Abstract
Cytomegaloviruses (CMVs) are members of the Betaherpesvirinae subfamily of the Herpesviridae, and their properties of latency, large DNA size, gene redundancy, and ability to be cloned as bacterial artificial chromosomes (BACs) suggest their utility as vaccine vectors. While the K181 strain of murine CMV (MCMV) is widely used to study MCMV biology, a BAC clone of this virus had not previously been produced. We report here the construction of a BAC clone of the K181Perth strain of MCMV. The in vivo and in vitro growth characteristics of virus derived from the K181 BAC were similar to those of wild-type K181. The utility of the K181 BAC as a method for the rapid production of vaccine vectors was assessed. A vaccine strain of BAC virus, expressing the self-fertility antigen, murine zona pellucida 3, was produced rapidly using standard bacterial genetics techniques and rendered female BALB/c mice infertile with a single intraperitoneal inoculation. In addition, attenuated vaccine strains lacking the open reading frames m07 to m12 exhibited no reduction in efficacy compared to the full-length vaccine strain. In conclusion, we describe the production of a K181-based BAC virus which behaved essentially as wild-type K181 and allowed the rapid production of effective viral vaccine vectors.
The use of viruses as vectors for the delivery of heterologous antigens has been considered because the immune system has evolved a sophisticated array of mechanisms to both detect and eliminate invading viruses. A viral vector also delivers the antigen directly into host cells, which allows for high-level intracellular expression. Hence, the viral vector acts as an adjuvant and as a delivery system. An effective viral vector should present the expressed antigen as an immune target and should remain in the host long enough to stimulate an effective response. A large range of viruses have been identified as potential vaccine vectors, including poliovirus (3, 27), vaccinia virus (40), rabies virus (31), adenovirus (47), and canarypox (7, 30). Viral vector-based vaccines that target infectious diseases (31, 47) as well as malignancies (27, 30) have been developed. The use of herpesviruses, such as herpes simplex virus, as vaccine vectors has also been explored (9, 36). The use of cytomegalovirus (CMV) has also been explored. Murine CMV (MCMV) has been used as viral vectored immunocontraceptive (VVIC) (24) and has been used in human dendritic cells to prime naïve CD8+ T cells to human immunodeficiency virus type 1 gp120 antigen (56).
CMVs are members of the Betaherpesvirinae subfamily of the Herpesviridae and contain a large double-stranded DNA genome of approximately 230,000 bp. CMVs cause acute benign infections in immunocompetent animals that persist in a latent state for the lifetime of the host. They are found in a broad range of mammalian species but exhibit strict species specificity (51). Their properties of latency, large DNA size, and strict species specificity make CMVs useful vaccine vectors.
Vaccine vectors are required to induce strong immune responses. CMVs induce strong and long-lived cytotoxic T-lymphocyte (CTL) responses (22) as well as long-lived antibody responses (2). It is likely that MCMV persistence and reactivation are involved in the long-lived immune responses to MCMV. We have previously shown that these long-lived responses to MCMV are also directed to MCMV-expressed antigen. Antibody to MCMV-expressed antigen was detected as late as day 108 after a single intraperitoneal (i.p.) inoculation of virus (24). Safety is also a requirement for a useful vaccine vector. CMVs are typically nonpathogenic in immunocompetent hosts, making them potentially safer than other vectors such as lentivirus. However, for use in humans, it may be important to produce vaccine vectors that are attenuated because of the number of immunocompromised individuals in the population. This should be technically feasible, as vaccine strains of MCMV that replicate poorly in vivo yet protect from challenge with virulent wild-type (wt) MCMV have been produced (26, 35). All these data suggest that CMV may make a useful, safe vaccine vector that can induce long-lived humoral and cell-mediated immune responses to heterologous antigen.
A potential advantage to the use of CMVs as viral vaccine vectors is the ability of these viruses to be cloned as bacterial artificial chromosomes (BACs). This has significantly enhanced the study and genetic manipulation of CMV allowing molecular tools designed for Escherichia coli to be used on herpesviruses (reviewed in reference 55). These techniques greatly enhance the ease of mutagenesis of herpesviruses and allow for rapid insertion of candidate antigens. The cloning of a herpesvirus (MCMV) as a BAC was first described by Messerle and colleagues in 1997 (33). BAC clones have subsequently been produced for a human CMV, as well as a guinea pig CMV and a rhesus monkey CMV (11, 15, 55). Other herpesviruses, such as murine gammaherpesvirus 68, pseudorabies virus, Epstein-Barr virus, Kaposi's sarcoma-associated herpesvirus, herpes simplex virus type 1, equine herpesvirus type 1, and Marek's disease virus, have also been cloned as BACs (42, 55, 59)
Two viral strains, Smith and K181, are widely used in the study of MCMV. However, only the Smith strain of MCMV has been cloned as a BAC (33, 54). The K181 strain of MCMV was first described as a Smith strain variant and was isolated from the salivary glands of mice after serial passage in vivo (34). Given the different restriction fragment length polymorphisms (RFLP) and growth kinetics reported for these strains, it is possible that K181 is a novel strain resulting from viral reactivation in a mouse inoculated with the Smith strain (19, 34). In vivo, the K181 strain of MCMV was originally reported to replicate to higher titers in the salivary glands of infected mice and to demonstrate enhanced mortality in newborn mice (34). In vitro, the K181 strain of MCMV was reported to cause smaller plaques than the Smith strain of MCMV and to grow to lower titers (19). However, there have been no recent comparisons of the virulence of these two strains of MCMV, and the Smith strain of virus used in the studies of Misra and Hudson in the 1980s (19, 34) gave different RFLP patterns that those reported by Ebeling et al. in 1983 (14) and Mercer et al. in 1983 (32). The origins of the Perth strain of K181 MCMV have been described elsewhere (8).
We and others have used the K181 strain of MCMV extensively for studies on MCMV host-virus interactions (5, 10, 35, 45, 49, 53). The K181 strain of MCMV has also been used in vaccination studies (35), and we have used the K181Perth strain of MCMV as a vaccine vector (24). Consequently, we set out to develop a K181 BAC that would facilitate all components of our research.
We describe here the production of a K181Perth-based BAC and its use as a vaccine vector. While MCMV BAC DNA has recently been used as a DNA vaccine for protection studies either as naked DNA (50, 52) or delivered in vivo by bacterial cells (12), the use of a BAC-derived virus as a vaccine vector has not been reported. We wished to test the efficacy of the K181 BAC as a vaccine vector and as a means of rapid vaccine production. The antigen chosen for study was murine zona pellucida 3 (mZP3), as we have previously shown that this antigen can be successfully used for MCMV-mediated VVIC (24). Originally, this antigen was chosen as part of an ongoing study into the potential use of a disseminating VVIC for control of free-living house mice (Mus musculus domesticus). House mice periodically erupt to plague proportions in Australia. However, mZP3 can also be seen as a model antigen, with female mouse fertility used as a readout of immunity. Finally, because persistence of CMV in immunocompromised hosts or the environment may be undesirable, we sought to determine whether attenuation of the MCMV vector would affect vaccine efficacy.
The K181 BAC-derived virus produced in these studies behaved in vitro and in vivo as wt K181 did. Additionally, a VVIC that effectively sterilized female BALB/c mice was produced. This is the first description of a K181-based BAC and its use as a VVIC. Finally, the attenuation of the vector had no effect on vaccine efficacy.
MATERIALS AND METHODS
Virus and cells.
The origins of the K181 strain of MCMV have been described previously (8). The RM427+ virus (a K181-based recombinant virus containing a LacZ cassette) was kindly provided by E. Mocarski (Stanford University, Stanford, Calif.). RM427+ is similar to the RM427 virus (49) with the exception that the integrity of the ssg1 gene is maintained (29). Virus stocks were propagated in mouse embryonic fibroblasts (MEFs) as previously described (13). Viral titers were determined in duplicate by plaque assay in MEFs (24). The bone marrow stromal cell line M2-10B4 (25) was used for the selection of recombinant viruses using mycophenolic acid and xanthine.
Animals.
Specific-pathogen-free BALB/c mice were obtained from the Animal Resource Centre (Murdoch, Perth, Western Australia, Australia) and housed under minimal disease conditions. Mouse care was based on the Australian Code of Practice and was approved by the University of Western Australia Animal Experimentation and Ethics Committee. Sentinel animals were found to be free of a suite of murine pathogens, including MCMV, after routine testing.
Isolation of viral and BAC DNA.
