Abstract
Vpr and selected mutants were used in a Saccharomyces cerevisiae two-hybrid screen to identify cellular interactors. We found Vpr interacted with 14-3-3 proteins, a family regulating a multitude of proteins in the cell. Vpr mutant R80A, which is inactive in cell cycle arrest, did not interact with 14-3-3. 14-3-3 proteins regulate the G2/M transition by inactivating Cdc25C phosphatase via binding to the phosphorylated serine residue at position 216 of Cdc25C. 14-3-3 overexpression in human cells synergized with Vpr in the arrest of cell cycle. Vpr did not arrest efficiently cells not expressing 14-3-3σ. This indicated that a full complement of 14-3-3 proteins is necessary for optimal Vpr function on the cell cycle. Mutational analysis showed that the C-terminal portion of Vpr, known to harbor its cell cycle-arresting activity, bound directly to the C-terminal part of 14-3-3, outside of its phosphopeptide-binding pocket. Vpr expression shifted localization of the mutant Cdc25C S216A to the cytoplasm, indicating that Vpr promotes the association of 14-3-3 and Cdc25C, independently of the presence of serine 216. Immunoprecipitations of cell extracts indicated the presence of triple complexes (Vpr/14-3-3/Cdc25C). These results indicate that Vpr promotes cell cycle arrest at the G2/M phase by facilitating association of 14-3-3 and Cdc25C independently of the latter's phosphorylation status.
The human immunodeficiency virus type 1 (HIV-1) accessory protein Vpr, a 15-kDa virion-associated molecule, is required for efficient viral propagation in vivo (14, 15, 49). Since Vpr is incorporated into the virion, it is thought to exert its effects at an early stage of the viral life cycle, possibly by helping to achieve optimal conditions for viral infection (6, 37). Several Vpr activities have been identified in vitro, such as facilitation of the nuclear translocation of the HIV-1 preintegration complex, coactivation of steroid hormone receptors, and modulation of apoptosis (20, 24, 39, 45, 48). Vpr also arrests cells at the G2/M boundary of the cell cycle (6, 15, 19, 22, 40), a function that has been proposed to facilitate viral propagation. Transition through the G2/M checkpoint in mammalian cells is strictly controlled by activation of a protein complex formed by a catalytic subunit, the cyclin-dependent kinase Cdc2, and its regulatory partner cyclinB1 through coordinated phosphorylation and dephosphorylation events (21, 36). The protein kinases Wee1 and Myt1 inactivate this complex by phosphorylating threonine residues at amino acids 14 and 15 of Cdc2, while the phosphatase Cdc25C activates it by dephosphorylating the same threonine residues (21, 35, 36). Vpr inactivates the Cdc2/cyclinB1 complex by keeping Cdc2 at a hyperphosphorylated state, possibly by modulating the function of a host protein(s), which acts upstream of Cdc2/cyclinB1, such as Wee1, Myt1, and Cdc25C (10, 19, 22, 31, 40, 42).
To identify Vpr partner proteins that support the cell cycle-arresting activity of Vpr, we performed several Saccharomyces cerevisiae two-hybrid screening assays using wild-type (WT) and mutant Vprs as baits. We found that human 14-3-3 proteins bind Vpr and contribute to its cell cycle-arresting activity. 14-3-3 proteins are key molecules in the regulation of cell cycle progression (1, 13, 34). The family consists of nine isotypes produced from at least seven distinct genes in vertebrates. 14-3-3 proteins bind phosphorylated serine/threonine residues at specific positions of their partner proteins and regulate their activities by changing their subcellular localization and/or stability. 14-3-3 proteins contain nine α-helical structures and form homo- and heterodimers through their N-terminal portion (41, 43, 51). The central third to fifth α-helixes create a binding pocket for a phosphorylated serine/threonine residue, and the C-terminal seventh to ninth helices determine the specificity to target peptide motifs (41, 51). 14-3-3 contains a nuclear export signal in the ninth helix (28, 41).
14-3-3 proteins regulate cell cycle progression by changing the activities of Cdc25C, Wee1, cyclin B1, and Chk1. DNA damage triggers Cdc25C phosphorylation at serine 216, providing the active binding site for 14-3-3 (8, 17, 33, 38, 44, 52). Binding of 14-3-3 to the phosphorylated Cdc25C causes translocation of the complex into the cytoplasm, separating the Cdc25C phosphatase from its nuclear substrates (28, 41, 52), such as Cdc2. In the absence of Cdc25C phosphatase activity, Cdc2 is kept in an inactive phosphorylated state and the cells no longer overcome the G2/M cell cycle checkpoint (38).
Our results indicate that Vpr acts as a bridging factor between 14-3-3 and Cdc25C, irrespective of the latter's phosphorylated states. This binding leads to removal of Cdc25C and contributes to the prolonged arrest at G2/M seen in the presence of Vpr.
MATERIALS AND METHODS
Plasmids and HIV-1 molecular clones.
Vpr-related plasmids pLexA-VPR, pCMV-FLAG-VPR, pCDNA3-VPR, and pLexA-Tat were described previously (24). pLexA-VPRL64A, VprL23A, VprL23,64,67A, and VprR80A were constructed by substituting the indicated amino acids of Vpr into the pLexA-VPR using a PCR-assisted in vitro mutagenesis reaction. pLexA-Vpr (64-96) was constructed by inserting the corresponding Vpr cDNA fragment into pLexA (Clontech, Palo Alto, Calif.). The HIV-1 molecular clone NL4-3GFP, which produces the green fluorescent protein (GFP)-expressing WT HIV-1 virus, was described previously (47). NL4-3GFPΔVpr was constructed by replacing the Nef coding sequence of the NL4-3GFPΔVpr obtained from the NIH AIDS Research and Reference Reagent Program (Rockville, Md.) with the Nef-GFP fusion sequence of NL4-3GFP. pCDNA3-14-3-3η and pGEX-14-3-3η were constructed by introducing 14-3-3η cDNA into pCDNA3 (Invitrogen, Carlsbad, Calif.) and pGEX-4T3 (Amersham Pharmacia Biotech, Piscataway, N.J.), respectively. pB42AD-14-3-3η (1-244), (110-244), (141-244), (168-244), (190-210) and (210-244) were constructed by inserting 14-3-3η cDNA fragments of the indicated portions of 14-3-3η into pB42AD (Clontech). pGAD424-14-3-3σ (1-270) and (106-270) were constructed by subcloning 14-3-3σ cDNA fragments of 1-270 or 106-270 of 14-3-3σ into pGAD424 (Clontech), respectively. Myc epitope-tagged Cdc25C-expressing plasmids, Myc-Cdc25C WT and S216A mutant, were kind gifts from J. A. De Caprio (Dana-Farber Cancer Institute and Harvard Medical School, Boston, Mass.). EGFP-Cdc25C fusion-expressing plasmids, pEGFP-C1-Cdc25C WT and S216A, were constructed by inserting cDNA from Myc-Cdc25C WT and S216A into pEGFP-C1, respectively (Clontech). pHook-1 was purchased from Invitrogen. Baculovirus-produced purified Vpr was obtained from insect cell lysates, infected with the virus containing His-Vpr sequence by using a histidine affinity column (Amersham Pharmacia Biotech) (32).
