Abstract
Nanotechnology is a rapidly expanding field seeking to utilize nano‐scale structures for a wide range of applications. Biologically derived nanostructures, such as viruses and virus‐like particles (VLPs), provide excellent platforms for functionalization due to their physical and chemical properties. Plant viruses, and VLPs derived from them, have been used extensively in biotechnology. They have been characterized in detail over several decades and have desirable properties including high yields, robustness, and ease of purification. Through modifications to viral surfaces, either interior or exterior, plant‐virus‐derived nanoparticles have been shown to support a range of functions of potential interest to medicine and nano‐technology. In this review we highlight recent and influential achievements in the use of plant virus particles as vehicles for diverse functions: from delivery of anticancer compounds, to targeted bioimaging, vaccine production to nanowire formation. WIREs Nanomed Nanobiotechnol 2017, 9:e1447. doi: 10.1002/wnan.1447
For further resources related to this article, please visit the WIREs website.
INTRODUCTION
Nanobiotechnology focusses on the use of biologically derived structures with at least one dimension smaller than 100 nm, which can be adapted to perform specific functions. In this review, we will highlight the key advancements in the use of synthetic plant virology as a basis for a number of nanobiotechnology and medical applications.
There are certain characteristics that determine the usefulness of a nanobiotechnology system. Ideally it should be possible to produce species (particles) of consistent size, structure and biophysical properties. Such particles should be amenable to the introduction of additional functional groups such as dyes, enzymes, peptides, or inorganic compounds. Also such technologies should have minimal toxicity and low impact to the environment, particularly in the case of medical applications. Additional factors that will ultimately affect the viability of a nano‐biotechnology platform include ease and cost of production, ease of containment and the low risk of cross‐contamination with mammalian pathogens.
Viruses, and noninfective virus‐like particles (VLPs) in particular, possess these desirable features. They are capable of self‐assembling into defined structures of known dimensions, show a degree of genetic flexibility to allow functionalization with proteinaceous species, and possess reactive amino acid side‐chains, that can be used for conjugation to inorganic or less amenable species. Using plant viruses reduces many of the risks associated with biological materials as a basis for medical nanotechnologies, and the use of noninfective VLPs results in low risk to the environment. These lower risks allow greater ease in handling, transportation and processing of viral nanoparticles (VNPs), making plant virus‐based particles particularly attractive platforms for a range of nanobiotechnological applications. Throughout this review, we will use VNP as a generic term to denote any particle derived primarily from viral proteins, and VLP to distinguish VNPs that do not contain viral genetic material required to be infectious.
Plant viruses have played a prominent role in biochemical and structural research, despite the chemical nature of viruses only being revealed in the mid‐1930s.1, 2 Plant viruses were an ideal system for early biochemical experiments as high viral titres, and simple purification protocols, meant high yields of reasonably pure material were easily achievable in an era preceding sophisticated recombinant technologies. These properties also led to plant viruses being in the vanguard of studies on viral architecture3 and were the first viruses for which detailed atomic structures were available through X‐ray crystallographic and fibre diffraction studies.4, 5, 6, 7 Concomitant with these structural studies was the determination of the nucleotide sequences of a number of plant viruses and the development of methods for the manipulation of their genomes (for a review, see Porta and Lomonossoff8). This solid foundation of viral research made plant viruses very attractive for exploitation as biotechnology platforms. This review seeks to introduce readers to some of the key concepts surrounding the ability to engineer plant viruses, either by genetic or chemical means, to generate synthetic nanoparticles for a wide range of applications.
Plant Virus Structures
The vast majority of plant virus genera are nonenveloped and have genomes consisting of one or more strands of positive‐sense RNA. In common with all viruses, the particles are composed of highly‐repetitive motifs of protein subunits that assemble around the genome to form large, well‐ordered macromolecular structures. Though the detailed nature of these repeating motifs differs between viruses, there are essentially two classes of symmetry—helical and icosahedral3 (Figure 1). The classic example of a helical virus is tobacco mosaic virus (TMV, Virgaviridae) in which 2130 identical protein subunits surround the genome to form rigid rods of 300 nm.4, 5 Other helical viruses include potato virus X (PVX, Alphaflexiviridae) and potato virus Y (PVY, Potyviridae), both of which form flexuous rods.9 In terms of icosahedral symmetry, some viral capsids, such as that of Cowpea chlorotic mottle virus (CCMV, Bromoviridae), are comprised of multiple copies of a single‐coat protein that form icosahedral cages around its genome.10 Other Icosahedral viruses, such as cowpea mosaic virus (CPMV, Secoviridae) are composed of more than one type of capsomeric protein, though typically in this case these are produced by proteolytic processing of a precursor, meaning that the mature capsid proteins are present in equal numbers in the assembled particle.10, 11 Assembly of mature viruses typically involves formation of smaller assembly intermediates before these associate to form the large multimeric viral structures; however, the precise details of how capsid proteins mature, how assembly intermediates are initiated, and how complete structures form varies between viruses.10, 12, 13, 14, 15, 16 Regardless, the periodicity of capsid proteins in mature virions allows accurate predictions and regulation of the degree of modification that can be introduced into particles.
Figure 1.
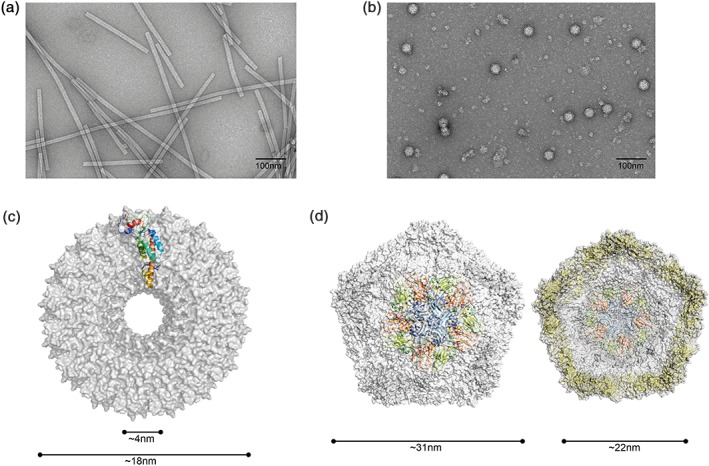
Structure and dimensions of plant viruses commonly used in nanobiotechnology. (a) Negative stain TEM of TMV showing rods of ~300 nm length. (b) Negative stain TEM of CPMV particles. (c) Cryo‐EM structure of a cross‐section of TMV (pdb 4udv). (d) Cryo‐EM structure of the external (left) and internal (right) surfaces of CPMV. Repeating motifs for cryo‐EM structures are shown as ribbons (monomer for TEM and pentamer for CPMV). [Correction added on 23 February 2017 after first online publication: labels (b) and (c) have been switched to match with the images.].
The precise size and highly symmetric nature of plant viruses makes them a powerful tool for structural biology, and has aided the development of structural techniques such as (cryo)‐electron microscopy. Improvements in electron microscopic techniques can be easily traced through studies into TMV structure, from development of negative stain transmission electron microscopy (TEM) methods in the late 1950s,17 to the advent of cryo‐EM structures defining helical arrangements 30 years later,18 and the rapid progression from near‐atomic resolution19 to atomic resolution structures that can now be produced without the need to crystallize proteins of interest.20 Such advances in structural biology are important for development of future biotechnological applications. Knowledge of the detailed three‐dimensional structure of virus particles resulted in identification of exposed loop regions of the coat proteins that permit genetic modification of the coat protein without interfering with the capsomeric interactions essential for assembly.21, 22
Production of Virus and VLPs
VNPs must be produced in plants in a different way, depending on whether they are infectious viruses or noninfectious VLPs. Indeed, infectious virus can be produced by infecting plants with the relevant virus; (whether it is wild‐type or genetically modified), while VLPs must be produced by the expression of just those proteins necessary for particle formation. The latter approach has been carried out in a variety of standard heterologous expression systems including Escherichia coli, 23, 24 yeast25, 26, 27 and insect cells.28 The infection approach results in the production of particles containing the viral genome which may require downstream processing to remove or inactivate the viral nucleic acid29, 30for reasons of safety or containment. However, using infectious virus usually has the advantage that it is possible to produce large quantities of material as the virus is able to spread within plants. The capsid expression approach often results in the production of assembled particles in vivo, but unlike infectious virus, these VLPs are either devoid of nucleic acid or encapsidate host RNA. Whichever method is chosen, the assembled particles can be further processed by, for example, in vitro disassembly and reassembly. The capsid expression approach has the advantage that it does not require the handling of infectious material and can be used to produce mutant coat proteins that are incompatible with a ‘live’ virus infection.
Recent work in the field of synthetic plant virology has led to the development of ‘deconstructed’ viral vectors, which can be used to produce high yields of plant virus‐derived VLPs in plants rather than in a heterologous system.31, 32 The term deconstructed here refers to the removal of viral genes not required for high levels of transcription and translation (such as viral coat proteins), resulting in vectors which contain only those viral elements that lead to enhanced protein yields, such as promoters and UTRs. The first example of deconstructed viral vector use was the production of VLPs based on CPMV.33 Though CPMV particles produced via infection have been extensively used in bio‐ and nanotechnology (see sections below), 90% of the particles contain the viral RNA and no in vitro disassembly/reassembly system has yet been devised for this virus; this has limited its potential as a system for encapsidation of cargo molecules. By transiently coexpressing the precursor of the two (L and S) viral coat proteins, VP60, and the protease necessary for its processing, Saunders et al.33 were able to produce particles which were morphologically similar to wild‐type CPMV but which were devoid of RNA and hence were termed empty VLPs or eVLPs. Structural studies by both cryo‐EM and crystallography34, 35 revealed the particles produced in this way to be identical to wild‐type CPMV apart from the absence of the genomic RNA. Moreover, the cryo‐EM structure allowed the visualization of a 24‐amino acid chain at the C‐terminus of the S coat protein.34 This 24‐amino acid region plays an important role in particle assembly and controls the permeability of the particles to small molecules,36, 37 with implications for loading desired compounds into CPMV VNPs (see Section Plant Viruses as Nanocarriers of Useful Cargo below). This strategy to produce eVLPs is by no means limited to CPMV; the related virus, grapevine fanleaf virus, has only a single type of subunit corresponding to uncleaved VP60, which Belval et al. were able to express to produce RNA‐free VLPs.38 These eVLPs could be produced using modified coat protein that presented a marker protein on either the external or internal particle surface. Furthermore, coexpression of two different versions of the coat protein resulted mosaic particles presenting different proteins on both surfaces. Such mosaics could be of interest to medical researchers looking to deliver encapsidated compounds to specific tissues, a theme further explored in ‘VNPs for biomedical delivery and bioimaging.’