MCMV DNA was extracted as previously described (54). Briefly, two T80 flasks of MEFs or M2-10B4 cells were infected with virus, and when the cytopathic effect (CPE) had reached 100%, the cells were harvested by freezing and scraping. Cell debris was pelleted at 850 × g for 10 min, and the supernatant was filtered through Whatman filter paper directly into centrifuge tubes. Virus was pelleted at 29,000 × g for 30 min at 4°C, and the pellet was resuspended in 500 μl of DNase I buffer (50 mM Tris HCl [pH 8.0], 5 mM MgCl2, 0.1 M sodium acetate, 100 μg of bovine serum albumin per ml). Extraviral DNA was digested for 1 h in 0.2 U of DNase I (Invitrogen, Carlsbad, Calif.) at room temperature, and the reaction was stopped with 20 μl of 500 mM EDTA (pH 8.0). Virus was treated with 500 μl of 1% sodium dodecyl sulfate (SDS) and 40 μl of proteinase K (20 mg/ml) for a minimum of 4 h at 56°C. Viral DNA was purified by phenol-chloroform extraction and precipitated in 1 volume of isopropanol. For the isolation of low-molecular-weight circular viral DNA, the method of Hirt was followed (17). Briefly, infected cells from a 10-cm-diameter dish were resuspended in 500 μl of 20 mM EDTA (pH 8.0) and then lysed with 500 μl of 1.2% SDS. Protein and high-molecular-weight DNA were precipitated by the addition of 660 μl of 5 M NaCl. The sample was incubated overnight at 4°C, and the proteins and high-molecular-weight DNA were removed by centrifugation at 15,000 × g at 4°C for 30 min. The supernatant was removed and extracted with phenol-chloroform. DNA was precipitated with 1 volume of isopropanol, washed twice with 70% ethanol, and resuspended in 50 μl of Tris-EDTA. BACs were isolated from E. coli by the alkaline lysis method (44) for screening and with the Nucleobond plasmid kit (Clontech, Palo Alto, Calif.) for purified DNA used in RFLP analysis and transfections.
Plasmids.
Plasmids pAJR02 and pAJR10 were produced using DNA from RM427+ virus. For construction of the recombination plasmid pAJR02, two PCR fragments were cloned into the plasmid pK18 (39). All sequence information and coordinates in this study were taken from the published Smith strain sequence (41) (accession number U68299). PCR primers were designed to amplify the right homology arm with EcoRI restriction enzyme (RE) sites on both primers. The RE sites are shown underlined (forward primer, 5′-GCA GAA TTC TCG ACC GCT TCA AAT GAT TGG GTT CG-3′; reverse primer, 5′-CGC GAA TTC CTT CGA GTT TAG ACA GCC GGT CAG TTG-3′). The region amplified spanned open reading frames (ORFs) m12 to m14 (nucleotides [nt] 12488 to 13576), and this region was cloned into pK18 to produce the plasmid pG4R. PCR primers were designed to amplify the left homology arm with a NotI site on the forward primer and an AvrII site on the reverse primer (forward primer, 5′-TCT GCG GCC GCT TGG AGC GAT ACG TTG ACA ATG-3′; reverse primer, 5′-CCC CCT AGG GTA CGC GAG CCA TAC GCA GAA CAC CA-3′). The PCR product containing the region from ORFs m06 to m07 (nt 5308 to 6522) was directionally cloned into pG4R to produce pG4RL. Finally, the recombination plasmid pAJR02 was produced by cloning the PacI fragment of pHA2 (1) containing the BAC vector, the guanosine phosphoribosyltransferase (gpt) gene from pKSO-gpt (33), and the enhanced green fluorescent protein (EGFP) gene from pEGFP-CI (Clontech) into the PacI site of pG4RL.
A repair plasmid, pAJR10, was constructed to replace the deleted region, nt 6522 to 12488, after insertion of the BAC cassette into the K181 virus. A new polylinker was inserted into pUC19 to create pAJR03. The polylinker was inserted into HindIII and EcoRI sites of pUC19 with destruction of these sites after ligation. The polylinker provided sites for BglII, DraI, PmeI, MluI, EagI, NotI, PacI, NcoI, DraI, PmeI, and BglII (5′-AAT TGA GAT CTG TTT AAA CAC GCG TCC GCG GCC GCT TAA TTA AGG GCC ATG GTT TAA ACA GAT CT-3′ and 5′-AGC TAG ATC TGT TTA AAC CAT GGC CCT TAA TTA AGC GGC CGC GGA CGC GTG TTT AAA CAG ATC TC-3′). The left homology arm for homologous recombination was provided by a BspHI and NotI double digest fragment of pHA2 containing the chloramphenicol resistance gene and the gpt gene which was cloned into the NotI and NcoI sites of pAJR03 to produce pAJR04. An additional polylinker (containing a SnaBI site and a MfeI site; 5′-CGC GCA ATT GGC CCC TAC GTA CCC GC-3′ and 5′-GGC CGC GGG TAC GTA GGG GCC AAT TG-3′) was cloned into the NotI and MluI sites of pAJR04 to produce pAJR04pl. To clone the MCMV component of the shuttle vector pAJR10, a 13-kb BglII fragment of the RM427+ virus was cloned into pUC19 to produce pAJR07. Plasmid pAJR08 was produced by subcloning a 8.6-kb fragment bound by the EagI site found at nt 6273 and the EcoRI site at nt 14964 of MCMV in pAJR07 into the NotI/MfeI site of PAJR04pl. Finally, the entire construct was removed from pAJR08 by BglII digestion and subcloned into the BamHI site of the shuttle vector pST76K-SR (18) to produce the repair vector pAJR10.
The vaccine strains of BAC-derived virus were produced by insertion of a mZP3 cDNA clone into the nonessential (10) immediate-early 2 (ie2) gene of MCMV as previously described (24). The pCMH492 plasmid containing the mZP3 cDNA under the control of the HCMV ie1 gene promoter and a simian virus 40 (SV40)-derived transcription termination signal inserted within the HindIII L fragment of MCMV strain K181 was produced as follows. A 1,302-bp BamHI/EcoRI DNA fragment of plasmid pZP3 (20) containing the entire mZP3 cDNA coding region (GenBank accession number M20026) was inserted into the expression vector pCMH411 (GenBank accession number AY122058) to produce pCMH412. Next, a 1,844-bp NotI DNA fragment of pCMH412 containing the mZP3 cDNA under the control of the HCMV ie1 gene promoter and the SV40-derived transcription termination signal was cloned into plasmid pH3LN to produce pCMH478. Plasmid pH3LN was derived from pK181-H3L (24). Plasmid pH3LN contains the entire HindIII L fragment (32) of K181 inserted into plasmid pUC9 with the 79-bp HpaI fragment replaced by a NotI linker. Finally, pCMH492 was produced by inserting a 8,934-bp HindIII DNA fragment of pCMH478 into the BamHI site of the plasmid pST76K-SR using HindIII (HindIII linker, 5′-AGC TTC TGG CGG TCG GGC-3′; universal linker, 5′-ACC GCC AGA-3′) and BamHI (BamHI linker, 5′-ATC TCG GCG GTG CCC G-3′; universal linker, 5′-ACC GCC AGA-3′) end-joining linkers.
Generation of recombinant viruses in tissue culture.
Cultures of MEFs that were 90% confluent were infected with K181 virus at a multiplicity of infection (MOI) of 3 and incubated for 2 h at 37°C in 5% CO2. Cells were harvested by trypsinization, washed, and resuspended in 400 μl of Optimem (GIBCO-BRL, Gaithersburg, Md.). For homologous recombination, 30 μg of the pAJR02 plasmid was digested with MluI and electroporated into infected MEFs. Cells were replated in a 6-cm-diameter plate with Dulbecco modified Eagle medium plus 10% fetal bovine serum and 24 h later were washed and given fresh medium. Cells were monitored daily for the development of plaques. Once the CPE reached 100%, the supernatant from infected cells was applied to the bone marrow stromal cell line M2-10B4. Selection for gpt gene expression was performed in 100 μM mycophenolic acid and 25 μM xanthine. M2-10B4 cells were more resistant to mycophenolic acid and xanthine than MEFs and thus more suitable for the selection procedure. Circular DNA was extracted from cells once 100% of the cells showing CPE were visibly expressing the EGFP gene. Extracted DNA was used to transform E. coli DH10B cells by electroporation, and the bacterial colonies resistant to chloramphenicol were selected. BAC clones were identified by RFLP analysis. One full-length clone, designated pARK14, was selected for repair of the deleted ORFs m07 to m12.
BAC mutagenesis and virus rescue.
All mutagenesis was performed by a two-step allele replacement method by homologous recombination in E. coli strain DH10B as previously described (54). The pAJR10 plasmid was used for repair of the BAC, pARK14, to produce the full-length K181 BAC pARK25. Insertion of the mZP3 antigen into the K181 BAC HindIII L fragment was accomplished with the pCMH492 plasmid by two-step allele replacement.
