Abstract
Exosomes are membranous extracellular nanovesicles of endocytic origin. Exosomes are known to carry host and pathogen-derived genomic, proteomic, lipidomic cargos and other extraneous molecules. Exosomes are secreted by diverse cell types into the extracellular milieu and are subsequently internalized by recipient neighboring or distal cells. Upon internalization, exosomes condition recipient cells by donating their cargos and/or activating various signal transduction pathways, consequently regulating physiological and pathophysiological processes. Exosomes facilitate intercellular communication, modulate cellular phenotype, and regulate microbial pathogenesis. We have previously shown that semen exosomes (SE) inhibit HIV-1 replication in various cell types. Here, we describe detailed protocols for characterizing SE. This protocol can be adapted or modified and used for evaluation of other extracellular vesicles of interest.
Keywords: Semen, Exosomes, Extracellular, Vesicles, Prostasomes, HIV
Background
Exosomes are membranous nanovesicles originating as a result of inward budding of endosomal membranes within the late endosomal compartment of a multitude of cell types (Simons and Raposo, 2009). Exosomes are released by many cell types ( Iglesias et al., 2012 ) into the extracellular milieu and are found in biological fluids including blood ( Kaur et al., 2014 ) urine ( Li et al., 2013 ) saliva ( Madison et al., 2015 ) and breast milk ( Madison et al., 2014 ; Naslund et al., 2014 ). Human semen contains a heterogenous population of nanovesicles ( Madison et al., 2014 ; Madison et al., 2015 ) produced by tissues of the male genital tract including prostate secretory acinar cells ( Sahlen et al., 2002 ) and epididymal epithelial cells ( Frenette et al., 2010 ) as well as cells of the vasa deferentia, testes, and the vesicular glands ( Renneberg et al., 1997 ; Sullivan et al., 2005 ). The variability in the cells that secret exosomes is reflected in the composition and function of exosomes. Thus, exosomal cargo composition and function are regulated by many factors including the type and condition of the originating cell (Raposo and Stoorvogel, 2013), cellular environment, and for in vivo derived exosomes; the condition of the donor ( Welch et al., 2017 ). Released exosomes when taken up by target cells transfer their cargo, including proteins ( Iglesias et al., 2012 ; Charrier et al., 2014 ), miRNA ( Shtam et al., 2013 ; Ong et al., 2014 ), and mRNA ( Tomasoni et al., 2013 ; Madison et al., 2014 ; Madison et al., 2015 ) to the target cells. As a result, exosomes are known to be involved in modulation of host immune response ( Kaur et al., 2014 ; Vojtech et al., 2014 ), and regulation of microbial pathogenesis ( Li et al., 2013 ; Arenaccio et al., 2014 ; Madison et al., 2014 ; Naslund et al., 2014 ; Vojtech et al., 2014 ; Madison et al., 2015 ).
While progress has been made in the field of exosome biology, many protocols are contradictory in the most effective and efficient method of characterizing exosomes (Taylor and Shah, 2015). Here, we provide a detailed protocol for evaluating the function and physical properties of semen exosomes ( Madison et al., 2014 ; Madison et al., 2015 ). This protocol lays the groundwork for evaluating other functional activities of semen exosomes, and for evaluating exosomes from other sources.
Materials and Reagents
Pipette tips (any brand)
15 polypropylene conical plastic tubes (DOT SCIENTIFIC, PerformR®, catalog number: 416-PG)
50 ml polypropylene conical plastic tubes (DOT SCIENTIFIC, PerformR®, catalog number: 451-PG)
12 well tissue culture plate (CELLTREAT Scientific Products, catalog number: 229112)
5 ml polystyrene round-bottom tubes (Corning, Falcon®, catalog number: 352052)
Microscope coverslip, 18 mm (Fisher Scientific, FisherbrandTM, catalog number: 12-545-100)
Coverslips (VWR, catalog number: 48382-041)
Microscope slides (Fisher Scientific, catalog number: 22-034-486)
1.7 m microcentrifuge tube (DOT SCIENTIFIC, catalog number: RN1700-GMT)
96 well solid white flat-bottom polystyrene microplates (Corning, catalog number: 3917)
96 well tissue culture plate (CELLTREAT Scientific Products, catalog number: 229196)
Disposable cuvettes (Eppendorf, catalog number: Z605050)
1 ml disposable syringes (BD, catalog number: 309659)
96 well tissue culture dishes
U937 human monocytic cell line
TZM-bl human vaginal epithelial cell line
Jurkat human T lymphocyte cell line
V428 (HPV-16 E6/E7 transformed human vaginal epithelial cell line)
VK2 (HPV-16 E6/E7 transformed human vaginal epithelial cell line)
Human semen
Cell-free HIV-1 virus stock, replication competent
ExoQuick (System Biosciences)
Phosphate buffered saline, DPBS 1x; without CaCl2 and MgCl2 (Thermo Fisher Scientific, GibcoTM)
Liposomes (Lipofectamine 2000) (Thermo Fisher Scientific, InvitrogenTM, catalog number: 11668019)
PKH67Green fluorescent kit (Sigma-Aldrich, catalog number: MINI67)
PKH26Red fluorescent kit (Sigma-Aldrich, catalog number: MINI26)
Fetal bovine serum (FBS) (Thermo Fisher Scientific, GibcoTM)
Quick Start Bradford Protein Assay Kit 1 (Bio-Rad Laboratories, catalog number: 500-0201)
Roswell Park Memorial Institute (RPMI) 1640 (Thermo Fisher Scientific, GibcoTM)
Penicillin/streptomycin (Thermo Fisher Scientific, GibcoTM)
Sodium pyruvate (Thermo Fisher Scientific, GibcoTM)
L-glutamine (Thermo Fisher Scientific, GibcoTM)
Keratinocyte serum free media (KSFM) (Thermo Fisher Scientific)
Human recombinant Epidermal Growth Factor 1-53 (Thermo Fisher Scientific)
