Abstract
Background:
We conducted this study to identify the influences and synergistic effects of smoking status and polymorphisms in hedgehog interacting protein (HHIP) on chronic obstructive pulmonary disease (COPD) and lung function decline.
Methods:
A cohort containing 306 COPD patients and 743 healthy subjects was recruited from 25,000 subjects. All selected subjects had chronic cough for over 2 years or a smoking history above 20 pack-years. After 8 years, all subjects were divided into 2 cohorts according to whether they had quit smoking or not. A follow-up of all patients was completed after another period of 10 years. Three variants in HHIP were genotyped to investigate the impacts of gene susceptibility on the development of COPD and lung function decline.
Results:
During the follow-up tests, forced expiratory volume in 1 s (FEV1) ratios decreased more significantly in COPD patients than in healthy subjects. For variant rs7654947, FEV1 decreased more significantly in CC and CT subjects than in TT subjects. FEV1 in COPD patients with a CC genotype from smoking cohorts reduced markedly when compared to ex-smoking cohorts (case, 30.75% vs. 35.5%; total, 28% vs. 32%).
Conclusions:
Our results showed that smoking and HHIP variant rs7654947 were associated with COPD development and lung function decline. Moreover, we found that cigarette smoking and gene susceptibility have cooperative effects on COPD risk and lung function decline.
Keywords: COPD, follow-up, HHIP, lung function, smoking
1. Introduction
Chronic obstructive pulmonary disease (COPD) is characterized by progressive airflow obstruction, airway inflammation, and systemic effects or comorbidities and affects approximately 10% of the worldwide population.[1] COPD is predicted to be the third-leading cause of death worldwide by 2030.[2] With the spread of air pollution, COPD has become one of the heaviest economic burdens associated with hospitalization, work absence, and disability.[3,4] There is compelling evidence supporting the hypothesis that COPD results from complex interactions between genetic factors and environmental exposure.[5–7] Genetic susceptibility is characterized by familial clustering; COPD risk in subjects with COPD family history is approximately 2 to 3 times higher than subjects from the general population.[8] Likewise, cigarette smoking is well recognized as a major environmental trigger and risk factor of COPD. In fact, cigarette smokers make up over 90% of COPD patients.[9] However, only approximately 20% of smokers are predisposed to the development of COPD,[10] for which the underlying factors remain unclear. Genetic backgrounds are likely involved.
Genome-wide association studies (GWASs) have been an important tool in identifying susceptible genes and loci for complex diseases. Since 2009, several loci have been identified to be associated with COPD.[11] One of these loci is hedgehog interacting protein (HHIP).[12] HHIP is an inhibitory protein for sonic hedgehog (SHH), which is crucial for the development of the lungs and other organs. Several single nucleotide polymorphisms (SNPs) in this gene have been demonstrated to be significantly associated with COPD risk.[12–14] Several replication studies have confirmed these associations.[15,16] However, most of these studies are cross-sectional studies or prospective studies with short follow-up periods. None of these reports analyzed the synergistic effects of cigarette smoking and gene susceptibility on COPD development and lung function decline. In our present study, we genotyped 3 SNPs in HHIP to expose the association between HHIP and COPD. Moreover, we also analyzed the combined effects of HHIP and cigarette smoking on COPD risks and lung function decline.
2. Methods
2.1. Study subjects
A total of 306 COPD patients and 743 age- and gender-matched healthy subjects were recruited in our study. All subjects were recruited from 15 villages in Haokou, Qianjiang, Hubei, China. The recruitment process and screening criteria have been described in detail in our previous study.[17] Briefly, we screened over 25,000 subjects to identify high-risk subjects and COPD patients. Of the screened subjects, 16,511 were over 15 years old and were selected for subsequent analyses. Among them, 3532 subjects had chronic cough for over 2 years or smoking history above 20 pack-years. These subjects were believed to be at risk of suffering from COPD. Pulmonary function tests were performed on these 3532 subjects. Of these patients, 306 subjects were diagnosed with COPD and were selected for the present study. An additional 743 age- and gender-matched healthy subjects were recruited as controls. The detailed inclusion criteria have been reported previously.[18] All subjects were followed-up for a period of 8 years. Afterward, the subjects were divided into 2 cohorts based on those who had quit smoking or not. During the follow up, 155 COPD patients and 307 controls quit smoking and were divided in the nonsmoking cohort. Meanwhile, 193 COPD patients and 379 controls remained smoking and were divided into the smoking group. All subjects were followed-up for another period of 10 years with lung function observations every 5 years.