Infectious virus was rescued from BAC DNA after transfection in MEFs by the calcium phosphate precipitation method (44). Recovered virus was monitored by fluorescence microscopy for loss of GFP expression (and hence BAC excision). Loss of expression was noted in the first passage. Virus from rescued BAC DNA was passaged five or six times for the production of stocks. Viruses are indicated by the “v” prefix. Thus, when the pARK25 BAC is rescued as a virus, it is designated vARK25. In all immunization experiments, tissue culture virus (TCV) was used, because the attenuation of recombinant MCMV with mZP3 DNA inserted (rMCMV-mZP3) precludes the production of salivary gland virus (SGV) stocks.
Southern blot analysis.
Viral DNA was subjected to RFLP analysis and Southern blotting according to standard methods (44). Probes were a 249-bp PCR amplicon from the pAJR02 plasmid containing the repeat region of pARK25 (forward primer, 5′-TCC GCA TCC CCG CGG ACG GTA AAG-3′; reverse primer, 5′-ACG TAC GCG AGC CAT ACG CAG AA-3′) or a 641-bp mZP3 PCR amplicon from the previously described (24) mZP3-containing virus, recombinant virus K181-ZP3 (forward primer, 5′-ACC CTC GCC CTG TGA GTG GCC-3′; reverse primer, 5′-AAT TAC TAC AGT TGC CAT GGC-3′). Probes were gel purified and digoxigenin labeled according to the manufacturer's instruction (DIG DNA labeling and detection kit; Roche, Indianapolis, Ind.).
Pathogenesis studies.
Viruses were derived from the appropriate BACs and used to infect female BALB/c mice (6 to 8 weeks of age). In all experiments, i.p. inoculations of 2 × 104 PFU of TCV or 1 × 104 PFU of SGV were used. All in vivo experiments were performed twice. Mice were sacrificed at 3, 5, 7, 10 and 18 days postinfection (n = 4 or 5 depending on the experiment). Serum was collected for the determination of immunoglobulin G1 (IgG1) and IgG2a antibodies to mZP3. The liver, lung, spleen, and salivary glands were removed and frozen (−80°C) for titration of infectious virus by plaque assay (24). Ovaries were collected and immediately fixed in Bouin's fixative, sectioned, and stained in hematoxylin. Sera from infected mice were tested by enzyme-linked immunosorbent assay (ELISA) for the presence of IgG1 and IgG2a antibodies to mZP3. For statistical analysis, all organ viral titer results were tested for normality, and the means were compared by a two-tailed t test.
ELISA.
Antibody levels were measured by adding sera from infected mice to ELISA plates coated with mZP3 antigen, prepared as previously described (24), with stepwise additions of biotinylated rat anti-mouse IgG1 or IgG2a (Southern Biotech Association, Inc., Birmingham, Ala.), streptavidin alkaline phosphatase (Amersham Biosciences, Piscataway, N.J.), and p-nitrophenyl phosphate (Sigma, St. Louis, Mo.). Since MCMV induces a polyclonal B-cell response (21, 38), the highest anti-mZP3 antibody level in mice infected with wt K181 virus was chosen as the cutoff for assessing specific anti-mZP3 antibody responses in mice infected with rMCMV-mZP3.
Immunocontraception.
Female BALB/c mice were injected i.p. with 2 × 104 PFU of TCV diluted in buffered saline with the osmolality of mouse serum using vARK25, vARK14, vARK25-mZP3, or vARK14-mZP3. Mice were allowed to mate immediately after infection. The mice were put into groups of three females per male and monitored for litters for 110 days. Cages containing unproductive females were either given new males, or the males were rotated between identical treatment groups to ensure the fertility of the male mice.
RESULTS
Strategy for cloning and mutagenesis of the MCMV genome.
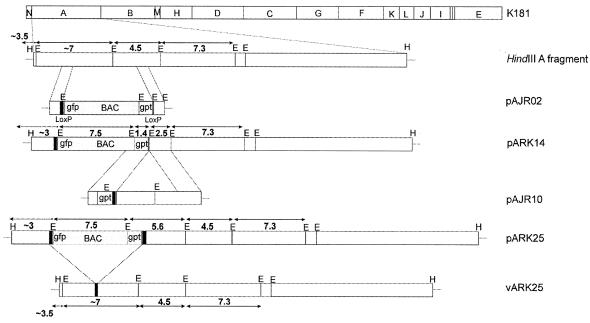
The strategy for the production of an MCMV BAC from K181 is shown in Fig. 1. The recombination vector pAJR02 contains homologous regions spanning nt 5308 to 6522 and nt 12488 to 13576 of the K181 genome. Homologous recombination resulted in the deletion of most of ORF m07 up to and including most of ORF m12 with the concomitant insertion of the BAC cassette. One clone, pARK14, was selected for further study. pARK14 BAC DNA was used to transfect MEFs, which resulted in the production of infectious viral progeny vARK14.
FIG. 1.
Cloning strategy for the production of the K181Perth-based BAC, pARK25. The BAC cassette contained within the pAJR02 plasmid was inserted into the K181 strain of MCMV in tissue culture by homologous recombination to produce the BAC pARK14 with a deletion in the genes m07 to m12 (nt 6522 to 12488). Repair of pARK14 was achieved by two-step allele replacement using the pAJR10 plasmid to produce the full-length BAC, pARK25. The EcoRI (E) and HindIII (H) RE sites, as well as the gpt and EGFP genes of the BAC cassette, are shown. Repair of pARK14 to pARK25 produces a 249-bp repeat region (black boxes) flanking the BAC cassette that allows for homologous recombination and reconstitution of wt genotype (vARK25) upon virus rescue. Fragment sizes (in kilobases) are shown in boldface type above or below the fragment lines.
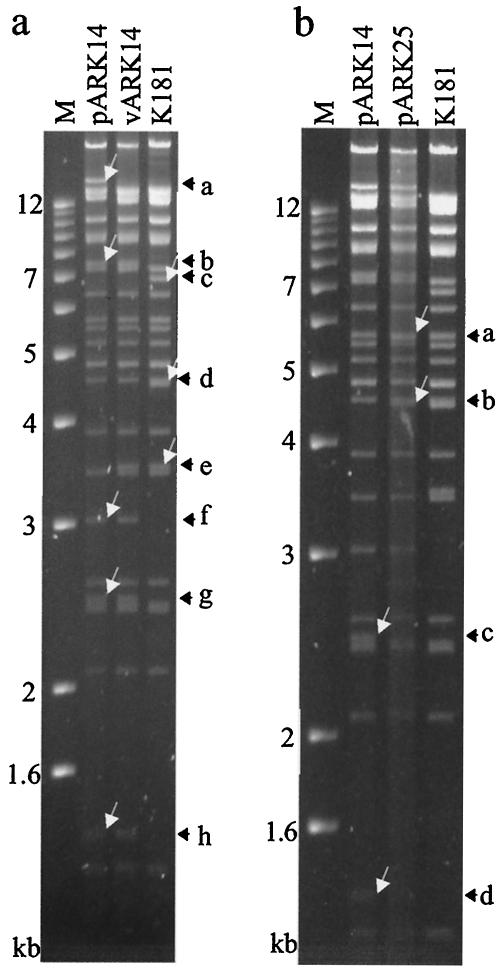
pARK14 BAC DNA and vARK14 viral DNA were purified and compared to parental K181 DNA by RFLP analysis (Fig. 2a). The resultant banding pattern was consistent with incorporation of the BAC cassette and retention of the K181 genome with the exception of the deleted regions between m07 to m12. In both pARK14 and vARK14, new EcoRI bands can be seen; these bands correspond to the expected fragments of 7,481 (fragment b) and 2,474 bp (fragment g). These fragments and an additional 1,367-bp fragment (h) were derived from the inserted BAC cassette. Likewise, there was loss of EcoRI U fragment of 4,487 bp (14) as a result of the insertion of BAC cassette, normally seen as a doublet in the K181 lane (fragment d). In comparison to the MCMV strain Smith, K181 has an additional EcoRI site approximately 3.5 kb from the left terminus of the virus that resulted in the substitution of the 10.5-kb EcoRI G fragment (14) with two fragments of 3.5 and 7 kb. Insertion of the BAC cassette disrupted the 7-kb fragment (c) and created a 3-kb fragment (f). Finally, circularization of the plasmid pARK14 causes fusion of the 3.5-kb (left arm) and 12.3-kb (right arm) terminal fragments of the virus to produce a 16-kb fragment with EcoRI digestion (fragment a). The left arm ∼3.5-kb fragment (e) reappears on linearization of pARK14 to vARK14. The 12.3-kb right terminal fragment was not discernible in Fig. 2a.
FIG. 2.
