Abstract
The 14.4-kDa hexon-associated protein IX (pIX) acts as a cement in the capsids of primate adenoviruses and confers a thermostable phenotype. Here we show that deletion of amino acids 100 to 114 of adenovirus type 5 pIX, which eliminates the conserved coiled-coil domain, impairs its capacity to self-associate. However, pIXΔ100-114 is efficiently incorporated into the viral capsid, and the resulting virions are thermostable. Deletion of the central alanine-rich domain, as in pIXΔ60-72, does not impair self-association, incorporation into the capsid, or the thermostable phenotype. These data demonstrate, first, that the self-association of pIX is dispensable for its incorporation into the capsid and generation of the thermostability phenotype and, second, that the increased thermostability results from pIX monomers binding to different hexon capsomers rather than capsid stabilization by pIX multimers.
The adenovirus (Ad) protein IX (pIX) is strongly conserved in primate mastadenoviruses. Cryoelectron microscopy image analyses of Ad particles reveal continuous trisymmetric densities between the nine-hexon capsomers that form the group-of-nine hexons on each facet of the icosahedral capsid structure (6, 18, 20). This suggested that there are 12 trimeric pIX proteins in each of the 20 facets. Although pIX is dispensable for viral replication, pIX-deficient particles are more thermolabile than wild-type (wt) particles (3). Therefore pIX is often described as capsid “cement.” pIX is required to package genomes with sizes greater than that of the wt Ad genome (7, 15). The location of pIX at or near the surface of the particle was confirmed by immune accessibility studies using pIX epitopes and heterologous epitopes linked to the pIX C terminus (1, 4, 10, 21). In addition, pIX has been implicated in regulating the expression of the Ad major late promoter (9, 14). Mutagenesis has pinpointed the central alanine-rich domain and the coiled-coil domain in the carboxyl-terminal part of pIX as being involved in transcription regulation (12). The coiled-coil domain contains a putative leucine zipper domain (16); it has been suggested that this domain is involved in the self-association and the presumed trimer formation of pIX molecules (12). Electron microscopy studies revealed that the integrity of the coiled-coil domain is required for formation of pIX-containing inclusion bodies (12, 13, 17). Further studies implicated the strongly conserved N-terminal domain (Fig. 1A) in capsid incorporation (12).
FIG. 1.
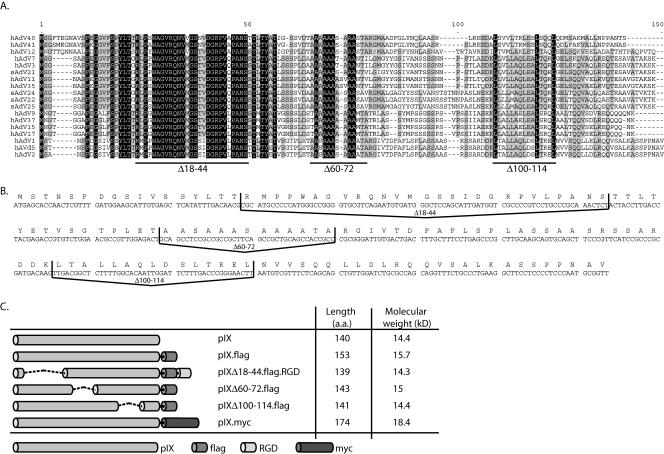
Conserved amino acid regions in pIX. (A) Amino acid sequence alignment of all available primate Ad pIX proteins (CLUSTAL W) (19). Fully conserved residues are shaded black. Residues that occur in more than 50% of the sequences are shaded gray. Lines under the aligned sequences indicate the deletions mentioned in the text. (B) DNA sequence of the human Ad type 5 pIX gene. Deletions of the three conserved domains were introduced by site-directed mutation PCR. The lines depict the DNA sequence deletions. (C) Schematic representation of the pIX variants used for coprecipitation assays in this study. Molecular weights are in thousands.