The study was approved by the institutional ethics committees of Tongji Hospital. Written informed consent was obtained from all participants.
2.2. SNP selection and genotyping
Three SNPs (rs7654947 and rs11100865 in the HHIP gene and rs12504628 near the HHIP gene) were selected for genotyping. The protocols for genomic DNA extraction and detection of genetic polymorphisms have been previously described.[17] Briefly, genomic DNA was extracted from 200 μL of peripheral venous blood by using a Blood Genomic DNA Purification Kit (Tiangen Biotech, Beijing, China) according to the manufacturer protocol. A TaqMan Genotyping system (Applied Biosystems, Foster City, CA) was used to genotype all participants for the selected SNPs. A GeneAmp PCR System 9700 (Applied Biosystems) was used for polymerase chain reactions. TaqMan Genotype software version 1.2 (Applied Biosystems) was used to analyze the reaction results. DNA samples of patients and controls were run in the same batches. To ensure intraplate and interplate genotype quality control, up to 10% of all genotypes were repeated to check for consistency.
2.3. Statistical analyses
The data were analyzed using SPSS 19.0 (SPSS Inc., Chicago, IL). For comparisons of baseline characteristics of participants, qualitative variables were tested using chi-squared tests. Continuous variables were expressed as the mean ± standard deviation and were compared by t test or 1-way analysis of variance. The distributions of genotypes were analyzed for deviations from the Hardy–Weinberg equilibrium (HWE) using chi-squared tests. Logistic regressions were used to test for genetic associations with and without adjusting for gender, age, body mass index, and smoking status. Haploview software version 4.0 (Daly Lab at the Broad Institute, Cambridge, MA) was used to assess linkage disequilibrium (LD) between SNPs. The figures were constructed with GraphPad Prism 5.01 software (GraphPad Software Inc., La Jolla, CA). All tests were 2-sided, and P values <.05 were considered statistically significant.
3. Results
3.1. General characteristics of study population
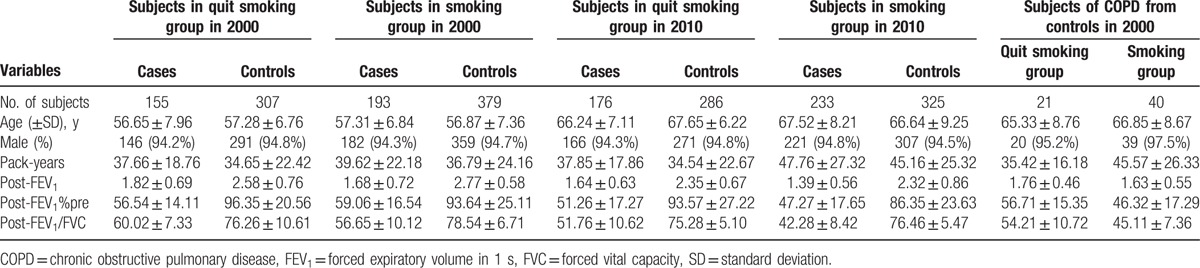
The general characteristics of the patients and controls have been reported in our previous study.[17] In brief, 306 COPD patients and 743 age- and gender-matched healthy subjects were recruited in 1992. All selected subjects had chronic cough over 2 years or smoking history above 20 pack-years. The average age was 50.81 in the patients and 49.19 in the controls, and the male percentage in the patients and controls were 93.5% and 92.1%, respectively. No significant differences were observed among these characteristics. The main characteristics of the selected subjects are shown in Table 1. By 2000, 348 COPD patients and 686 healthy subjects remained in our analysis. Among them, 155 patients and 307 control subjects had quit smoking. Meanwhile, 193 COPD patients and 379 controls continued smoking. We followed these subjects for another period of 10 years with lung function observations every 5 years. No significant differences were observed between the separated cohorts about these characteristics. After the additional 10 years, 21 control subjects developed COPD in the ex-smoking groups, and 40 control subjects developed COPD in the smoking groups, as shown in Table 2.
Table 1.
Characters of study subjects.