Bovine pituitary extract (BPE) (Thermo Fisher Scientific)
0.25% trypsin-EDTA, phenol red dissociation reagent (Thermo Fisher Scientific, GibcoTM, catalog number: 25200-056)
Collagen, type 1 from rat tail (Sigma-Aldrich, catalog number: C3867)
Paraformaldehyde (2%) (Fisher Scientific, catalog number: T353-500)
Vectashield antifade reagent with DAPI (Vector Laboratories, catalog number: H-1200)
Exosome-human CD63 Isolation/Detection (Thermo Fisher Scientific, InvitrogenTM, catalog number: 10606D)
Albumin, bovine fraction V (BSA) (RPI, catalog number: A30075-100.0)
Anti-human CD63-FITC (BioLegend, catalog number: 353005)
Aldehyde/Sulfate latex beads (Thermo Fisher Scientific, Molecular ProbesTM, catalog number: A37301)
MHC-II monoclonal antibody
Isotype control antibody (mouse IgG1)
Glycine (RPI, catalog number: G36050-500.0)
Anti-human CD63-PE (BioLegend, catalog number: 353003)
Triton-X-100 (Sigma-Aldrich, catalog number: X100)
Acetylthiocholine chloride (Sigma-Aldrich, catalog number: A5626)
5,5’-Dithiobis 2-nitrobenzoic acid (Sigma-Aldrich, catalog number: D8130)
Sodium carbonate, anhydrous (RPI, catalog number: S25025-500.0)
NuPAGE LDS sample buffer (4x) (Thermo Fisher Scientific, NovexTM, catalog number: NP0008)
NuPAGETM NovexTM 4-12% Bis-Tris Protein Gels, 1.5 mm, 10-well (Thermo Fisher Scientific, InvitrogenTM, catalog number: NP0335PK2)
NuPAGE MOPS SDS running buffer (20x) (Thermo Fisher Scientific, NovexTM, catalog number: NP0001)
Pierce Silver Stain Kit (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 24612)
RNeasy Mini Kit (QIAGEN, catalog number: 74104)
RNase-Free DNase Set (QIAGEN, catalog number: 79254)
High-capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific, Applied BiosystemsTM, catalog number: 4368814)
QuantiFast SYBR Green PCR Kit (QIAGEN, catalog number: 204054)
Agarose (RPI, catalog number: A20090)
Ethidium bromide solution (Bio-Rad Laboratories, catalog number: 1610433)
1x TAE buffer
Steady-Glo (Promega, catalog number: E2510)
NuPAGE sample reducing agent (10x) (Thermo Fisher Scientific, NovexTM, catalog number: NP0004)
MES (Sigma-Aldrich, catalog number: M2933)
1 N NaOH (Avantor Performance Materials®, J.T.Baker®, catalog number: 563502)
Distilled water (any brand)
Exosome-depleted FBS (Thermo Fisher Scientific, GibcoTM, catalog number: 26140079)
MTT reagent (Thermo Fisher Scientific, Molecular ProbesTM, catalog number: M6494)
Nonidet P-40 substitute (RPI, catalog number: N59000)
Hydrochloric acid (HCl), ACS reagent, 37% (Sigma-Aldrich, catalog number: 258148)
Isopropanol (Fisher Scientific, catalog number: A416-4)
Dulbecco’s modified Eagle medium (DMEM) (Thermo Fisher Scientific, GibcoTM)
Exosome-depleted FBS (see Recipes)
Lysing sample buffer for protein footprint (see Recipes)
Storage buffer (see Recipes)
MES buffer (see Recipes)
FACS wash buffer (see Recipes)
MTT reagent (see Recipes)
MTT solvent (see Recipes)
Equipment
Pipettes (any brand)
Sterile tweezers (any brand)
Centrifuge (Eppendorf, model: 5415 D)
Ultracentrifuge (Beckman Coulter, model: Optima L-90K)
SW40Ti rotor (Beckman Coulter, catalog number: 331362)
Polyallomer centrifuge tubes 14 x 89 mm for SW41Ti rotor (Beckman Coulter, catalog number: 344059)
37 °C, 5% CO2 cell culture incubator (NuAire, model: NU-5510)
FACSCalibur flow cytometer (BD) (see Notes for laser and filter specifications)
FACSVerse flow cytometer (BD) (see Notes for laser and filter specifications)
FACSAria flow cytometer (BD) (see Notes for laser and filter specifications)
Laser scanning confocal microscope (Nikon Instruments, model: Eclipse TE2000)
Magnetic separator for 1.7 ml tubes
Rotating mixer
Microplate reader (Tecan Trading, model: Infinite® M200 Pro)
DynaPro Nanostar (Wyatt Technologies, model: DynaPro Nanostar)
NanoSight LM10 (Malvern Instruments, model: NanoSight LM10)
NanoDrop spectrophotometer (Thermo Fisher Scientific)
SW60 Ti rotor (Beckman Coulter, catalog number: 335649)
Polyallomer centrifuge tubes 11 x 60 mm for SW60Ti rotor (Beckman Coulter, catalog number: 355636)
7500 fast real-time PCR system (Thermo Fisher Scientific, model: 7500 Fast Real-time PCR System)
Luminometer (BioTek Instruments, model: Synergy H1 Hybrid Reader)
Laminar flow hood
SW32 Ti rotor (Beckman Coulter, model: 369650)
Polyallomer centrifuge tubes 25 x 89 mm for SW32Ti rotor (Beckman Coulter, catalog number: 344058)
pH meter (any brand)
Gel running tank(Thermo Fisher Scientific, NovexTM, model: XCell SureLock® Mini-Cell, catalog number: EI0001)
Sephacryl S300-HR 16/60 gel filtration prepacked column (GE Healthcare catalog number: 17-1167-01)
BioLogic DuoFlow Workstation (Bio-Rad Laboratories)
BioLogic BioFrac fraction collector (Bio-Rad Laboratories)
Gel electrophoresis horizontal apparatus (Bio-Rad Laboratories, model: Wide Mini-Sub Cell GT Cell, catalog number: 1704468EDU)
Dry Block Heater for microcentrifuge tubes (Thermo Fisher Scientific)
Software
FlowJo analysis software (TreeStar)
BioLogic DuoFlow software version 5.3 (Bio-Rad Laboratories)
Dynamics software (Wyatt Technology)
NTA software (Malvern Instruments)
Procedure
-
Acquisition of human semen samples
This study utilized existing human specimens (semen) and therefore is not human subjects’ research. The samples were discarded from routine examinations and not linked to any identifiers.
Collect semen by dry manual stimulation and ejaculation into sterile 15 ml polypropylene conical tubes.
Store samples at room temperature for 30 min to promote liquefaction and then centrifuge for 10 min at 1,000 × g at 4 °C to pellet spermatozoa.
