Abstract
The human cytomegalovirus (HCMV) glycoprotein US11 diverts class I major histocompatibility complex (MHC) heavy chains (HC) from the endoplasmic reticulum (ER) to the cytosol, where HC are subjected to proteasome-mediated degradation. In mouse embryonic fibroblasts that are deficient for X-box binding protein 1 (XBP-1), a key transcription factor in the unfolded protein response (UPR) pathway, we show that degradation of endogenous mouse HC is impaired. Moreover, the rate of US11-mediated degradation of ectopically expressed HLA-A2 is reduced when XBP-1 is absent. In the human astrocytoma cell line U373, turning on expression of US11, but not US2, is sufficient to induce a UPR, as manifested by upregulation of the ER chaperone Bip and by splicing of XBP-1 mRNA. In the presence of dominant-negative versions of XBP-1 and activating transcription factor 6, the kinetics of class I MHC HC degradation were delayed when expression of US11 was turned on. The magnitude of these effects, while reproducible, was modest. Conversely, in cells that stably express high levels of US11, the degradation of HC is not affected by the presence of the dominant negative effectors of the UPR. An infection of human foreskin fibroblasts with human cytomegalovirus induced XBP-1 splicing in a manner that coincides with US11 expression. We conclude that the contribution of the UPR is more pronounced on HC degradation shortly after induction of US11 expression and that US11 is sufficient to induce such a response.
The human cytomegalovirus (HCMV) glycoproteins US2 and US11 specifically target class I major histocompatibility complex (MHC) heavy chains (HC) for dislocation from the endoplasmic reticulum (ER) membrane to the cytosol, where they are degraded by the proteasome. Many similarities exist between the series of events catalyzed by these viral glycoproteins and disposal of misfolded proteins in the ER. Although the exact mechanism by which class I HC are rerouted for dislocation by US2 and US11 is still unknown, studies using mutants of US2, US11, and class I HC show that dislocation by US11 and US2 involves distinct mechanisms. One of these distinctions is the unique role of the US11 transmembrane (TM) domain, which mediates the interaction with the transmembrane protein Derlin-1 (15). This interaction is necessary for class I MHC dislocation, since a single amino acid replacement, Q192L in the TM of US11 (Q192L) abolishes this interaction and, with it, the ability to dislocate class I HC (15, 16).
ER stress is a state in which accumulation of misfolded protein in the ER is not matched with the folding capacity of the ER. Persistent ER stress eventually leads to cell death (12). To prevent intoxication by unfolded proteins, eukaryotes developed at least two mechanisms to deal with such ER stress (3). The unfolded protein response (UPR) is an ER-to-nucleus signaling pathway. The outcome of this signaling ultimately coordinates the protein load in the ER with the folding capacity of the ER (7); concomitantly, terminally misfolded proteins are destined for degradation by dislocation from the ER to the cytoplasm, where proteolysis by the ubiquitin-proteasome machinery occurs (1). Both mechanisms, the UPR and degradation of ER proteins, ensure that only properly folded proteins are transported to their site of action.
In Saccharomyces cerevisiae, the two pathways are interconnected. Target genes of the UPR include those that encode products participating in the degradation of ER proteins (29). Strains with deletions of the UPR genes cannot properly dispose of misfolded ER proteins (2). Inhibition of ER degradation by elimination of E2 and E3 enzymes, which participate in the ubiquitination of misfolded ER proteins, strongly induces the UPR (4). In fact, the genetic link between degradation of ER proteins and the UPR was exploited as a screening tool to isolate alleles of the Sec61 translocon and the Bip chaperone that are protein translocation proficient and protein dislocation deficient, demonstrating biochemical distinctions between translocation into the ER and dislocation from the ER (11, 35).
