Supplemental Digital Content is available in the text
Keywords: differentially expressed genes, hepatocellular carcinoma, KEGG pathway, miRNA-target genes, protein-protein interaction network
Abstract
Objective:
We aimed to identify some pivotal genes and pathways for hepatocellular carcinoma (HCC) transformation from cirrhosis and explore potential targets for treatment of the disease.
Methods:
The GSE17548 microarray data were downloaded from Gene Expression Omnibus database, and 37 samples (20 cirrhosis and 17 HCC samples) were used for analysis. The differentially expressed genes (DEGs) in HCC tissues were compared with those in cirrhosis tissues and analyzed using the limma package. Gene ontology-biological process and Kyoto encyclopedia of genes and genomes (KEGG) pathway enrichment analyses were performed using ClueGO and CluePedia tool kits, and the key KEGG pathway was analyzed using the R package pathview. The regulatory factor miRNA of DEGs was extracted from 3 verified miRNAs-target databases using the multiMiR R package. Moreover, a protein-protein interaction (PPI) network was constructed using the Cytoscape software.
Results:
DEGs including cyclin-dependent Kinase 1 (CDK1), PDZ-binding kinase (PBK), ribonucleotide reductase M2 (RRM2), and abnormal spindle homolog, and microcephaly-associated drosophila (ASPM) were the hub proteins with higher degrees in the PPI network. The cell cycle pathway (CDK1 enriched) and p53 signaling pathway (CDK1 and RRM2 enriched) were significantly enriched by DEGs.
Conclusion:
CDK1, PBK, RRM2, and ASPM may be key genes for HCC transformation from cirrhosis. Furthermore, cell cycle and p53 signaling pathways may play vital mediatory roles; CDK1 may play crucial roles in HCC transformed from cirrhosis via cell cycle and p53 signaling pathways, and RRM2 might be involved in HCC transformed from cirrhosis via the p53 signaling pathway.
1. Introduction
Hepatocellular carcinoma (HCC), caused by specific risk factors including aflatoxin exposure and hepatitis virus infections, is the third major cause of cancer-related mortality all over the world.[1,2] The incidence of HCC increases with aging and reaches the peak at age of 70 years.[3] Besides, HCC is 2.4 times more likely to occur in men than in women.[4] It is estimated that >600,000 new HCC cases are diagnosed worldwide every year, and 55% of these cases are in China.[4] Therefore, it is urgent to identify effective treatments for HCC, and additional studies on the molecular mechanisms of HCC may provide new options for the treatment of this disease.
Some reports on the molecular mechanisms of HCC have been published recently. Several studies showed that stathmin could promote cell proliferation and suppress cell apoptosis by regulating the expression of tumor proliferation-related genes, and that it is involved in the development of HCC.[5–8] Furthermore, Jiang et al found that signal transducer and activator of transcription 4 (STAT4) and α β heterodimer of type MHC Class II (HLA-DQ) were 2 genetic susceptibility loci for hepatitis B virus-related HCC.[9] However, AT rich interactive domain 2 (ARID2) is a tumor suppressor gene of HCC,[10] and tumor protein P53 (TP53) and β-catenin are the most frequently mutated tumor suppressor gene and oncogene in HCC, respectively.[11] The Janus Kinase (JAK)/STAT pathway and Wnt/β-catenin pathway may play roles in HCC acting as 2 major oncogenic drivers,[11] and Wnt/β-catenin pathway may cooperate with oxidative stress metabolism in HCC and ras/mitogen-activated protein kinase (MAPK) pathways.[12] Furthermore, microRNA-7 (miR-7), a tumor suppressor, can reverse the metastasis and inhibit the tumorigenesis of HCC by phosphoinositide 3-kinase (PI3K)/protein kinase B (Akt)/mammalian target of rapamycin (mTOR)-signaling pathway.[13] microRNA-26a (MiR-26a) can suppress tumor growth and metastasis of HCC via targeting the interleukin-6-Stat3 pathway.[14] However, the molecular mechanisms leading to the development and progression of HCC have not been fully demonstrated.
Using the microarray data of GSE17548, a previous study indicated that senescence bypass played a central role in HCC engendering systematic changes,[15] and 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase (ATIC), cell cyclin D1 (CCND1), and amino acid metabolism pathways were important genes and pathways related to HCC, respectively.[2] In the present study, using the microarray data of GSE17548, we analyzed the differentially expressed genes (DEGs) in HCC tissues and compared them with those in cirrhosis tissues. Thereafter, pathway enrichment analyses for DEGs were performed and miRNA-target genes were predicted. Furthermore, a protein-protein interaction (PPI) network was constructed. We aimed to identify some pivotal genes and pathways for HCC transformed from cirrhosis, and explore potential targets for the treatment of the disease.