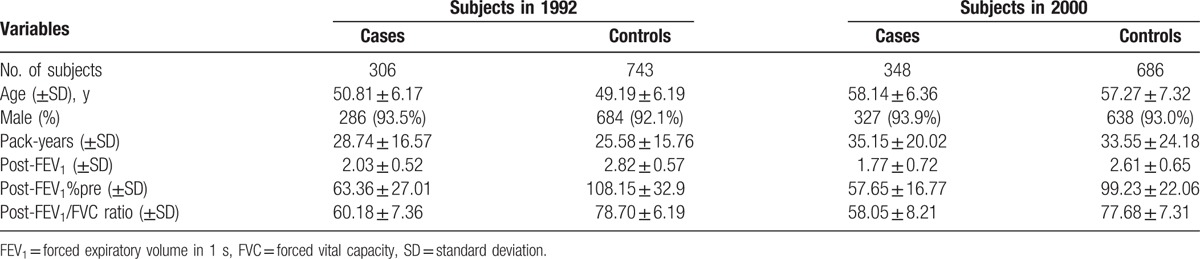
Table 2.
Characteristics of subjects in different smoking status groups.
3.2. Effects of cigarette smoking on lung function decline
The overall forced expiratory volume in 1 s (FEV1) ratio decreased by 35.06%. As shown in Figure 1A, the FEV1 declined both in the COPD patients and in the healthy controls during the follow-up period. When compared with the control subjects, the FEV1 ratio in the COPD patients decreased more significantly (41.45% vs. 31.90%, P < .05). As shown in Figure 1B, both in the ex-smoking cohorts and the smoking cohorts, the FEV1 ratio in the COPD patients decreased more significantly than in the control patients. As shown in Figure 1C, when compared with the ex-smoking cohorts, the FEV1 ratio in the smoking cohorts declined more significantly.
Figure 1.
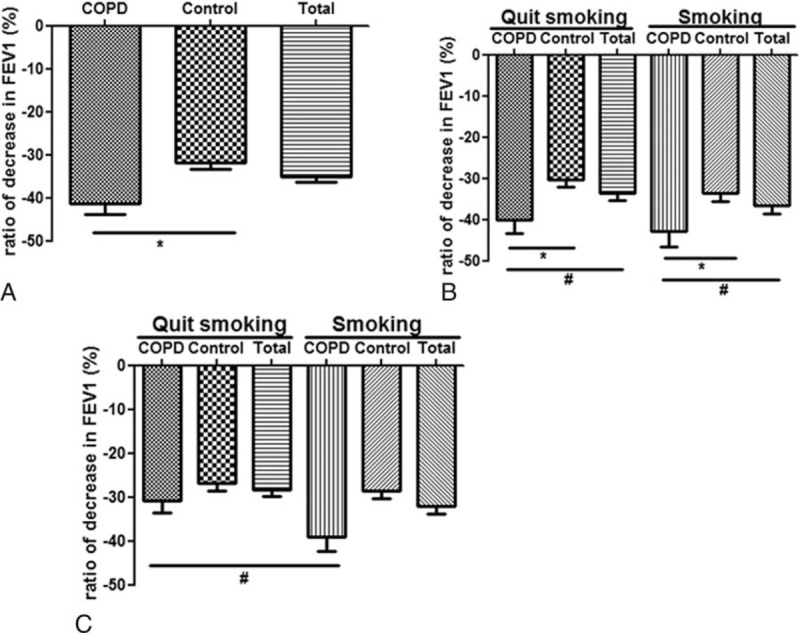
The FEV1 decline in different groups during the follow-up. (A) FEV1 decline both in COPD and controls. (B) FEV1 decline in quit smoking and smoking groups. (C) FEV1 decline in quit smoking and smoking groups. ∗P < .01; #P < .05. COPD = chronic obstructive pulmonary disease, FEV1 = forced expiratory volume in 1 s.
3.3. Association between polymorphisms and lung function decline
In our analysis, 3 SNPs were genotyped. The rs7654947 and rs11100865 were located in the intron of the HHIP gene, and rs12504628 was located upstream. All SNPs were in accordance with the HWE in the patient and control groups (P > .05). No LD was indicated by the Haploview4.0 software for any pair-wise combination or among all 3 SNPs. After adjusting for age, sex, and pack-years smoking, multivariate unconditional logistic regression analyses showed that rs11100865 and rs7654947 were associated with increased risks of COPD in 2010.
Next, we analyzed the influence of the variants on FEV1 decline; the results were shown in Figure 2. For the rs7654947 SNP, the FEV1 in the COPD patients with CC and CT genotypes decreased more significantly than the TT genotype during the follow-up period from 1992 to 2000. The difference remained significant when the COPD patients and healthy controls were combined. For the rs11100865 and rs12504628 SNPs, the FEV1 decline in the different genotypes were not significantly different, as shown in Figure 2B and C. During the follow-up period from 2000 to 2010, the results of the FEV1 decline were consistent with the findings from the first follow-up period from 1992 to 2000, as shown in Figure 2D–F. Briefly, during the follow-up period from 2000 to 2010, the FEV1 in the CT and CC subjects decreased more significantly when compared with the TT subjects for rs7654947 in both patients and overall. There were no significant differences in the other 2 SNP genotypes on lung function decline.