Remove seminal plasma from spermatozoa-containing pellets. Pellets can be discarded or stored for downstream analysis.
Store seminal plasma samples at -80 °C until required for exosome purification by ExoQuick or ultracentrifugation. We have found no discernable difference between the two methods of purification.
-
ExoQuick purification of exosomes
Thaw seminal plasma samples before centrifuging at 2,000 × g for 15 min at 4 °C in a 50 ml conical tube. Transfer supernatant to a new tube and centrifuge again at 10,000 × g for 30 min at 4 °C in a 50 ml conical tube to pellet cellular debris.
Place clarified seminal plasma in a fresh 50 ml conical tube and add ExoQuick at a ratio of 4:1 (clarified seminal plasma/ExoQuick). Mix by inversion, and incubate at 4 °C overnight (12-24 h).
Centrifuge the clarified seminal plasma/ExoQuick mixture at 1,500 × g for 30 min at 4 °C.
Remove the supernatant (exosome free clarified seminal plasma and ExoQuick) and repeat centrifugation at 1,500 × g for 10 min at 4 °C without resuspension of the pellet.
Remove the residual supernatant.
Resuspend the exosome pellet in PBS to 1/10 of the original volume of seminal plasma, quantify protein concentration by Bradford assay, and aliquot.
-
Ultracentrifugation purification of exosomes
Dilute clarified seminal plasma 50% in PBS, and ultracentrifuge at 100,000 × g for 2 h at 4 °C to pellet exosomes using an SW41Ti rotor.
Wash exosome pellets in PBS three times with ultracentrifugation at 100,000 × g for 30 min per wash (1.5 h). A volume of PBS should be used that completely fills the ultracentrifuge tubes to avoid collapse of the tubes.
Resuspend exosomes in PBS to 1/10 of the original volume of seminal plasma, quantify protein concentration by Bradford assay, and aliquot.
-
Fluorescent labeling of exosomes, liposome controls or PBS controls
Fluorescently label PBS control, Lipofectamine 2000 derived liposome control, or exosomes using PKH67Green or PKH26Red kits according to manufacturer’s instructions with the following modifications: Add 1 mg of purified exosomes or liposome control or an equivalent volume of PBS to 250 µl of PBS and mix with 250 µl of Diluent C in SW41Ti ultracentrifuge tubes.
Add 4 µl of PKH67Green or PKH26Red lipophilic dye to 500 µl of Diluent C in a separate tube at room temperature (RT) in the dark and perform all subsequent steps at RT in the dark.
Combine the tube containing dye and Diluent C with the tube containing exosomes or controls in PBS and Diluent C in the SW41Ti ultracentrifuge tube. Rapidly mix by manual pipetting. Incubate for 5 min in the dark at RT and vortex twice during the 5-min incubation.
After the 5 min of incubation, add exosome-free FBS in a 1:1 ratio to the ultracentrifuge tube containing dye, exosomes or controls, and Diluent C. Incubate for 1 min to occupy unbound dye.
Ultracentrifuge the solution at 100,000 × g for 30 min. Discard the supernatant.
Resuspend the pellet containing dye-labelled exosomes or controls in PBS, transfer to another SW41Ti ultracentrifuge tube and wash three times by ultracentrifugation at 100,000 × g for 30 min per wash. After each wash, transfer the PBS resuspended pellet to a new SW41Ti ultracentrifuge tube to minimize transfer of unbound dye. A volume of PBS should be used that completely fills the ultracentrifuge tube to avoid collapse of the tube.
After the final wash, remove the supernatant, and resuspend the pellet in PBS to the original volume of the exosomes. Quantify dye-labelled exosomes or controls using Bradford Protein Assay Kit, aliquot, and store in the dark at -80 °C until use.
-
Cell culture preparation for internalization of fluorescent exosomes
Grow human monocytic and lymphocytic cell lines in tissue culture dishes in RPMI supplemented with 10% exosome depleted FBS, 100 U/ml penicillin, 100 μg/ml streptomycin, 1 mM sodium pyruvate, and 0.3 mg/ml L-glutamine in a 5% CO2 incubator at 37 °C.
Grow TZM-bl vaginal epithelial cell lines in tissue culture dishes in DMEM supplemented with 10% exosome depleted FBS, 100 U/ml penicillin, 100 μg/ml streptomycin, 1 mM sodium pyruvate, and 0.3 mg/ml L-glutamine in a 5% CO2 incubator at 37 °C.
Grow VK2 and V428 vaginal epithelial cell lines in tissue culture dishes in keratinocyte serum free media supplemented with 0.1 ng/ml prequalified human recombinant Epidermal Growth Factor 1-53, 0.05 mg/ml BPE, 100 U/ml penicillin and 100 μg/ml streptomycin.
Grow all cell types in 12 well tissue culture dishes. Seed cells at 1 x 105 cells per well in 1 ml volume of the correct cell culture media for each cell type (see Note 1).
Expose cells to 25-100 μg/ml of red or green fluorescent exosomes or controls for 0, 3, 6, 9, 12 or 24 h incubation periods in a 5% CO2 incubator at 37 °C. The incubation of SE with cells should be completed in cell culture media. Final volume of SE incubation with cells will depend on the concentration of exosomes after fluorescent labeling.
Wash TZM-bl, VK2 or V428 adherent cells three times in 1 ml PBS in the plate and detach the cells from the plate using 0.5 ml of 0.25% trypsin-EDTA dissociation reagent. Neutralize the trypsin with 0.5 ml 10% exosome free FBS in DMEM. Collect the cells from the wells and transfer to 5 ml polystyrene round-bottom tubes. Wash the cells three times in 1 ml PBS with 5 min centrifugation at 500 × g at 4 °C.
Collect monocytic and lymphocytic suspension cells in 5 ml polystyrene round-bottom tubes. Treat with trypsin as described in step E5, and neutralize with 10% exosome free FBS in RPMI. Wash the cells three times in 1 ml PBS with 5 min centrifugation at 500 × g at 4 °C.
-
Assess cellular uptake and exosome internalization kinetics utilizing FACS analysis or confocal microscopy.
-
For FACS analysis:
Fix cells for 15 min on ice with 300 μl 2% paraformaldehyde.
Wash the cells two times in PBS with 5 min centrifugation at 500 × g in between each wash. Resuspend the cell pellets in 300 μl PBS.