To get more insight in the structure and function of pIX and its role in capsid stabilization, we generated a series of hAd5 mutants with deletions in the pIX gene. The modified pIX proteins were tested for their capacity to self-associate and to be incorporated into the capsid. Subsequently we tested virus particles carrying the mutant pIX for their thermostability. We employed site-directed mutagenesis (QuikChange site-directed mutagenesis kit; Stratagene) of the expression plasmid pAd5pIX (21) to generate the following deletion mutant proteins: pIXΔ18-44, which has a deletion of amino acids (aa) 18 to 44 in the N-terminal region; pIXΔ60-72, in which aa 60 to 72 of the alanine-rich stretch have been removed; and pIXΔ100-114, which lacks aa 100 to 114, which comprise the central part of the coiled-coil domain (Fig. 1B and Table 1). The deletions were chosen on the basis of both the sequence conservation among the serotypes and the previously characterized domains (12). For the self-association assay we generated C-terminal Flag epitope-tagged versions of our mutant pIX proteins and a MYC epitope-tagged version of wt pIX. An additional RGD motif was fused to the pIXΔ18-44 construct, generating a pIX molecule with a molecular weight similar to those of the others (Fig. 1C). To examine whether the deletion mutant proteins were capable of being incorporated into the viral capsid of hAd5dl313, which lacks a functional pIX (3), this virus was propagated on 911 helper cells (5) that transiently expressed the mutant pIX proteins as described previously (21). All pIX proteins were produced in the 911 helper cells (Fig. 2A). After CsCl banding and purification, the viruses were assayed for the presence of pIX by Western blot analysis. Both the pIXΔ60-72 and pIXΔ100-114 mutant proteins were incorporated efficiently (Fig. 2B). In contrast, pIXΔ18-44 was not incorporated into the capsid, implying a role for this domain in capsid incorporation (12).
TABLE 1.
Primer list
| Primer, orientationa | Sequence (5′-3′) |
|---|---|
| N-ter-Del, For | TCATATTTGACAACGACTACCTTGACCTAC |
| N-ter-Del, Rev | GTAGGTCAAGGTAGTCGTTGTCAAATATGA |
| Ala-Del, For | ACGCCGTTGGAGACTCGCGGGATTGTGACT |
| Ala-Del, Rev | AGTCACAATCCCGCGAGTCTCCAACGGCGT |
| Leu-Del, For | GCCCGCGATGACAAGAATGTCGTTTCTCAG |
| Leu-Del, Rev | CTGAGAAACGACATTCTTGTCATCGCGGGC |
| PCRdeletion, For | GGTCTTATGTAGTTTTGTATC |
| PCRdeletion, Rev | CGCTGGTCCCGGGCCTACCG |
| Fib-FWD-PstI, For | CAGCACTGCAGACCATGAAGCGCGCAAGACCGTC |
| Fib-REV-XhoI, Rev | CGACACTCGAGCTATCATTCTTGGGCAATGTATGAAAAAGTG |
For, forward; Rev, reverse.
FIG. 2.
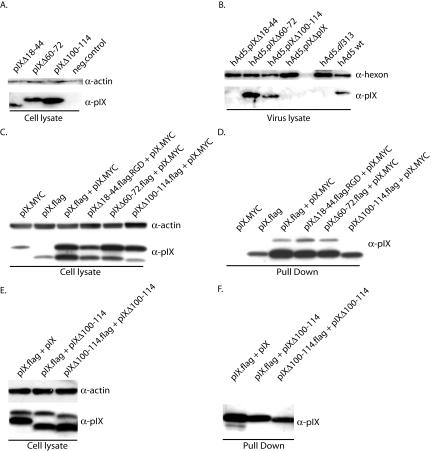
Assay to measure pIX incorporation into the capsid. (A) Cultured 911 cells were transfected with the various pIX expression plasmids. Twenty hours posttransfection, the cells were infected with hAd5dl313 viruses. Two days later, protein extracts were prepared and analyzed by Western blotting. The blots were probed with anti-pIX or antiactin sera. The negative control (neg.control) is a lysate from untransfected 911 cells. (B) From parallel cultures of transfected and pIX-expressing cells, the progeny hAd5dl313 viruses were harvested, purified, and assayed for the presence of the pIX variants in the capsids by Western analysis. The negative control is hAd5dl313 virus lysate propagated on normal 911 cells; the positive control is wt hAd5. (C) pIX self-association assay. To test the self-association of pIX molecules, an expression plasmid containing the pIX.MYC gene and a plasmid containing a gene encoding one of the Flag-tagged pIX deletion mutants were cotransfected into 911 cells. The cell lysates of these cells were used for coprecipitation with Flag-beads. As a negative control we used 911 cells that were transfected with the pIX.MYC expression plasmid only. A protein lysate from 9ll cells transfected with the pIX.Flag plasmid was used as a positive control. (D) Western blot analysis of the pIX proteins associating with pIX-Flag. After coprecipitation the samples were analyzed by Western blotting with anti-pIX serum for detection of the pIX variants. (E) Western blot analysis of 911 cells transfected with plasmids coding for pIX.Flag, wt pIX, pIXΔ100-114, and pIXΔ100-114-Flag. The blots were probed with anti-pIX or antiactin sera. (F) Western blot analysis of the pIX proteins associating with the Flag-tagged pIX and pIXΔ100-114. The blots were probed with anti-pIX serum.