Figure 2.
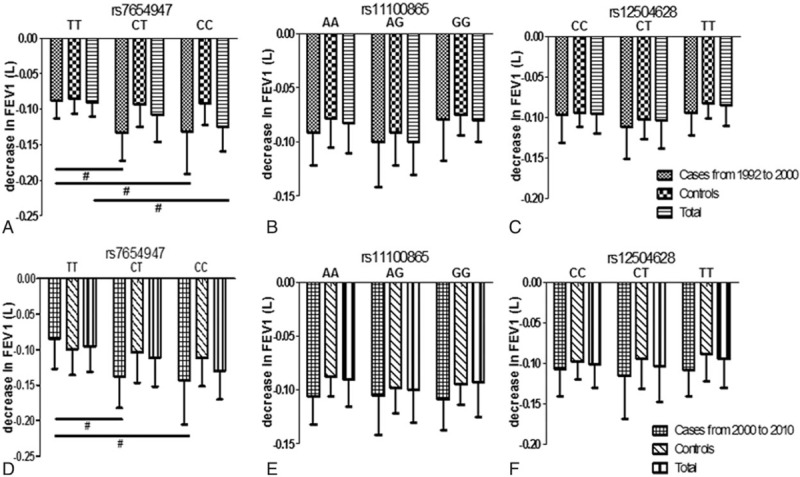
The FEV1 decline in different genotypes of subjects during the follow-up. (A) FEV1 decline in different genotypes of rs7654947 from 1992 to 2000. (B) FEV1 decline in different genotypes of rs11100865 from 1992 to 2000. (C) FEV1 decline in different genotypes of rs12504628 from 1992 to 2000. (D) FEV1 decline in different genotypes of rs7654947 from 2000 to 2010. (E) FEV1 decline in different genotypes of rs11100865 from 2000 to 2010. (F) FEV1 decline in different genotypes of rs12504628 from 2000 to 2010. ∗P < .01; #P < .05. FEV1 = forced expiratory volume in 1 s.
3.4. Cooperative effect of smoking status and gene susceptibility on lung function decline
To further determine the influence of rs7654947 on lung function under different smoking statuses, we separately analyzed the influence of rs7654947 in the ex-smoking and smoking cohorts. In the ex-smoking cohorts, the FEV1 declined more significantly in the CC and CT subjects compared with the TT subjects, as shown in Figure 3A. Similar results were observed in the smoking cohorts. When we analyzed the patients and controls separately, the difference remained significant. The FEV1 declined more significantly in the CC and CT subjects when compared with the TT subjects, which further indicated that the rs7654947 loci was associated with lung function decline, as shown in Figure 3B. Moreover, the FEV1 in the CC subjects from the smoking cohorts reduced more markedly than that of the CC subjects from the ex-smoking cohorts in both the COPD patients and overall (case, 35.5% vs. 30.75%; total, 32% vs. 28%).
Figure 3.
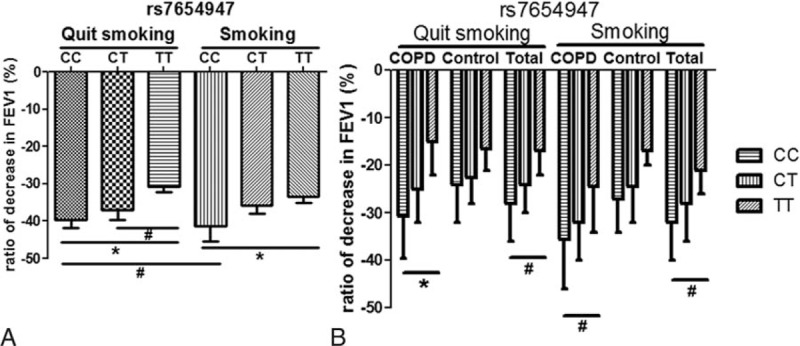
The FEV1 decline in different genotypes of rs7654947 among different groups during the follow-up. (A) FEV1 decline in different genotypes of rs7654947 among quit smoking and smoking groups. (B) FEV1 decline in different genotypes of rs7654947 among COPD and smoking groups. ∗P < .01; #P < .05. COPD = chronic obstructive pulmonary disease, FEV1 = forced expiratory volume in 1 s.