The mammalian UPR differs in a number of aspects from the yeast UPR. Instead of the single sensor (Ire1p) of the UPR found in yeast, the mammalian UPR includes at least three distinct transducers: PERK, activating transcription factor 6 (ATF6), and IRE1. Upon imposition of ER stress in mammalian cells, PERK is rapidly activated to phosphorylate the α subunit of eukaryotic initiation factor 2 (eIF-2α). Phosphorylation of eIF-2α attenuates translation and thereby reduces the protein load in the secretory system. ATF6, once properly engaged, undergoes regulated intramembrane proteolysis by S1P and S2P proteases to liberate an ATF6 cytosolic fragment from the membrane for transport to the nucleus. ATF6 activates transcription of several target genes, including X-box binding protein 1 (XBP-1). IRE1, in a mechanism similar to that used by Ire1p/Hac1 in yeast, splices XBP-1 mRNA. The spliced XBP-1 mRNA gives rise to a 371-residue polypeptide that comprises the original N-terminal DNA binding domain and an additional transactivation domain in the C terminus (14). The cytosolic fragment of ATF6 and spliced XBP-1 can heterodimerize to form potent activators of UPR target genes (13, 34).
In mammalian cells, physiological links between degradation of ER misfolded proteins and the UPR are becoming apparent. An ER lectin with significant homology to α1,2-mannosidase, called ER degradation-enhancing α-mannosidase-like protein (EDEM), is required for the degradation of misfolded null Hong Kong variant of α1-anti-trypsin (α1-AT NHK). EDEM is strongly induced by the UPR. In fact, mouse embryonic fibroblasts (MEFs) express EDEM under the control of the IRE1α/XBP-1 arm of the mammalian UPR. Therefore, expression of EDEM in MEFs is thus far the only example of a link between the UPR and the ER degradation machinery in mammalian cells (33). Mere expression of murine HC in yeast results in the presence of the misfolded HC in the yeast ER. While its degradation is Ire1 dependent, the misfolded HC itself induces the UPR only weakly. Interestingly, RNA interference-mediated attenuation in the expression of the Caenorhabditis elegans homologue of Derlin-1, the newly identified protein implicated in dislocation, activates the UPR. This observation suggests a possible link between US11-mediated dislocation and the UPR (32). Therefore, the link between the UPR and protein degradation from the ER deserves closer scrutiny, especially with mammalian cells.
Here, we explore the role of the UPR in the US11-mediated degradation of class I HC. We show that MEFs deficient in XBP-1 support the degradation of HC, but that they do so with reduced efficiency compared to wild-type (wt) MEFs. Overall, US11-mediated degradation of HC is less sensitive to modulation of the UPR than is degradation of α1-AT NHK. We further demonstrate that US11, but not the Q192L mutant, is by itself an inducer of the UPR. Upon HCMV infection, UPR is induced in a manner that coincides with US11 expression. We propose that this trait facilitates the dislocation of HC early after viral infection, when US11 levels may be limiting.
MATERIALS AND METHODS
Cell lines, antibodies and chemicals.
U373-MG astrocytoma cells transfected with US11 have been described previously (23). All astrocytoma cell lines were cultured in Dulbecco's modified Eagle's medium (DMEM) as described previously (27). Tet-On systems (Clontech, Palo Alto, Calif.) for US2 and US11 in the U373 cell lines were constructed according to the manufacturer's guidelines. Single-cell clones were screened by reduction of HLA-A surface expression upon doxycycline (DOX) treatment, as measured by fluorescence-activated cell sorter by using W6/32 monoclonal antibodies. XBP-1−/− MEFs have been described previously (13). Human foreskin fibroblasts (HFF) were cultures in DMEM supplemented with 10% fetal bovine serum, HEPES, and antibiotics. The antibodies used in this study have been described previously (19, 28). Goat anti-human α1-antitrypsin was purchased from ICN Biomedicals. Polyclonal goat anti-β-actin was purchased from Santa Cruz Biotechnology. Mouse monoclonal anti-KDEL (Stressgen, Victoria, Canada) was used to immunoprecipitate Bip.
Dual-luciferase assay.