Yeast two-hybrid screening assay.
The yeast two-hybrid screening was performed in a HeLa cell cDNA library with the LexA system (16). To study the specificity of the identified interactions, we used a yeast mating assay (18). For a yeast two-hybrid assay, yeast strain EGY48 (Clontech) was transformed with the lacZ reporter plasmid p8OP-LacZ (Clontech), pLexA-VPR (WT or mutants), or pLexA-Tat and the indicated pB42AD-14-3-3η plasmids (12). The cells were grown in a selective medium to the early stationary phase and permeabilized with CHCl3-sodium dodecyl sulfate (SDS) treatment, and β-galactosidase activity was measured in the cell suspension by using Galactolight PLUS (Tropix, Bedford, Mass.).
In vitro binding assay.
35S-labeled Vpr WT and mutants were generated by in vitro translation using the wheat germ extract from pCDNA3-VPRs. They were tested for interaction with glutathione-S-transferase (GST)-14-3-3η, immobilized on glutathione-Sepharose beads in buffer containing 50 mM Tris-HCl (pH 8.0), 50 mM NaCl, 1 mM EDTA, 0.1% NP-40, 10% glycerol, and 0.1 mg of bovine serum albumin/ml at 4°C for 1.5 h. After vigorous washing with the buffer, proteins were eluted and separated on 16% SDS-polyacrylamide gel electrophoresis (SDS-PAGE) gels. Gels were fixed and exposed to films.
Cell cultures and transfections.
HCT116 WT and 14-3-3σ knockout (KO) cells were kindly provided by B. Vogelstein (Johns Hopkins University, Baltimore, Md.) (4). 293, HeLa, and HCT116 cells were maintained in Dulbecco's modified eagle's medium or McCoy's 5A medium containing 10% fetal bovine serum and 100 U of penicillin and 1 μg of streptomycin/ml. Cells were transfected using Lipofectin and CaPO4 methods, as described previously (11, 23).
Cell cycle analysis.
293, HeLa, HCT116 WT, and 14-3-3σ KO cells were transfected with the indicated plasmids. pHook-1 was included in experiments with HeLa cells to identify transfection-positive cells. At 48 h after transfection, cells were stained for 1 h with 10 μg of Hoechst 33342 (Molecular Probes, Eugene, Oreg.)/ml, and their DNA content was analyzed in an LSR flow cytometer (BD Biosciences, San Jose, Calif.) using the CellQuest software (BD Biosciences) for data acquisition. Cell cycle analysis was performed with the cell cycle platform provided by FlowJo (Tree Star Inc., San Carlos, Calif.). Transfected HeLa cells were enriched before DNA staining using the pHook-1 method following the company's recommendation.
VSV-G-pseudotyped HIV-1 molecular clones and infection of HCT116 cells.
Stocks of infectious pseudotyped HIV-1 molecular clones were generated by cotransfection of either 10 μg of NL4-3GFP or NL4-3GFPΔvpr and 0.5 μg of vesicular stomatitis virus G protein (VSV-G) DNA in 293 cells. Forty-eight hours after transfection, the supernatants were harvested, spun for 10 min at 1,200 rpm (300 × g) aliquoted, and stored at −80°C.
For infection, HCT116 WT and 14-3-3σ KO cells were incubated for 2 h at 37°C with 2 ml of supernatant containing the VSV-G-pseudotyped HIV-1 molecular clones. Thirty-six hours after the infection, the cells were stained for 1 h with Hoescht 33342 and their DNA content and cell cycle profile were analyzed as described above.
Detection and localization of GFP-fused Cdc25C proteins.
HCT116 WT and 14-3-3σ KO cells were cultured on poly-lysine-coated cover slides and transfected with the indicated plasmids. They were fixed with 4% paraformaldehyde and subsequently mounted on glass slides with Vectashield/DAPI (4,6-diamidino-2-phenylindole; Vector Laboratories, Inc., Burlingame, Calif.). Emitted signals were recorded with the Zeiss LSM510 confocal microscope (Carl Zeiss, Thornwood, New York, N.Y.) at the Microscopy & Imaging Core (National Institute of Child Health and Human Development, Bethesda, Md.) with the assistance of Vincent Schram. Signal intensity of enhanced green fluorescent protein (EGFP) in the nucleus and the cytoplasm was measured in over 20 cells by using the LSM5 image analyzer (Carl Zeiss), and the mean ± standard error (SE) values of their ratios were calculated after subtracting background signal intensity in the same image field.
Coimmunoprecipitation experiments.
293 cells were grown in 175-cm2 flasks and transfected with 15 μg of the indicated plasmids. At 48 h after transfection, cell were lysed with buffer containing 50 mM Tris-HCl (pH 8.0), 120 mM NaCl, 0.5% NP-40, and 1 tablet of Complete tablets (Roche Molecular Biochemicals, Indianapolis, Ind.)/50 ml, and extracts were cleared by centrifugation at 12,000 × g for 15 min at 4°C. Supernatants were harvested and incubated with the indicated antibody for 3 h at 4°C. The protein-antibody complexes were subsequently collected by adding protein A/G agarose (Santa Cruz Biotechnology, Inc.) and washed three times with the buffer containing 20 mM Tris-HCl (pH 8.0), 100 mM NaCl, 0.5% NP-40, and 1 tablet of Complete/50 ml. Samples were separated on 12 or 16% SDS-PAGE gels and blotted on nitrocellulose membranes, which were subsequently treated with the indicated antibody. Blotted proteins were visualized with the enhanced chemiluminescence reaction (Amersham Pharmacia Biotech).
RESULTS
Identification of 14-3-3 as a Vpr-interacting protein.