Next we evaluated the ability of our mutant pIX proteins to self-associate. The plasmids encoding the pIX.MYC and pIX.Flag mutant proteins were coexpressed in 911 cells as described previously (21) in the absence of Ad. After 48 h the cells were washed with phosphate-buffered saline, followed by lysis in radioimmunoprecipitation assay lysis buffer (50 mM Tris-Cl [pH 7.5], 150 mM NaCl, 0.1% sodium dodecyl sulfate [SDS], 0.5% deoxycholate, 1% NP-40). Expression of these pIX derivatives was confirmed by Western blotting. Protein concentrations were measured with a protein assay kit (BCA protein assay kit; Pierce, Rockford, Ill.). After SDS-polyacrylamide gel electrophoresis (PAGE) of 15 μg of cell extract, the proteins were transferred to an Immobilon-P transfer membrane (Millipore, Etten-Leur, The Netherlands). The pIX proteins were detected with rabbit anti-pIX serum (2) (1:2,000), followed by a horseradish peroxidase-labeled goat anti-rabbit serum (1:5,000; Santa Cruz Biotechnology, Inc., Santa Cruz, Calif.) as described previously (Fig. 2C) (21). As a loading control a mouse antibody recognizing human actin (1:500; clone C4; ICN Biomedicals, Inc., Zoetermeer, The Netherlands) was used. To determine whether the proteins associated with each other, we performed a coimmunoprecipitation assay. Fifty micrograms of cell lysate was incubated with 1 ml of a Flag-bead suspension (ANTI-flag M2 freezer-safe affinity gel; Sigma Aldrich Chemie, Zwijndrecht, The Netherlands) for 2 h at 4°C. Subsequently the beads were washed with Tris-buffered saline (50 mM Tris-Cl, 150 mM NaCl [pH 7.4]) and resuspended in 2× sample buffer (10% glycerol, 4% SDS, 120 mM Tris-Cl [pH 6.7], 5% bromophenol blue). After size fractionation by SDS-15% PAGE and blotting, the pIX proteins were detected with anti-pIX serum.
In the positive-control sample both the pIX.Flag and pIX.MYC proteins were detected (Fig. 2D), indicating that pIX.MYC associates with pIX.Flag. Also, the deletion mutant proteins pIXΔ18-44 and pIXΔ60-72 associated with pIX.MYC, demonstrating that neither the conserved N-terminal domain nor the alanine stretch is involved in pIX self-association. The latter observation is contrary to an earlier study that had implicated the alanine stretch in pIX self-association (12). The difference may be explained by the presence of a large tag at the N terminus of pIX in the earlier study, rather than at the C terminus in our study. In agreement with previous studies (12), deletion of the conserved domain in the coiled-coil region (pIXΔ100-114) impaired association with pIX.MYC. To study whether the Flag-tagged pIXΔ100-114 can associate with other pIXΔ100-114 molecules, a plasmid encoding a nontagged pIXΔ100-114 was coexpressed with the Flag-tagged pIXΔ100-114 in 911 cells (Fig. 2E and F). As a positive control the plasmids coding for pIX and pIX.Flag were used. Whereas the tagged wt pIX could coprecipitate the untagged wt pIX protein, neither the tagged wt pIX nor the tagged pIXΔ100-114 could precipitate the untagged pIXΔ100-114. This demonstrates that pIXΔ100-114 associates neither with wt pIX nor with pIXΔ100-114.