Transient expression has also been used to produce VLPs of the unrelated member of the Tombusviridae family, turnip crinkle virus39 (Tombusviridae). Expression of the full‐length coat protein led to the formation of particles closely resembling the native virus particle with 180 coat protein subunits, and these particles encapsidated host RNA. Deletion of the N‐terminal RNA‐binding domain of the coat protein resulted in the production of small RNA‐free particles containing only 60 subunits. Attempts to display foreign proteins fused to all copies of the coat protein resulted in significantly reduced yields. However, by incorporating wild‐type subunits to produce mosaic particles, this effect can be alleviated (Saunders, K., Castells‐Graells, R. and Lomonossoff, G.P., in preparation).
In Vitro Assembly of Structures
The coat proteins of a number of plant viruses have been shown to be able to self‐assemble to form VLPs in vitro, including the well‐studied TMV.40 In this case, the TMV origin of assembly sequence (OAS) from the genomic RNA alone can direct particle formation by the coat protein in vitro under appropriate buffer conditions. By this means the assembly of rod‐shaped particles with modified coat protein conferring desirable properties to the resulting rod‐like particles has been attempted.41 Here, Eiben et al. note that mutant rod formation was possible only by mixing mutant coat protein synthesized in E. coli with wild‐type coat protein derived from a plant TMV infection in the assembly reaction. Similar in vitro assembly has been demonstrated for flexuous rod and icosahedral plant viruses42, 43, 44, 45 allowing the encapsidation of a range of foreign species, discussed further in Section Plant Viruses as Nanocarriers of Useful Cargo.
In addition to in vitro assembly to produce particles with wild‐type morphologies, filamentous viruses can be modified to produce novel structures, altering the biophysical properties of the resulting nanoparticles. Most dramatic is the production of nanospheres of various dimensions from the rod virus TMV, which maintain high thermal stability.46, 47
Of potential interest to nanobiotechnological applications, nonnative TMV structures such as ‘kinked nanoboomerangs’ can be made by introducing RNA molecules with multiple OASs, with ‘multipods’ produced in a similar manner.48 Such structure may allow the development of biocatalysis nanostructures with high surface area for efficient enzyme display. Metal‐nucleic acid conjugates with multiple TMV OASs results in the formation of ‘nanostars,’48 and similarly large networks of Turnip mosaic virus ‘Nanonets’ bound to Candida antarctica Lipase B resulted in catalytically active supra‐macromolecular complexes49 (Figure 2).
Figure 2.
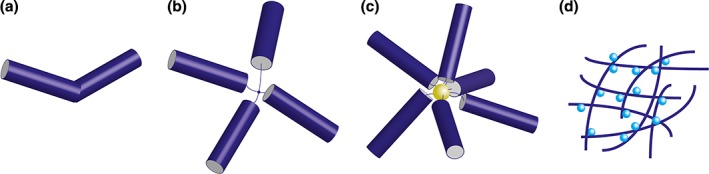
Representations of novel VNP structures for functionalization. (a) TMV‐derived ‘nano‐boomerang’ (b) TMV tetrapod, both derived by in vitro formation mediated by either two or four OAS on a single RNA. (c) Nano‐star formed by conjugating multiple TMV OAS to a gold nanoparticle. (d) Catalytically active TuMV nanonet formed by conjugation to C. antarctica Lipase B.
Such hybrid architectures may find favor in future nano‐technical applications such as nanowires and nanobiocatalysis assemblies. Not only can changes in nanoparticles structure alter physical properties such as surface area and thus for example reaction efficacy, but different architectures also affects in vivo behavior,50, 51 raising the possibility of choosing specific architectures with different pharmokinetic profiles. Although heterologous expression of TMV may be appealing for biotechnology, wild‐type TMV coat protein expressed in yeast and bacteria led to the formation of rod‐like particles of indeterminate length without the necessity of the OAS nucleotide region in the template RNA.26 Thus compared to in vitro rod formation, the heterogeneous nature of the particles formed, and lack of control over assembly normally mediated by RNA means that heterologous expression systems appears to be of limited use in this instance.
Once a particle of desired physical properties has been selected, both icosahedral and filamentous viruses structures can be functionalized via both surface modifications and use of their internal cavities.
Functionalizing Viral Surfaces
The advent of modern recombinant technologies has allowed greater functionalization of the outer surface of viruses either by the direct genetic insertion of functional enzymes or peptides into predetermined loci, or the addition of nonnative amino acids to facilitate chemical conjugation using reactive side chains, such as those of cysteine, lysine and glutamate.
Genetic insertion of a peptide or protein may appeal to researchers with knowledge of the target virus’ structure as the position of such inserts should be predictable, and once purified the VNPs may not require additional processing for functionalization. One of the problems encountered with this approach, however, is that the presence of the inserted sequence often adversely affects the yield of virus that can be obtained, particularly if the insert is large or positively charged.52, 53 For CPMV, structural studies have been useful in rationalizing such limitations, and improving insertion sites to maximize VNP activity22, 52, 54; however, such data are not available for every prospective VNP. A potential solution to this problem is to use specific antibodies or antibody fragments to mediate interactions with antigens or other proteins of interest. For example, mosaic PVX VNPs have been produced in planta, which display engineered antibody fragments capable of antigen‐binding, despite such insertions typically preventing VNP formation.55, 56 This was achieved by including a short peptide that resulted in a mixed population of wild‐type and modified proteins via a ribosomal stutter mechanism. By choosing different antibodies for fusion to PVX coat protein in this system, it should be possible to produce VNPs functionalized with a wider range of species than would otherwise be possible.
Alternatively, researchers may choose to modify the outside of wild‐type particles by chemical conjugation in order to avoid the potentially destabilizing effect of genetic insertion. This process is not always straightforward however. For example TMV lacks exposed reactive cysteine and lysine amino acids, and so modification of wild‐type particles relies on the less commonly used azo‐coupling through existing tyrosine residues.57 Nevertheless, mutant TMV particles have been synthesized that include nonnative amino acids to allow thiol‐ or amine‐selective chemistry.58, 59 Similarly, modified TMV possessing a surface‐exposed cysteine residue has been coupled with sensor enzymes such as glucose oxidase and horseradish peroxidase, resulting in multivalent nanoscale platform for the ordered presentation of bioactive proteins.60 The scope for genetic fusions to the termini of TMV capsid proteins is limited, as this can inhibit virus formation, with wild‐type morphologies requiring mosaics of wild‐type and mutant protein.41
Chemical modifications to the solvent‐exposed external surface of icosahedral particle can carried out using either virus particles or VLPs, while modification of the internal surface is usually confined to empty VLPs as the presence of genomic RNA tends to occlude access to the reactive amino acid side chains.37 Multiple examples of surface modifications of icosahedral particles exist, using the carboxyl groups of aspartic and glutamic acid,61 the ε‐amino group of lysine,62, 63, 64, 65 the thiol group of cysteine65, 66 and the phenol side chain of tyrosine.67 It is also possible to genetically introduce or remove amino acids with reactive side changes in order to better control the levels of modification, or facilitate alternative chemistries for conjugation. For example, Wang et al.66 and Gillitzer et al.68 introduced cysteine residues to the surface of CPMV and CCMV, respectively while Chatterji et al.69 successively reduced the number of lysine residues on the surface of CPMV. Once a desirable chemistry has been identified (using either wild‐type or mutant VNPs), conjugation can facilitate functionalization with a wide range of species including fluorescent dyes, electroactive compounds, drug molecules, quantum dots, specific metals, and even active enzymes.63, 70, 71, 72
The interior surface of certain VNPs is also amenable to modification: the interior surface of CPMV VLPs can be chemically modified with fluorescent dyes via naturally occurring cysteines.37 Modification of the interior surface of VLPs derived from CCMV has also been achieved: the wild‐type coat protein is highly positively charged due to the presence of the basic amino acids lysine and arginine. This positively charged interior surface was used to promote mineralization within the preformed capsid to produce defined inorganic nanoparticles.73 By producing CCMV VLPs with an altered interior charge, it was possible to alter the range of materials that could be encapsulated.74
Modifications to both the solvent‐exposed surfaces of VLPs and to the internal cavity of many plant viruses allow synthetic viruses to interact with a variety of organic and inorganic substrates for a range of biotechnology applications, and to act as nanocarriers for both medical and bioimaging use.
Plant Viruses as Nanocarriers of Useful Cargo
Plant viruses have a strong track record of being used as carriers of useful cargo for a plethora of nanotechnological and biomedical applications,75, 76 which we will divide here into three broad categories: delivery of therapeutics, bioimaging, and metallization. Cargo carrying utilizes the interior viral cavity as a vehicle for specific molecules; however, the introduction of defined cargoes into viruses is nontrivial. There are two main strategies for loading plant viral particles with foreign cargo: the infusion technique seeks to allow diffusion of a cargo of interest into the preformed viral particle, whereas the caging strategy aims to trigger particle formation around the cargo of interest (Figure 3). Each strategy has been used successfully with different viruses for different types of cargo, described below.
Figure 3.
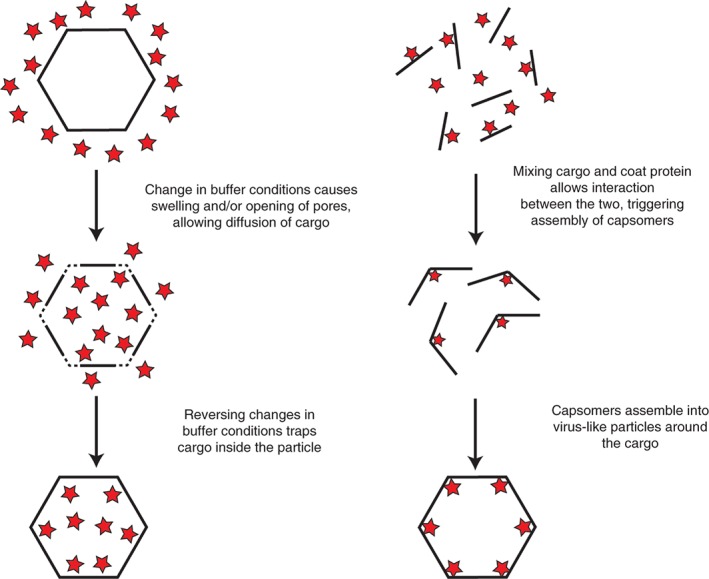
Schematic of the key methods used to encapsidate specific cargoes into VNPs. Left shows swelling‐mediated infusion of nanoparticles. Right demonstrates cargo caging.