2. Methods
2.1. Microarray data
The microarray data of GSE17548 were downloaded from the Gene Expression Omnibus (GEO) database, which was based on the platform of GPL570 ([HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array, Affymetrix Inc, Ankara, Turkey) and deposited by Ozturk et al.[15] A total of 37 samples, including 20 cirrhosis samples and 17 HCC samples, were used for the analysis. Samples were derived from patients aged between 41 and 69 years, comprising more males than females. The etiology of these samples was hepatitis B virus (HBV), hepatitis C virus (HCV), or HBV + HDV (hepatitis D virus). Tissues in HCC samples were resected from inside the tumor, whereas those for cirrhosis samples were resected from adjacent tissue surrounding the tumor.
2.2. DEG analysis
The raw expression data were preprocessed by robust multiarray average (RMA)[16] algorithm using the Affy package in Bioconductor. After the preprocessing of the raw expression data, the DEGs in HCC tissues were compared with those in cirrhosis tissues and analyzed using the unpaired t test provided by limma package[17] in R/Bioconductor. |logFC| ≥1 and P value <.05 were considered as threshold values.
2.3. Pathway enrichment analysis
The Kyoto Encyclopedia of Genes and Genomes (KEGG) is a database used to map associated genes to their respective pathways.[18] ClueGO,[19] a Cytoscape plug-in, can decipher functionally grouped Gene Ontology (GO) and pathway annotation networks with a hypergeometric test, calculate the kappa coefficient[20] of pathways, and study functional correlations among pathways. CluePedia,[21] another Cytoscape plug-in, is a tool used to search new markers potentially related with pathways. Furthermore, ClueGO combined with CluePedia can derive better results. In the present study, KEGG pathway enrichment analysis was performed using ClueGO and CluePedia tool kits to study the functions of DEGs. A P value of <.05 and kappa coefficient of 0.4 were considered as threshold values. Furthermore, the key KEGG signaling pathway was analyzed using the R package pathview.[22]
2.4. Predictions of miRNA-target genes
The multiMiR R package[23] integrates miRNAs-target interactions with their disease and drug associations. The results of 14 databases were synthesized and merged into 3 categories including 3 verified miRNAs-target databases (miRecords, miRTarBase, and TarBase), 8 predicted miRNAs-target databases (DIANA-microT, ElMMo, MicroCosm, miRanda, miRDB, PicTar, PITA, and TargetScan), and 3 disease-/drug-related miRNA databases (miR2Disease, PharmacomiR, and PhenomiR) in this multiMiR R package.
The regulatory factor miRNA of DEGs was extracted from 3 verified miRNAs-target databases with multiMiR R package. We selected at least 2 verified relationship pairs to construct miRNA-DEGs regulatory networks. Furthermore, the gene ontology-biological process (GO-BP) was performed for target genes of key miRNA using ClueGO and CluePedia tool kits. A P value of <.05 was considered as the threshold value.
2.5. Construction of PPI network
The Search Tool for the Retrieval of Interacting Genes (STRING) database can provide predicted and experimental information of protein interactions.[24] The DEGs were mapped into PPIs and a combined score of >0.7 was identified as the cutoff standard. The PPI network was constructed using Cytoscape software.[25]
3. Results
3.1. Data preprocessing
After background correction and normalization, a total of 20,108 genes were obtained in 37 HCC and cirrhosis tissue samples (Fig. 1).
Figure 1.
Boxplots of sample data. The lateral axis represents the names of samples, and the longitudinal axis represents the genes. Red represents hepatocellular carcinoma samples and blue represents cirrhosis samples.
3.2. DEGs and pathway enrichment analyses
As shown in Figure 2, a total of 224 DEGs including 77 downregulated and 147 upregulated genes (supplementary Table 1) were identified in HCC tissues compared to in cirrhosis tissues, using |logFC| ≥1 and P value <.05 as threshold values. The function annotation of DEGs is shown in Figure 3. The significantly enriched pathways, by DEGs, included the cell cycle pathway, p53 signaling pathway, caffeine metabolism pathway, primary bile acid biosynthesis pathway, bile secretion pathway, and retinol metabolism pathway. Furthermore, cyclin-dependent kinase 1 (CDK1) was enriched in the cell cycle pathway and p53 signaling pathway, and ribonucleotide reductase M2 (RRM2) was enriched in the p53 signaling pathway.