U373 cells were transfected by using FuGene6 reagent (Roche) according to the manufacturer's instructions. After transfection, cells were incubated for 2 days in the presence or absence of doxycycline (1 μg/ml). For each transfection, 4 μg of CMV-driven ATF6-dominant negative (DN) (30) or XBP-1-DN (13) were cotransfected with 1 μg of 5× ATF6GL3 and 100 ng of the Renilla luciferase reporter pRL-TK as an internal control. Plasmid pcDNA3.1 was used to bring the total amount of DNA to 5 μg for untreated controls. Where indicated, tunicamycin (1 μg/ml) was added in the last 12 h of incubation. Cells were then lysed and assayed for firefly and Renilla luciferase activities by using a Dual-Luciferase Assay kit (Promega, Madison, Wis.). The data shown are the averages from four independent experiments ± standard deviations.
Plasmid construction, transient transfection, and retrovirus production.
XBP-1-DN and ATF6-DN were cloned into the pMiG murine stem cell virus vector harboring an internal ribosome entry site-green fluorescent protein (IRES-GFP) element to sort infected cells. Viral particles were made in 293T cells by a triple transfection of the retroviral vector (2 μg), pMD-gag-pol (2 μg), and pVSV-G (2 μg) by using Effectene (QIAGEN). Cells were infected as previously described (16). MEFs were transfected by Effectene (QIAGEN) and U373 cells were transfected by FuGene6 (Roche) according to the corresponding manufacturer's specifications.
Reverse transcriptase (RT)-PCR analysis.
Total RNA was isolated by using TRIzol (Gibco-BRL, Carlsbad, Calif.). RNAs were used for first-strand synthesis with Superscript reverse transcriptase (Invitrogen, Carlsbad, Calif.). PCR primers 5′-ACACGCTTGGGAATGGACAC-3′ and 5′-CCATGGGAAGATGTTATGGG-3′, encompassing the missing sequences in XBP-1, were used for the PCR amplification with Platinum PCR Supermix (Invitrogen). Cycling conditions were as follows: 95°C for 3 min and 58°C for 40 s, 35 cycles of 72°C for 45 s, and 95°C for 45 s. A PCR for GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was performed to validate cDNA synthesis. We separated PCR products by 11% polyacrylamide gel electrophoresis (PAGE) gel and visualized them by ethidium bromide staining.
Metabolic labeling, pulse-chase analysis, and immunoprecipitation.
Cells were detached by trypsin treatment, followed by starvation in methionine- and cysteine-free DMEM for 1 h. Cells were metabolically labeled with 500 μCi/ml of [35S]methionine-cysteine (1,200 Ci/mmol; PerkinElmer Life Sciences, Boston, Mass.) at 37°C for the times indicated. Pulse-chase experiments, cell lysis, and immunoprecipitation were performed as described previously (23). The immunoprecipitates were analyzed by SDS-PAGE followed by fluorography. Densitometry was performed by phosphorimager by using ImageQuant 1.0 software (Molecular Dynamics, Sunnyvale, Calif.).
HCMV infection.
The HCMV strain AD169 was obtained from the American Type Culture Collection (Rockville, Md.). HFF cells were seeded onto 162-cm2 flasks and allowed to grow to 80% confluency. Cells were infected with purified HCMV at a multiplicity of infection (MOI) of 10 in 8 ml of the growth medium. Cells were incubated at 37°C for 2 h and virus-containing medium was replaced with fresh growth medium. The time course was initiated by the addition of the fresh medium. At the indicated times, adherent cells were harvested by trypsin treatment. Ten percent of the cells were used to prepare total cell extract for Western blot analysis, and RNA was extracted by TRIzol from the remaining sample.
RESULTS
Stability of class I MHC HC is enhanced in XBP-1−/− cells.