Characterization of BAC pARK14 with ORFs m07 to m12 deleted. (a) Insertion of the BAC cassette into the K181Perth strain of MCMV was achieved by homologous recombination. The EcoRI restriction digest patterns of the pARK14 BAC, rescued virus vARK14, and parental virus K181 are shown. The 16-kb band caused by fusion of the EcoRI terminal fragments in the pARK14 plasmid (fragment a) was lost in vARK14 and replaced by the left terminal fragment at ∼3.5 kb (fragment e) and a 12.3-kb right terminal fragment (not discernible in this figure). In both pARK14 and vARK14, new EcoRI fragments of 7.5 kb (fragment b), 2.5 kb (fragment g), and 1.4 kb (fragment h) are a result of the inserted BAC cassette (Fig. 1). Likewise, the EcoRI U fragment at 4.5 kb was lost as a result of the insertion of BAC cassette; it is normally seen as a doublet in the K181 lane (fragment d). The 4.5-kb EcoRI U band was replaced by a truncated 2.5-kb band (fragment g). K181 has an additional EcoRI site (compared to the Smith strain of MCMV) located approximately 3.5 kb from the left terminus of the virus that resulted in the substitution of the 10.5-kb EcoRI G fragment with two fragments of 3.5 and 7 kb. Insertion of the BAC cassette resulted in the loss of the 7-kb fragment (fragment c) and created a 3.1-kb fragment (fragment f). (b) Production of the full-length K181Perth BAC pARK25. Two-step allele replacement was used to restore the m07 to m12 genes in pARK14. The EcoRI restriction digest patterns of the BAC pARK14, the repaired BAC pARK25, and the parental virus K181 are shown. Repair of pARK14 to pARK25 results in the insertion of a 5.6-kb fragment (fragment a), seen as a doublet in the pARK25 lane. The 4.5-kb EcoRI U is restored (fragment b), also seen as a doublet (pARK25 and K181 lanes). Restoration of the 4.5-kb EcoRI U fragment resulted in the loss of the truncated 2.5-kb fragment (fragment c). The 1.4-kb BAC cassette fragment (fragment d) is lost on repair of pARK25. Lane M contains a 1-kb DNA molecular size ladder.
Truncated pARK14 BAC was restored to full length by reinserting ORFs m07 to m12 by two-step allele replacement as outlined in Fig. 1. Successful repair was monitored by RFLP analysis. The EcoRI sites of the wt virus and the K181 BAC are shown in Fig. 1. Figure 2b demonstrates the RFLP pattern obtained with one repaired clone, pARK25, compared to pARK14 and K181 DNA. The BAC pARK25 RFLP was as expected. The BAC pARK25 resembles pARK14 with the exception that both the 2.5-kb band (fragment c) and the 1.4-kb band (fragment d) were lost, with a gain of a 5.6-kb band, seen as a doublet (fragment a) and restoration of the 4.5-kb U band (fragment b), also seen as a doublet in the pARK25 lane. The BAC pARK25 resembles K181 viral DNA with the exception that the BAC cassette bands are present and the circularization band as detailed in Fig. 2a are present.
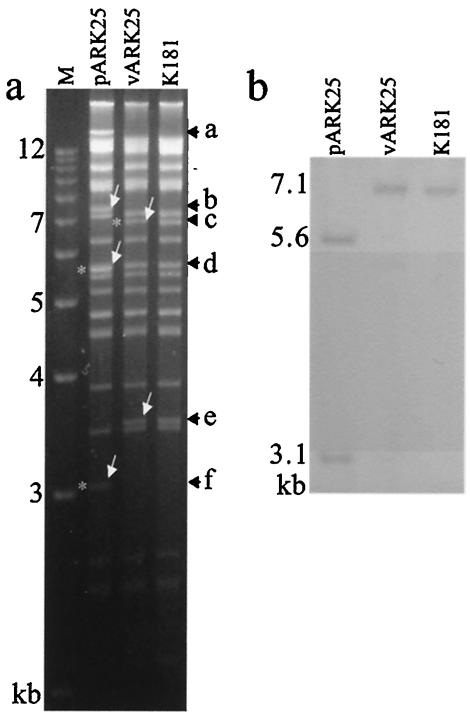
Construction of the pARK25 BAC utilizes a previously published method for restoring the BAC to wt virus by excising the bacterial sequences (54). In this instance, the repair of pARK14 to produce pARK25 resulted in a 249-bp MCMV sequence repeat, flanking the BAC cassette (solid black box in Fig. 1). Once transfected into eukaryotic cells, viral packaging restrictions preferentially lead to homologous recombination between the repeat sequences and exclusion of the BAC sequence. Restoration of the virus to wt genotype was monitored by the loss of GFP expression and was confirmed by RFLP analysis (Fig. 3a). Loss of GFP expression was rapid, beginning in the first passage. By passage 2, GFP expression in cells demonstrating CPE was less than 1% (data not shown). Once linear, DNA from vARK25-infected cells was indistinguishable from K181 DNA. RFLP analysis was performed with EcoRI (Fig. 3a). Southern blot analysis (Fig. 3b) shows the expected band size of 7.1 kb when the 249-bp repeat region was used as a probe. Other enzymes tested include BamHI, XbaI, HpaI, and PstI (data not shown). In some instances, homologous recombination occurred between the SV40-derived transcription termination signal that was present twice in the BAC cassette. In these instances, the gpt gene remained in the viral genome. An additional BAC not reported in this study has been produced; in this BAC, the gpt gene and its SV40-derived transcription termination signal were removed to prevent this from occurring.
FIG. 3.
Rescue of wild-type K181Perth genotype from pARK25 BAC in tissue culture. (a) The full-length BAC pARK25 was transfected into MEFs to produce the virus stock vARK25. Repeated passage of vARK25 (n = 5) resulted in the production of viral DNA indistinguishable from parental K181 MCMV viral DNA when digested with EcoRI. The 16-kb band caused by fusion of the EcoRI terminal fragments in the pARK25 plasmid (fragment a) was lost in vARK25 and replaced by the left terminal fragment at ∼3.5 kb (fragment e) and a 12.3-kb fragment that is not discernible in this figure. Homologous recombination between the MCMV repeat sequences (black boxes in Fig. 1) resulted in the loss of the BAC cassette fragments at 7.5 kb (fragment b) and the 5.6-kb fragment (fragment d). Restoration of the 7-kb fragment of MCMV (fragment c) was seen with concomitant loss of the 3.1-kb fragment (fragment f). Fragments d and f contain the MCMV repeat sequence in pARK25, the single “repeat” region in vARK25 is present in the 7-kb fragment (fragment c) in vARK25 (asterisks). Lane M contains a 1-kb DNA molecular size ladder. (b) Southern blot analysis of Fig. 3a using the MCMV repeat region (either side of the BAC cassette) as a probe demonstrated the expected bands at 5.6 and 3.1 kb from pARK25. As expected, a single fragment of 7.1 kb was present in both the K181 and vARK25 DNA samples.
Growth characteristics of viral BAC.
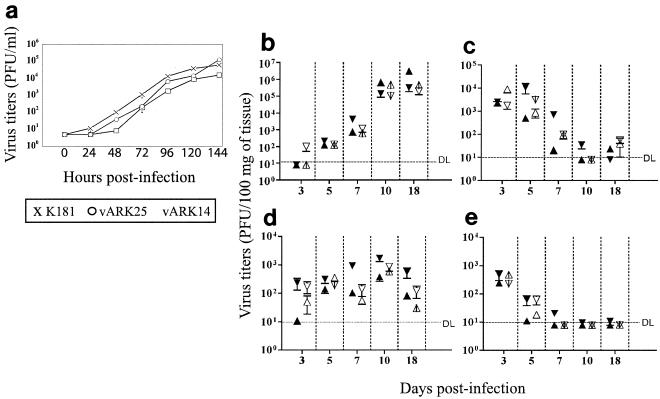
The in vitro growth characteristics of vARK25 and vARK14 viruses were similar to those of the parental K181 strain of MCMV as shown by multistep growth curves (Fig. 4a). These data indicate that the ORFs m07 to m12, deleted in vARK14, are not essential for the replication of K181 in vitro.
FIG. 4.
Full-length BAC-derived virus vARK25 and truncated virus vARK14 retain normal in vitro growth characteristics. (a) In vitro replication of vARK25 and vARK14 was assessed in comparison to the in vitro replication of parental K181 by a multistep growth curve (MOI of 0.01). Supernatants of infected cells were harvested at the indicated time points, and the viral titers were determined in duplicate on MEFs by standard plaque assay. The detection limit of the plaque assay was 25 PFU/ml. (b to e) In vivo replication. Full-length BAC-derived virus vARK25 retains in vivo infectivity. Mice were infected i.p. with 104 PFU of SGV of either the K181Perth strain of MCMV or vARK25, and the viral titer in the salivary gland (b), spleen (c), lung (d), and liver (e) was assessed over an 18-day period. The results of two independent experiments are shown. Viral titers in the organs of vARK25-infected mice (black symbols) and K181-infected mice (white symbols) are shown. The means ± standard errors of the means (error bars) are depicted. One experiment with five mice per group (▵) and a repeat experiment with four mice per group (▿) are shown. The growth characteristics of BAC-derived virus vARK25 were similar to those of parental K181Perth strain of virus in all tissues tested. Significant differences in the values for vARK25 and K181 were tested by two-tailed t test and recorded only when evident in both experiments. The limit of detection (DL) is indicated by the broken line. When not shown, error bars are smaller than the symbols.