Infusion
The infusion strategy relies on causing viral particles to swell in such a way that pores open in the capsid, allowing diffusion of small cargo. Reversing the swelling then causes these pores to close, thus trapping the cargo inside. The method required to achieve such reversible swelling in tomato bushy stunt virus (TBSV, Tombusviridae) was described by Perez et al.,77 and relies on the chelation then addition of divalent cations to cause opening then closing of pores. This method has been used to load TBSV virions with ethidium bromide,78 and a similar strategy was used by Loo et al.79 as well as Lockney et al.80 to load the chemotherapy drug doxorubicin into particles of red clover necrotic mosaic virus (RCNMV, Tombusviridae) thanks to the interaction between the drug and viral nucleic acid. The same technique was used by Zeng et al. to load doxorubicin into particles of the distantly related cucumber mosaic virus (CMV, Bromoviridae).81 This technique has also found a use in agronomy, where the soil mobility of RCNMV led Cao et al.82 to load the nematicide Abamectin into the viral particles through infusion. This encapsulated form of Abamectin had similar bioavailability to nematodes as the free form of the pesticide, but with improved soil mobility, making it more effective at protecting crop roots from nematode infection.
In some cases, infusion does not even require swelling of the viral particles to allow ingress of cargo: it is possible to use the native nucleic acid content of the virions as an electrostatic sponge to attract and retain positively charged cargo. This is the strategy that was used by Yildiz et al.83 as well as Wen et al.84 to load particles of CPMV with imaging agents and therapeutic molecules. This relies the natural affinity for these molecules with nucleic acid, and these experiments demonstrate that potential to use infused CPMV particles as imaging and therapy vehicles. A similar method was employed to load the platinum‐containing anticancer drug candidate Phenanthriplatin into the channel of the rod‐shaped TMV through a one‐step loading protocol that also exploited the electrostatic interaction between the positively charged cargo and the negatively charged interior of the viral particle.85
Caging
The caging strategy relies on assembling viral particles around the cargo of interest, either in vivo or after disassembling virions in vitro. The plant virus that has been most often used for caging of foreign cargo is almost certainly CCMV. The conditions required to disassemble the virus particles were described by Adolph,86 and had the advantage of being very simple: shifts in pH and ionic strength allowed for easily controlled swelling, and eventually complete disassembly of the viral particles. This mechanism has been extensively studied,87 and the reversibility of this process allowed later groups to use the disassembly–reassembly mechanism to remove native nucleic acid from the inside of the particles and replace it with cargo of interest. In one example, CCMV particles have been used as gene therapy candidates: the native viral RNA is removed after disassembly of the virions, and the capsid dimers are reassembled around heterologous RNA originating from the mammalian Sindbis virus.88 The authors even demonstrated that the heterologous RNA was released and expressed in mammalian cells upon transfection with the chimeric viral particles. Proteins can also be specifically packaged inside CCMV particles by using the caging strategy in concert with some targeted genetic engineering of the CCMV capsid protein. Indeed, the N‐terminus of the CCMV capsid protein can be fused to the K‐coil of a leucine zipper, while the C‐terminus of a protein cargo is fused to the E. coli, allowing noncovalent binding of the cargo to the capsid dimers, and self‐assembly of VLPs around the protein cargo.89
More stable covalent binding of cargo has also been induced through the use of bacterial Sortase A: The C‐terminus of the CCMV capsid protein is modified to end in a glycine residue, and covalent binding can take place with any cargo (small molecule of protein) fused to an leucine proline glutamate threonine glycine (LPETG) amino acid motif via the action of trans‐acting Sortase A.90 Encapsidation of whole proteins into CCMV VLPs allows the creation of nanoreactors; nanoscale cages with enzymatic activity. This was demonstrated by Comellas‐Aragones et al.,91 who disassembled CCMV virions and reassembled them around individual molecules of horseradish peroxidase, with the enzymatic substrate and product capable of diffusing in and out of the nanoreactors. This has huge implications for enzyme‐based therapeutics, as demonstrated by Sanchez‐Sanchez et al.,92 who used a similar method for the encapsulation of a bacterial cytochrome p450 inside CCMV VLPs. These VLPs were enzymatically functional and could process prodrugs into cytotoxic active forms, which has important implications for targeted drug delivery. The closely related virus CMV can also be used for caging of cargo by functionalization of a nucleic acid intermediate. Disassembled CMV capsids can be made to reassemble around heterologous DNA, which can be used directly as cargo of interest, but if the DNA is functionalized with dyes or protein (through biotin–streptavidin interactions), the capsid–DNA interaction allows encapsulation of a wide range of cargoes.93
Two areas which are attracting a considerable amount of attention with regards to loading cargo inside plant viral particles are the fields of biodelivery and mineralization of nano‐scale structures. To achieve their goals, researchers in both of these fields have made use of both the infusion as well as the caging strategies, each with great success.
VNPs for Biomedical Delivery and Bioimaging
Efficient drug delivery is greatly affected by the biochemical properties of the target compound and its interactions with the host. For compounds that are quickly eliminated from the body, are poorly soluble, or may not be able to efficiently pass cell membranes, the use of carrier molecules can improve drug delivery and thus efficacy. Such carriers must be small enough to move through the bloodstream, nontoxic, biocompatible and able to enter cells. Plant viruses have all of these properties, and have been the subject of much research as potential biomedical vehicles.94 With distinct surface and interior residues, VNPs are good candidates for manipulation to protect poorly soluble drugs while maintaining good biocompatibility.
In addition to drug biocompatibility, efficient transport to target tissues is a major challenge in pharmacology. This is a particular issue in the case of anticancer drugs which usually discriminate between cancerous and normal cells by the fact that the cancer cells are dividing more rapidly and differentially certain cell‐surface receptors.95, 96 However, anticancer drugs are toxic to all cells and thus often have severe side effects.
Nanotechnology‐based drug delivery is aided by physiological differences between tumorous and healthy tissue. Tumors have been shown to retain more nanoparticles than healthy tissue and display increased permeability to larger nanomaterials on the basis of disrupted tight junctions. This results in preferential penetration of nanoparticles into tumorous tissue, known as the enhanced permeability and retention effect. Viruses specifically are useful for biomedical delivery due to their bioavailability, biodistribution, and persistence in a mammalian organism. Studies to ascertain these characteristics have been undertaken with CPMV,97, 98 CCMV,99 TMV,100 and PVX.51, 101, 102 Taken together, these studies reveal that plant viral particles are generally safe, well tolerated, with high bioavailability, broad biodistribution, and relatively short persistence in animal models.
Many wild‐type plant viruses display nonspecific cell entry, or at least entry into a wide range of cells, which may be deleterious for delivery of cytotoxic drugs. Untargeted uptake can be reduced by conjugation of surface residues to polyethylene glycol (PEGylation),103 and more targeted delivery facilitated by specific surface modifications. CPMV has been shown to interact with the intermediate filament protein vimentin,104 and this interaction facilitates CPMV uptake into a range of cells (including macrophages and certain cancerous cells). Using this natural uptake, Wen et al.75 were able to decorate CPMV with a photosensitizing agent to target macrophages and tumorous cells for destruction. Similarly, the loading of TMV with Zn‐EpPor has been shown to enhance photosensitizer uptake into melanoma cells.105
Integrins are another example of cell‐surface receptors that are upregulated in many cancers.106 Specific peptide motifs, such as the RGD motif seen in Adenovirus are recognized by subtypes of integrins, and when this peptide is introduced to the surface of CPMV, either by genetic fusion to exposed loops or via chemical conjugation, uptake of CPMV into multiple cancer cell lines can be increased.107 Similar approaches have been used to target ovarian cancer cells. Folic acid receptors (FR) are overexpressed in ovarian cancer cells, and so FR‐mediated endocytosis is a potential strategy for cell‐specific drug delivery.108 This strategy was successfully employed by Ren et al.109 and Zeng et al.81: modification of the surfaces of two unrelated viruses with folic acid for cell targeting, and infusion of the internal cavity of the VNPs with doxorubicin resulted in a significant increase in drug delivery into cancer cells.
The ability to incorporate custom cargoes into protective protein shells, and targeting of these to specific tissue types allows the use of VNPs as bioimaging agents. Using NHS ester chemistry both flexuous rod virus PVX and the icosahedral virus CPMV can be functionalized with a number of commercial fluorescent dyes for imaging in cell cultures and to mark embryo vasculature and tumor tissue in chick and mouse systems.37, 110, 111, 112, 113 The use of more complex two‐step bioconjugation methods allows further functionalization of PVX to target fluorescent particle uptake into cancer cells.114 In addition to commonly used single‐photon fluorescent markers, two‐photon dyes can be preferable for bioimaging as such dyes give lower background fluorescence115 and can reduce radiation damage to surrounding tissue.116, 117 Recently the coupling of TMV particles to the two‐photon dye BF3‐NCS has been demonstrated to allow visualization of diseased brain vasculature in mice.118 Imaging using nonoptical methods such as MRI is also achievable by loading metals such as gadolinium into VNPs.119, 120 Such metal‐VNP‐specific interactions, referred to as mineralization, are further discussed in Section Mineralization of Viral Scaffolds.
VNPs provide a good basis for biomedical technology, not only as chassis capable of transporting specific cargo, but also as scaffolds for antigen presentation and vaccination. With the development of plant‐virus based technologies capable of producing a wide range of non‐infectious VLPs, plant‐derived VLPs is an area of considerable interest.
Synthetic Plant Virology for Vaccine Production
Vaccines provide acquired immunity to the infection of a particular micro‐organism by stimulating the immune system against it. Vaccines against viral diseases are commonly produced from lab‐cultured pathogens which are then attenuated or inactivated. This comes with the risk of residual pathogenic activity due to incomplete inactivation, attenuation, or reversion.121 A potentially safer approach is therefore the recombinant production of specific components of pathogens, which can mimic the immunological properties of the original virus without its pathogenic properties.