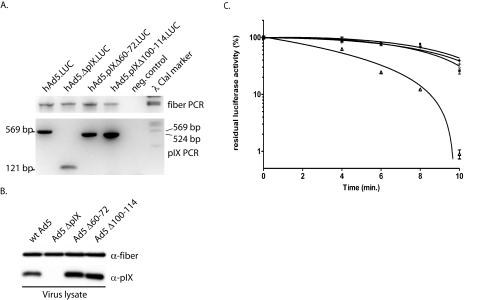
Next we tested the pIX mutant proteins for their capacity to function as capsid cement in a thermostability assay. To this end, we introduced the Δ60-72 and Δ100-114 deletions into the viral backbone by site-directed mutation PCR of the pTrackCMV-Luc shuttle plasmid (21). In addition, a mutant protein, called hAd5ΔpIX. LUC, from which all codons for pIX were deleted, was generated. The Δ18-44 mutant protein was omitted from this study since this mutant pIX is not incorporated into the viral capsid and therefore capsid stability would be expected to be the same as that for an Ad lacking the pIX gene (21). Replication-incompetent hAd5CMV-Luc viruses carrying mutant pIX and ΔpIX were generated as described previously (8). The CsCl-banded and purified viruses were tested for the presence of the deletion in the pIX gene based on product sizes by PCR (Fig. 3A) with the primers listed in Table 1. The PCR products have been verified by sequencing to further confirm the integrity of the deletions in the viral backbone (data not shown). PCR of the fiber gene in the viral backbone was used as an internal control. The viruses carrying the Δ60-72, Δ100-114, and ΔpIX mutations could be propagated with kinetics and titers similar to those for the hAd5.Luc virus that carries the wt pIX gene (data not shown). Western analysis of the purified viruses showed incorporation of pIXΔ60-72 and pIXΔ100-114 equal to that of wt pIX (Fig. 3B). The deletion of the central alanine stretch (Δ60-72) as well as the deletion of a large part of the coiled-coil region (Δ100-114) did not have a negative effect on the incorporation of pIX into the virus capsid. To test for capsid stability, 150-μl aliquots of each of the viruses were incubated at 45°C for 0, 4, 6, 8, and 10 min and subsequently rapidly cooled on ice for 5 min. Next U2OS cells were infected with 100 μl of virus suspension from each aliquot. After 24 h viral titers were determined by measuring the luciferase activity, as described previously (11). The virus lacking pIX, hAd5 ΔpIX.LUC, rapidly decreased in titer in response to incubation at 45°C, whereas vectors encoding wt pIX were thermostable (Fig. 3C) (3). The viruses with the central alanine stretch deleted (Δ60-72) and the coiled-coil (Δ100-114) region were as thermostable as wt virus. These results show that the coiled-coil region is essential for self-association but does not mediate pIX-dependent thermostabilization of the viral particle. Similarly, we found that, contrary to previously reported evidence (12), the alanine stretch is dispensable for self-association and induction of the thermostable phenotype.
FIG. 3.