To identify cellular proteins interacting with Vpr, we performed a yeast two-hybrid screen of a HeLa cell cDNA library using WT Vpr as bait. In addition to several proteins already known to interact with Vpr, such as HHR23A and Rch1 (importin α), we identified several molecules, shown in Table 1, which might interact with Vpr in vivo and contribute to its activities. To examine whether any of them might affect cell cycle-arresting activity of Vpr, we tested their interaction with WT and mutant Vprs in a yeast mating assay. Many Vpr mutants are known, affecting one or several Vpr activities (9, 16, 24, 29). Examples of such Vpr mutants are VprL64A (replacing leucine at amino acid 64 with alanine) and R80A (replacing arginine at amino acid 80 with alanine): the former is active in cell cycle arrest but deficient in the nuclear receptor coactivator activity, whereas the latter has the reverse phenotype (16, 24, 46). We also used a Vpr fragment comprising amino acids 64 to 96. This fragment contains the C-terminal acidic domain that is necessary but not sufficient for cell cycle arrest by Vpr. All candidate gene products interacted with WT Vpr as well as the R80A mutant, while neither of them was associated with VprL64A (Table 1). Their interaction with Vpr (64-96) was variable. Since the candidates interacted with a Vpr mutant inactive in cell cycle arrest, the results indicated that the identified molecules are not likely to be involved in the cell cycle-arresting activity of Vpr. Since we observed that the VprL64A mutant showed a greatly reduced number of interactors upon screening of the library, we hypothesized that use of this mutant might help in reducing nonspecific interactions and facilitate the identification of specific host partner proteins for Vpr's cell cycle-arresting activity. We thus performed a second round of yeast two-hybrid screening using VprL64A as bait. In addition to several other candidates, we found that 14-3-3 protein family members η and θ, interacted with VprL64A, with 14-3-3η being the more frequently identified interactor (Table 2). 14-3-3η also showed strong interaction with the WT Vpr as well as with VprL64A in a yeast mating assay (data not shown).
TABLE 1.
Summary of the yeast two-hybrid screening using WT Vpr as baita
| Potential interactor | GenBank accession no. | No. of clones | Interaction with Vprs
|
|||
|---|---|---|---|---|---|---|
| WT | L64A | R80A | 64-96 | |||
| D123 | D14878 | 5 | + | − | + | + |
| HHR23A | D21235 | 4 | + | − | + | − |
| Rch1 (importin α) | U09559 | 3 | + | − | + | + |
| Tre oncogene | X63546 | 3 | + | − | + | + |
| Cullin 1 | AF062536 | 2 | + | − | + | + |
| ATP-binding protein | AJ01084 | 1 | + | − | + | − |
| Chromosome-associated polypeptide (HCAP) | AF020043 | 1 | + | − | + | − |
| C-terminal binding protein 2 | AF016507 | 1 | + | − | + | − |
| MAD-3 (I-κB) | M69043 | 1 | + | − | + | + |
| PDCD (programmed cell death-2/Rp8 homolog) | S78085 | 1 | + | − | + | − |
| Putative SMC-like protein | AJ005015 | 1 | + | − | + | − |
| Retinoblastoma-binding protein (RbAp46) | U35143 | 1 | + | − | + | − |
| RNA polymerase II subunit hsRBP7 | U20659 | 1 | + | − | + | − |
| Signal transducer and activator of transcription 5B | U47686 | 1 | + | − | + | + |
| TOM-1-like protein | AJ010071 | 1 | + | − | + | − |
Total clones sequenced = 159.
TABLE 2.
Summary of the 14-3-3 clones identified using VprL64A as bait in yeast two-hybrid screensa
| Potential interactor | GenBank accession no. | No. of clones | Interaction with Vprs
|
||
|---|---|---|---|---|---|
| WT | L64A | R80A | |||
| 14-3-3η | X78138 | 5 | + | + | − |
| 14-3-3θ | X56468 | 2 | + | + | − |
Total clones sequenced = 96.
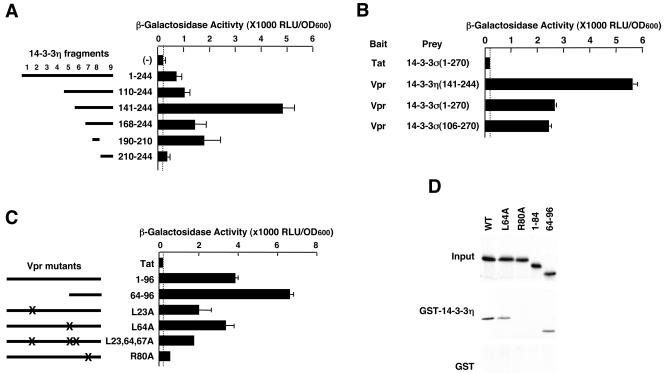
Using 14-3-3 fragments, we next studied the region of 14-3-3 required for Vpr binding in a yeast two-hybrid assay (Fig. 1A). The results indicated that Vpr bound most strongly to 14-3-3η amino acids 141 to 244. The smallest 14-3-3η fragment that supported binding to Vpr was amino acids 190 to 210, which contain the eighth α-helix of this protein (41, 51). The interaction of Vpr with full-length 14-3-3η was weaker in this assay. One reason for this result may be toxicity after expression of the full-length molecule in yeast cells grown in galactose- and raffinose-containing but histidine-, tryptophan-, and uracil-depleted selective medium (data not shown). This result was consistent with our yeast two-hybrid screens, in which all Vpr-interacting 14-3-3η clones contained the last 100 amino acids, which correspond to the sixth to the ninth α-helices of the protein (data not shown). We also examined binding of Vpr to 14-3-3σ in the same yeast two-hybrid assay. Vpr bound to both full-length and the C-terminal half of 14-3-3σ (Fig. 1B). We next tested a panel of Vpr mutants in a yeast two-hybrid assay to determine the portion of Vpr that supports binding to 14-3-3η (Fig. 1C). Mutant R80A, which is known to abolish Vpr's cell-cycle arresting activity, inactivated the binding to 14-3-3η, whereas deletion or point mutations in the N-terminal portion of Vpr were still active for binding.
FIG. 1.
Direct Interaction of Vpr with members of the 14-3-3 family. (A) Vpr interacts with the C-terminal portion of 14-3-3η in a yeast two-hybrid assay. Numbers (1 to 9) indicate the positions of the nine α-helical regions in 14-3-3. (B) Vpr interacts with 14-3-3σ in addition to 14-3-3η in a yeast two-hybrid assay. (C) Identification of the Vpr region interacting with 14-3-3η. In panels A to C, plasmids expressing indicated bait and prey molecules were cotransfected with p8OP-LacZ in the EGY48 yeast strain, and their interaction was detected by measuring the produced β-galactosidase activity. (D) Direct interaction of Vpr with 14-3-3η in a GST pull-down assay. In vitro-translated and -labeled Vpr-related molecules were incubated with bacterially produced GST-fused 14-3-3η. Samples were loaded and run on 16% SDS-PAGE gels and detected by autoradiography. Vpr mutants R80A and Vpr1-84 did not show interaction with 14-3-3η.