Heterologous production and purification of immunogenic regions, for example solvent‐exposed surface peptides, that can be introduced into hosts can result in immune responses. The use of nanoparticles such as VLPs as display scaffolds, enhances the immunogenicity of the antigen by presenting a quasi‐crystalline ordered repeat antigen structure to the immune system122, 123 (further explored in Plummer and Manchester124). Because the point of an antigen‐carrier VNP is that the VNP moiety is incapable of causing an infection in the animal host, replicating plant viruses used for antigen display can be considered antigen‐carrier VLPs, even though they are not technically VLPs. There are numerous examples of replicating plant viruses used to display immunogenic epitopes of animal pathogens for vaccine purposes. Chimaeric CPMV particles displaying a foot and mouth disease virus epitope, human rhinovirus epitope, or HIV epitope fused to the S coat protein have been produced in cowpea plants, and these particles could stimulate an immune response against the target epitope in test animals.125, 126, 127 Moreover, this technique also permitted the development of a vaccine against mink enteritis virus that provides protective immunity in target animals.128 TMV particles have also been used to display heterologous epitopes: a leaky stop codon strategy was used to produce chimaeric TMV particles in which the C‐terminus of some of the coat proteins is fused to epitopes from the malaria parasite,129 influenza virus, or HIV.130 It has subsequently shown that TMV particles presenting a short epitope from murine hepatitis virus was able to stimulate protective immunity in mice.131
Chemical conjugation of antigens provides a further alternative to genetic fusion, with tyrosine‐mediated linkage of the weakly immunogenic hapten estriol to the surface of TMV leading to a significant immune response in mice.132
Although VLP‐mediated antigen display can increase immune protection, this is not always the case.133, 134, 135 Furthermore, the use of a single VLP scaffold for antigen delivery may not be sustainable as exposure to the scaffold may lead to an immune response against the VLP moiety. Instead, recent work in the field of synthetic plant virology has used deconstructed viral vectors to produce high yields of pharmaceutically interesting proteins (for a recent review, see Peyret & Lomonossoff32).
Although beyond the scope of this review to cover in detail, the use of such vectors and heterologous plant systems such as Nicotiana benthamiana to produce nonplant VLPs for vaccines is an exciting area of research that cannot go without mention. In this approach the structural proteins for a target virus are cloned into a high‐expression vector and transiently expressed in hosts such as N. benthamiana. Protein expression results in particles that morphologically resemble their infectious counterparts, and thus can be expected to act as an effective vaccine. Similar methods can be used in traditional mammalian and insect cell‐culture; however, the use of plants has a number of significant benefits. This approach has been used successfully to produce a number of different VLPs as vaccine candidates (Table 1). It is possible to produce VLPs composed of a small number of capsid proteins, such as Hepatitis B core‐like particles and Human Papillomavirus VLPs,142, 149 and more complex nonenveloped proteins such as Bluetongue virus.148 The transient expression in plants of VLPs from a variety of animal viruses is further reviewed in Marsian and Lomonossoff, 2016.150
Table 1.
Key Vaccine Candidates Produced Using Deconstructed Viral Vectors. Further Details Can Be Found in Marsian and Lomonossoff 2016.
| Virus | Summary | Reference |
|---|---|---|
| Hepatitis B | Tabletized transgenic lettuce containing HBsAg VLPs is orally immunogenic in mice | Pniewski 2011136 |
| Hepatitis C | Cucumber mosaic virus nanoparticles carrying a Hepatitis C virus‐derived epitope, orally immunogenic in rabbits | Nuzzaci 2010137 |
| Hepatitis C | Papaya mosaic virus‐like particles fused to a hepatitis C virus epitope: evidence for the critical function of multimerization, mixed response in mice | Denis 2007138 |
| Influenza | Influenza virus‐like particles induce a protective immune response against a lethal viral challenge in mice, produced for H7N9 outbreak virus | D'Aoust 2008139 |
| Papillomavirus | HPV‐16 L1 VLPs via agroinfiltration‐mediated transient expression or via transplastomic expression | Maclean 2007,140 Fernandez‐San 2008141 |
| Papillomavirus | Expression of HPV‐8 L1 VLPs | Matic 2012142 |
| Papillomavirus | transient expression of chimaeric L1::L2 VLPs and proof of increased breadth of immune response | Pineo 2013143 |
| Bovine papillomavirus | Transient expression of BPV L1 VLPs | Love 2012144 |
| HIV | Expression of Gag VLPs in transgenic tobacco chloroplasts | Scotti 2009145 |
| Human norovirus | NaVCP VLPs in which generate a mucosal and serum antibody response | Mathew 2014146 |
| Rotavirus | Immunogenic rotavirus‐like particles in transgenic plants | Yang 2011147 |
| Bluetongue virus | Protective bluetongue virus‐like particles | Thuenemann 2013148 |
Mineralization of Viral Scaffolds
Many VNP technological and medical applications, such MRI, are dependent on the ability to form complexes between VNPs and specific metals and metal‐based compounds. In the case of mineralization, inorganic nanoparticles and microstructures can be deposited and assembled on virus structures that act as biological templates or scaffolds.151, 152 Mineralization has been reported for both the interior and exterior surfaces of icosahedral and rod‐shaped virus particles (Figure 4). VNPs present several advantages like their nanoscale size, symmetry, polyvalence and monodispersity.75, 152 Hybrid organic–inorganic species such as metallized VNPs can be useful for a broad range of applications such as catalysis, semiconductors, drugs and contrast agents.73, 75
Figure 4.
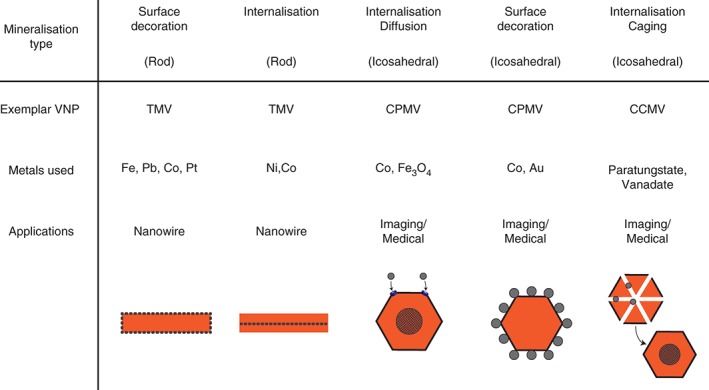
Summary of different VNP‐metal interactions. Both icosahedral and rod‐shaped viruses can be used to either cage metals, or act as scaffolds to decorate with a variety of metals for a range of biomedical and nano‐biotechnological functions.
Icosahedral viruses such as CCMV and CPMV have been demonstrated to be able to interact with a variety of metals, with the positively charged internal cavity of CCMV allowing inorganic crystal nucleation.153 pH‐dependent swelling, as described above, results in the formation of 60 pores in the capsid, which can then be used to load paratungstate and vanadate.73 CPMV shows similarities to CCMV in its ability be loaded with metals cobalt or iron‐oxide154; however, in order to load CCMV with iron, nine basic amino acids at the N‐terminus of the coat protein must be substituted with glutamate residues to alter the charge of the inner cavity to allow oxidative mineralization.74, 155 The surface of CPMV can be decorated directly with ferrocene derivatives.154 Alternatively, custom decoration with cobalt–platinum, iron–platinum or zinc sulfide can be achieved by conjugating mineralizing peptides to the viral surface, or genetic fusion of hexa‐histidine motifs to the C‐terminus of the small coat protein subunit.36, 156
Filamentous viruses such as TMV can also be modified to facilitate mineralization, either externally or internally. Immobilization of both silica and a range of metals on the surface of TMV has been demonstrated,151, 152, 157, 158 and the inner cavity can likewise be utilized for mineralization of nickel and cobalt.159, 160 With a range of available VNPs with different physical characteristics (icosahedral, rigid, or flexuous rod), and by identifying the appropriate modifications required for specific metal interactions, VNPs appear to be a viable platform for the generation of a number of organic–inorganic hybrids.
CONCLUSION
Plant viruses are an integral tool for nanobiotechnology. Much of this is due to the extensive foundational work regarding plant virus structure, genetics, and biochemistry. Viruses have many properties making them ideal chassis for biotechnological functions, with plant viruses having additional benefits with regards to ease of production and purification. Production is one of the main limiting factors for widespread use of plant‐derived VNPs, as current research can be relatively labor‐intensive; activities such as pricking out seedlings for growth are typically done manually. This issue is by no means insurmountable, as automation is feasible for most aspects of plant growth, and such issues must be weighed against the benefits of using simple media and growth conditions. The success of companies such as North America‐based Medicago using plants as an expression system demonstrate the viability of industrial‐scale use of plants for protein expression.
As both wild‐type viruses and noninfectious VLPs pose little threat to human health, they are a good choice for nanomedical uses. As with any protein‐based system there are risks of inducing an immune response to VNPs which may limit their long‐term usefulness in both antigen display and live‐animal bioimaging; however, to what extent this will prevent their clinical usefulness is uncertain. With very promising preclinical data regarding VNP toxicity, retention, and biodistribution, a key area of research will be the demonstration of clinical efficacy. If VNPs can be demonstrated to be viable clinically, improved methods for cell targeting will be crucial for widespread use of VNPs for drug delivery and gene therapy. The development of resources such as TumorHOPE,161 a database for tumor homing peptides, will play a vital role in developing novel targeting methods that may be ultimately beneficial where existing drug treatments show little discrimination between diseased and healthy tissue.
Functionalization of external surfaces combined with manipulation of viral morphologies opens a large number of potential applications in nanotechnology, from nanowires to nanoreactors. Increasing our understanding of virus tolerance to manipulation, generation of more novel structures, and methods of functionalizing with active enzymes without affecting activity is going to be key in expanding the range of VNPs with custom properties.
Currently there are no plant‐based nanobiotechnology products on the market. We believe that as researchers are able to demonstrate greater success in producing VNPs with desirable properties, increased interest from both private and public bodies will drive investment in both fundamental research and infrastructure required to produce economically viable technologies. Part of this is likely to be due to economies of scale: despite low running costs for plant production, high upfront costs for industrial‐scale expression means that very few sites are capable of producing industrial levels of plant‐derived nanotech materials. Specialized medium‐scale production facilities that will allow researchers to test the feasibility of novel VNPs will be an important step in demonstrating viability of plants as an expression platform, thus hopefully stimulating wider interest in the field.
Furthermore, new technologies are likely to be a driving force in plant‐based VNP production, and plant‐based heterologous expression in general. Cell‐based methods such as the BY2 cell‐pack method for transient expression162 that allow medium‐ to high‐throughput screening will be essential to allow candidate screening analogous to existing E. coli based methods. Although the production timescales are unlikely to match those of E. coli (expression typically requiring several days as opposed to several hours), transient expression screens are still a viable means to facilitate construct design and optimization for enhanced yield, and improved particle characteristics.
ACKNOWLEDGMENTS
This work was supported by the UK Biotechnological and Biological Sciences Research Council (BBSRC) Institute Strategic Program Grant ‘Understanding and Exploiting Plant and Microbial Secondary Metabolism’ (BB/J004596/1) and the John Innes Foundation.
Conflict of interest: The authors have declared no conflicts of interest for this article.
The copyright line for this article was changed on 27 April 2017 after original online publication.