4. Discussion
In this study, we analyzed the influences of smoking status and 3 SNPs on COPD development and lung function decline. We found that rs7654947 and cigarette smoking significantly impacted lung function in both the COPD patients and healthy subjects. For rs7654947, the FEV1 of the CC subjects from the smoking cohorts declined more markedly than the ex-smoking cohorts, implying cigarette smoking and the rs7654947 loci likely had co-operative effects on the development of COPD and lung function decline.
HHIP is a member of the HHIP family. The hedgehog (HH) protein families were conserved during evolution. They play important roles in a wide range of human developmental processes, including anteroposterior patterns of limbs and the regulation of left–right asymmetry in embryonic development. HHIP is a highly conserved, vertebrate-specific inhibitor of HH signaling. It interacts with all 3 HH family members, SHH, IHH, and DHH. Three variants (rs1828591,[12] rs13118928,[19] and rs13141641[13]) in this gene have been demonstrated to significantly associate with COPD through GWAS.[20] The relationship between rs12504628 and COPD has also been reported in a case–control study.[14] However, whether the variants of HHIP and cigarette smoking have cooperative effects on the development of COPD and lung function decline has remained unclear. In this study, we analyzed 3 variants in the HHIP gene to further explore their relationship with COPD development and lung function decline. Our longitudinal study demonstrated that the variant rs7654947 had an enormous impact on COPD development and lung function decline. Cigarette smoking is one of the most important causes of morbidity and mortality in the general population and has also been implicated as a significant risk factor for COPD.[21] Hence, we further explored the impact of smoking status on COPD development and lung function decline. After 8 years of follow-up, we divided the selected subjects into 2 cohorts based on whether they had or had not quit smoking. Ten years later, the FEV1 declined in both the ex-smoking and smoking cohorts. The smoking cohorts showed a more significant decline when compared with the ex-smoking cohorts. The FEV1 of the CC subjects from the smoking cohorts reduced much more markedly than that from the ex-smoking cohorts in both the COPD patients and overall. These results confirmed that cigarette smoking and the rs7654947 loci may have synergistic effects on the development of COPD and lung function decline.
Although we conducted a long-term prospective study over the course of 18 years, several limitations should be noted. First, we only identified 3 SNPs in our study. The incomplete coverage may not provide a full representation of the entire gene and therefore, may not have fully described the contribution of HHIP. Future systematic studies with other SNPs in HHIP are warranted to evaluate the role of this gene in COPD development and lung function decline. Second, the number of subjects examined in this study was relatively small and partly weakened our statistical power. So further analyses with larger subject numbers to verify these results might be needed. Third, some of the patients in the control population may develop COPD in the future beyond the follow-up term of this study. Meantime, 14 control subjects suffered from cancer or died with unknown causes during the follow up. So these loss to follow-up might influence our results.
5. Conclusion
Our results showed that the rs7654947 variant of HHIP was associated with COPD development and lung function decline. Moreover, our study also demonstrated that both gene susceptibility and cigarette smoking were involved in the development of COPD and lung function decline. More importantly, our results also suggested that cigarette smoking and the rs7654947 loci had a combined effect on the development of COPD and lung function decline.
Acknowledgments
The authors thank the colleagues who contributed to collection and phenotypic characterization of the clinical sampling and genotyping. The authors especially thank those who kindly agreed to participate in the studies.
Footnotes
Abbreviations: COPD = chronic obstructive pulmonary disease, FEV1 = forced expiratory volume in 1 s, GWAS = genome-wide association study, HHIP = hedgehog interacting protein, HWE = Hardy–Weinberg equilibrium, LD = linkage disequilibrium, SNP = single nucleotide polymorphism.
This study was approved by the ethics committee of Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology. All procedures performed in this study involving human participants were in accordance with the ethical standards of the institutional research committee.
Conceived and designed the experiments: JX and JY. Analyzed the data: JZ, ML, and XW. Contributed reagents/materials/analysis tools: JZ and YX. Wrote the paper: JZ. Designed the study: QN. All authors read and approved the final manuscript.
This study was supported by the National Natural Science Foundation of China (No. 81570033, 81570047, 81470227, 81370145, and 81370156), National Key Basic Research and Development Program (973 Program, No. 20l5CB553403), Chinese Medical Association Research Project (No. 2013BAI09B00), and Changjiang Scholars and Innovative Research Team in University (No. IRT_14R20).
