scholar]
- 29.Lindow JC, Kuwano M, Moriya S, Grossman AD. Subcellular localization of the Bacillus subtilis structural maintenance of chromosomes (SMC) protein. Mol Microbiol. 2002;46:997–1009. doi: 10.1046/j.1365-2958.2002.03235.x. [DOI] [PubMed] [Google Scholar]
- 30.Lin DC, Levin PA, Grossman AD. Bipolar localization of a chromosome partition protein in Bacillus subtilis. Proc Natl Acad Sci U S A. 1997;94:4721–4726. doi: 10.1073/pnas.94.9.4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rudner DZ, Fawcett P, Losick R. A family of membrane-embedded metalloproteases involved in regulated proteolysis of membrane-associated transcription factors. Proc Natl Acad Sci U S A. 1999;96:14765–14770. doi: 10.1073/pnas.96.26.14765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levin PA, Losick R. Transcription factor Spo0A switches the localization of the cell division protein FtsZ from a medial to a bipolar pattern in Bacillus subtilis. Genes & Development. 1996;10:478–488. doi: 10.1101/gad.10.4.478. [DOI] [PubMed] [Google Scholar]
- 33.Fujita M. Temporal and selective association of multiple sigma factors with RNA polymerase during sporulation in Bacillus subtilis. Genes Cells. 2000;5:79–88. doi: 10.1046/j.1365-2443.2000.00307.x. [DOI] [PubMed] [Google Scholar]
- 34.El Sayyed H, et al. Mapping Topoisomerase IV Binding and Activity Sites on the E. coli Genome. PLoS Genet. 2016;12:e1006025. doi: 10.1371/journal.pgen.1006025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nolivos S, et al. MatP regulates the coordinated action of topoisomerase IV and MukBEF in chromosome segregation. Nature communications. 2016;7:10466. doi: 10.1038/ncomms10466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Teytelman L, Thurtle DM, Rine J, van Oudenaarden A. Highly expressed loci are vulnerable to misleading ChIP localization of multiple unrelated proteins. Proc Natl Acad Sci U S A. 2013;110:18602–18607. doi: 10.1073/pnas.1316064110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McGhee JD, von Hippel PH. Formaldehyde as a probe of DNA structure. r. Mechanism of the initial reaction of Formaldehyde with DNA. Biochemistry. 1977;16:3276–3293. doi: 10.1021/bi00634a002. [DOI] [PubMed] [Google Scholar]
- 38.Ireton K, Gunther NW, Grossman AD. spo0J is required for normal chromosome segregation as well as the initiation of sporulation in Bacillus subtilis. J Bacteriol. 1994;176:5320–5329. doi: 10.1128/jb.176.17.5320-5329.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Graham TG, et al. ParB spreading requires DNA bridging. Genes Dev. 2014;28:1228–1238. doi: 10.1101/gad.242206.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berlatzky IA, Rouvinski A, Ben-Yehuda S. Spatial organization of a replicating bacterial chromosome. Proc Natl Acad Sci U S A. 2008;105:14136–14140. doi: 10.1073/pnas.0804982105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Broedersz CP, et al. Condensation and localization of the partitioning protein ParB on the bacterial chromosome. Proc Natl Acad Sci U S A. 2014;111:8809–8814. doi: 10.1073/pnas.1402529111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