REFERENCES
- 1. Stanley WM. Isolation of a crystalline protein possessing the properties of tobacco‐mosaic virus. Science 1935, 81:644–645. [DOI] [PubMed] [Google Scholar]
- 2. Bawden FC, Pirie NW, Bernal JD, Fankuchen I. Liquid crystalline substances from virus‐infected plants. Nature 1936, 138:1051–1052. [Google Scholar]
- 3. Caspar DL, Klug A. Physical principles in the construction of regular viruses. Cold Spring Harb Symp Quant Biol 1962, 27:1–24. [DOI] [PubMed] [Google Scholar]
- 4. Stubbs G, Warren S, Holmes K. Structure of RNA and RNA binding site in tobacco mosaic virus from 4‐A map calculated from X‐ray fibre diagrams. Nature 1977, 267:216–221. [DOI] [PubMed] [Google Scholar]
- 5. Bloomer AC, Champness JN, Bricogne G, Staden R, Klug A. Protein disk of tobacco mosaic virus at 2.8 A resolution showing the interactions within and between subunits. Nature 1978, 276:362–368. [DOI] [PubMed] [Google Scholar]
- 6. Harrison SC, Olson AJ, Schutt CE, Winkler FK, Bricogne G. Tomato bushy stunt virus at 2.9 A resolution. Nature 1978, 276:368–373. [DOI] [PubMed] [Google Scholar]
- 7. Abad‐Zapatero C, Abdel‐Meguid SS, Johnson JE, Leslie AG, Rayment I, Rossmann MG, Suck D, Tsukihara T. Structure of southern bean mosaic virus at 2.8 A resolution. Nature 1980, 286:33–39. [DOI] [PubMed] [Google Scholar]
- 8. Porta C, Lomonossoff GP. Use of viral replicons for the expression of genes in plants. Mol Biotechnol 1996, 5:209–221. [DOI] [PubMed] [Google Scholar]
- 9. Varma A, Gibbs AJ, Woods RD, Finch JT. Some observations on structure of filamentous particles of several plant viruses. J Gen Virol 1968, 2:107–114. [DOI] [PubMed] [Google Scholar]
- 10. Rossmann MG, Johnson JE. Icosahedral RNA virus structure. Annu Rev Biochem 1989, 58:533–573. [DOI] [PubMed] [Google Scholar]
- 11. Lomonossoff GP, Johnson JE. The synthesis and structure of comovirus capsids. Prog Biophys Mol Biol 1991, 55:107–137. [DOI] [PubMed] [Google Scholar]
- 12. Bancroft JB. The self‐assembly of spherical plant viruses. Adv Virus Res 1970, 16:99–134. [DOI] [PubMed] [Google Scholar]
- 13. Casjens S, King J. Virus assembly. Annu Rev Biochem 1975, 44:555–611. [DOI] [PubMed] [Google Scholar]
- 14. Klug A. The tobacco mosaic virus particle: structure and assembly. Philos Trans R Soc Lond B Biol Sci 1999, 354:531–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Atabekov J, Dobrov E, Karpova O, Rodionova N. Potato virus X: structure, disassembly and reconstitution. Mol Plant Pathol 2007, 8:667–675. [DOI] [PubMed] [Google Scholar]
- 16. Zhang W, Olson NH, Baker TS, Faulkner L, Agbandje‐McKenna M, Boulton MI, Davies JW, McKenna R. Structure of the Maize streak virus geminate particle. Virology 2001, 279:471–477. [DOI] [PubMed] [Google Scholar]
- 17. Brenner S, Horne RW. A negative staining method for high resolution electron microscopy of viruses. Biochim Biophys Acta 1959, 34:103–110. [DOI] [PubMed] [Google Scholar]
- 18. Jeng TW, Crowther RA, Stubbs G, Chiu W. Visualization of alpha‐helices in tobacco mosaic virus by cryo‐electron microscopy. J Mol Biol 1989, 205:251–257. [DOI] [PubMed] [Google Scholar]
- 19. Sachse C, Chen JZ, Coureux PD, Stroupe ME, Fandrich M, Grigorieff N. High‐resolution electron microscopy of helical specimens: a fresh look at tobacco mosaic virus. J Mol Biol 2007, 371:812–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fromm SA, Bharat TA, Jakobi AJ, Hagen WJ, Sachse C. Seeing tobacco mosaic virus through direct electron detectors. J Struct Biol 2015, 189:87–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lomonossoff GP, Johnson JE. Use of macromolecular assemblies as expression systems for peptides and synthetic vaccines. Curr Opin Struct Biol 1996, 6:176–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lin T, Porta C, Lomonossoff G, Johnson JE. Structure‐based design of peptide presentation on a viral surface: the crystal structure of a plant/animal virus chimera at 2.8 A resolution. Fold Des 1996, 1:179–187. [DOI] [PubMed] [Google Scholar]
- 23. Bragard C, Duncan GH, Wesley SV, Naidu RA, Mayo MA. Virus‐like particles assemble in plants and bacteria expressing the coat protein gene of Indian peanut clump virus. J Gen Virol 2000, 81:267–272. [DOI] [PubMed] [Google Scholar]
- 24. Zhao X, Fox JM, Olson NH, Baker TS, Young MJ. In vitro assembly of cowpea chlorotic mottle virus from coat protein expressed in Escherichia coli and in vitro‐transcribed viral cDNA. Virology 1995, 207:486–494. [DOI] [PubMed] [Google Scholar]
- 25. Brumfield S, Willits D, Tang L, Johnson JE, Douglas T, Young M. Heterologous expression of the modified coat protein of Cowpea chlorotic mottle bromovirus results in the assembly of protein cages with altered architectures and function. J Gen Virol 2004, 85:1049–1053. [DOI] [PubMed] [Google Scholar]
- 26. Kadri A, Wege C, Jeske H. In vivo self‐assembly of TMV‐like particles in yeast and bacteria for nanotechnological applications. J Virol Methods 2013, 189:328–340. [DOI] [PubMed] [Google Scholar]
- 27. Mueller A, Kadri A, Jeske H, Wege C. In vitro assembly of tobacco mosaic virus coat protein variants derived from fission yeast expression clones or plants. J Virol Methods 2010, 166:77–85. [DOI] [PubMed] [Google Scholar]
- 28. Young M, Willits D, Uchida M, Douglas T. Plant viruses as biotemplates for materials and their use in nanotechnology. Annu Rev Phytopathol 2008, 46:361–384. [DOI] [PubMed] [Google Scholar]
- 29. Langeveld JP, Brennan FR, Martinez‐Torrecuadrada JL, Jones TD, Boshuizen RS, Vela C, Casal JI, Kamstrup S, Dalsgaard K, Meloen RH, et al. Inactivated recombinant plant virus protects dogs from a lethal challenge with canine parvovirus. Vaccine 2001, 19:3661–3670. [DOI] [PubMed] [Google Scholar]
- 30. Ochoa WF, Chatterji A, Lin T, Johnson JE. Generation and structural analysis of reactive empty particles derived from an icosahedral virus. Chem Biol 2006, 13:771–778. [DOI] [PubMed] [Google Scholar]
- 31. Sainsbury F, Lomonossoff GP. Transient expressions of synthetic biology in plants. Curr Opin Plant Biol 2014, 19:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Peyret H, Lomonossoff GP. When plant virology met Agrobacterium: the rise of the deconstructed clones. Plant Biotechnol J 2015, 13:1121–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Saunders K, Sainsbury F, Lomonossoff GP. Efficient generation of cowpea mosaic virus empty virus‐like particles by the proteolytic processing of precursors in insect cells and plants. Virology 2009, 393:329–337. [DOI] [PubMed] [Google Scholar]
- 34. Hesketh EL, Meshcheriakova Y, Dent KC, Saxena P, Thompson RF, Cockburn JJ, Lomonossoff GP, Ranson NA. Mechanisms of assembly and genome packaging in an RNA virus revealed by high‐resolution cryo‐EM. Nat Commun 2015, 6:10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Huynh NT, Hesketh EL, Saxena P, Meshcheriakova Y, Ku YC, Hoang LT, Johnson JE, Ranson NA, Lomonossoff GP, Reddy VS. Crystal structure and proteomics analysis of empty virus‐like particles of cowpea mosaic virus. Structure 2016, 24:567–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sainsbury F, Saunders K, Aljabali AA, Evans DJ, Lomonossoff GP. Peptide‐controlled access to the interior surface of empty virus nanoparticles. Chembiochem 2011, 12:2435–2440. [DOI] [PubMed] [Google Scholar]
- 37. Wen AM, Shukla S, Saxena P, Aljabali AA, Yildiz I, Dey S, Mealy JE, Yang AC, Evans DJ, Lomonossoff GP, et al. Interior engineering of a viral nanoparticle and its tumor homing properties. Biomacromolecules 2012, 13:3990–4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Belval L, Hemmer C, Sauter C, Reinbold C, Fauny JD, Berthold F, Ackerer L, Schmitt‐Keichinger C, Lemaire O, Demangeat G, et al. Display of whole proteins on inner and outer surfaces of grapevine fanleaf virus‐like particles. Plant Biotechnol J 2016, 14:2288–2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Saunders K, Lomonossoff GP. The generation of turnip crinkle virus‐like particles in plants by the transient expression of wild‐type and modified forms of its coat protein. Front Plant Sci 2015, 6:1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Butler PJ. Self‐assembly of tobacco mosaic virus: the role of an intermediate aggregate in generating both specificity and speed. Philos Trans R Soc Lond B Biol Sci 1999, 354:537–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Eiben S, Stitz N, Eber F, Wagner J, Atanasova P, Bill J, Wege C, Jeske H. Tailoring the surface properties of tobacco mosaic virions by the integration of bacterially expressed mutant coat protein. Virus Res 2014, 180:92–96. [DOI] [PubMed] [Google Scholar]
- 42. Arkhipenko MV, Petrova EK, Nikitin NA, Protopopova AD, Dubrovin EV, Yaminskii IV, Rodionova NP, Karpova OV, Atabekov JG. Characteristics of artificial virus‐like particles assembled in vitro from potato virus X coat protein and foreign viral RNAs. Acta Naturae 2011, 3:40–46. [PMC free article] [PubMed] [Google Scholar]
- 43. Chen C, Daniel MC, Quinkert ZT, De M, Stein B, Bowman VD, Chipman PR, Rotello VM, Kao CC, Dragnea B. Nanoparticle‐templated assembly of viral protein cages. Nano Lett 2006, 6:611–615. [DOI] [PubMed] [Google Scholar]