Analyze fluorescence using a FACSCalibur or FACSVerse flow cytometer (BD) to detect the PKH67Green (FL-1 or FITC channel, respectively) or PKH26Red (FL-2 or PE channel, respectively) transferred from exosomes to cells during fusion and uptake.
Determine cellular frequency and fluorescence intensity using FlowJo analysis software (TreeStar) (Figure 1).
-
For confocal microscopy analysis of exosome internalization:
Coat coverslips with collagen by adding sterile, round 18 mm coverslips to the wells of a 12 well plate. Add 0.5 ml of 50 μg/ml collagen solution to the wells. Incubate the plate at 37 °C for 1 h, after which you move the plate to 4 °C for overnight incubation. After overnight incubation, remove the collagen solution and gently wash the coverslips in the wells three times with PBS. After the final wash, aspirate all the PBS and immediately proceed to step E8b.ii.
Grow TZM-bl, VK2 or V428 adherent cells in 12 well tissue culture plates on top of collagen coated microscope cover slips, expose to fluorescent exosomes as described in step E4 and wash three times in PBS in the plate.
Fix cells on the coverslips in the plate with 2% paraformaldehyde for 15 min with the plate on ice.
Remove coverslips from the plate and add a drop of Vectashield antifade reagent to each coverslip.
Mount coverslips down on microscope slides.
Assess fusion, uptake and internalization kinetics of fluorescent red or green exosomes using laser scanning confocal microscopy. Representative images are shown in Madison et al., 2015 .
-
-
Detection of surface exosomal markers
Here we will describe detection of human CD63 in SE, but this protocol may be modified for detection of other common exosomal surface markers in SE. Human CD63 in SE is detected per the manufacturer’s instructions using the exosome-human CD63 isolation/detection kit from Invitrogen where after acquiring SE:
Resuspend 25 μg of SE to a total volume of 100 μl in 0.1% BSA.
Resuspend anti-CD63 coated magnetic beads by vortexing for 30 sec. Transfer 20 μl of beads to a microcentrifuge tube and wash the beads by adding 200 μl of 0.1% BSA and vortexing.
Place the tube containing the beads on a magnet separator for 1 min. Discard the supernatant before removing the tube from the magnet.
Add the SE solution to the washed beads and mix by pipetting.
Incubate the SE/beads solution at 4 °C overnight (18-20 h) in a rotating mixer.
Centrifuge the SE/beads solution for 3-5 sec.
Wash the SE bound beads in 300 μl of 0.1% BSA and mix by pipetting before placing the tube on the magnet for 1 min and discarding the supernatant.
Remove the tube from the magnet. Wash the SE bound beads in 400 μl of 0.1% BSA and mix by pipetting.
Place the tube on the magnet for 1 min and discard the supernatant before removal from the magnet.
Resuspend the SE bound beads in 300 μl of 0.1% BSA.
Transfer 100 μl of the SE bound beads to a new microcentrifuge tube. Add 5 μl of anti-human CD63-FITC (BioLegend) and mix by pipetting.
Incubate the SE-bounds beads and antibody for 60 min at room temperature in the dark on an oscillating plate.
Wash the antibody stained SE-bound beads in 300 μl of 0.1% BSA and mix by pipetting. Place the tube on the magnet for 1 min, and discard the supernatant before removal of the tube from the magnet.
-
Repeat the washing step twice before resuspending the SE-bound beads in 300 μl of 0.1% BSA. Transfer the resuspended SE-bound beads to a 5 ml polystyrene round-bottom tube before analysis on FACSVerse instrument and FlowJo (Tree Star) software (Figure 2).
Alternatively, common exosomal markers in SE may also be detected with the use of non-magnetic beads. Here we provide instructions for the use of latex beads to detect CD63 in SE.
Per manufacturer instructions, 2.5 ml of resuspended, surfactant-free, 4-μm diameter, aldehyde/sulphate, latex beads (Invitrogen, Molecular Probes, hereafter referred to as latex beads) were washed twice in 10 ml of 0.025 M, pH 6.0 2-(N-morpholino) ethenesulfonic acid (MES) buffer with centrifugation at 3,000 × g for 20 min at 4 °C.
Following the second and final wash, the latex beads were resuspended in 5 ml MES buffer.
Incubate 100 µl latex beads with 100 µl of anti MHC-II MAb or isotype control antibody prepared in MES at a concentration of 1 mg/ml at room temperature overnight with gentle agitation.
Sediment latex beads with conjugated antibody by centrifugation at 3,000 × g for 20 min at 4 °C.
Remove supernatant (unbound antibody).
Wash latex beads with conjugated antibody thrice in 1 ml PBS (0.1 M, pH 7.2) at 3,000 × g for 20 min at 4 °C.
Resuspend latex beads in 100 µl of storage buffer (see Recipes).
-
ExoQuick purified exosomes (100 μg) were incubated with *2 x 105 anti MHC-II or isotype control coated latex beads in a final volume of 100 μl PBS (0.1 M, pH 7.2) first for 15 min at room temperature followed by overnight at 4 °C with gentle agitation.
*Note: Concentration of beads/ml differs by lot number.
The reaction was stopped by 30 min incubation with PBS (0.1 M, pH 7.2) including 0.2% glycine to saturate any vacant sites on the latex beads.
The exosome and antibody bound latex bead preparation was then washed thrice in FACS wash buffer (see Recipes).
Exosomes coated beads were then incubated with the appropriate concentration of anti-CD63 conjugated to PE (BioLegend) or isotype control antibody for 1 h at room temperature in the absence of light followed by three washes in FACS buffer.
The resulting immunofluorescence was analyzed by use of a FACSAria flow cytometer (BD) and FlowJo analysis software (TreeStar) (Figure 2).
-
SE acetylcholinesterase activity
Lyse 50 μg of SE in 2% Triton-X-100 at a 1:1 volumetric ratio.
Add 5 μl of SE/Triton-X reaction to a 96-well flat bottom clear plate in replicates of three.
Combine 1.25 mM acetylthiocholine chloride (Sigma-Aldrich) and 0.1 mM 5,5’-dithiobis 2-nitrobenzoic acid (Sigma-Aldrich) in a 1:1 ratio to reach a final volume of 100 μl per well. Add 100 μl of the mixed solutions to each of the exosome containing wells. Be sure the microplate reader has reached 37 °C before adding this solution to the wells as the reaction will start immediately after addition.