Virus stability assay. For the thermostability assay deletions were made in the conserved alanine stretch (Δ60-72) and in the conserved coiled-coil domain (Δ100-114) of the viral backbone. Viruses were propagated on 911 cells and purified by CsCl gradient centrifugation. (A) The presence of the deletions was tested by PCR with primers that flanked the pIX gene in the virus backbone. PCR on hAd5.LUC, which contains wt pIX, should reveal a band of 569 bp, PCR on hAd5.ΔpIX.LUC viruses should reveal a band of 171 bp, PCR on hAd5.pIXΔ60-72.LUC should reveal a band of 528 bp, and PCR on hAd5.pIXΔ100-114.LUC should reveal a band of 525 bp. As internal control, a PCR on the fiber gene was performed with the same samples. Water was used in both PCRs as a negative control. (B) The incorporation levels of the mutant pIX were assayed by Western blotting. As positive and negative controls, wt Ad5 and Ad5 ΔpIX, respectively, were included. (C) Thermostability of wt and mutant viruses. Aliquots of the vectors were incubated in a water bath at 45°C for 4, 6, 8, or 10 min. Residual infectious virus titers were estimated by determining the capacity of the virus to induce luciferase activity in U2OS cell 24 h after infection. The results are presented as percentages of residual luciferase activity. ○, ▵, +, and *, wt hAd5.LUC, hAd5.ΔpIX.LUC, hAd5.pIXΔ60-72.LUC, and hAd5.pIXΔ100-114.LUC, respectively. Each bar represents the cumulative mean ± standard deviation of triplicate analyses.
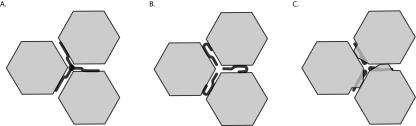
The mechanism by which pIX cements the capsid into a thermostable virion is still unclear. Our data show that pIX self-association is not required. This excludes the model in which three pIX molecules form a trimer that simultaneously binds three different hexon capsomers, thereby stabilizing the complex via the self-associated pIX molecules (Fig. 4A). This model would require pIX to interact with a single hexon capsomer only. Our data demonstrate that pIX stabilizes the capsid independently of self-association. This suggests a model in which the pIX N-terminal domain binds simultaneously to two different hexon capsomers (Fig. 4B). The individual pIX molecules then provide links between neighboring capsomers. Alternatively, a mechanism in which single pIX molecules strongly bind to single hexon capsomers to induce a stability-enhancing conformational change cannot yet be excluded (Fig. 4C). Mapping the interaction domains of hexon and pIX, as well as high-resolution structural analyses of the adenovirus particle, should facilitate the resolution of this issue. Nevertheless, our observation that the self-interaction domain is dispensable for capsid stability, together with the recent observations that pIX only moderately affects replication and transcription of Ads (14), leaves the precise function of the conserved C-terminal domain of pIX enigmatic.
FIG. 4.
Models for pIX-hexon interaction. (A) Three hexon capsomers are kept together via a pIX trimer. In this model a pIX molecule binds to a single hexon capsomer and two other pIX molecules. (B) Three hexon capsomers are kept together via hexon capsomer-pIX-hexon capsomer interactions; there is no multimerization of pIX molecules required. (C) Three hexon capsomers bind more strongly to each other as result of a conformational change induced by pIX.
Acknowledgments
We thank Keith N. Leppard for supplying the anti-pIX serum, members of the FP6 GIANT consortium for the stimulating discussions, and David Baker for critically reading the manuscript.
This work was supported in part by the Technology Foundation STW (program LGN 66.3977).
REFERENCES
- 1.Akalu, A., H. Liebermann, U. Bauer, H. Granzow, and W. Seidel. 1999. The subgenus-specific C-terminal region of protein IX is located on the surface of the adenovirus capsid. J. Virol. 73:6182-6187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caravokyri, C., and K. N. Leppard. 1995. Constitutive episomal expression of polypeptide IX (pIX) in a 293-based cell line complements the deficiency of pIX mutant adenovirus type 5. J. Virol. 69:6627-6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Colby, W. W., and T. Shenk. 1981. Adenovirus type 5 virions can be assembled in vivo in the absence of detectable polypeptide IX. J. Virol. 39:977-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dmitriev, I. P., E. A. Kashentseva, and D. T. Curiel. 2002. Engineering of adenovirus vectors containing heterologous peptide sequences in the C terminus of capsid protein IX. J. Virol. 76:6893-6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fallaux, F. J., O. Kranenburg, S. J. Cramer, A. Houweling, H. van Ormondt, R. C. Hoeben, and A. J. van der Eb. 1996. Characterization of 911: a new helper cell line for the titration and propagation of early region 1-deleted adenoviral vectors. Hum. Gene Ther. 7:215-222. [DOI] [PubMed] [Google Scholar]