To demonstrate a direct interaction of 14-3-3 and Vpr, we used a GST pull-down assay with bacterially produced GST-14-3-3η and in vitro-translated and -labeled Vpr or Vpr mutants. We found that Vpr was able to bind to 14-3-3 in vitro, demonstrating a direct interaction (Fig. 1D). Interestingly, use of the mutants VprR80A and Vpr (1-84) eliminated this interaction, whereas VprL64A and Vpr (1-64) were active, in agreement with the results of the yeast two-hybrid assay.
14-3-3 enhanced Vpr's effect on cell cycle arrest.
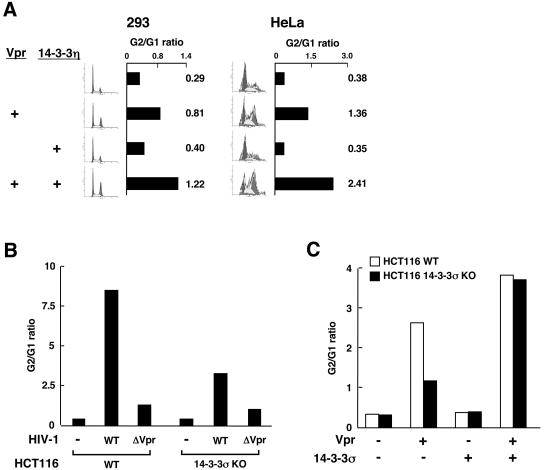
To address the functional relevance of 14-3-3 and Vpr association on the cell cycle, we coexpressed these two proteins in HeLa and 293 cells and examined the cell cycle profile. We found that coexpression of 14-3-3η potentiated Vpr's cell cycle arrest, whereas transfection of 14-3-3η alone had almost no effect in either cell line (Fig. 2A).
FIG. 2.
(A) Effect of Vpr and 14-3-3η on the cell cycle profile and G2/G1 ratio in HeLa and 293 cells. Cells were transfected with plasmids encoding Vpr and/or 14-3-3η, and their cell cycle profile presented as the G2/G1 ratio (shown as bars and numbers) was examined by fluorescence-activated cell sorter (FACS) analysis. (B) 14-3-3σ is important for cell cycle arrest induced by Vpr in HCT116 cells. HCT116 WT or 14-3-3σ KO cells were infected with the VSV-G-pseudotyped HIV-1 molecular clone NL4-3GFP or NL4-3GFPΔvpr. The cell cycle profiles of infected cells are indicated as the G2/G1 ratio after FACS analysis. (C) Supplementation of 14-3-3σ in 14-3-3σ KO cells restores cell cycle arrest by Vpr. HCT116 WT or 14-3-3σ KO cells were transfected with pCDNA3-Vpr in the presence or absence of plasmid expressing 14-3-3σ. The cell cycle profiles of transfected cells are indicated as the G2/G1 ratio determined by FACS analyses.
14-3-3 isoform elimination decreased the cell cycle arrest induced by infection with HIV-1 molecular clones.
As an alternative method to examine the role of 14-3-3 proteins in the cell cycle arrest induced by HIV-1, we used the 14-3-3σ-deficient HCT116 human cell line (14-3-3σ KO cells) (4) as a target for infection with HIV-1 molecular clones. These cells are the only ones available with knockouts of any members of the 14-3-3 family of proteins. Infectious stocks of NL4-3GFP (47) and the Vpr-deficient molecular clone NL4-3GFPΔvpr were generated by cotransfection of either molecular clone together with VSV-G DNA into 293 cells. VSV-G allows infection of HCT116 cells, which lack HIV surface receptors. Two days after transfection of 293 cells, the supernatants were harvested and, after low-speed centrifugation, used to infect the WT HCT116 cell line and a 14-3-3σ KO derivative cell line. Two days after infection the cells were stained with Hoechst 33342 and their cell cycle profiles were analyzed by flow cytometry. NL4-3GFP induced G2/M cell cycle arrest in the WT HCT116 cells, whereas the Vpr-deficient virus showed a dramatically lower effect, indicating that Vpr expressed from the WT molecular clone was mainly responsible for the cell cycle arrest (Fig. 2B). In contrast to these results, the frequency of cells arrested in G2/M was significantly decreased in 14-3-3σ KO HCT116 cells infected with NL4-3GFP, indicating that the presence of 14-3-3σ was important for Vpr's effects on cell cycle. The frequency of infected cells in G2/M was similar in both WT and 14-3-3σ KO HCT116 cells infected with NL4-3GFPΔvpr. Supplementation of 14-3-3σ protein by transfection back into 14-3-3σ KO cells almost completely reversed the observed Vpr-generated defect in the cell cycle (Fig. 2C). Taken together, these results suggest that Vpr effects on the cell cycle are severely diminished in cells not expressing a full set of 14-3-3 proteins.
Vpr sequestered Cdc25C in the cytoplasm independently of serine 216.
To address the mechanism underlying the effect of 14-3-3 on Vpr's cell cycle-arresting activity, we focused on the Cdc25C molecule. Interaction of 14-3-3 with this phosphatase is important and has been well examined among 14-3-3 partner proteins that are functional in cell cycle progression (8, 17, 33, 38, 44, 52, 53).