- 44. Hu Y, Zandi R, Anavitarte A, Knobler CM, Gelbart WM. Packaging of a polymer by a viral capsid: the interplay between polymer length and capsid size. Biophys J 2008, 94:1428–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sun J, DuFort C, Daniel MC, Murali A, Chen C, Gopinath K, Stein B, De M, Rotello VM, Holzenburg A, et al. Core‐controlled polymorphism in virus‐like particles. Proc Natl Acad Sci U S A 2007, 104:1354–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hart RG. Morphological changes accompanying thermal denaturation of tobacco mosaic virus. Biochim Biophys Acta 1956, 20:388–389. [DOI] [PubMed] [Google Scholar]
- 47. Atabekov J, Nikitin N, Arkhipenko M, Chirkov S, Karpova O. Thermal transition of native tobacco mosaic virus and RNA‐free viral proteins into spherical nanoparticles. J Gen Virol 2011, 92:453–456. [DOI] [PubMed] [Google Scholar]
- 48. Eber FJ, Eiben S, Jeske H, Wege C. RNA‐controlled assembly of tobacco mosaic virus‐derived complex structures: from nanoboomerangs to tetrapods. Nanoscale 2015, 7:344–355. [DOI] [PubMed] [Google Scholar]
- 49. Cuenca S, Mansilla C, Aguado M, Yuste‐Calvo C, Sanchez F, Sanchez‐Montero JM, Ponz F. Nanonets derived from turnip mosaic virus as scaffolds for increased enzymatic activity of immobilized candida antarctica lipase B. Front Plant Sci 2016, 7:464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shukla S, Eber FJ, Nagarajan AS, DiFranco NA, Schmidt N, Wen AM, Eiben S, Twyman RM, Wege C, Steinmetz NF. The impact of aspect ratio on the biodistribution and tumor homing of rigid soft‐matter nanorods. Adv Healthc Mater 2015, 4:874–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shukla S, Ablack AL, Wen AM, Lee KL, Lewis JD, Steinmetz NF. Increased tumor homing and tissue penetration of the filamentous plant viral nanoparticle Potato virus X. Mol Pharm 2013, 10:33–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Porta C, Spall VE, Findlay KC, Gergerich RC, Farrance CE, Lomonossoff GP. Cowpea mosaic virus‐based chimaeras. Effects of inserted peptides on the phenotype, host range, and transmissibility of the modified viruses. Virology 2003, 310:50–63. [DOI] [PubMed] [Google Scholar]
- 53. Lico C, Capuano F, Renzone G, Donini M, Marusic C, Scaloni A, Benvenuto E, Baschieri S. Peptide display on Potato virus X: molecular features of the coat protein‐fused peptide affecting cell‐to‐cell and phloem movement of chimeric virus particles. J Gen Virol 2006, 87:3103–3112. [DOI] [PubMed] [Google Scholar]
- 54. Taylor KM, Lin T, Porta C, Mosser AG, Giesing HA, Lomonossoff GP, Johnson JE. Influence of three‐dimensional structure on the immunogenicity of a peptide expressed on the surface of a plant virus. J Mol Recognit 2000, 13:71–82. [DOI] [PubMed] [Google Scholar]
- 55. Smolenska L, Roberts IM, Learmonth D, Porter AJ, Harris WJ, Wilson TM, Santa CS. Production of a functional single chain antibody attached to the surface of a plant virus. FEBS Lett 1998, 441:379–382. [DOI] [PubMed] [Google Scholar]
- 56. Cruz SS, Chapman S, Roberts AG, Roberts IM, Prior DA, Oparka KJ. Assembly and movement of a plant virus carrying a green fluorescent protein overcoat. Proc Natl Acad Sci U S A 1996, 93:6286–6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Schlick TL, Ding Z, Kovacs EW, Francis MB. Dual‐surface modification of the tobacco mosaic virus. J Am Chem Soc 2005, 127:3718–3723. [DOI] [PubMed] [Google Scholar]
- 58. Demir MaS MH. A chemoselective biomolecular template for assembling diverse nanotubular materials. Nanotechnology 2002, 13:541. [Google Scholar]
- 59. Yi H, Nisar S, Lee SY, Powers MA, Bentley WE, Payne GF, Ghodssi R, Rubloff GW, Harris MT, Culver JN. Patterned assembly of genetically modified viral nanotemplates via nucleic acid hybridization. Nano Lett 2005, 5:1931–1936. [DOI] [PubMed] [Google Scholar]
- 60. Koch C, Wabbel K, Eber FJ, Krolla‐Sidenstein P, Azucena C, Gliemann H, Eiben S, Geiger F, Wege C. Modified TMV particles as beneficial scaffolds to present sensor enzymes. Front Plant Sci 2015, 6:1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Steinmetz NF, Lomonossoff GP, Evans DJ. Cowpea mosaic virus for material fabrication: addressable carboxylate groups on a programmable nanoscaffold. Langmuir 2006, 22:3488–3490. [DOI] [PubMed] [Google Scholar]
- 62. Wang Q, Kaltgrad E, Lin T, Johnson JE, Finn MG. Natural supramolecular building blocks. Wild‐type cowpea mosaic virus. Chem Biol 2002, 9:805–811. [DOI] [PubMed] [Google Scholar]
- 63. Aljabali AA, Barclay JE, Steinmetz NF, Lomonossoff GP, Evans DJ. Controlled immobilisation of active enzymes on the cowpea mosaic virus capsid. Nanoscale 2012, 4:5640–5645. [DOI] [PubMed] [Google Scholar]
- 64. Sen Gupta S, Kuzelka J, Singh P, Lewis WG, Manchester M, Finn MG. Accelerated bioorthogonal conjugation: A practical method for the Ligation of diverse functional molecules to a polyvalent virus scaffold. Bioconjug Chem 2005, 16:1572–1579. [DOI] [PubMed] [Google Scholar]
- 65. Gonzalez MJ, Plummer EM, Rae CS, Manchester M. Interaction of Cowpea mosaic virus (CPMV) nanoparticles with antigen presenting cells in vitro and in vivo. PLoS One 2009, 4:e7981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wang Q, Lin T, Johnson JE, Finn MG. Natural supramolecular building blocks. Cysteine‐added mutants of cowpea mosaic virus. Chem Biol 2002, 9:813–819. [DOI] [PubMed] [Google Scholar]
- 67. Meunier S, Strable E, Finn MG. Crosslinking of and coupling to viral capsid proteins by tyrosine oxidation. Chem Biol 2004, 11:319–326. [DOI] [PubMed] [Google Scholar]
- 68. Gillitzer E, Willits D, Young M, Douglas T. Chemical modification of a viral cage for multivalent presentation. Chem Commun (Camb) 2002, 20:2390–2391. [DOI] [PubMed] [Google Scholar]
- 69. Chatterji A, Ochoa WF, Paine M, Ratna BR, Johnson JE, Lin T. New addresses on an addressable virus nanoblock; uniquely reactive Lys residues on cowpea mosaic virus. Chem Biol 2004, 11:855–863. [DOI] [PubMed] [Google Scholar]
- 70. Steinmetz NF, Lin T, Lomonossoff GP, Johnson JE. Structure‐based engineering of an icosahedral virus for nanomedicine and nanotechnology. Curr Top Microbiol Immunol 2009, 327:23–58. [DOI] [PubMed] [Google Scholar]
- 71. Lomonossoff GP, Evans DJ. Applications of plant viruses in bionanotechnology. Curr Top Microbiol Immunol 2014, 375:61–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Shah SN, Steinmetz NF, Aljabali AA, Lomonossoff GP, Evans DJ. Environmentally benign synthesis of virus‐templated, monodisperse, iron‐platinum nanoparticles. Dalton Trans 2009, 40:8479–8480. [DOI] [PubMed] [Google Scholar]
- 73. Douglas TY. M Host–guest encapsulation of materials by assembled virus protein cages. Nature 1998, 393:152–155. [Google Scholar]
- 74. Douglas T, Strable E, Willits D, Aitouchen A, Libera M, Young M. Protein engineering of a viral cage for constrained nanomaterials synthesis. Advanced Materials 2002, 14:415–418. [Google Scholar]
- 75. Wen AM, Steinmetz NF. Design of virus‐based nanomaterials for medicine, biotechnology, and energy. Chem Soc Rev 2016, 45:4074–4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. van Kan‐Davelaar HE, van Hest JC, Cornelissen JJ, Koay MS. Using viruses as nanomedicines. Br J Pharmacol 2014, 171:4001–4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Perez J, Defrenne S, Witz J, Vachette P. Detection and characterization of an intermediate conformation during the divalent ion‐dependent swelling of tomato bushy stunt virus. Cell Mol Biol (Noisy‐le‐Grand) 2000, 46:937–948. [PubMed] [Google Scholar]
- 78. Grasso S, Lico C, Imperatori F, Santi L. A plant derived multifunctional tool for nanobiotechnology based on Tomato bushy stunt virus. Transgenic Res 2013, 22:519–535. [DOI] [PubMed] [Google Scholar]
- 79. Loo L, Guenther RH, Lommel SA, Franzen S. Infusion of dye molecules into Red clover necrotic mosaic virus. Chem Commun (Camb) 2008, 88‐90. DOI: 10.1039/B714748A. [DOI] [PubMed] [Google Scholar]
- 80. Lockney DM, Guenther RN, Loo L, Overton W, Antonelli R, Clark J, Hu M, Luft C, Lommel SA, Franzen S. The Red clover necrotic mosaic virus capsid as a multifunctional cell targeting plant viral nanoparticle. Bioconjug Chem 2011, 22:67–73. [DOI] [PubMed] [Google Scholar]
- 81. Zeng Q, Wen H, Wen Q, Chen X, Wang Y, Xuan W, Liang J, Wan S. Cucumber mosaic virus as drug delivery vehicle for doxorubicin. Biomaterials 2013, 34:4632–4642. [DOI] [PubMed] [Google Scholar]
- 82. Cao J, Guenther RH, Sit TL, Lommel SA, Opperman CH, Willoughby JA. Development of abamectin loaded plant virus nanoparticles for efficacious plant parasitic nematode control. ACS Appl Mater Interfaces 2015, 7:9546–9553. [DOI] [PubMed] [Google Scholar]
- 83. Yildiz I, Lee KL, Chen K, Shukla S, Steinmetz NF. Infusion of imaging and therapeutic molecules into the plant virus‐based carrier cowpea mosaic virus: cargo‐loading and delivery. J Control Release 2013, 172:568–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Wen AM, Wang YM, Jiang K, Hsu GC, Gao HY, Lee KL, Yang AC, Yu X, Simon DI, Steinmetz NF. Shaping bio‐inspired nanotechnologies to target thrombosis for dual optical‐magnetic resonance imaging. J Mater Chem B 2015, 3:6037–6045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Czapar AE, Zheng YR, Riddell IA, Shukla S, Awuah SG, Lippard SJ, Steinmetz NF. Tobacco mosaic virus delivery of phenanthriplatin for cancer therapy. ACS Nano 2016, 10:4119–4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Adolph KW. The conformation of the RNA in cowpea chlorotic mottle virus: dye‐binding studies. Eur J Biochem 1975, 53:449–455. [DOI] [PubMed] [Google Scholar]