The authors have no conflicts of interest to disclose.
References
- [1].Lopez AD, Mathers CD. Measuring the global burden of disease and epidemiological transitions: 2002–2030. Ann Trop Med Parasitol 2006;100:481–99. [DOI] [PubMed] [Google Scholar]
- [2].Herse F, Kiljander T, Lehtimaki L. Annual costs of chronic obstructive pulmonary disease in Finland during 1996–2006 and a prediction model for 2007–2030. NPJ Prim Care Respir Med 2015;25:15015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Restrepo RD. Year in review 2014: COPD. Respir Care 2015;60:1057–60. [DOI] [PubMed] [Google Scholar]
- [4].Hu G, Zhong N, Ran P. Air pollution and COPD in China. J Thorac Dis 2015;7:59–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Coyle M, Shergis JL, Liu S, et al. Safety of Chinese herbal medicine for chronic obstructive pulmonary disease. Evid Based Complement Alternat Med 2015;2015:380678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].De Soyza A, Calverley PM. Large trials, new knowledge: the changing face of COPD management. Eur Respir J 2015;45:1692–703. [DOI] [PubMed] [Google Scholar]
- [7].Hassett DJ, Borchers MT, Panos RJ. Chronic obstructive pulmonary disease (COPD): evaluation from clinical, immunological and bacterial pathogenesis perspectives. J Microbiol (Seoul, Korea) 2014;52:211–26. [DOI] [PubMed] [Google Scholar]
- [8].Soto FJ, Varkey B. Evidence-based approach to acute exacerbations of COPD. Curr Opin Pulm Med 2003;9:117–24. [DOI] [PubMed] [Google Scholar]
- [9].COPD: improving prevention and care. Lancet (Lond, Engl) 2015;385:830. [DOI] [PubMed] [Google Scholar]
- [10].Zuo L, He F, Sergakis GG, et al. Interrelated role of cigarette smoking, oxidative stress, and immune response in COPD and corresponding treatments. Am J Physiol Lung Cell Mol Physiol 2014;307:L205–18. [DOI] [PubMed] [Google Scholar]
- [11].Hobbs BD, Hersh CP. Integrative genomics of chronic obstructive pulmonary disease. Biochem Biophys Res Commun 2014;452:276–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Pillai SG, Ge D, Zhu G, et al. A genome-wide association study in chronic obstructive pulmonary disease (COPD): identification of two major susceptibility loci. PLoS Genet 2009;5:e1000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Cho MH, Castaldi PJ, Wan ES, et al. A genome-wide association study of COPD identifies a susceptibility locus on chromosome 19q13. Hum Mol Genet 2012;21:947–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Soler Artigas M, Wain LV, Repapi E, et al. Effect of five genetic variants associated with lung function on the risk of chronic obstructive lung disease, and their joint effects on lung function. Am J Respir Crit Care Med 2011;184:786–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kim WJ, Oh YM, Lee JH, et al. Genetic variants in HHIP are associated with FEV1 in subjects with chronic obstructive pulmonary disease. Respirology (Carlton, VIC) 2013;18:1202–9. [DOI] [PubMed] [Google Scholar]
- [16].Wang B, Zhou H, Yang J, et al. Association of HHIP polymorphisms with COPD and COPD-related phenotypes in a Chinese Han population. Gene 2013;531:101–5. [DOI] [PubMed] [Google Scholar]
- [17].Xie J, Wu H, Xu Y, et al. Gene susceptibility identification in a longitudinal study confirms new loci in the development of chronic obstructive pulmonary disease and influences lung function decline. Respir Res 2015;16:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Xie J, Zhao J, Xiao C, et al. Reduced heat shock protein 70 in airway smooth muscle in patients with chronic obstructive pulmonary disease. Exp Lung Res 2010;36:219–26. [DOI] [PubMed] [Google Scholar]
- [19].Cho MH, Boutaoui N, Klanderman BJ, et al. Variants in FAM13A are associated with chronic obstructive pulmonary disease. Nat Genet 2010;42:200–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Nakamura H. Genetics of COPD. Allergol Int 2011;60:253–8. [DOI] [PubMed] [Google Scholar]
- [21].Boudewijn IM, Postma DS, Telenga ED, et al. Effects of ageing and smoking on pulmonary computed tomography scans using parametric response mapping. Eur Respir J 2015;46:1193–6. [DOI] [PMC free article] [PubMed] [Google Scholar]