The replication kinetics of salivary gland-derived vARK25 in adult BALB/c mice were similar to those of parental K181 SGV (Fig. 4b to e). Stocks of vARK25 SGV were produced to assess the pathogenicity of vARK25 SGV compared to K181 SGV. After six passages in tissue culture, vARK25 was passaged two more times in weanling BALB/c female mice. Passage 2 SGV stocks were made and compared to passage 2 K181 SGV. BALB/c female mice were inoculated i.p. with 104 PFU of SGV in two independent experiments (four and five mice per group, respectively). There were minor experimental variations between the means of each group in respective experiments (Fig. 4b to e). However, there was no consistent statistically significant difference between the organ viral titers of vARK25- and K181-infected mice. Titers of vARK25 were similar to parental K181 virus in the salivary glands with both reaching viral titers of more than 105 PFU/100 mg of tissue by day 18 (Fig. 4b). Additionally, the kinetics of salivary gland infection were similar for both vARK25 and K181 SGV (Fig. 4b). In addition, no differences in the replication of vARK25 compared to K181 in the spleen, lungs, and livers of infected mice were detected (Fig. 4c to e).
Production of vaccine strains of MCMV BACs.
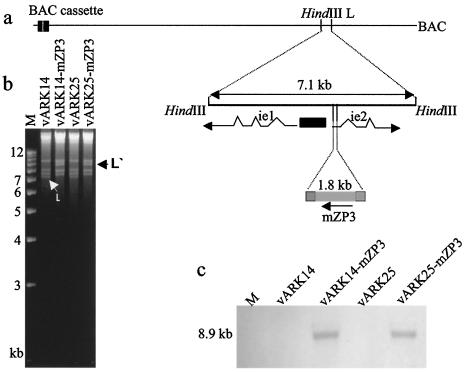
Disruption of the ie2 gene does not inhibit in vitro growth (28) or in vivo growth (10). Consequently, we chose this site for the insertion of mZP3 into the pARK25 and pARK14 BACs. Two-step allele replacement was performed as described for the repair of pARK14. Figure 5a outlines the cloning strategy employed. Correct insertion was confirmed by RFLP and Southern blot analyses (Fig. 5b and c). Note the change in the size of the HindIII L fragment from 7.1 to 8.9 kb in vaccine strains of virus. Western blotting confirmed the expression of mZP3 protein in both vARK14-mZP3- and vARK25-mZP3-infected MEFs (data not shown).
FIG. 5.
Production of vaccine strains of BAC virus. Truncated pARK14 and full-length pARK25 were used to produce mZP3-expressing vaccine strains of virus by two-step allele replacement. The mZP3 cDNA was inserted into the ie2 gene of MCMV (a). Correct insertion was assessed by HindIII RFLP analysis and demonstrated by an increase in the size of the HindIII L fragment (b) and was confirmed by Southern blotting and hybridization with an mZP3 probe (c). M lanes contain 1-kb DNA molecular size ladder.
Growth characteristics of vaccine strains in vitro and in vivo.
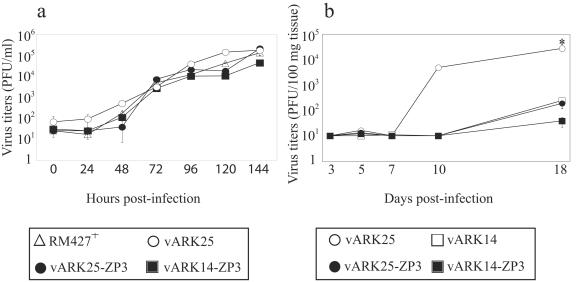
The insertion of mZP3 cDNA had no effect on the in vitro growth characteristics of either vARK14-mZP3 or vARK25-mZP3 compared to the LacZ-expressing virus RM427+ or to parental vARK25 by multistep growth curves (Fig. 6a).
FIG. 6.
Growth characteristics of vaccine strains of virus. (a) The in vitro growth characteristics of vaccine strains of virus are similar to those of vARK25 and the K181-based LacZ-expressing virus RM427+. The means ± standard errors of the means (error bars) of viral titers in a multistep growth curve (MOI of 0.01) are shown. The minimum level of sensitivity was 25 PFU/ml. When not shown, error bars are smaller than the symbols. (b) Truncated virus vARK14 and vaccine strains of virus were attenuated in vivo. In vivo viral replication was assessed after i.p. infection of female BALB/c mice with 104 PFU of TCV. The means ± standard error of the means (error bars) of viral titers in the salivary gland (five mice per time point). Only vARK25 virus was detected in the salivary glands of infected mice on day 10, and the viral titers of vARK25 virus were significantly higher than viral titers in vARK14-infected mice on day 18 (P = 0.0052). Titers of vaccine strains of virus were similar to or lower than that of vARK14 in the salivary gland at all time points. Similar data were obtained in a repeat experiment. Significant differences (two-tailed t test) between vARK25 and vARK14 are indicated by an asterisk.
Previous experience has shown that SGV stocks cannot normally be produced from vaccine viruses expressing fertility antigen (24; unpublished observations). For this reason, in vivo growth characteristics of mZP3-expressing vaccine strains were compared to those of parental viruses using TCV stocks. TCV stocks were injected i.p. at a dose of 2 × 104 PFU in adult female BALB/c mice, and the viral titers were assessed in the spleen, liver, lungs, and salivary gland of infected mice over an 18-day period. Vaccine strains of virus and vARK14 were severely attenuated in vivo compared to vARK25 (Fig. 6b). Only vARK25 could be detected in the salivary glands at day 10, and the vARK25 titer was significantly higher than the vARK14 titer at day 18 (P = 0.0052). All viruses were detected in the spleen on day 3 but were rapidly cleared by day 5 (data not shown). Parental and mZP3-expressing recombinant virus titers were below the level of detection in the liver and lungs of infected mice (data not shown). Similar results were observed in a repeat experiment with the exception that vARK25 could be detected at low titers in the lungs of infected mice.
A surprising finding was that the degree of attenuation induced by insertion of the mZP3 cDNA into the ie2 gene of MCMV was similar to the degree induced by the deletion of ORFs m07 to m12 and the retention of the large BAC cassette. This was most evident in the salivary glands where the titers of vARK14 were similar to those of vARK25-mZP3 (Fig. 6b). In a repeat experiment, attenuation of vARK25-mZP3 in the salivary gland was more pronounced (data not shown). Hence, the effect of losing six ORFs and retaining a large 8.8-kb fragment of bacterial DNA on salivary gland replication was similar to the effect of expressing a single mammalian cDNA.
BAC-derived virus expressing mZP3 induces sterility in female mice.
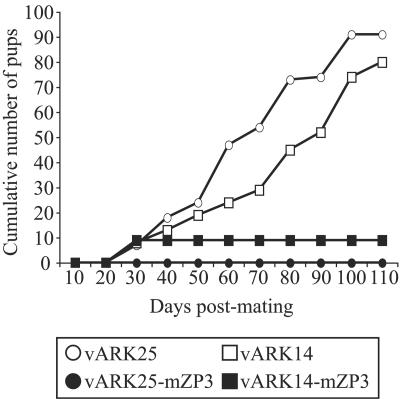
Tissue culture-derived virus stocks of vARK14-mZP3 and vARK25-mZP3 were tested for their ability to induce an immunocontraceptive response in female BALB/c mice. Mice (six mice per group) were inoculated i.p. with 2 × 104 TCV and were immediately allowed to mate with male BALB/c mice. Control mice received parental vARK14 or vARK25 virus lacking the mZP3 cDNA. vARK25-mZP3 produced a powerful immunocontraceptive response in female BALB/c mice, and no litters were produced for the 110 days of the experiment (Fig. 7). Similarly, only one of six mice infected with vARK14-mZP3 had pups after infection and then only in the first round of breeding. These results were seen despite the fact that viral titers of mZP3-expressing recombinant viruses were low in infected animals. Similar data were seen in a repeat experiment with time points taken at 30, 50, and 70 days (data not shown). Histological examination of the ovaries from mice infected with mZP3-expressing viruses revealed almost complete follicle depletion with no overt inflammatory cell infiltrate (data not shown).
FIG. 7.