- 6.Furcinitti, P. S., J. van Oostrum, and R. M. Burnett. 1989. Adenovirus polypeptide IX revealed as capsid cement by difference images from electron microscopy and crystallography. EMBO J. 8:3563-3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghosh-Choudhury, G., Y. Haj-Ahmad, and F. L. Graham. 1987. Protein IX, a minor component of the human adenovirus capsid, is essential for the packaging of full length genomes. EMBO J. 6:1733-1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He, T. C., S. Zhou, L. T. da Costa, J. Yu, K. W. Kinzler, and B. Vogelstein. 1998. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. USA. 95:2509-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lutz, P., M. Rosa-Calatrava, and C. Kedinger. 1997. The product of the adenovirus intermediate gene IX is a transcriptional activator. J. Virol. 71:5102-5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meulenbroek, R. A., K. L. Sargent, J. Lunde, B. J. Jasmin, and R. J. Parks. 2004. Use of adenovirus protein IX (pIX) to display large polypeptides on the virion—generation of fluorescent virus through the incorporation of pIX-GFP. Mol. Ther. 9:617-624. [DOI] [PubMed] [Google Scholar]
- 11.Rademaker, H. J., M. A. Abou El Hassan, G. A. Versteeg, M. J. Rabelink, and R. C. Hoeben. 2002. Efficient mobilization of E1-deleted adenovirus type 5 vectors by wild-type adenoviruses of other serotypes. J. Gen. Virol. 83:1311-1314. [DOI] [PubMed] [Google Scholar]
- 12.Rosa-Calatrava, M., L. Grave, F. Puvion-Dutilleul, B. Chatton, and C. Kedinger. 2001. Functional analysis of adenovirus protein IX identifies domains involved in capsid stability, transcriptional activity, and nuclear reorganization. J. Virol. 75:7131-7141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosa-Calatrava, M., F. Puvion-Dutilleul, P. Lutz, D. Dreyer, H. de The, B. Chatton, and C. Kedinger. 2003. Adenovirus protein IX sequesters host-cell promyelocytic leukaemia protein and contributes to efficient viral proliferation. EMBO Rep. 4:969-975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sargent, K. L., R. A. Meulenbroek, and R. J. Parks. 2004. Activation of adenoviral gene expression by protein IX is not required for efficient virus replication. J. Virol. 78:5032-5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sargent, K. L., P. Ng, C. Evelegh, F. L. Graham, and R. J. Parks. 2004. Development of a size-restricted pIX-deleted helper virus for amplification of helper-dependent adenovirus vectors. Gene Ther. 11:504-511. [DOI] [PubMed] [Google Scholar]
- 16.Shu, W., H. Ji, and M. Lu. 1999. Trimerization specificity in HIV-1 gp41: analysis with a GCN4 leucine zipper model. Biochemistry 38:5378-5385. [DOI] [PubMed] [Google Scholar]
- 17.Souquere-Besse, S., E. Pichard, O. Filhol, V. Legrand, M. Rosa-Calatrava, A. G. Hovanessian, C. Cochet, and F. Puvion-Dutilleul. 2002. Adenovirus infection targets the cellular protein kinase CK2 and RNA-activated protein kinase (PKR) into viral inclusions of the cell nucleus. Microsc. Res. Tech. 56:465-478. [DOI] [PubMed] [Google Scholar]
- 18.Stewart, P. L., S. D. Fuller, and R. M. Burnett. 1993. Difference imaging of adenovirus: bridging the resolution gap between X-ray crystallography and electron microscopy. EMBO J. 12:2589-2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Oostrum, J., and R. M. Burnett. 1985. Molecular composition of the adenovirus type 2 virion. J. Virol. 56:439-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vellinga, J., M. J. Rabelink, S. J. Cramer, D. J. van den Wollenberg, H. Van der Meulen, K. N. Leppard, F. J. Fallaux, and R. C. Hoeben. 2004. Spacers increase the accessibility of peptide ligands linked to the carboxyl terminus of adenovirus minor capsid protein IX. J. Virol. 78:3470-3479. [DOI] [PMC free article] [PubMed] [Google Scholar]