- 87. Tama F, Brooks CL 3rd.. The mechanism and pathway of pH induced swelling in cowpea chlorotic mottle virus. J Mol Biol 2002, 318:733–747. [DOI] [PubMed] [Google Scholar]
- 88. Azizgolshani O, Garmann RF, Cadena‐Nava R, Knobler CM, Gelbart WM. Reconstituted plant viral capsids can release genes to mammalian cells. Virology 2013, 441:12–17. [DOI] [PubMed] [Google Scholar]
- 89. Minten IJ, Ma Y, Hempenius MA, Vancso GJ, Nolte RJ, Cornelissen JJ. CCMV capsid formation induced by a functional negatively charged polymer. Org Biomol Chem 2009, 7:4685–4688. [DOI] [PubMed] [Google Scholar]
- 90. Schoonen L, Pille J, Borrmann A, Nolte RJ, van Hest JC. Sortase A‐mediated N‐terminal modification of cowpea chlorotic mottle virus for highly efficient cargo loading. Bioconjug Chem 2015, 26:2429–2434. [DOI] [PubMed] [Google Scholar]
- 91. Comellas‐Aragones M, Engelkamp H, Claessen VI, Sommerdijk NA, Rowan AE, Christianen PC, Maan JC, Verduin BJ, Cornelissen JJ, Nolte RJ. A virus‐based single‐enzyme nanoreactor. Nat Nanotechnol 2007, 2:635–639. [DOI] [PubMed] [Google Scholar]
- 92. Sanchez‐Sanchez L, Cadena‐Nava RD, Palomares LA, Ruiz‐Garcia J, Koay MS, Cornelissen JJ, Vazquez‐Duhalt R. Chemotherapy pro‐drug activation by biocatalytic virus‐like nanoparticles containing cytochrome P450. Enzyme Microb Technol 2014, 60:24–31. [DOI] [PubMed] [Google Scholar]
- 93. Lu X, Thompson JR, Perry KL. Encapsidation of DNA, a protein and a fluorophore into virus‐like particles by the capsid protein of cucumber mosaic virus. J Gen Virol 2012, 93:1120–1126. [DOI] [PubMed] [Google Scholar]
- 94. Koudelka KJ, Pitek AS, Manchester M, Steinmetz NF. Virus‐Based Nanoparticles as versatile nanomachines. Annu Rev Virol 2015, 2:379–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Rusch V, Baselga J, Cordon‐Cardo C, Orazem J, Zaman M, Hoda S, McIntosh J, Kurie J, Dmitrovsky E. Differential expression of the epidermal growth factor receptor and its ligands in primary non‐small cell lung cancers and adjacent benign lung. Cancer Res 1993, 53:2379–2385. [PubMed] [Google Scholar]
- 96. Ross JF, Chaudhuri PK, Ratnam M. Differential regulation of folate receptor isoforms in normal and malignant tissues in vivo and in established cell lines. Physiologic and clinical implications. Cancer 1994, 73:2432–2443. [DOI] [PubMed] [Google Scholar]
- 97. Rae CS, Khor IW, Wang Q, Destito G, Gonzalez MJ, Singh P, Thomas DM, Estrada MN, Powell E, Finn MG, et al. Systemic trafficking of plant virus nanoparticles in mice via the oral route. Virology 2005, 343:224–235. [DOI] [PubMed] [Google Scholar]
- 98. Singh P, Prasuhn D, Yeh RM, Destito G, Rae CS, Osborn K, Finn MG, Manchester M. Bio‐distribution, toxicity and pathology of cowpea mosaic virus nanoparticles in vivo. J Control Release 2007, 120:41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Kaiser CR, Flenniken ML, Gillitzer E, Harmsen AL, Harmsen AG, Jutila MA, Douglas T, Young MJ. Biodistribution studies of protein cage nanoparticles demonstrate broad tissue distribution and rapid clearance in vivo. Int J Nanomedicine 2007, 2:715–733. [PMC free article] [PubMed] [Google Scholar]
- 100. Bruckman MA, Randolph LN, VanMeter A, Hern S, Shoffstall AJ, Taurog RE, Steinmetz NF. Biodistribution, pharmacokinetics, and blood compatibility of native and PEGylated tobacco mosaic virus nano‐rods and ‐spheres in mice. Virology 2014, 449:163–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Shukla S, Wen AM, Ayat NR, Commandeur U, Gopalkrishnan R, Broome AM, Lozada KW, Keri RA, Steinmetz NF. Biodistribution and clearance of a filamentous plant virus in healthy and tumor‐bearing mice. Nanomedicine (Lond) 2014, 9:221–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Lee KL, Shukla S, Wu M, Ayat NR, El Sanadi CE, Wen AM, Edelbrock JF, Pokorski JK, Commandeur U, Dubyak GR, et al. Stealth filaments: polymer chain length and conformation affect the in vivo fate of PEGylated potato virus X. Acta Biomater 2015, 19:166–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Steinmetz NF, Manchester M. PEGylated viral nanoparticles for biomedicine: the impact of PEG chain length on VNP cell interactions in vitro and ex vivo. Biomacromolecules 2009, 10:784–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Koudelka KJ, Destito G, Plummer EM, Trauger SA, Siuzdak G, Manchester M. Endothelial targeting of cowpea mosaic virus (CPMV) via surface vimentin. PLoS Pathog 2009, 5:e1000417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Lee KL, Carpenter BL, Wen AM, Ghiladi RA, Steinmetz NF. High aspect ratio nanotubes formed by tobacco mosaic virus for delivery of photodynamic agents targeting melanoma. ACS Biomat Sci Eng 2016, 2:838–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Koistinen P, Heino J. Integrins in Cancer Cell Invasion. Madam Curie Bioscience Austin, TX: Landes Bioscience; 2000‐2013. [Google Scholar]
- 107. Hovlid ML, Steinmetz NF, Laufer B, Lau JL, Kuzelka J, Wang Q, Hyypia T, Nemerow GR, Kessler H, Manchester M, et al. Guiding plant virus particles to integrin‐displaying cells. Nanoscale 2012, 4:3698–3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Bareford LM, Swaan PW. Endocytic mechanisms for targeted drug delivery. Adv Drug Deliv Rev 2007, 59:748–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Ren Y, Wong SM, Lim LY. Folic acid‐conjugated protein cages of a plant virus: a novel delivery platform for doxorubicin. Bioconjug Chem 2007, 18:836–843. [DOI] [PubMed] [Google Scholar]
- 110. Steinmetz NF, Mertens ME, Taurog RE, Johnson JE, Commandeur U, Fischer R, Manchester M. Potato virus X as a novel platform for potential biomedical applications. Nano Lett 2010, 10:305–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Lewis JD, Destito G, Zijlstra A, Gonzalez MJ, Quigley JP, Manchester M, Stuhlmann H. Viral nanoparticles as tools for intravital vascular imaging. Nat Med 2006, 12:354–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Cho CF, Ablack A, Leong HS, Zijlstra A, Lewis J. Evaluation of nanoparticle uptake in tumors in real time using intravital imaging. J Vis Exp 2011, 21:2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Leong HS, Steinmetz NF, Ablack A, Destito G, Zijlstra A, Stuhlmann H, Manchester M, Lewis JD. Intravital imaging of embryonic and tumor neovasculature using viral nanoparticles. Nat Protoc 2010, 5:1406–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Chariou PL, Lee KL, Wen AM, Gulati NM, Stewart PL, Steinmetz NF. Detection and imaging of aggressive cancer cells using an epidermal growth factor receptor (EGFR)‐targeted filamentous plant virus‐based nanoparticle. Bioconjug Chem 2015, 26:262–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Kim D, Moon H, Baik SH, Singha S, Jun YW, Wang T, Kim KH, Park BS, Jung J, Mook‐Jung I, et al. Two‐photon absorbing dyes with minimal autofluorescence in tissue imaging: application to in vivo imaging of amyloid‐beta plaques with a negligible background signal. J Am Chem Soc 2015, 137:6781–6789. [DOI] [PubMed] [Google Scholar]
- 116. Denk W, Strickler JH, Webb WW. Two‐photon laser scanning fluorescence microscopy. Science 1990, 248:73–76. [DOI] [PubMed] [Google Scholar]
- 117. Squirrell JM, Wokosin DL, White JG, Bavister BD. Long‐term two‐photon fluorescence imaging of mammalian embryos without compromising viability. Nat Biotechnol 1999, 17:763–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Niehl A, Appaix F, Bosca S, van der Sanden B, Nicoud JF, Bolze F, Heinlein M. Fluorescent tobacco mosaic virus‐derived bio‐nanoparticles for intravital two‐photon imaging. Front Plant Sci 2015, 6:1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Shriver LP, Plummer EM, Thomas DM, Ho S, Manchester M. Localization of gadolinium‐loaded CPMV to sites of inflammation during central nervous system autoimmunity. J Mater Chem B 2013, 1:5256–5263. [DOI] [PubMed] [Google Scholar]
- 120. Bruckman MA, Yu X, Steinmetz NF. Engineering Gd‐loaded nanoparticles to enhance MRI sensitivity via T(1) shortening. Nanotechnology 2013, 24:462001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Rezapkin GV, Chumakov KM, Lu Z, Ran Y, Dragunsky EM, Levenbook IS. Microevolution of Sabin 1 strain in vitro and genetic stability of oral poliovirus vaccine. Virology 1994, 202:370–378. [DOI] [PubMed] [Google Scholar]
- 122. Clarke BE, Newton SE, Carroll AR, Francis MJ, Appleyard G, Syred AD, Highfield PE, Rowlands DJ, Brown F. Improved immunogenicity of a peptide epitope after fusion to hepatitis B core protein. Nature 1987, 330:381–384. [DOI] [PubMed] [Google Scholar]
- 123. Francis MJ, Hastings GZ, Brown AL, Grace KG, Rowlands DJ, Brown F, Clarke BE. Immunological properties of hepatitis B core antigen fusion proteins. Proc Natl Acad Sci U S A 1990, 87:2545–2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Plummer EM, Manchester M. Viral nanoparticles and virus‐like particles: platforms for contemporary vaccine design. WIREs Nanomed Nanobiotechnol 2011, 3:174–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Usha R, Rohll JB, Spall VE, Shanks M, Maule AJ, Johnson JE, Lomonossoff GP. Expression of an animal virus antigenic site on the surface of a plant virus particle. Virology 1993, 197:366–374. [DOI] [PubMed] [Google Scholar]