Figure 2.
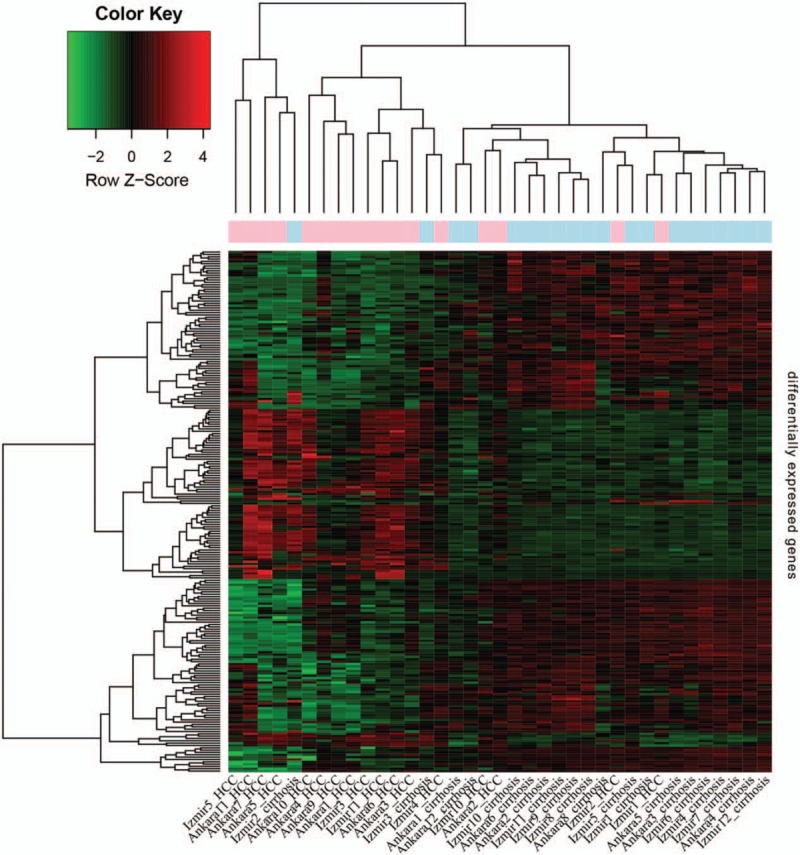
Heat maps of the differentially expressed genes in hepatocellular carcinoma tissues compared to in cirrhosis tissues. The lateral axis represents the names of samples, and the longitudinal axis represents the genes. Red represents hepatocellular carcinoma samples, whereas blue represents cirrhosis samples.
Figure 3.
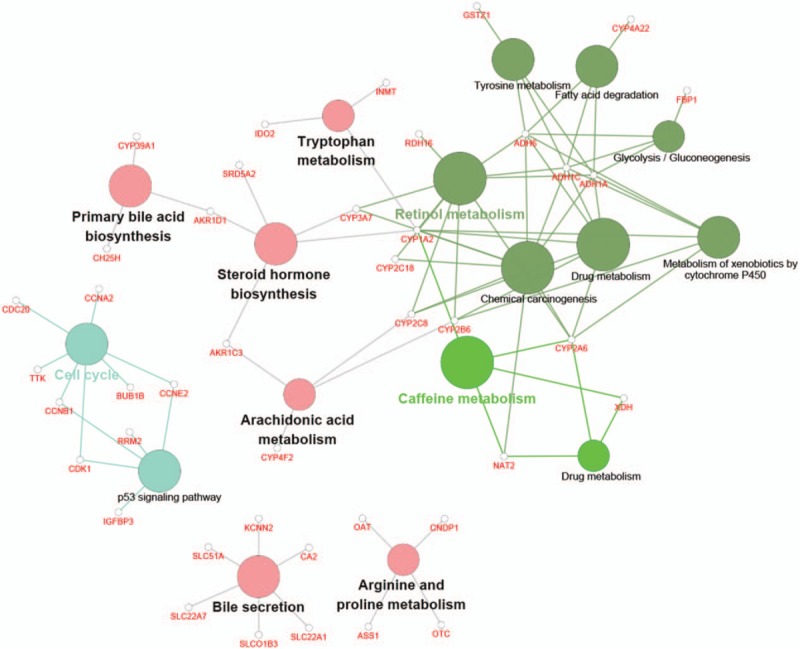
Function annotation of differentially expressed genes. Each node is a Kyoto Encyclopedia of Genes and Genomes pathway item. The node size reflects pathway significance (fdr): the smaller the fdr value, the larger the node size is. Edge between nodes reflects shared or common genes: the wider the edge, the larger the overlap is. Different node colors represent different functional groups. The most significant pathway of each group has a highlighted label.