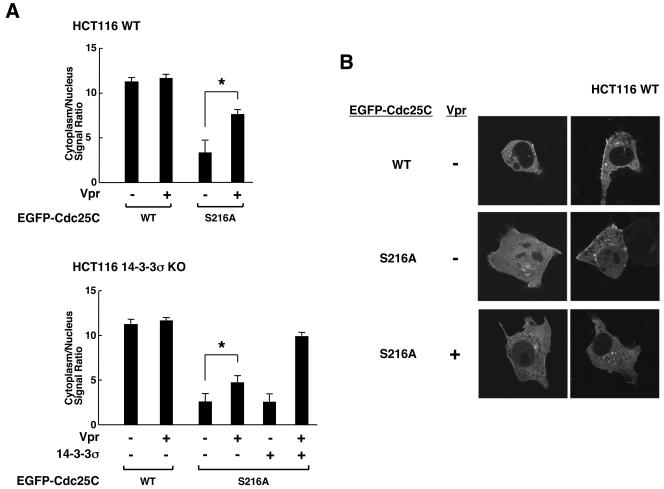
We examined the distribution of EGFP-Cdc25C fusion proteins in the presence or absence of Vpr in HCT116 WT and 14-3-3σ KO cells (Fig. 3). Signal intensity of EGFP-Cdc25C in the nucleus and the cytoplasm of over 20 cells cotransfected with Vpr-expressing plasmid was measured, and the mean ± SE values of their ratios are shown in Fig. 3A after subtracting background signal intensity. Almost all cells of both cell lines had the WT EGFP-Cdc25C in the cytoplasm in the absence or presence of Vpr. We thus used the mutant EGFP-Cdc25C S216A, in which the serine residue at amino acid 216 was replaced with alanine. In HCT116 WT cells, the intracellular distribution of this mutant moderately shifted to the nucleus when compared to the WT Cdc25C, as previously reported (Fig. 3A, upper panel) (8, 28, 34, 53). Coexpression of Vpr shifted EGFP-Cdc25CS216A toward the cytoplasm, although Cdc25CS216A did not have a serine residue at amino acid 216, which is required for the cytoplasmic localization of the WT Cdc25C by 14-3-3 (8, 38). In 14-3-3σ KO cells, Vpr's ability to localize EGFP-Cdc25CS216A in the cytoplasm was decreased, and coexpression of 14-3-3σ reversed this defect (Fig. 3A, lower panel). Representative images showing the effect of Vpr on the subcellular localization of EGFP-Cdc25C WT and S216A are shown in Fig. 3B.
FIG. 3.
Vpr facilitates redistribution of Cdc25CS216A into the cytoplasm of HCT116 WT and 14-3-3σ KO cells. (A) HCT116 WT and 14-3-3σ KO cells were transfected with EGFP-Cdc25C or the S216A mutant-expressing plasmids in the presence or absence of pCDNA3-Vpr. Cdc25C and Cdc25CS216A localization was examined in over 20 cells. Signal intensity of EGFP-Cdc25Cs in the cytoplasm and the nucleus was evaluated, and the mean ± SE values of their ratios are shown. (B) Representative images of subcellular localization of EGFP-Cdc25C in the absence or presence of Vpr expression are shown. *, P < 0.01 (t test) for the indicated two conditions.
Vpr supports triple complex formation with 14-3-3 and Cdc25C.
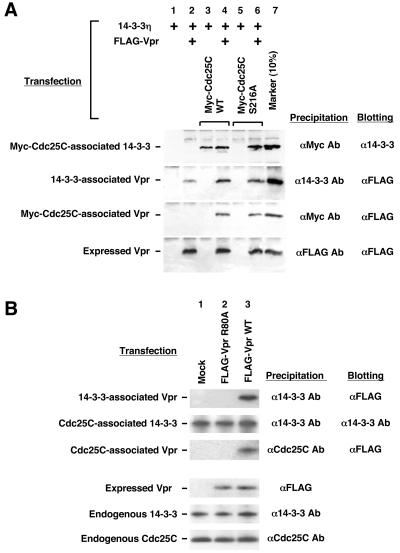
The above results indicated that, in the presence of Vpr, 14-3-3 appeared to cause translocation of Cdc25C into the cytoplasm even in the absence of serine 216. Since phosphorylation of this serine residue is important for binding of 14-3-3 to Cdc25C (38, 44), these results suggested that Vpr might somehow bridge 14-3-3 and Cdc25C independently of the latter's phosphorylation status. Therefore, we examined the in vivo interaction of 14-3-3 and Cdc25C in the presence of Vpr in a coimmunoprecipitation assay (Fig. 4A). For this, we transfected 293 cells with combinations of FLAG-tagged Vpr, 14-3-3, and Myc-tagged Cdc25C, as indicated in Fig. 4A. The epitope-tagged molecules facilitated the quantitative immunoprecipitation of the molecular complexes containing the tagged molecules from 293 cell extracts. The precipitated complexes were separated on SDS-PAGE gels, and the various proteins were identified after immunoblotting with either anti-14-3-3 or anti-FLAG antibody.
FIG. 4.
Vpr forms a trimeric complex with 14-3-3 and Cdc25C. (A) Vpr supports complex formation between 14-3-3 and Cdc25C independently of a serine residue at amino acid 216 of Cdc25C. 293 cells were transfected with plasmids expressing FLAG-Vpr, 14-3-3η, and Myc-tagged Cdc25C WT or S216A mutant. The complexes were precipitated with the indicated antibodies and run on SDS-12 or 16% PAGE gels. After blotting on a nitrocellulose membrane, 14-3-3 or FLAG-Vpr was detected with anti-FLAG (M2) antibody. (B) WT Vpr, but not the R80A mutant, was associated with endogenous Cdc25C and 14-3-3. 293 cells were transfected with WT Vpr- or R80A mutant-expressing plasmid. The complexes were precipitated with the indicated antibodies and run on 12 or 16% SDS-PAGE gels. After blotting on a nitrocellulose membrane, FLAG-Vpr, endogenous 14-3-3, and Cdc25C were detected with the appropriate antibodies, as indicated.
In the absence of Vpr, 14-3-3 was specifically coprecipitated with Cdc25C (lane 3), but not with Cdc25C S216A (lane 5). In contrast, in the presence of Vpr, 14-3-3 was coprecipitated also with Cdc25C S216A (lane 6), which does not normally associate with 14-3-3. Since two of three proteins were mutually coprecipitated by their specific antibodies, we concluded that Vpr helped formation of 14-3-3/Vpr/Cdc25C complexes.
Since the interactions observed were obtained upon 14-3-3 overexpression, we examined similar coimmunoprecipitations of Vpr with endogenous 14-3-3 and Cdc25C molecules in the same 293 cells in the absence of transfected 14-3-3 (Fig. 4B). As expected, WT Vpr was associated with 14-3-3 and Cdc25C (lane 3), whereas the 14-3-3-binding-defective VprR80A failed to interact with these molecules (lane 2), suggesting that Vpr was coprecipitated with Cdc25C through its binding to 14-3-3. Taken together, these results strongly indicate that Vpr promotes the association of 14-3-3 and Cdc25C S216A, bypassing the requirement for phosphorylation at serine 216 of Cdc25C.