Read absorbance at 450 nm on a microplate reader for a total of 30 min in 5 min intervals at 37 °C (Figure 3).
-
Dynamic Light Scattering (DLS) of SE
Dilute 0.1 mg/ml of SE in 200 μl of PBS and analyze size by DynaPro NanoStar DLS (Wyatt Technologies) using a total volume of 150 μl in plastic cuvettes.
Complete data analysis using Dynamics software.
Use an average of ten measurements per exosome sample to determine radius, diameter, and %intensity (Figure 4).
-
SE NanoSight nanoparticle tracking analysis (NTA)
Prepare control suspension containing uniformly sized (100 or 200 nm) polystyrene particles.
Use the suspension to align the foci of the laser and microscope.
Make serial dilutions of each SE specimen in PBS to a final volume of 0.5 ml.
Inject 0.5 ml of diluted SE into NanoSight LM10 NTA using 1 ml disposable syringe.
Analyzed individual members of the prepared serial dilutions until the raw concentration detected is within the recommended range for the instrument.
Using the identified dilution, record three 30 sec videos for each specimen.
Repeat the analysis 3 times using the same settling to ensure repeatable and accurate measurement.
Complete post-acquisition analysis with NTA software to determine size and concentration of SE.
View the results tab for i) total number of particles traced, ii) average number of particles, and iii) particle concentration.
Calculate total concentration of exosomes per ml of semen accounting for the dilution factor used for NanoSight analysis (Figure 4). Obtain error bars by analyzing standard deviation of the 3 measurements of each sample.
-
Fractionation of SE into membrane and luminal contents
Treat 100 μg SE with 5-10x volume of 0.1 M sodium carbonate pH = 11.5. Mix by vortexing.
Incubate for 30 min to 1 h at 4 °C. At this point, total protein from the lysed exosomes can be measured by NanoDrop spectrophotometer at 280 nm. Complete the following steps to separate lysed SE into membrane and luminal fractions.
Transfer SE/sodium carbonate mix to SW60 Ti ultracentrifuge tubes. Bring up the volume with PBS to fill the tube.
Ultracentrifuge at 150,000 x g for 1.5-2 h at 4 °C. The pellet contains the membrane fraction while the supernatant contains the luminal fraction.
Remove the luminal ‘supernatant’ fraction. This can be concentrated by MW filter cutoffs before protein quantification by Bradford assay and downstream analysis or storage at -80 °C until use.
Resuspend the membrane fraction in PBS to the original volume of SE before lysis. Quantify protein by Bradford assay, aliquot, and store at -80 °C or use in downstream analysis (Figure 5).
-
SE protein footprint
Lyse 5 μg of SE in 20 μl of PBS at a ratio of 3 μl NuPAGE lysing sample buffer (see Recipes) per 7 μl SE.
Heat SE in sample buffer at 90 °C for 10 min.
Load a maximum volume of 40 μl on a 10 well 1.5 mm NuPAGE 4-12% Bis-Tris gel.
Run the gel at 200 V for ~50 min in NuPAGE 20x MOPS SDS running buffer diluted to 1x.
Silver stain the gel following the manufacturer’s protocol (Pierce silver stain kit Thermo Scientific).
-
Examination of RNA integrity and content of SE
Use a starting concentration of at least 12 μg of SE for RNA extraction.
Extract SE RNA using the Qiagen RNeasy kit per the manufacturer’s instructions and complete the optional DNase treatment using Qiagen RNase-free DNase set.
Determine RNA concentration by NanoDrop spectrophotometer. To evaluate RNA integrity of SE, RNA can be analyzed by an Agilent BioAnalysis run using RNA 6000 Pico chips if the RNA concentration is < 50 ng/μl. If the RNA concentration is > 50 but < 500 ng/μl, the RNA can be evaluated using RNA 6000 Nano chips (Figure 6).
Use equivalent concentrations of RNA for cDNA synthesis using High-Capacity cDNA Reverse Transcription Kit (ABI).
Use human gene specific primers to amplify CD9, CD63, and GAPDH or other genes of interest by Quantifast Sybr green technology (QIAGEN) and 7500 fast real-time machine.
Visualize PCR amplicons on a 2% agarose gel by ethidium bromide staining. Representative images are shown in Madison et al., 2014 .
-
SE inhibition of HIV-1 infectivity
Grow TZM-bl cells in 96 well tissue culture dishes in DMEM supplemented with 10% exosome depleted FBS, 100 U/ml penicillin, 100 μg/ml streptomycin, 1 mM sodium pyruvate, and 0.3 mg/ml L-glutamine in a 5% CO2 incubator at 37 °C.
Preincubate 100 μg/ml SE or PBS vehicle with 8 RT units of HIV-1 virus for 1 h at 37 °C in DMEM supplemented with 10% exosome depleted FBS, 100 U/ml penicillin, 100 μg/ml streptomycin, 1 mM sodium pyruvate, and 0.3 mg/ml L-glutamine. Preincubate supplemented DMEM with PBS vehicle as uninfected control. Mix before incubation by vortexing.
Remove DMEM media from TZM-bl cells. Add 100 μl per well in triplicate of SE/HIV-1, PBS vehicle/HIV-1, or PBS vehicle/DMEM to TZM-bl cells.
Incubate in a 5% CO2 incubator at 37 °C for 24 h.
-
Access cell viability by MTT assay and HIV-1 infectivity by Steady-Glo luciferase assay (see Note 5; Figure 7)
-
For MTT assay:
Add 20 μl 5 mg/ml of MTT reagent to each well in replicates of three. Incubate for 3.5 h in a 5% CO2 incubator at 37 °C.
Remove the media and MTT solution from the wells. Add 150 μl off MTT solvent (see Recipes) to each well and incubate for 15 min in the dark on an oscillating plate.
Read absorbance at 590 nm on a microplate reader.
-
For Steady-Glo luciferase assay:
Remove DMEM media from TZM-bl cells. Add 100 μl of Steady-Glo luciferase reagent to each well in replicates of three, including uninfected control. Allow 5 min for cell lysis.
Transfer 90 μl of lysed cells/Steady-Glo mixture to a solid white 96 well plate, avoiding the formation of bubbles.
Read luciferase activity in a microplate luminometer.