3.3. Regulatory networks of miRNA-DEGs
Most of the DEGs were regulated by hsa-miRNA-193b-3p, hsa-miRNA-192–5p, hsa-miRNA-26b-5p, and hsa-miRNA-335–5p (Fig. 4; supplementary Table 2). The hsa-miRNA-26b-5p and hsa-miRNA-335–5p targeted DEGs were not significantly enriched in any GO-BP terms. The hsa-miRNA-192–5p targeted DEGs played a role in cytokinesis and sister chromatid segregation. The hsa-miRNA-193b-3p targeted DEGs also influenced microtube cytoskeleton organization involved in mitosis and sister chromatid segregation.
Figure 4.
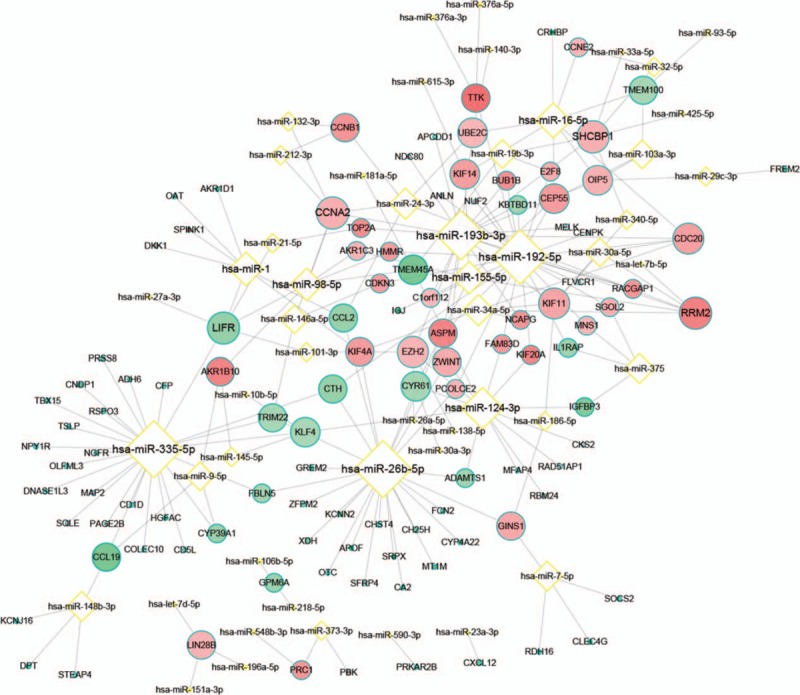
Regulatory networks of miRNA-differentially expressed genes. Yellow border diamond node represents miRNA, whereas blue green border round node represents genes. Red nodes represent upregulation and green nodes represent downregulation. The lighter the nodes, the larger the degrees of differences and |logFC| are. The higher the degree, the bigger the size is.
3.4. PPI network analysis
As shown in Figure 5, a total of 796 edges and 94 nodes were obtained based on STRING database, and a group of upregulated DEGs was gathered into a cluster. Furthermore, CDK1, PDZ-binding kinase (PBK), RRM2, and abnormal spindle homolog, microcephaly associated Drosophila (ASPM) were found to be the hub proteins with higher degrees in the PPI network.
Figure 5.