DISCUSSION
Here we present several lines of evidence indicating that the identified interaction of Vpr to 14-3-3 proteins has functional significance for the cell cycle regulation of HIV-1 Vpr. To identify proteins involved in Vpr's cell cycle-arresting activity, we performed a yeast two-hybrid screening using VprL64A mutant as bait, which is defective in the transcriptional activity but retains the Vpr ability to induce cell cycle arrest. We reasoned that this approach would reduce the number of interactors without eliminating the ones responsible for cell cycle arrest. This approach successfully eliminated binding to many cellular proteins interacting with Vpr and allowed the identification as Vpr partners of several 14-3-3 proteins known to affect the cell cycle at several different steps. Recent genetic analysis employing fission yeast, Schizosaccharomyces pombe, also indicated involvement of rad24 on Vpr's cell cycle activity, where rad24 encodes a member of the 14-3-3 protein family (31). Among the 14-3-3-encoding clones that we found in the yeast two-hybrid assay, 14-3-3η was the most frequently represented. In functional assays, 14-3-3η cooperatively enhanced Vpr's cell cycle-arresting activity. Furthermore, in HCT116 cells, endogenous 14-3-3σ appears to facilitate cell cycle arrest induced by Vpr expressed after infection by an HIV-1 molecular clone as well as following transfection of a Vpr-expressing plasmid. These results indicate that a full complement of 14-3-3 proteins is necessary for optimal function of Vpr and that elimination of 14-3-3σ severely affected Vpr function. We found that Vpr induces translocation of Cdc25C into the cytoplasm independently of serine 216 in HCT116 cells. The ability of Vpr to translocate Cdc25CS216A into the cytoplasm was blunted in 14-3-3σ KO cells, and supplementation of 14-3-3σ reversed this defect. Using Vpr and 14-3-3 mutants, we further characterized the direct interaction of these molecules. We found that the C-terminal acidic domain of Vpr binds to the C-terminal portion of 14-3-3, outside of the binding pocket for phosphopeptides. Thus, Vpr binds 14-3-3 proteins and causes cell cycle arrest by inducing association between 14-3-3 and Cdc25C independently of the latter's phosphorylation status at serine 216. Vpr appears to function as an adaptor molecule, bridging these two cell cycle-regulating molecules.
Cellular events promoting cell cycle arrest at the G2/M checkpoint lead to phosphorylation of Cdc25C on serine 216 (7, 38). 14-3-3 binds the phosphorylated serine and promotes Cdc25 translocation into the cytoplasm, thus preventing the dephosphorylation and inactivation of the nuclear Cdc2 (8, 44, 52). Our data support the conclusion that Vpr bypasses the phosphorylation step of Cdc25C; upon Vpr binding, 14-3-3 interacts with Cdc25C and induces its translocation into the cytoplasm. This interaction is independent of the serine 216 phosphorylation status, since the mutant Cdc25C S216A also forms complexes and is transported into the cytoplasm.
14-3-3 proteins play a significant role in cell cycle progression at several different stages. First, they regulate Cdc25C activity (8, 17, 28, 33, 38, 41, 44, 52). Second, they bind Wee1 kinase and increase the stability and enhance the activity of this protein (27, 50). Third, 14-3-3σ also binds and activates Chk1 and Cdk2 kinases by appropriately localizing these molecules inside the nucleus (5). Activation of Chk1 causes phosphorylation of Cdc25C, producing a binding site for 14-3-3, leading to inactivation of its phosphatase activity (44, 52). Fourth, 14-3-3 binds both phosphorylated Cdc2 and cyclinB1 and inactivates their complex by exporting it into the cytoplasm (4, 26). Therefore, it is possible that Vpr also modulates activities of the above-described kinases and phosphatases by changing their binding specificity to 14-3-3. Since Vpr binds 14-3-3 at the C-terminal part, a region that plays an important role in its binding specificity to phosphopeptides (41), binding of Vpr may alter the binding affinity of 14-3-3 to its partner proteins. Alternatively, Vpr might facilitate or inhibit the association of 14-3-3 to its partner molecules with additional interactions. Furthermore, association of Vpr to 14-3-3 may also have consequences for the regulation of apoptosis (3, 30). It is known that 14-3-3 may affect apoptosis by interacting with BAD, a member of the Bcl-2 family of proteins that regulate mitochondrial permeability (2, 3, 25). The presence of Vpr might influence the association between 14-3-3 and BAD resulting in increased apoptotic signals triggered by Bcl-2.
In conclusion, we have found that Vpr binds 14-3-3 and promotes the formation of a trimolecular complex with Cdc25C even in the absence of a phosphorylated serine residue at position 216. Vpr may act as a bridge between 14-3-3 and Cdc25C or, alternatively, change the conformation of 14-3-3, resulting in increased binding affinity to nonphosphorylated molecules. This complex formation may promote the cytoplasmic accumulation of Cdc25C and thus subsequent cell cycle arrest.
Acknowledgments
We thank B. Vogelstein for cell lines, J. A. De Caprio for plasmids, and P. Carney and K. Zachman for superb technical assistance.
REFERENCES
- 1.Aitken, A. 1996. 14-3-3 and its possible role in co-ordinating multiple signaling pathways. Trends Cell Biol. 6:341-347. [DOI] [PubMed] [Google Scholar]
- 2.Bernardi, P., V. Petronilli, F. Di Lisa, and M. Forte. 2001. A mitochondrial perspective on cell death. Trends Biochem. Sci. 26:112-117. [DOI] [PubMed] [Google Scholar]
- 3.Boya, P., B. Roques, and G. Kroemer. 2001. New EMBO members' review: viral and bacterial proteins regulating apoptosis at the mitochondrial level. EMBO J. 20:4325-4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan, T. A., H. Hermeking, C. Lengauer, K. W. Kinzler, and B. Vogelstein. 1999. 14-3-3Sigma is required to prevent mitotic catastrophe after DNA damage. Nature 401:616-620. [DOI] [PubMed] [Google Scholar]
- 5.Chen, L., T. H. Liu, and N. C. Walworth. 1999. Association of Chk1 with 14-3-3 proteins is stimulated by DNA damage. Genes Dev. 13:675-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen, E. A., E. F. Terwilliger, Y. Jalinoos, J. Proulx, J. G. Sodroski, and W. A. Haseltine. 1990. Identification of HIV-1 vpr product and function. J. Acquir. Immune Defic. Syndr. 3:11-18. [PubMed] [Google Scholar]
- 7.Conklin, D. S., K. Galaktionov, and D. Beach. 1995. 14-3-3 proteins associate with cdc25 phosphatases. Proc. Natl. Acad. Sci. USA 92:7892-7896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dalal, S. N., C. M. Schweitzer, J. Gan, and J. A. DeCaprio. 1999. Cytoplasmic localization of human cdc25C during interphase requires an intact 14-3-3 binding site. Mol. Cell. Biol. 19:4465-4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Di Marzio, P., S. Choe, M. Ebright, R. Knoblauch, and N. R. Landau. 1995. Mutational analysis of cell cycle arrest, nuclear localization, and virion packaging of human immunodeficiency virus type 1 Vpr. J. Virol. 69:7909-7916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elder, R. T., M. Yu, M. Chen, S. Edelson, and Y. Zhao. 2000. Cell cycle G2 arrest induced by HIV-1 Vpr in fission yeast (Schizosaccharomyces pombe) is independent of cell death and early genes in the DNA damage checkpoint. Virus Res. 68:161-173. [DOI] [PubMed] [Google Scholar]
- 11.Felber, B. K., M. Hadzopoulou-Cladaras, C. Cladaras, T. Copeland, and G. N. Pavlakis. 1989. Rev protein of human immunodeficiency virus type 1 affects the stability and transport of the viral mRNA. Proc. Natl. Acad. Sci. USA 86:1495-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finley, R. L., and R. Brent. 1995. Interaction trap cloning with yeast, p. 169-203. In B. D. Hames and D. M. Glover (ed.), DNA cloning, expression systems: a practical approach. Oxford University Press, Oxford, England.