-
Figure 1. Internalization of fluorescent exosomes by FACS.
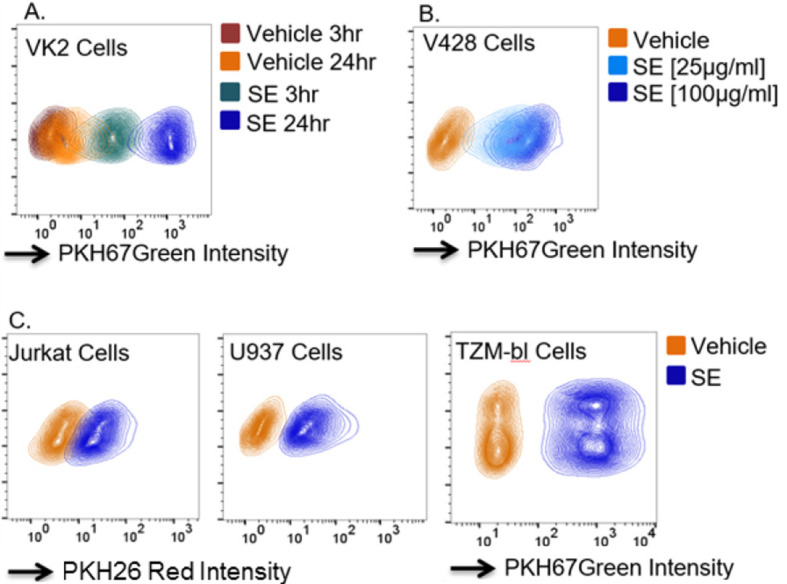
A. VK2 cellular uptake of PKH67 green labeled PBS vehicle or 100 μg/ml semen exosomes (SE) at 3- and 24-h post exposure. B. V428 cellular uptake of PK67 green labeled PBS vehicle or 25 or 100 μg/ml SE at 24-h post exposure. C. Jurkat, U937, and TZM-bl cellular uptake of PKH26 red or PKH67 green labeled PBS vehicle or 100 μg/ml SE at 24-h post exposure. The y-axis shows the forward scattering value (FSC) of the cell populations.
Figure 2. CD63 expression on SE.
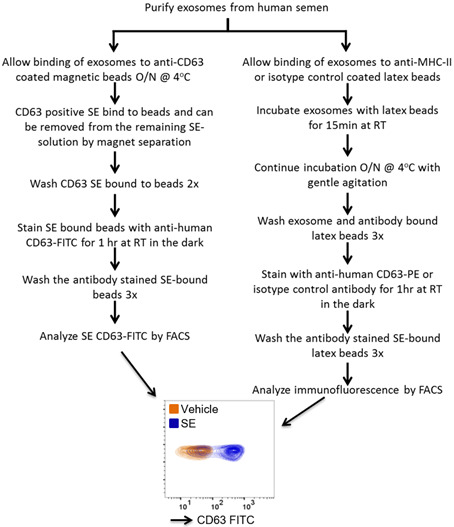
25 μg SE or PBS vehicle were incubated overnight with α-CD63 coated magnetic beads to facilitate binding. Unbound SE was removed before staining of bead-bound SE with α-CD63-FITC and FACS analysis. O/N = overnight. The y-axis shows the forward scattering value (FSC) of the beads.
Figure 3. Acetylcholine esterase activity of SE.
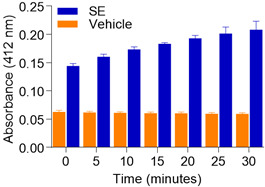
50 μg SE or PBS vehicle were lysed in Triton-X-100. AChE activity was measured at 5 min intervals for a total of 30 min. Error bars represent standard deviation.
Figure 4. SE size and concentration estimation.
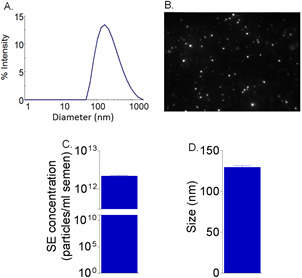
A. Dynamic light scattering indicates approximate diameter of SE. 0.1 mg/ml of SE was used to measure the diameter of SE. Shown is the range of exosome diameters in the population. B. Representative image from NanoSight NTA video clip showing SE particles. C. Approximation of SE particles per ml of semen calculated from NanoSight estimation of concentration. D. Approximation of SE size by NanoSight. N = refers to the number of donors that were combined for exosome purification.
Figure 5. Separation of SE into different fractions.
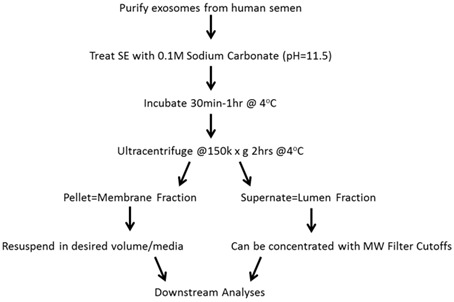
SE can be fractionated into membrane and luminal components.
Figure 6. RNA integrity of SE.
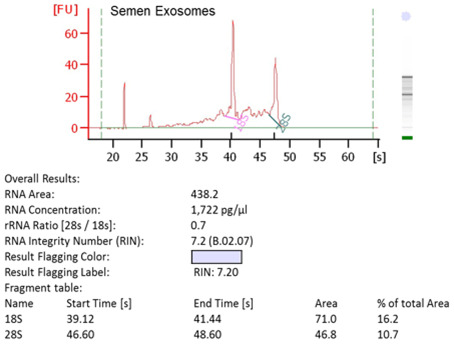
RNA was extracted from SE and analyzed by Agilent Bioanalyzer.
Figure 7. SE inhibition of HIV-1.