Vaccine strains of virus induce immunocontraception in recipient female mice. Adult female BALB/c mice (six mice per group) were inoculated i.p. with 2 × 104 PFU of TCV and paired with fertile male mice on the same day. Fertility was monitored over a 110-day period. Mice infected with the vaccine strain of virus either had no litters or had an initial litter and remained infertile for the duration of the experiment. The full-length vaccine strain of virus vARK25-mZP3 was no more effective than vARK14-mZP3. Similar data were seen in a repeat experiment (data not shown).
The production of serum antibody to mZP3 was assessed by ELISAs. Only mice inoculated with recombinant MCMVs expressing mZP3 produced detectable levels of anti-mZP3 antibody. Considerably more anti-mZP3 antibody was detected in the mice treated with vARK25-mZP3 virus than in the mice infected with vARK14-mZP3 virus. This was reflected in the percentage of animals that were seropositive as well as in the earlier kinetics of antibody production. In vARK25-mZP3 virus-infected mice, IgG1 antibody to mZP3 was detected as early as day 7 with 25% of mice seropositive and with 100% of mice seropositive by day 18. Similar data were seen with IgG2a antibody (50% positive at day 7 and 100% positive at day 18). In contrast, seroconversion occurred later in vARK14-mZP3-infected mice, and only 50% of mice had IgG1 and IgG2a antibody by day 18. By day 30, all mice infected with mZP3-expressing recombinant viruses were seropositive for anti-mZP3 antibodies (data not shown).
DISCUSSION
We report here the construction of a new MCMV BAC based on the K181Perth strain of the virus. This BAC, with a self-excisable BAC cassette, was indistinguishable from the parental K181 virus when examined by RFLP analysis and multistep growth curves and retained similar pathogenicity in vivo. From the full-length (wt) BAC, pARK25, we have created a recombinant MCMV encoding the self-fertility antigen mZP3 that was capable of breaking self-tolerance and inducing an immunocontraceptive response. In addition, an attenuated vaccine strain of recombinant virus expressing mZP3 and lacking ORFs m07 to m12 was produced and was as effective at inducing immunocontraception as the full-length virus expressing mZP3. Hence, attenuation of MCMV by deletion of viral genes can be used to produce a vaccine strain of virus without necessarily affecting the efficacy of the vaccine.
The previously described MCMV BAC was based on the Smith strain of MCMV (33, 54). However, the K181 strain of MCMV is used by a number of investigators worldwide and is more virulent than the Smith strain of MCMV (34). The K181 strain has been widely used to study innate immunity to MCMV, especially interactions with NK cells (5, 53) and was the strain of MCMV used to describe the Cmv1r locus that defines NK cell-mediated resistance to MCMV in C57BL/6 mice (45, 46). The K181 strain has also been used extensively in studies of dendritic cell interactions (4, 6) and host-mediated inflammatory and immunopathological responses (23). However, in the past, changes to the K181 genome had to be made using the older and more time-consuming methods of homologous recombination in tissue culture. The K181 BAC described here should allow the rapid progress made in the study of the Smith strain of MCMV to be applied to the K181 strain of MCMV.
The original method of BAC cassette excision relied on homologous recombination between MCMV sequences flanking the BAC cassette, with rescue of wt virus after serial passage in tissue culture (54). Since then, Smith and Enquist have described the production of a self-excisable BAC cassette system that relied on Cre recombinase-mediated excision of the BAC cassette via flanking LoxP sites (48). This technique has subsequently been applied to the construction of a human CMV BAC (57) and a rhesus monkey CMV BAC (11). In these studies, the insertion site of the BAC cassette was chosen so that the remnant LoxP sequence did not affect viral function. However, because it is possible that the remnant LoxP site could affect a gene or splice variant not known at this time, we chose to use the originally described method of BAC cassette excision (54), in which no extraneous sequences remain in the viral genome. In 1999, Wagner and colleagues (54) used a repeat region of 529 bp, while we used a shorter repeat region of 249 bp which still allowed for rapid recovery of a wt K181 DNA genotype. In our hands, loss of the BAC cassette from the K181 BAC was rapid and was detected within the first passage. While this method relies on repeated rounds of passage to recover wt DNA, our experience was that less than 1% of infected cells were GFP positive (and thus contained the BAC cassette) by passage two. In addition, serial passage was required in any case to amplify sufficient virus to make stocks for subsequent experiments. However, as a cautionary note, we did find that the repeat SV40-derived transcription termination signal (125 bp) within the BAC cassette was also a site for homologous recombination which left the gpt gene within the viral genome. Once recombination occurred via the SV40 repeats, this genotype was fixed, even in vivo. It is likely that most virus preparations of vARK25 had some level of chimerism. However, this chimerism appeared to have no effect on in vitro or in vivo growth of vARK25. This problem was circumvented in a subsequent version of pARK25 that had the gpt gene and its SV40 sequences removed (not detailed in this study).
In recent studies, we have tested more than 10 candidate fertility antigens for use in VVIC, most of which were ineffective (16; unpublished data). Consequently, there was a need for a rapid construction method for testing future antigens. Prior VVICs were constructed by standard homologous recombination in mammalian cells. This method has the disadvantages of being laborious, error prone, and time-consuming. Therefore, we sought to test the efficacy of the K181 BAC as a rapid method for the production of viral vaccine vectors. We chose to use the previously successful self-antigen, mZP3, as a test antigen (24). The K181 BAC described in this study was used to successfully, and rapidly, construct two effective VVICs. More rapid methods of gene insertion have since been developed (reviewed in reference 55) and should allow faster vaccine construction in the future.
Successful immunocontraception with the full-length K181-based BAC expressing the mZP3 cDNA in female mice was anticipated (24). However, it was important to show that the BAC-derived VVIC described in this report was as effective as the previously described VVIC and that the K181 virus had not undergone any adventitious mutations during construction that precluded its use as a vaccine.
In this study, the effect of attenuation of the virus on vaccine efficacy was assessed. Although others have demonstrated that attenuated vaccine strains of MCMV can be used to afford protection from wt MCMV (26, 35), we did not know whether a single inoculation of an attenuated MCMV vaccine vector would be sufficient to break tolerance to a self-antigen. Immunization with the VVIC vARK14-mZP3 was no less effective at inducing sterility in female mice as the full-length VVIC vARK25-mZP3. This was despite the finding that removal of these ORFs (and retention of the BAC cassette) attenuated the in vivo growth of deletion mutant virus vARK14. The m07 to m12 ORFs form part of the m02 gene family, which contains the known immune evasion genes, m04 and m06, and other genes that have been linked to resistance to natural killer cell control (37). However, the exact functions of the ORFs m07, m08, m09, m10, m11, and m12 are not known, although it is known that the m09 ORF can be removed without affecting MCMV growth in BALB/c mice (58). These studies show that putative immune evasion genes can be removed from viral vaccine vectors without necessarily affecting immunogenicity of the expressed antigen.
A surprising finding in these studies was that the expression of a single fertility antigen, mZP3 (vARK25-mZP3) led to a level of in vivo attenuation similar to the level seen with the deletion of ORFs m07 to m12 and the retention of the BAC cassette (vARK14). This effect was most pronounced in the salivary gland. Two possible reasons for this attenuation can be suggested. Disruption of the ie2 gene by the insertion of the mZP3 cDNA causes attenuation of the virus. Alternatively, attenuation could be immunological in nature, i.e., the strong immune response to the inserted antigen, mZP3, could allow for better immune control of the virus. Others have reported that insertional inactivation of the ie2 gene does not affect in vivo replication of the K181 strain of MCMV and allows dissemination to the salivary gland (10, 43). The reason for this attenuation therefore remains unclear; however, the development of a K181 BAC should facilitate the study of this attenuation in the future.
In conclusion, we have produced a fully functional BAC of the K181Perth strain of MCMV. The efficacy of the BAC-derived virus was tested as a vaccine. BAC-derived K181 virus expressing a self-fertility antigen was capable of inducing infertility in female mice. An attenuated vaccine virus missing ORFs m07 to m12 also functioned as a vaccine vector with an efficacy equal to that of the full-length virus. These studies demonstrate the utility of the MCMV BAC system for developing viral vectored vaccines and may allow for a targeted appraisal of the roles of various viral genes, such as immune evasion genes, in the immune response to a heterologously expressed antigen.
Acknowledgments
This work was funded in part by the Pest Animal Control Cooperative Research Centre. M.M. was supported by a grant from the NBL3 programme of the Bundesministerium für Bildung und Forschung (BMBF). U.H.K. was supported by grants from the Deutsche Forschungsgemeinschaft and by the State of Bavaria.
We thank Elizabeth Williams for technical assistance.