- 126. Porta C, Spall VE, Loveland J, Johnson JE, Barker PJ, Lomonossoff GP. Development of cowpea mosaic virus as a high‐yielding system for the presentation of foreign peptides. Virology 1994, 202:949–955. [DOI] [PubMed] [Google Scholar]
- 127. McLain L, Porta C, Lomonossoff GP, Durrani Z, Dimmock NJ. Human immunodeficiency virus type 1‐neutralizing antibodies raised to a glycoprotein 41 peptide expressed on the surface of a plant virus. AIDS Res Hum Retroviruses 1995, 11:327–334. [DOI] [PubMed] [Google Scholar]
- 128. Dalsgaard K, Uttenthal A, Jones TD, Xu F, Merryweather A, Hamilton WD, Langeveld JP, Boshuizen RS, Kamstrup S, Lomonossoff GP, et al. Plant‐derived vaccine protects target animals against a viral disease. Nat Biotechnol 1997, 15:248–252. [DOI] [PubMed] [Google Scholar]
- 129. Turpen TH, Reinl SJ, Charoenvit Y, Hoffman SL, Fallarme V, Grill LK. Malarial epitopes expressed on the surface of recombinant tobacco mosaic virus. Biotechnology (N Y) 1995, 13:53–57. [DOI] [PubMed] [Google Scholar]
- 130. Sugiyama Y, Hamamoto H, Takemoto S, Watanabe Y, Okada Y. Systemic production of foreign peptides on the particle surface of tobacco mosaic virus. FEBS Lett 1995, 359:247–250. [DOI] [PubMed] [Google Scholar]
- 131. Koo M, Bendahmane M, Lettieri GA, Paoletti AD, Lane TE, Fitchen JH, Buchmeier MJ, Beachy RN. Protective immunity against murine hepatitis virus (MHV) induced by intranasal or subcutaneous administration of hybrids of tobacco mosaic virus that carries an MHV epitope. Proc Natl Acad Sci U S A 1999, 96:7774–7779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Zhao X, Chen L, Luckanagul JA, Zhang X, Lin Y, Wang Q. Enhancing antibody response against small molecular hapten with tobacco mosaic virus as a polyvalent carrier. Chembiochem 2015, 16:1279–1283. [DOI] [PubMed] [Google Scholar]
- 133. Arora U, Tyagi P, Swaminathan S, Khanna N. Virus‐like particles displaying envelope domain III of dengue virus type 2 induce virus‐specific antibody response in mice. Vaccine 2013, 31:873–878. [DOI] [PubMed] [Google Scholar]
- 134. Middelberg AP, Rivera‐Hernandez T, Wibowo N, Lua LH, Fan Y, Magor G, Chang C, Chuan YP, Good MF, Batzloff MR. A microbial platform for rapid and low‐cost virus‐like particle and capsomere vaccines. Vaccine 2011, 29:7154–7162. [DOI] [PubMed] [Google Scholar]
- 135. Mazeike E, Gedvilaite A, Blohm U. Induction of insert‐specific immune response in mice by hamster polyomavirus VP1 derived virus‐like particles carrying LCMV GP33 CTL epitope. Virus Res 2012, 163:2–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Pniewski T, Kapusta J, Bociag P, Wojciechowicz J, Kostrzak A, Gdula M, Fedorowicz‐Stronska O, Wojcik P, Otta H, Samardakiewicz S, et al. Low‐dose oral immunization with lyophilized tissue of herbicide‐resistant lettuce expressing hepatitis B surface antigen for prototype plant‐derived vaccine tablet formulation. J Appl Genet 2011, 52:125–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Nuzzaci M, Vitti A, Condelli V, Lanorte MT, Tortorella C, Boscia D, Piazzolla P, Piazzolla G. In vitro stability of Cucumber mosaic virus nanoparticles carrying a Hepatitis C virus‐derived epitope under simulated gastrointestinal conditions and in vivo efficacy of an edible vaccine. J Virol Methods 2010, 165:211–215. [DOI] [PubMed]
- 138. Denis J, Majeau N, Acosta‐Ramirez E, Savard C, Bedard MC, Simard S, Lecours K, Bolduc M, Pare C, Willems B, et al. Immunogenicity of papaya mosaic virus‐like particles fused to a hepatitis C virus epitope: evidence for the critical function of multimerization. Virology 2007, 363:59–68. [DOI] [PubMed] [Google Scholar]
- 139. D'Aoust MA, Lavoie PO, Couture MM, Trepanier S, Guay JM, Dargis M, Mongrand S, Landry N, Ward BJ, Vezina LP. Influenza virus‐like particles produced by transient expression in Nicotiana benthamiana induce a protective immune response against a lethal viral challenge in mice. Plant Biotechnol J 2008, 6:930–940. [DOI] [PubMed] [Google Scholar]
- 140. Maclean J, Koekemoer M, Olivier AJ, Stewart D, Hitzeroth II, Rademacher T, Fischer R, Williamson AL, Rybicki EP. Optimization of human papillomavirus type 16 (HPV‐16) L1 expression in plants: comparison of the suitability of different HPV‐16 L1 gene variants and different cell‐compartment localization. J Gen Virol 2007, 88:1460–1469. [DOI] [PubMed] [Google Scholar]
- 141. Millan AFS, Ortigosa SM, Hervas‐Stubbs S, Corral‐Martinez P, Segui‐Simarro JM, Gaetan J, Coursaget P, Veramendi J. Human papillomavirus L1 protein expressed in tobacco chloroplasts self‐assembles into virus‐like particles that are highly immunogenic. Plant Biotechnol J 2008, 6:427–441. [DOI] [PubMed] [Google Scholar]
- 142. Matic S, Masenga V, Poli A, Rinaldi R, Milne RG, Vecchiati M, Noris E. Comparative analysis of recombinant Human Papillomavirus 8 L1 production in plants by a variety of expression systems and purification methods. Plant Biotechnol J 2012, 10:410–421. [DOI] [PubMed] [Google Scholar]
- 143. Pineo CB, Hitzeroth II, Rybicki EP. Immunogenic assessment of plant‐produced human papillomavirus type 16 L1/L2 chimaeras. Plant Biotechnol J 2013, 11:964–975. [DOI] [PubMed] [Google Scholar]
- 144. Love AJ, Chapman SN, Matic S, Noris E, Lomonossoff GP, Taliansky M. In planta production of a candidate vaccine against bovine papillomavirus type 1. Planta 2012, 236:1305–1313. [DOI] [PubMed] [Google Scholar]
- 145. Scotti N, Alagna F, Ferraiolo E, Formisano G, Sannino L, Buonaguro L, De Stradis A, Vitale A, Monti L, Grillo S, et al. High‐level expression of the HIV‐1 Pr55gag polyprotein in transgenic tobacco chloroplasts. Planta 2009, 229:1109–1122. [DOI] [PubMed] [Google Scholar]
- 146. Mathew LG, Herbst‐Kralovetz MM, Mason HS. Norovirus Narita 104 virus‐like particles expressed in Nicotiana benthamiana induce serum and mucosal immune responses. Biomed Res Int 2014, 2014:807539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Yang Y, Li X, Yang H, Qian Y, Zhang Y, Fang R, Chen X. Immunogenicity and virus‐like particle formation of rotavirus capsid proteins produced in transgenic plants. Sci China Life Sci 2011, 54:82–89. [DOI] [PubMed] [Google Scholar]
- 148. Thuenemann EC, Meyers AE, Verwey J, Rybicki EP, Lomonossoff GP. A method for rapid production of heteromultimeric protein complexes in plants: assembly of protective bluetongue virus‐like particles. Plant Biotechnol J 2013, 11:839–846. [DOI] [PubMed] [Google Scholar]
- 149. Mechtcheriakova IA, Eldarov MA, Nicholson L, Shanks M, Skryabin KG, Lomonossoff GP. The use of viral vectors to produce hepatitis B virus core particles in plants. J Virol Methods 2006, 131:10–15. [DOI] [PubMed] [Google Scholar]
- 150. Marsian J, Lomonossoff GP. Molecular pharming – VLPs made in plants. Curr Opin Biotechnol 2016, 37:201–206. [DOI] [PubMed] [Google Scholar]
- 151. Shenton W, Douglas T, Young M, Stubbs G, Mann S. Inorganic‐organic nanotube composites from template mineralization of tobacco mosaic virus. Adv Mater 1999, 11:253–256. [Google Scholar]
- 152. Pokorski JK, Steinmetz NF. The art of engineering viral nanoparticles. Mol Pharm 2011, 8:29–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Douglas T, Young M. Virus particles as templates for materials synthesis. Adv Mater 1999, 11:679–681. [Google Scholar]
- 154. Aljabali AA, Sainsbury F, Lomonossoff GP, Evans DJ. Cowpea mosaic virus unmodified empty viruslike particles loaded with metal and metal oxide. Small 2010, 6:818–821. [DOI] [PubMed] [Google Scholar]
- 155. Heddle JG. Protein cages, rings and tubes: useful components of future nanodevices? Nanotechnol Sci Appl 2008, 1:67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Aljabali AA, Lomonossoff GP, Evans DJ. CPMV‐polyelectrolyte‐templated gold nanoparticles. Biomacromolecules 2011, 12:2723–2728. [DOI] [PubMed] [Google Scholar]
- 157. Tseng RJ, Tsai CL, Ma LP, Ouyang JY. Digital memory device based on tobacco mosaic virus conjugated with nanoparticles. Nat Nanotechnol 2006, 1:72–77. [DOI] [PubMed] [Google Scholar]
- 158. Gorzny ML, Walton AS, Wnek M, Stockley PG, Evans SD. Four‐probe electrical characterization of Pt‐coated TMV‐based nanostructures. Nanotechnology 2008, 19:165704. [DOI] [PubMed] [Google Scholar]
- 159. Knez M, Bittner AM, Boes F, Wege C, Jeske H, Maiss E, Kern K. Biotemplate synthesis of 3‐nm nickel and cobalt nanowires. Nano Lett 2003, 3:1079–1082. [Google Scholar]
- 160. Tsukamoto R, Muraoka M, Seki M, Tabata H, Yamashita I. Synthesis of CoPt and FePt3 nanowires using the central channel of tobacco mosaic virus as a biotemplate. Chem Mater 2007, 19:2389–2391. [Google Scholar]
- 161. Kapoor P, Singh H, Gautam A, Chaudhary K, Kumar R, Raghava GP. TumorHoPe: a database of tumor homing peptides. PLoS One 2012, 7:e35187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Rademacher T. Method for the generation and cultivation of a plant cell pack. EP 2012‐2623603 A1: 20130807, Fraunhofer Institute, 2013.