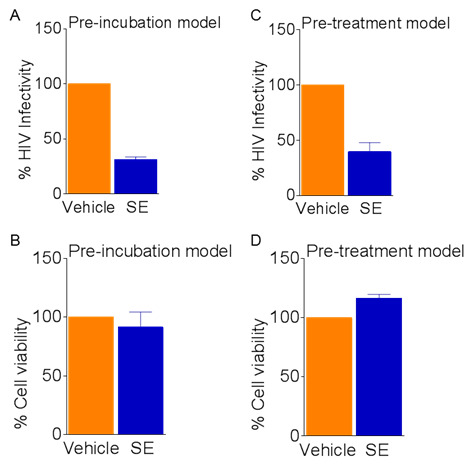
Exosomes (100 μg/ml) or vehicle PBS were preincubated with 8 RT units HIV-1 NL4.3 virus for 1 h at 37 °C before infection of TZM-bl cells for 24 h. A. Infectivity measured by luciferase units; B. Viability is determined by MTT. TZM-bl cells were pretreated with exosomes (100 μg/ml) or vehicle PBS for 24 h before infection with 8 RT units HIV-1 NL4.3 virus for an additional 24 h. C. Infectivity read by luciferase units; D. Viability determined by MTT. Vehicle set as reference at 100% for infectivity and viability. Error bars are standard deviations.
Data analysis
Each assay should be experimentally repeated at least three times with replicates of three per experiment to verify reproducibility.
Average replicates of independent experiments to evaluate statistical significance. Where appropriate, compare results to vehicle control.
Plot data using graphing software, such as GraphPad Prism. For analyses that are not represented graphically such as Bioanalyzer, protein footprint, and FACS analysis representative images may be shown.
Notes
-
Cell culture
When growing cells, to minimize evaporation, only plate cells in the inner wells of the plate and fill the outermost wells with PBS. We recommend plating cells the day before they will be used to allow the cells to normalize. Because SE alters cell viability at differing concentrations and in a cell-type dependent manner, it is important to evaluate cell cytotoxicity in all experiments involving cell treatments with SE ( Madison et al., 2014 ; Madison et al., 2015 ). This is also important when evaluating HIV-1 infectivity as high concentrations of virus or SE may be cytotoxic to cells and influence analysis of results.
-
Storage of exosomes
We found that repeated freeze-thawing of exosomes decreases functional activity. We recommend that after isolation, to aliquot exosomes into individual microcentrifuge tubes (< 100 μg) before freezing for storage to retain functional activity.
-
Discarding of supernatant
When discarding supernatant after pelleting or washing exosomes, we recommend removal by pipette aspiration rather than inversion to ensure complete removal. However, exosome pellets may not always be easily observed, depending on the concentration used, and caution should be used to not dislodge the pellet.
-
Sterility
As often as feasibly possible, all experimental steps should be completed under a laminar flow hood to ensure an aseptic environment. Contamination may influence downstream analyses.
-
HIV-1 infectivity
Because TZM-bl cells contain background fluorescence, we recommend plating 10,000 cells per well in a 96 well format to minimize background luciferase expression. Depending on the cell line used, the HIV-1 isolate used may vary depending on the receptor/co-receptors expressed on that cell line. If evaluating infection in cells that are not a reporter cell line, infection may be evaluated by qRT-PCR using HIV-1 gene specific primers. Data may be confirmed by measuring intracellular or extracellular HIV-1 RT activity ( Madison et al., 2014 ; Madison et al., 2015 ). All HIV-1 experiments must be carried out in accordance with biosafety training and laboratory environment requirements.
-
SE-mediated inhibition of HIV-1
SE also inhibits HIV-1 infection during a pretreatment model of infection. In this model, after growing the cells, SE is added to cells 24 h before virus infection, and remains inhibitory to infection. The HIV-1 inhibitory characteristic of SE is upheld in both the preincubation and pretreatment models with other cells lines such as: Jurkat, SUPT1, U937, PM1, THP-1, CEM, and PBLs ( Madison et al., 2014 ; Madison et al., 2015 ).
-
Flow cytometer specifications
Laser and filter specifications of flow cytometer systems used in this protocol are included below.
FACSCalibur flow cytometer (BD)
Lasers: Air-cooled, argon-ion, 488 nm, 15 mW
Emission detection:
FL1 530/30
FL2 585/42
FL3 670 LP
FL4 661/16
SSC 488/10
FSC 488/10
FACSVerse flow cytometer (BD)
Lasers: Blue laser, 488 nm, 20 mW, beam spot size 9 x 63 μm
Red laser, 640 nm, 40 mW, beam spot size 9 x 63 μm
Violet laser, 405 nm, 40 mW, beam spot size 9 x 63 μm
Emission detection:
FITC 527/32
PE 586/42
PerCp 700/54
APC 660/10
SSC 488/15
FSC 488/10
FACSAria flow cytometer (BD)
Lasers: Coherent Sapphire, solid state, 488 nm, 20mW
JDS Uniphase HeNe, air-cooled, 633 nm, 18mW
Point Source Violet, solid state, 405 nm, 15 mW
Emission detection:
FITC 530/30
PE 576/26
PerCp 695/40
APC 660/20
SSC 488/10
FSC 488/10
Recipes
-
Exosome-depleted FBSUltracentrifuge FBS at 100,000 × g for 2 h at 4 °C in SW32Ti ultracentrifuge tubes using SW32Ti rotor
Collect supernatant and store at 4 °C for up to 1 week or at -80 °C for longer periods of time
-
Lysing sample buffer for protein footprint
Mix 250 μl NuPAGE 4x LDS sample buffer with 100 μl NuPAGE 10x reducing agent
Add mixed buffer to SE samples at 3 μl buffer per 7 μl SE
-
Storage buffer
PBS (0.1 M, pH 7.2) and 0.1% glycine
-
MES buffer
Dissolve MES in distilled water for a concentration of 0.025 M. Determine pH with a pH meter, and adjust the pH with 1 N NaOH to 6.0
-
FACS wash buffer
1% exosome-depleted FBS in PBS
-
MTT reagent
Resuspend MTT reagent in PBS for final concentration of 5 mg/ml, per manufacturer’s instructions
-
MTT solvent
0.1% NP-40 and 4 mM HCl in isopropanol
Acknowledgments
This work was supported by the National Institute on Drug Abuse (NIDA) grant 1R01DA042348-01 (to CMO), National Institutes of Health (NIH) 5T32AI007533-18 (to JLW), and NIH T32 postdoctoral training grant in Infectious Diseases (to MNM), shared Instrumentation Grants 1S10RR025439-01 to the University of Iowa Central Microscopy Core facility, and Holden Comprehensive Cancer Center support grant P30CA086862. The authors are thankful to Aloysius Klingelhutz of the University of Iowa for providing V428 cells, to Bartholomey Konan and Melanie Freeman of the Reproductive Specialty Laboratory of Middle Tennessee and to Amy E.T. Sparks of the University of Iowa Hospitals and Clinics (UIHC) In Vitro Fertilization and Reproductive Testing Laboratory for providing pre-existing, de-identified human donor semen samples. We acknowledge the support of University of Iowa core facilities, including Central Microscopy, X-ray crystallography, and DNA core. We thank Sankar Baruah and Lokesh Gakhar of University of Iowa Crystallography Core Facility for help with Dynamic light scattering. The authors declare that they have no competing interests. MNM, JLW, and CMO wrote the paper. MNM and JLW contributed equally to this manuscript and are thus co-first authors. All authors reviewed the manuscript and approved the final version. Protocols described herein are adapted from our previously published works ( Madison et al., 2014 and 2015; Welch et al., 2017 ).