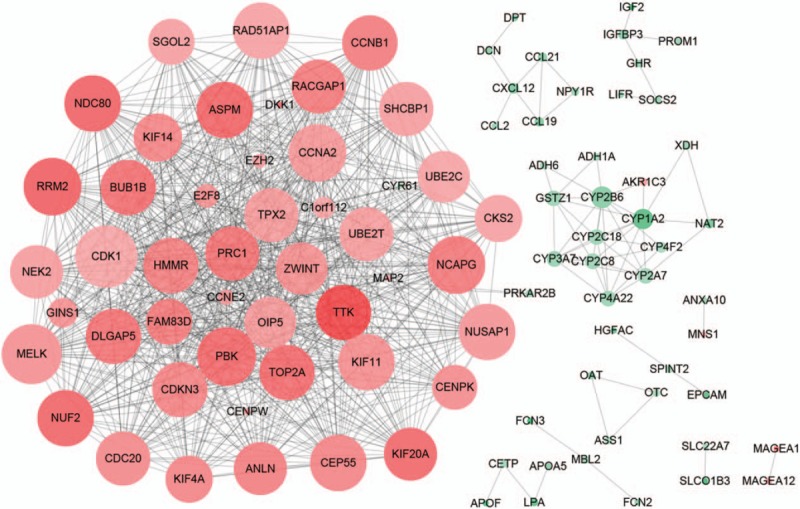
Protein-protein interaction network of differentially expressed genes. Red nodes represent upregulation, whereas green nodes represent downregulation. The lighter the nodes, the larger the degree of difference is. The higher the degree, the bigger the size is.
4. Discussion
In the present study, a total of 224 DEGs, including 77 downregulated and 147 upregulated genes, were identified in HCC tissues compared to in cirrhosis tissues. Our results showed that CDK1, PBK, RRM2, and ASPM were the hub proteins with higher degrees in the PPI network. Furthermore, the cell cycle and p53 signaling pathways were significantly enriched by DEGs. In addition, CDK1 was enriched in the cell cycle and p53 signaling pathways, and RRM2 was enriched in the p53 signaling pathway.
A previous study showed that CDK1 plays a role in the development of HCC by regulating the subcellular localization of apoptin.[26] Zhang et al[27] suggested that miR-582–5p regulated the development of HCC by inhibiting the expression of CDK1. A separate study also showed that PBK can promote tumor cell proliferation via regulation of the DNA damage response and p38 mitogen-activated protein kinase activity.[28] It is also known that high cell proliferation is closely associated with tumor recurrence in HCC.[29] SCY1-like 1-binding protein 1 (SCYL1-BP1) affects cell cycle arrest in HCC cells through RRM2 and Cyclin F.[30] Small-interfering RNA-mediated knockdown of RRM2 can depress HCC cell proliferation.[31] Furthermore, Lee et al[32] indicated that high expression of RRM2 could be a useful marker to predict early recurrence of HCC following curative hepatectomy. ASPM plays important roles in cell proliferation and neurogenesis, and enhanced invasive/metastatic potential in HCC can be predicted by ASPM overexpression.[33] Wurmbach et al[34] indicated that ASPM could be as a potential marker for early HCC. Therefore, our results are in line with previous findings and suggest that CDK1, PBK, RRM2, and ASPM may be key genes associated with the development of HCC.
In this study, cell cycle and p53 signaling pathways were significantly enriched by DEGs. The p110γ, one subclass of PI3K, can promote cell proliferation in HCC by controlling the cell cycle checkpoint.[35] Furthermore, miR-138 is a frequently downregulated miRNA in HCC, and can cause cell cycle arrest in HCC by targeting cyclin D3.[36] Besides, oleanolic acid can play inhibitory effects on HCC via cell cycle arrest and apoptosis mediated by extracellular signal–regulated kinase-p53 (ERK-p53).[37] Thus, the cell cycle pathway plays important mediatory roles in the development of HCC. The p53 signaling pathway is a potent barrier for tumor progression, and p53 plays important roles in hepatocarcinogenesis.[38,39] Moreover, p53 is a significant correlative regulatory factor of the α-fetoprotein (AFP) expression in HCC,[40] and Tu et al[41] found that the activation of F-box and WD repeat domain-containing 7 (Fbxw7) via adenoviral delivery of p53 caused increased proteasomal degradation of cyclin E and c-Myc, and thus recombinant human adenovirus-p53 injection could be a possible therapeutic agent for HCC. Thus, the p53 signaling pathway also plays significant mediatory roles for the progression of HCC. Therefore, our results are in line with those from previous studies and suggest that cell cycle and p53 signaling pathways play vital mediatory roles in the development of HCC.
As stated above, CDK1 and RRM2 may be key genes associated with the development of HCC, and cell cycle and p53 signaling pathways play vital roles in the development of HCC. Furthermore, in the present study, CDK1 was enriched in cell cycle and p53 signaling pathways, whereas RRM2 was enriched in the p53 signaling pathway. This suggests that CDK1 affects HCC via its roles in the cell cycle and p53 signaling pathways, and RRM2 affects HCC via its role in the p53 signaling pathway.