REFERENCES
- 1.Adler, H., M. Messerle, M. Wagner, and U. H. Koszinowski. 2000. Cloning and mutagenesis of the murine gammaherpesvirus 68 genome as an infectious bacterial artificial chromosome. J. Virol. 74:6964-6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alford, C. A., and W. J. Britt. 1990. Cytomegalovirus, p. 1981-2010. In B. N. Fields and D. M. Knipe (ed.), Fields virology, vol. 2. Raven Press, New York, N.Y. [Google Scholar]
- 3.Andino, R., D. Silvera, S. D. Suggett, P. L. Achacoso, C. J. Miller, D. Baltimore, and M. B. Feinberg. 1994. Engineering poliovirus as a vaccine vector for the expression of diverse antigens. Science 265:1448-1451. [DOI] [PubMed] [Google Scholar]
- 4.Andrews, D. M., C. E. Andoniou, F. Granucci, P. Ricciardi-Castagnoli, and M. A. Degli-Esposti. 2001. Infection of dendritic cells by murine cytomegalovirus induces functional paralysis. Nat. Immunol. 2:1077-1084. [DOI] [PubMed] [Google Scholar]
- 5.Andrews, D. M., H. E. Farrell, E. H. Densley, A. A. Scalzo, G. R. Shellam, and M. A. Degli-Esposti. 2001. NK1.1+ cells and murine cytomegalovirus infection: what happens in situ? J. Immunol. 166:1796-1802. [DOI] [PubMed] [Google Scholar]
- 6.Andrews, D. M., A. A. Scalzo, W. M. Yokoyama, M. J. Smyth, and M. A. Degli-Esposti. 2003. Functional interactions between dendritic cells and NK cells during viral infection. Nat. Immunol. 4:175-181. [DOI] [PubMed] [Google Scholar]
- 7.Berencsi, K., Z. Gyulai, E. Gonczol, S. Pincus, W. I. Cox, S. Michelson, L. Kari, C. Meric, M. Cadoz, J. Zahradnik, S. Starr, and S. Plotkin. 2001. A canarypox vector-expressing cytomegalovirus (CMV) phosphoprotein 65 induces long-lasting cytotoxic T cell responses in human CMV-seronegative subjects. J. Infect. Dis. 183:1171-1179. [DOI] [PubMed] [Google Scholar]
- 8.Booth, T. W., A. A. Scalzo, C. Carrello, P. A. Lyons, H. E. Farrell, G. R. Singleton, and G. R. Shellam. 1993. Molecular and biological characterization of new strains of murine cytomegalovirus isolated from wild mice. Arch. Virol. 132:209-220. [DOI] [PubMed] [Google Scholar]
- 9.Brockman, M. A., and D. M. Knipe. 2002. Herpes simplex virus vectors elicit durable immune responses in the presence of preexisting host immunity. J. Virol. 76:3678-3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cardin, R. D., G. B. Abenes, C. A. Stoddart, and E. S. Mocarski. 1995. Murine cytomegalovirus IE2, an activator of gene expression, is dispensable for growth and latency in mice. Virology 209:236-241. [DOI] [PubMed] [Google Scholar]
- 11.Chang, W. L., and P. A. Barry. 2003. Cloning of the full-length rhesus cytomegalovirus genome as an infectious and self-excisable bacterial artificial chromosome for analysis of viral pathogenesis. J. Virol. 77:5073-5083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cicin-Sain, L., W. Brune, I. Bubic, S. Jonjic, and U. H. Koszinowski. 2003. Vaccination of mice with bacteria carrying a cloned herpesvirus genome reconstituted in vivo. J. Virol. 77:8249-8255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis-Poynter, N. J., D. M. Lynch, H. Vally, G. R. Shellam, W. D. Rawlinson, B. G. Barrell, and H. E. Farrell. 1997. Identification and characterization of a G protein-coupled receptor homolog encoded by murine cytomegalovirus. J. Virol. 71:1521-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ebeling, A., G. M. Keil, E. Knust, and U. H. Koszinowski. 1983. Molecular cloning and physical mapping of murine cytomegalovirus DNA. J. Virol. 47:421-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hahn, G., H. Khan, F. Baldanti, U. H. Koszinowski, M. G. Revello, and G. Gerna. 2002. The human cytomegalovirus ribonucleotide reductase homolog UL45 is dispensable for growth in endothelial cells, as determined by a BAC-cloned clinical isolate of human cytomegalovirus with preserved wild-type characteristics. J. Virol. 76:9551-9555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hardy, C. M., G. Clydesdale, K. J. Mobbs, J. Pekin, M. L. Lloyd, C. Sweet, G. R. Shellam, and M. A. Lawson. 2004. Assessment of contraceptive vaccines based on recombinant mouse sperm protein PH20. Reprod. Fertil. Dev. 127:325-334. [DOI] [PubMed] [Google Scholar]
- 17.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365-369. [DOI] [PubMed] [Google Scholar]
- 18.Hobom, U., W. Brune, M. Messerle, G. Hahn, and U. H. Koszinowski. 2000. Fast screening procedures for random transposon libraries of cloned herpesvirus genomes: mutational analysis of human cytomegalovirus envelope glycoprotein genes. J. Virol. 74:7720-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hudson, J. B., D. G. Walker, and M. Altamirano. 1988. Analysis in vitro of two biologically distinct strains of murine cytomegalovirus. Arch. Virol. 102:289-295. [DOI] [PubMed] [Google Scholar]
- 20.Jackson, R. J., D. J. Maguire, L. A. Hinds, and I. A. Ramshaw. 1998. Infertility in mice induced by a recombinant ectromelia virus expressing mouse zona pellucida glycoprotein 3. Biol. Reprod. 58:152-159. [DOI] [PubMed] [Google Scholar]
- 21.Karupiah, G., T. E. Sacks, D. M. Klinman, T. N. Fredrickson, J. W. Hartley, J. H. Chen, and H. C. Morse III. 1998. Murine cytomegalovirus infection-induced polyclonal B cell activation is independent of CD4+ T cells and CD40. Virology 240:12-26. [DOI] [PubMed] [Google Scholar]
- 22.Khan, N., N. Shariff, M. Cobbold, R. Bruton, J. A. Ainsworth, A. J. Sinclair, L. Nayak, and P. A. Moss. 2002. Cytomegalovirus seropositivity drives the CD8 T cell repertoire toward greater clonality in healthy elderly individuals. J. Immunol. 169:1984-1992. [DOI] [PubMed] [Google Scholar]
- 23.Lawson, C. M., H. L. O'Donoghue, and W. D. Reed. 1992. Mouse cytomegalovirus infection induces antibodies which cross-react with virus and cardiac myosin: a model for the study of molecular mimicry in the pathogenesis of viral myocarditis. Immunology 75:513-519. [PMC free article] [PubMed] [Google Scholar]
- 24.Lloyd, M. L., G. R. Shellam, J. M. Papadimitriou, and M. A. Lawson. 2003. Immunocontraception is induced in BALB/c mice inoculated with murine cytomegalovirus expressing mouse zona pellucida 3. Biol. Reprod. 68:2024-2032. [DOI] [PubMed] [Google Scholar]
- 25.Lutarewych, M. A., M. R. Quirk, B. A. Kringstad, W. Li, C. M. Verfaillie, and M. C. Jordan. 1997. Propagation and titration of murine cytomegalovirus in a continuous bone marrow-derived stromal cell line (M2-10B4). J. Virol. Methods 68:193-198. [DOI] [PubMed] [Google Scholar]
- 26.MacDonald, M. R., X. Y. Li, R. M. Stenberg, A. E. Campbell, and H. W. Virgin IV. 1998. Mucosal and parenteral vaccination against acute and latent murine cytomegalovirus (MCMV) infection by using an attenuated MCMV mutant. J. Virol. 72:442-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mandl, S., L. J. Sigal, K. L. Rock, and R. Andino. 1998. Poliovirus vaccine vectors elicit antigen-specific cytotoxic T cells and protect mice against lethal challenge with malignant melanoma cells expressing a model antigen. Proc. Natl. Acad. Sci. USA 95:8216-8221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manning, W. C., and E. S. Mocarski. 1988. Insertional mutagenesis of the murine cytomegalovirus genome: one prominent alpha gene (ie2) is dispensable for growth. Virology 167:477-484. [PubMed] [Google Scholar]
- 29.Manning, W. C., C. A. Stoddart, L. A. Lagenaur, G. B. Abenes, and E. S. Mocarski. 1992. Cytomegalovirus determinant of replication in salivary glands. J. Virol. 66:3794-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marshall, J. L., M. J. Hawkins, K. Y. Tsang, E. Richmond, J. E. Pedicano, M. Z. Zhu, and J. Schlom. 1999. Phase I study in cancer patients of a replication-defective avipox recombinant vaccine that expresses human carcinoembryonic antigen. J. Clin. Oncol. 17:332-337. [DOI] [PubMed] [Google Scholar]