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1.Arenaccio C., Chiozzini C., Columba-Cabezas S., Manfredi F., Affabris E., Baur A. and Federico M.(2014). Exosomes from human immunodeficiency virus type 1(HIV-1)-infected cells license quiescent CD4+ T lymphocytes to replicate HIV-1 through a Nef- and ADAM17-dependent mechanism . J Virol 88(19): 11529-11539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Charrier A., Chen R., Chen L., Kemper S., Hattori T., Takigawa M. and Brigstock D. R.(2014). Exosomes mediate intercellular transfer of pro-fibrogenic connective tissue growth factor(CCN2) between hepatic stellate cells, the principal fibrotic cells in the liver. Surgery 156(3): 548-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frenette G., Girouard J., D'Amours O., Allard N., Tessier L. and Sullivan R.(2010). Characterization of two distinct populations of epididymosomes collected in the intraluminal compartment of the bovine cauda epididymis. Biol Reprod 83(3): 473-480. [DOI] [PubMed] [Google Scholar]
- 4.Iglesias D. M., El-Kares R., Taranta A., Bellomo F., Emma F., Besouw M., Levtchenko E., Toelen J., van den Heuvel L., Chu L., Zhao J., Young Y. K., Eliopoulos N. and Goodyer P.(2012). Stem cell microvesicles transfer cystinosin to human cystinotic cells and reduce cystine accumulation in vitro . PLoS One 7: e42840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaur S., Singh S. P., Elkahloun A. G., Wu W., Abu-Asab M. S. and Roberts D. D.(2014). CD47-dependent immunomodulatory and angiogenic activities of extracellular vesicles produced by T cells. Matrix biology 37: 49-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li J., Liu K., Liu Y., Xu Y., Zhang F., Yang H., Liu J., Pan T., Chen J., Wu M., Zhou X. and Yuan Z.(2013). Exosomes mediate the cell-to-cell transmission of IFN-alpha-induced antiviral activity. Nat Immunol 14: 793-803. [DOI] [PubMed] [Google Scholar]
- 7.Madison M. N., Jones P. H. and Okeoma C. M.(2015). Exosomes in human semen restrict HIV-1 transmission by vaginal cells and block intravaginal replication of LP-BM5 murine AIDS virus complex. Virology 482: 189-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Madison M. N., Roller R. J., Okeoma C. M.(2014). Human semen contains exosomes with potent anti-HIV-1 activity. Retrovirology 11: 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Naslund T. I., Paquin-Proulx D., Paredes P. T., Vallhov H., Sandberg J. K. and Gabrielsson S.(2014). Exosomes from breast milk inhibit HIV-1 infection of dendritic cells and subsequent viral transfer to CD4+ T cells . AIDS 28(2): 171-180. [DOI] [PubMed] [Google Scholar]
- 10.Ong S. G., Lee W. H., Huang M., Dey D., Kodo K., Sanchez-Freire V., Gold J. D. and Wu J. C.(2014). Cross talk of combined gene and cell therapy in ischemic heart disease: role of exosomal microRNA transfer. Circulation 1): S60-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raposo G. and Stoorvogel W.(2013). Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol 200(4): 373-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Renneberg H., Konrad L., Dammshauser I., Seitz J. and Aumuller G.(1997). Immunohistochemistry of prostasomes from human semen. Prostate 30(2): 98-106. [DOI] [PubMed] [Google Scholar]
- 13.Sahlen G. E., Egevad L., Ahlander A., Norlen B. J., Ronquist G. and Nilsson B. O.(2002). Ultrastructure of the secretion of prostasomes from benign and malignant epithelial cells in the prostate. Prostate 53(3): 192-199. [DOI] [PubMed] [Google Scholar]
- 14.Shtam T. A., Kovalev R. A., Varfolomeeva E. Y., Makarov E. M., Kil Y. V. and Filatov M. V.(2013). Exosomes are natural carriers of exogenous siRNA to human cells in vitro . Cell Commun Signal 11: 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simons M. and Raposo G.(2009). Exosomes--vesicular carriers for intercellular communication. Curr opin cell biolo 21(4): 575-581. [DOI] [PubMed] [Google Scholar]
- 16.Sullivan R., Saez F., Girouard J. and Frenette G.(2005). Role of exosomes in sperm maturation during the transit along the male reproductive tract. Blood Cells Mol Dis 35(1): 1-10. [DOI] [PubMed] [Google Scholar]
- 17.Taylor D. D. and Shah S.(2015). Methods of isolating extracellular vesicles impact down-stream analyses of their cargoes. Methods 87: 3-10. [DOI] [PubMed] [Google Scholar]
- 18.Tomasoni S., Longaretti L., Rota C., Morigi M., Conti S., Gotti E., Capelli C., Introna M., Remuzzi G. and Benigni A.(2013). Transfer of growth factor receptor mRNA via exosomes unravels the regenerative effect of mesenchymal stem cells. Stem Cells Dev 22(5): 772-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vojtech L., Woo S., Hughes S., Levy C., Ballweber L., Sauteraud R. P., Strobl J., Westerberg K., Gottardo R., Tewari M. and Hladik F.(2014). Exosomes in human semen carry a distinctive repertoire of small non-coding RNAs with potential regulatory functions. Nucleic Acids Res 42(11): 7290-7304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Welch J. L., Madison M. N., Margolick J. B., Galvin S., Gupta P., Martínez-Maza O., Dash C. and Okeoma C. M.(2017). Effect of prolonged freezing of semen on exosome recovery and biologic activity. Sci Rep 7: 45034. [DOI] [PMC free article] [PubMed] [Google Scholar]