There were few limitations in the present study. Previous studies showed that the risk factors of HCC included hepatitis B and C infections, cirrhosis, and alcohol intake,[42] and cirrhosis was the largest risk factor, accounting for 80% to 90% of the total number of HCC cases.[43] In total, 1% to 1.5% of cirrhosis cases are transformed to HCC every year,[44] indicating the potential to identify molecular mechanisms associated with HCC via comparisons between HCC and cirrhosis. In this study, we performed bioinformatics analysis for microarray data, including 20 cirrhosis samples and 17 HCC samples. Several key genes and pathways were found to be important for HCC transformed from cirrhosis. In this study, we did not perform the analysis of the normal expression data, which, in addition to HCC and cirrhosis data, could provide more convincing findings. As such, we will carry out a comprehensive analysis including normal expression data, cirrhosis data, and HCC data in the future. Furthermore, we did not analyze other publicly available HCC data from GEO in parallel with adjacent normal tissues. A step-wise progressive analysis including other publicly available HCC data from normal to cirrhosis to HCC is needed in future studies.
5. Conclusions
In conclusion, CDK1, PBK, RRM2, and ASPM may be key genes for HCC transformed from cirrhosis. Furthermore, cell cycle and p53 signaling pathways play crucial roles in the development of HCC from cirrhosis. CDK1 may play important roles in HCC transformed from cirrhosis via cell cycle and p53 signaling pathways, and RRM2 may play a role in HCC transformed from cirrhosis via the p53 signaling pathway. However, owing to lack of experimental verification, normal expression data, and adequate sample size, further studies on the molecular mechanisms of HCC are needed.
Supplementary Material
Supplementary Material
Footnotes
Abbreviations: Akt = protein kinase B, ASPM = Abnormal Spindle Homolog, Microcephaly Associated Drosophila, ATIC = 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase, CCND1 = cell cyclin D1, CDK1 = cyclin-dependent kinase 1, DEGs = differentially expressed genes, GEO = Gene Expression Omnibus, GO-BP = gene ontology-biological process, HCC = hepatocellular carcinoma, HLA-DQ = α β heterodimer of type MHC Class II, JAK = Janus kinase, KEGG = Kyoto Encyclopedia of Genes and Genomes, miR-26a = microRNA-26a, mTOR = mammalian target of rapamycin, PBK = PDZ-binding kinase, PI3K = phosphoinositide 3-kinase, RRM2 = ribonucleotide reductase M2, STAT4 = signal transducer and activator of transcription 4.
Declarations: Ethics approval: All studies have been approved by the Scientific and Ethical Committee of the second affiliated hospital of Nantong University and performed in accordance with the ethical standards.
Written informed consent was obtained from participants before the study.
Authors’ contributions: B.S.H. and W.B.D. conceived and designed the study; Y.H., S.C.G., B.S.H., and W.X.S. performed the experiments; J.B.Y., B.S.H., and J.X. wrote the article; J.H.G. and W.B.D. reviewed and edited the manuscript; all authors read and approved the manuscript.
This article was supported by the social development fund of Nantong (Contract grant number: HS2014063), the 12th Talent Summit of Top Six Industries in Jiangsu Province (Contract grant number: WSW080), and the Project of Enhancing Medicine with Science and Education under the 13th Five-Year Plan of Nantong (Contract grant number: Nantong MED Sci & Edu [2016] 23).
All authors declare that they have no conflicts of interest.
Supplemental Digital Content is available for this article.