- 13.Fu, H., R. R. Subramanian, and S. C. Masters. 2000. 14-3-3 proteins: structure, function, and regulation. Annu. Rev. Pharmacol. Toxicol. 40:617-647. [DOI] [PubMed] [Google Scholar]
- 14.Gibbs, J. S., A. A. Lackner, S. M. Lang, M. A. Simon, P. K. Sehgal, M. D. Daniel, and R. C. Desrosiers. 1995. Progression to AIDS in the absence of a gene for vpr or vpx. J. Virol. 69:2378-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goh, W. C., M. E. Rogel, C. M. Kinsey, S. F. Michael, P. N. Fultz, M. A. Nowak, B. H. Hahn, and M. Emerman. 1998. HIV-1 Vpr increases viral expression by manipulation of the cell cycle: a mechanism for selection of Vpr in vivo. Nat. Med. 4:65-71. [DOI] [PubMed] [Google Scholar]
- 16.Gragerov, A., T. Kino, G. Ilyina-Gragerova, G. P. Chrousos, and G. N. Pavlakis. 1998. HHR23A, the human homologue of the yeast repair protein RAD23 interacts specifically with Vpr protein and prevents cell cycle arrest but not the transcriptional effects of Vpr. Virology 245:323-330. [DOI] [PubMed] [Google Scholar]
- 17.Graves, P. R., C. M. Lovly, G. L. Uy, and H. Piwnica-Worms. 2001. Localization of human Cdc25C is regulated both by nuclear export and 14-3-3 protein binding. Oncogene 20:1839-1851. [DOI] [PubMed] [Google Scholar]
- 18.Gyuris, J., E. Golemis, H. Chertkov, and R. Brent. 1993. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell 75:791-803. [DOI] [PubMed] [Google Scholar]
- 19.He, J., S. Choe, R. Walker, P. Di Marzio, D. O. Morgan, and N. R. Landau. 1995. Human immunodeficiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J. Virol. 69:6705-6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heinzinger, N. K., M. I. Bukinsky, S. A. Haggerty, A. M. Ragland, V. Kewalramani, M. A. Lee, H. E. Gendelman, L. Ratner, M. Stevenson, and M. Emerman. 1994. The Vpr protein of human immunodeficiency virus type 1 influences nuclear localization of viral nucleic acids in nondividing host cells. Proc. Natl. Acad. Sci. USA 91:7311-7315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jackman, M. R., and J. N. Pines. 1997. Cyclins and the G2/M transition. Cancer Surv. 29:47-73. [PubMed] [Google Scholar]
- 22.Jowett, J. B., V. Planelles, B. Poon, N. P. Shah, M. L. Chen, and I. S. Chen. 1995. The human immunodeficiency virus type 1 vpr gene arrests infected T cells in the G2 + M phase of the cell cycle. J. Virol. 69:6304-6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kino, T., and G. P. Chrousos. 2003. Tumor necrosis factor α receptor- and Fas-associated FLASH inhibits transcriptional activity of the glucocorticoid receptor by binding to and interfering with its interaction with p160 type nuclear receptor coactivators. J. Biol. Chem. 278:3023-3029. [DOI] [PubMed]
- 24.Kino, T., A. Gragerov, J. B. Kopp, R. H. Stauber, G. N. Pavlakis, and G. P. Chrousos. 1999. The HIV-1 virion-associated protein Vpr is a coactivator of the human glucocorticoid receptor. J. Exp. Med. 189:51-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korsmeyer, S. J. 1999. BCL-2 gene family and the regulation of programmed cell death. Cancer Res. 59:1693s-1700s. [PubMed] [Google Scholar]
- 26.Laronga, C., H. Y. Yang, C. Neal, and M. H. Lee. 2000. Association of the cyclin-dependent kinases and 14-3-3 sigma negatively regulates cell cycle progression. J. Biol. Chem. 275:23106-23112. [DOI] [PubMed] [Google Scholar]
- 27.Lee, J., A. Kumagai, and W. G. Dunphy. 2001. Positive regulation of Wee1 by Chk1 and 14-3-3 proteins. Mol. Biol. Cell 12:551-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lopez-Girona, A., B. Furnari, O. Mondesert, and P. Russell. 1999. Nuclear localization of Cdc25 is regulated by DNA damage and a 14-3-3 protein. Nature 397:172-175. [DOI] [PubMed] [Google Scholar]
- 29.Mahalingam, S., V. Ayyavoo, M. Patel, T. Kieber-Emmons, and D. B. Weiner. 1997. Nuclear import, virion incorporation, and cell cycle arrest/differentiation are mediated by distinct functional domains of human immunodeficiency virus type 1 Vpr. J. Virol. 71:6339-6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marzo, I., C. Brenner, N. Zamzami, S. A. Susin, G. Beutner, D. Brdiczka, R. Remy, Z. H. Xie, J. C. Reed, and G. Kroemer. 1998. The permeability transition pore complex: a target for apoptosis regulation by caspases and bcl-2-related proteins. J. Exp. Med. 187:1261-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Masuda, M., Y. Nagai, N. Oshima, K. Tanaka, H. Murakami, H. Igarashi, and H. Okayama. 2000. Genetic studies with the fission yeast Schizosaccharomyces pombe suggest involvement of wee1, ppa2, and rad24 in induction of cell cycle arrest by human immunodeficiency virus type 1 Vpr. J. Virol. 74:2636-2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mirani, M., I. Elenkov, S. Volpi, N. Hiroi, G. P. Chrousos, and T. Kino. 2002. HIV-1 protein Vpr suppresses IL-12 production from human monocytes by enhancing glucocorticoid action: potential implications of Vpr coactivator activity for the innate and cellular immunity deficits observed in HIV-1 infection. J. Immunol. 169:6361-6368. [DOI] [PubMed] [Google Scholar]