- 31.McGettigan, J. P., R. J. Pomerantz, C. A. Siler, P. M. McKenna, H. D. Foley, B. Dietzschold, and M. J. Schnell. 2003. Second-generation rabies virus-based vaccine vectors expressing human immunodeficiency virus type 1 Gag have greatly reduced pathogenicity but are highly immunogenic. J. Virol. 77:237-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mercer, J. A., J. R. Marks, and D. H. Spector. 1983. Molecular cloning and restriction endonuclease mapping of the murine cytomegalovirus genome (Smith strain). Virology 129:94-106. [DOI] [PubMed] [Google Scholar]
- 33.Messerle, M., I. Crnkovic, W. Hammerschmidt, H. Ziegler, and U. H. Koszinowski. 1997. Cloning and mutagenesis of a herpesvirus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA 94:14759-14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Misra, V., and J. B. Hudson. 1980. Minor base sequence differences between the genomes of two strains of murine cytomegalovirus differing in virulence. Arch. Virol. 64:1-8. [DOI] [PubMed] [Google Scholar]
- 35.Morley, P. J., P. Ertl, and C. Sweet. 2002. Immunisation of Balb/c mice with severely attenuated murine cytomegalovirus mutants induces protective cellular and humoral immunity. J. Med. Virol. 67:187-199. [DOI] [PubMed] [Google Scholar]
- 36.Murphy, C. G., W. T. Lucas, R. E. Means, S. Czajak, C. L. Hale, J. D. Lifson, A. Kaur, R. P. Johnson, D. M. Knipe, and R. C. Desrosiers. 2000. Vaccine protection against simian immunodeficiency virus by recombinant strains of herpes simplex virus. J. Virol. 74:7745-7754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oliveira, S. A., S. H. Park, P. Lee, A. Bendelac, and T. E. Shenk. 2002. Murine cytomegalovirus m02 gene family protects against natural killer cell-mediated immune surveillance. J. Virol. 76:885-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Price, P., S. D. Olver, A. E. Gibbons, and G. R. Shellam. 1993. B-cell activation following murine cytomegalovirus infection: implications for autoimmunity. Immunology 78:14-21. [PMC free article] [PubMed] [Google Scholar]
- 39.Pridmore, R. D. 1987. New and versatile cloning vectors with kanamycin-resistance marker. Gene 56:309-312. [DOI] [PubMed] [Google Scholar]
- 40.Ramirez, J. C., M. M. Gherardi, and M. Esteban. 2000. Biology of attenuated modified vaccinia virus Ankara recombinant vector in mice: virus fate and activation of B- and T-cell immune responses in comparison with the Western Reserve strain and advantages as a vaccine. J. Virol. 74:923-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rawlinson, W. D., H. E. Farrell, and B. G. Barrell. 1996. Analysis of the complete DNA sequence of murine cytomegalovirus. J. Virol. 70:8833-8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rudolph, J., D. J. O'Callaghan, and N. Osterrieder. 2002. Cloning of the genomes of equine (EHV-1) strains KyA and racL11 as bacterial artificial chromosomes (BAC). J. Vet. Med. B 49:31-36. [DOI] [PubMed] [Google Scholar]
- 43.Saederup, N., S. A. Aguirre, T. E. Sparer, D. M. Bouley, and E. S. Mocarski. 2001. Murine cytomegalovirus CC chemokine homolog MCK-2 (m131-129) is a determinant of dissemination that increases inflammation at initial sites of infection. J. Virol. 75:9966-9976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 45.Scalzo, A. A., N. A. Fitzgerald, A. Simmons, A. B. La Vista, and G. R. Shellam. 1990. Cmv-1, a genetic locus that controls murine cytomegalovirus replication in the spleen. J. Exp. Med. 171:1469-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scalzo, A. A., N. A. Fitzgerald, C. R. Wallace, A. E. Gibbons, Y. C. Smart, R. C. Burton, and G. R. Shellam. 1992. The effect of the Cmv-1 resistance gene, which is linked to the natural killer cell gene complex, is mediated by natural killer cells. J. Immunol. 149:581-589. [PubMed] [Google Scholar]
- 47.Shiver, J. W., T. M. Fu, L. Chen, D. R. Casimiro, M. E. Davies, R. K. Evans, Z. Q. Zhang, A. J. Simon, W. L. Trigona, S. A. Dubey, L. Huang, V. A. Harris, R. S. Long, X. Liang, L. Handt, W. A. Schleif, L. Zhu, D. C. Freed, N. V. Persaud, L. Guan, K. S. Punt, A. Tang, M. Chen, K. A. Wilson, K. B. Collins, G. J. Heidecker, V. R. Fernandez, H. C. Perry, J. G. Joyce, K. M. Grimm, J. C. Cook, P. M. Keller, D. S. Kresock, H. Mach, R. D. Troutman, L. A. Isopi, D. M. Williams, Z. Xu, K. E. Bohannon, D. B. Volkin, D. C. Montefiori, A. Miura, G. R. Krivulka, M. A. Lifton, M. J. Kuroda, J. E. Schmitz, N. L. Letvin, M. J. Caulfield, A. J. Bett, R. Youil, D. C. Kaslow, and E. A. Emini. 2002. Replication-incompetent adenoviral vaccine vector elicits effective anti-immunodeficiency-virus immunity. Nature 415:331-335. [DOI] [PubMed] [Google Scholar]
- 48.Smith, G. A., and L. W. Enquist. 2000. A self-recombining bacterial artificial chromosome and its application for analysis of herpesvirus pathogenesis. Proc. Natl. Acad. Sci. USA 97:4873-4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stoddart, C. A., R. D. Cardin, J. M. Boname, W. C. Manning, G. B. Abenes, and E. S. Mocarski. 1994. Peripheral blood mononuclear phagocytes mediate dissemination of murine cytomegalovirus. J. Virol. 68:6243-6253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suter, M., A. M. Lew, P. Grob, G. J. Adema, M. Ackermann, K. Shortman, and C. Fraefel. 1999. BAC-VAC, a novel generation of (DNA) vaccines: a bacterial artificial chromosome (BAC) containing a replication-competent, packaging-defective virus genome induces protective immunity against herpes simplex virus 1. Proc. Natl. Acad. Sci. USA 96:12697-12702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sweet, C. 1999. The pathogenicity of cytomegalovirus. FEMS Microb. Rev. 23:457-482. [DOI] [PubMed] [Google Scholar]
- 52.Tischer, B. K., D. Schumacher, M. Beer, J. Beyer, J. P. Teifke, K. Osterrieder, K. Wink, V. Zelnik, F. Fehler, and N. Osterrieder. 2002. A DNA vaccine containing an infectious Marek's disease virus genome can confer protection against tumorigenic Marek's disease in chickens. J. Gen. Virol. 83:2367-2376. [DOI] [PubMed] [Google Scholar]
- 53.Voigt, V., C. A. Forbes, J. N. Tonkin, M. A. Degli-Esposti, H. R. Smith, W. M. Yokoyama, and A. A. Scalzo. 2003. Murine cytomegalovirus m157 mutation and variation leads to immune evasion of natural killer cells. Proc. Natl. Acad. Sci. USA 100:13483-13488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wagner, M., S. Jonjic, U. H. Koszinowski, and M. Messerle. 1999. Systematic excision of vector sequences from the BAC-cloned herpesvirus genome during virus reconstitution. J. Virol. 73:7056-7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wagner, M., Z. Ruzsics, and U. H. Koszinowski. 2002. Herpesvirus genetics has come of age. Trends Microbiol. 10:318-324. [DOI] [PubMed] [Google Scholar]
- 56.Wang, X., M. Messerle, R. Sapinoro, K. Santos, P. K. Hocknell, X. Jin, and S. Dewhurst. 2003. Murine cytomegalovirus abortively infects human dendritic cells, leading to expression and presentation of virally vectored genes. J. Virol. 77:7182-7192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu, D., G. A. Smith, L. W. Enquist, and T. Shenk. 2002. Construction of a self-excisable bacterial artificial chromosome containing the human cytomegalovirus genome and mutagenesis of the diploid TRL/IRL13 gene. J. Virol. 76:2316-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhan, X. Y., M. Lee, J. Q. Xiao, and F. Y. Liu. 2000. Construction and characterization of murine cytomegaloviruses that contain transposon insertions at open reading frames m09 and M83. J. Virol. 74:7411-7421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou, F. C., Y. J. Zhang, J. H. Deng, X. P. Wang, H. Y. Pan, E. Hettler, and S. J. Gao. 2002. Efficient infection by a recombinant Kaposi's sarcoma-associated herpesvirus cloned in a bacterial artificial chromosome: application for genetic analysis. J. Virol. 76:6185-6196. [DOI] [PMC free article] [PubMed] [Google Scholar]