References
- [1].Jemal A, Bray F, Center MM, et al. Global cancer statistics. Cancer J Clin 2011;61:69–90. [DOI] [PubMed] [Google Scholar]
- [2].Shangguan H, Tan S, Zhang J. Bioinformatics analysis of gene expression profiles in hepatocellular carcinoma. Eur Rev Med Pharmacol Sci 2015;19:2054–61. [PubMed] [Google Scholar]
- [3].El-Serag HB, Mason AC. Rising incidence of hepatocellular carcinoma in the United States. N Engl J Med 1999;340:745–50. [DOI] [PubMed] [Google Scholar]
- [4].Parkin DM, Bray F, Ferlay J, et al. Global cancer statistics, 2002. Cancer J Clin 2005;55:74–108. [DOI] [PubMed] [Google Scholar]
- [5].Yuan RH, Jeng YM, Chen HL, et al. Stathmin overexpression cooperates with p53 mutation and osteopontin overexpression, and is associated with tumour progression, early recurrence, and poor prognosis in hepatocellular carcinoma. J Pathol 2006;209:549–58. [DOI] [PubMed] [Google Scholar]
- [6].Li C, Tan YX, Zhou H, et al. Proteomic analysis of hepatitis B virus-associated hepatocellular carcinoma: identification of potential tumor markers. Proteomics 2005;5:1125–39. [DOI] [PubMed] [Google Scholar]
- [7].Chen Y, Lin MC, Yao H, et al. Lentivirus-mediated RNA interference targeting enhancer of zeste homolog 2 inhibits hepatocellular carcinoma growth through down-regulation of stathmin. Hepatology 2007;46:200–8. [DOI] [PubMed] [Google Scholar]
- [8].Singer S, Ehemann V, Brauckhoff A, et al. Protumorigenic overexpression of stathmin/Op18 by gain-of-function mutation in p53 in human hepatocarcinogenesis. Hepatology 2007;46:759–68. [DOI] [PubMed] [Google Scholar]
- [9].Jiang D-K, Sun J, Cao G, et al. Genetic variants in STAT4 and HLA-DQ genes confer risk of hepatitis B virus-related hepatocellular carcinoma. Nat Genet 2013;45:72–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Li M, Zhao H, Zhang X, et al. Inactivating mutations of the chromatin remodeling gene ARID2 in hepatocellular carcinoma. Nat Genet 2011;43:828–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Kan Z, Zheng H, Liu X, et al. Whole-genome sequencing identifies recurrent mutations in hepatocellular carcinoma. Genome Res 2013;23:1422–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Guichard C, Amaddeo G, Imbeaud S, et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet 2012;44:694–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Fang Y, Xue JL, Shen Q, et al. MicroRNA-7 inhibits tumor growth and metastasis by targeting the phosphoinositide 3-kinase/Akt pathway in hepatocellular carcinoma. Hepatology 2012;55:1852–62. [DOI] [PubMed] [Google Scholar]
- [14].Yang X, Liang L, Zhang XF, et al. MicroRNA-26a suppresses tumor growth and metastasis of human hepatocellular carcinoma by targeting interleukin-6-Stat3 pathway. Hepatology 2013;58:158–70. [DOI] [PubMed] [Google Scholar]
- [15].Yildiz G, Arslan-Ergul A, Bagislar S, et al. Genome-wide transcriptional reorganization associated with senescence-to-immortality switch during human hepatocellular carcinogenesis. PLoS One 2013;8:e64016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Gautier L, Cope L, Bolstad BM, et al. affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 2004;20:307–15. [DOI] [PubMed] [Google Scholar]
- [17].Smyth G. Limma: linear models for microarray data Bioinformatics and Computational Biology Solutions using R and Bioconductor 2005. New York: Springer; 2005. [Google Scholar]
- [18].Altermann E, Klaenhammer TR. PathwayVoyager: pathway mapping using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. BMC Genom 2005;6:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Bindea G, Mlecnik B, Hackl H, et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009;25:1091–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ramos H, Shannon P, Aebersold R. The protein information and property explorer: an easy-to-use, rich-client web application for the management and functional analysis of proteomic data. Bioinformatics 2008;24:2110–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Bindea G, Galon J, Mlecnik B. CluePedia Cytoscape plugin: pathway insights using integrated experimental and in silico data. Bioinformatics 2013;29:661–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Luo W, Brouwer C. Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 2013;29:1830–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ru Y, Kechris KJ, Tabakoff B, et al. The multiMiR R package and database: integration of microRNA–target interactions along with their disease and drug associations. Nucleic Acids Res 2014;42:e133–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Szklarczyk D, Franceschini A, Kuhn M, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res 2011;39suppl 1:D561–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Kohl M, Wiese S, Warscheid B. Cytoscape: software for visualization and analysis of biological networks. Methods Mol Biol 2011;696:291–303. [DOI] [PubMed] [Google Scholar]