- 33.Morris, M. C., A. Heitz, J. Mery, F. Heitz, and G. Divita. 2000. An essential phosphorylation-site domain of human cdc25C interacts with both 14-3-3 and cyclins. J. Biol. Chem. 275:28849-28857. [DOI] [PubMed] [Google Scholar]
- 34.Muslin, A. J., and H. Xing. 2000. 14-3-3 proteins: regulation of subcellular localization by molecular interference. Cell Signal. 12:703-709. [DOI] [PubMed] [Google Scholar]
- 35.Nebreda, A. R., and I. Ferby. 2000. Regulation of the meiotic cell cycle in oocytes. Curr. Opin. Cell Biol. 12:666-675. [DOI] [PubMed] [Google Scholar]
- 36.Ohi, R., and K. L. Gould. 1999. Regulating the onset of mitosis. Curr. Opin. Cell Biol. 11:267-273. [DOI] [PubMed] [Google Scholar]
- 37.Paxton, W., R. I. Connor, and N. R. Landau. 1993. Incorporation of Vpr into human immunodeficiency virus type 1 virions: requirement for the p6 region of gag and mutational analysis. J. Virol. 67:7229-7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peng, C. Y., P. R. Graves, R. S. Thoma, Z. Wu, A. S. Shaw, and H. Piwnica-Worms. 1997. Mitotic and G2 checkpoint control: regulation of 14-3-3 protein binding by phosphorylation of Cdc25C on serine-216. Science 277:1501-1505. [DOI] [PubMed] [Google Scholar]
- 39.Popov, S., M. Rexach, G. Zybarth, N. Reiling, M. A. Lee, L. Ratner, C. M. Lane, M. S. Moore, G. Blobel, and M. Bukrinsky. 1998. Viral protein R regulates nuclear import of the HIV-1 pre-integration complex. EMBO J. 17:909-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Re, F., D. Braaten, E. K. Franke, and J. Luban. 1995. Human immunodeficiency virus type 1 Vpr arrests the cell cycle in G2 by inhibiting the activation of p34cdc2-cyclin B. J. Virol. 69:6859-6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rittinger, K., J. Budman, J. Xu, S. Volinia, L. C. Cantley, S. J. Smerdon, S. J. Gamblin, and M. B. Yaffe. 1999. Structural analysis of 14-3-3 phosphopeptide complexes identifies a dual role for the nuclear export signal of 14-3-3 in ligand binding. Mol. Cell 4:153-166. [DOI] [PubMed] [Google Scholar]
- 42.Rogel, M. E., L. I. Wu, and M. Emerman. 1995. The human immunodeficiency virus type 1 vpr gene prevents cell proliferation during chronic infection. J. Virol. 69:882-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rosenquist, M., P. Sehnke, R. J. Ferl, M. Sommarin, and C. Larsson. 2000. Evolution of the 14-3-3 protein family: does the large number of isoforms in multicellular organisms reflect functional specificity? J. Mol. Evol. 51:446-458. [DOI] [PubMed] [Google Scholar]
- 44.Sanchez, Y., C. Wong, R. S. Thoma, R. Richman, Z. Wu, H. Piwnica-Worms, and S. J. Elledge. 1997. Conservation of the Chk1 checkpoint pathway in mammals: linkage of DNA damage to Cdk regulation through Cdc25. Science 277:1497-1501. [DOI] [PubMed] [Google Scholar]
- 45.Stewart, S. A., B. Poon, J. B. Jowett, and I. S. Chen. 1997. Human immunodeficiency virus type 1 Vpr induces apoptosis following cell cycle arrest. J. Virol. 71:5579-5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Subbramanian, R. A., A. Kessous-Elbaz, R. Lodge, J. Forget, X. J. Yao, D. Bergeron, and E. A. Cohen. 1998. Human immunodeficiency virus type 1 Vpr is a positive regulator of viral transcription and infectivity in primary human macrophages. J. Exp. Med. 187:1103-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Valentin, A., W. Lu, M. Rosati, R. Schneider, J. Albert, A. Karlsson, and G. N. Pavlakis. 1998. Dual effect of interleukin 4 on HIV-1 expression: implications for viral phenotypic switch and disease progression. Proc. Natl. Acad. Sci. USA 95:8886-8891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vodicka, M. A., D. M. Koepp, P. A. Silver, and M. Emerman. 1998. HIV-1 Vpr interacts with the nuclear transport pathway to promote macrophage infection. Genes Dev. 12:175-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang, B., Y. C. Ge, P. Palasanthiran, S. H. Xiang, J. Ziegler, D. E. Dwyer, C. Randle, D. Dowton, A. Cunningham, and N. K. Saksena. 1996. Gene defects clustered at the C-terminus of the vpr gene of HIV-1 in long-term nonprogressing mother and child pair: in vivo evolution of vpr quasispecies in blood and plasma. Virology 223:224-232. [DOI] [PubMed] [Google Scholar]
- 50.Wang, Y., C. Jacobs, K. E. Hook, H. Duan, R. N. Booher, and Y. Sun. 2000. Binding of 14-3-3β to the carboxyl terminus of Wee1 increases Wee1 stability, kinase activity, and G2-M cell population. Cell Growth Differ. 11:211-219. [PubMed] [Google Scholar]
- 51.Yaffe, M. B., K. Rittinger, S. Volinia, P. R. Caron, A. Aitken, H. Leffers, S. J. Gamblin, S. J. Smerdon, and L. C. Cantley. 1997. The structural basis for 14-3-3:phosphopeptide binding specificity. Cell 91:961-971. [DOI] [PubMed] [Google Scholar]
- 52.Zeng, Y., K. C. Forbes, Z. Wu, S. Moreno, H. Piwnica-Worms, and T. Enoch. 1998. Replication checkpoint requires phosphorylation of the phosphatase Cdc25 by Cds1 or Chk1. Nature 395:507-510. [DOI] [PubMed] [Google Scholar]
- 53.Zeng, Y., and H. Piwnica-Worms. 1999. DNA damage and replication checkpoints in fission yeast require nuclear exclusion of the Cdc25 phosphatase via 14-3-3 binding. Mol. Cell. Biol. 19:7410-7419. [DOI] [PMC free article] [PubMed] [Google Scholar]