- [26].Zhao J, Han S-X, Ma J-L, et al. The role of CDK1 in apoptin-induced apoptosis in hepatocellular carcinoma cells. Oncol Rep 2013;30:253–9. [DOI] [PubMed] [Google Scholar]
- [27].Zhang Y, Huang W, Ran Y, et al. miR-582-5p inhibits proliferation of hepatocellular carcinoma by targeting CDK1 and AKT3. Tumor Biol 2015. 1–8. [DOI] [PubMed] [Google Scholar]
- [28].Ayllon V, O’connor R. PBK/TOPK promotes tumour cell proliferation through p38 MAPK activity and regulation of the DNA damage response. Oncogene 2007;26:3451–61. [DOI] [PubMed] [Google Scholar]
- [29].Van Nhieu JT, Renard CA, Wei Y, et al. Nuclear accumulation of mutated (-catenin in hepatocellular carcinoma is associated with increased cell proliferation. Am J Pathol 1999;155:703–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Wang Y, Zhi Q, Ye Q, et al. SCYL1-BP1 affects cell cycle arrest in human hepatocellular carcinoma cells via Cyclin F and RRM2. Anticancer Agents Med Chem 2016;16:440–6. [DOI] [PubMed] [Google Scholar]
- [31].Satow R, Shitashige M, Kanai Y, et al. Combined functional genome survey of therapeutic targets for hepatocellular carcinoma. Clin Cancer Res 2010;16:2518–28. [DOI] [PubMed] [Google Scholar]
- [32].Lee B, Ha SY, Song DH, et al. High expression of ribonucleotide reductase subunit M2 correlates with poor prognosis of hepatocellular carcinoma. Gut Liver 2014;8:662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Lin S-Y, Pan H-W, Liu S-H, et al. ASPM is a novel marker for vascular invasion, early recurrence, and poor prognosis of hepatocellular carcinoma. Clin Cancer Res 2008;14:4814–20. [DOI] [PubMed] [Google Scholar]
- [34].Wurmbach E, Chen Yb, Khitrov G, et al. Genome-wide molecular profiles of HCV-induced dysplasia and hepatocellular carcinoma. Hepatology 2007;45:938–47. [DOI] [PubMed] [Google Scholar]
- [35].Dituri F, Mazzocca A, Lupo L, et al. PI3K class IB controls the cell cycle checkpoint promoting cell proliferation in hepatocellular carcinoma. Int J Cancer 2012;130:2505–13. [DOI] [PubMed] [Google Scholar]
- [36].Wang W, Zhao L-J, Tan Y-X, et al. MiR-138 induces cell cycle arrest by targeting cyclin D3 in hepatocellular carcinoma. Carcinogenesis 2012;33:1113–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Wang X, Bai H, Zhang X, et al. Inhibitory effect of oleanolic acid on hepatocellular carcinoma via ERK–p53-mediated cell cycle arrest and mitochondrial-dependent apoptosis. Carcinogenesis 2013;34:1323–30. [DOI] [PubMed] [Google Scholar]
- [38].Edamoto Y, Hara A, Biernat W, et al. Alterations of RB1, p53 and Wnt pathways in hepatocellular carcinomas associated with hepatitis C, hepatitis B and alcoholic liver cirrhosis. Int J Cancer 2003;106:334–41. [DOI] [PubMed] [Google Scholar]
- [39].Yang Y, Xia T, Li N, et al. Combined effects of p53 and MDM2 polymorphisms on susceptibility and surgical prognosis in hepatitis B virus-related hepatocellular carcinoma. Protein Cell 2013;4:71–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Peng SY, Chen WJ, Lai PL, et al. High (-fetoprotein level correlates with high stage, early recurrence and poor prognosis of hepatocellular carcinoma: significance of hepatitis virus infection, age, p53 and (-catenin mutations. Int J Cancer 2004;112:44–50. [DOI] [PubMed] [Google Scholar]
- [41].Tu K, Zheng X, Zhou Z, et al. Recombinant human adenovirus-p53 injection induced apoptosis in hepatocellular carcinoma cell lines mediated by p53-Fbxw7 pathway, which controls c-Myc and cyclin E. PLoS One 2013;8:e68574. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [42].Sherman M. Thieme Medical Publishers, Hepatocellular Carcinoma: Epidemiology, Risk Factors, and Screening. Seminars in Liver Disease. New York, NY: 2005. [DOI] [PubMed] [Google Scholar]
- [43].Nordenstedt H, White DL, El-Serag HB. The changing pattern of epidemiology in hepatocellular carcinoma. Dig Liver Dis 2010;42:S206–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Liu C-J, Kao J-H. Hepatitis B virus-related hepatocellular carcinoma: epidemiology and pathogenic role of viral factors. J Chinese Med Assoc 2007;70:141–5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