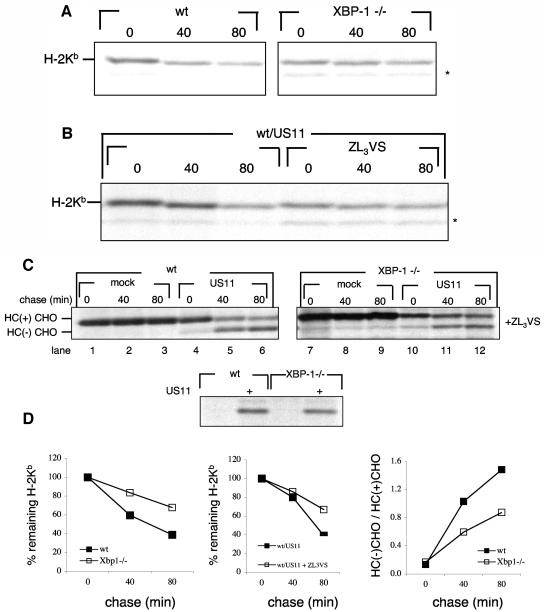
To study the role of UPR in US11-mediated degradation of class I MHC HC, we compared the stability of endogenous HC in MEFs devoid of XBP-1 to that in wt MEF cells. Cells were pulse-labeled for 20 min and chased at 37°C. Endogenous class I MHC (H-2Kb) was retrieved by immunoprecipitation with an antiserum raised against the cytoplasmic tail of class I MHC. Regardless of the presence of US11, class I MHC is unstable in wt MEFs. However, in XBP-1−/− cells, H-2Kb is more stable (Fig. 1A and D, left panel) suggesting the involvement of the IRE1α/XBP-1 pathway in its degradation. Introduction of US11 to wt MEFs did not result in accumulation of deglycosylated H-2Kb heavy chains when cells were chased in the presence of a proteasome inhibitor, although H-2Kb heavy chains were stabilized (Fig. 1B and D, middle panel). Moreover, US11 did not enhance the degradation of H-2Kb compared to mock transfectants in either wt or XBP-1−/− MEFs (data not shown). We conducted similar experiments at 25°C. At this temperature, class I MHC products are stable regardless of peptide loading (17) and US11 retains its capacity to exert its function (31; B. Tirosh and H. Ploegh, unpublished observation). When cells were chased at room temperature, class I MHC products were markedly stabilized, suggesting that a defect in peptide loading may well account for the instability of HC measured at 37°C (data not shown). At reduced temperatures, the expression of US11 did not significantly affect the decay of H-2Kb, and we did not observe a deglycosylated intermediate of the endogenous HC in the presence of a proteasome inhibitor at 37°C or at 25°C (Fig. 1B and results not shown). This is most likely due to the reduced capacity of US11 to interact with mouse class I alleles compared to the interaction of US11 with human HLA class I molecules (18). Therefore, to investigate US11-mediated degradation of class I HC, we transfected HLA-A2 either alone or in conjunction with US11 into XBP-1−/− and wt cells. In this situation, US11-mediated degradation of HC displays the characteristics normally seen in human cells, in which the glycosylated HC is converted to a deglycosylated intermediate when proteasomal activity is blocked (Fig. 1C, lanes 1 to 3 versus lanes 4 to 6) (31). To compare the rate of dislocation between the cell lines, we determined the ratio of the deglycosylated HC intermediate to the glycosylated HC precursor. The higher this ratio, the more extensive is the dislocation of HC. Although dislocation of HLA-A2 was clearly seen with XBP-1−/− cells (Fig. 1C, lanes 10 to 12) its rate was slow in comparison to wt cells (Fig. 1D, right panel) despite comparable levels of US11 expression (Fig. 1C, lower panel). Interestingly, when US2 was introduced to wt or XBP-1−/− MEFs in conjunction with HLA-A2, the latter was not dislocated (data not shown). The lack of US2 activity in the MEFs is most likely attributable to the exclusive interaction of US2 with correctly folded, peptide-loaded class I MHC (5, 6). Peptide loading in MEFs appears to be defective. Therefore, MEFs are ill-suited to study the role of XBP-1 in US2-mediated dislocation. We conclude that maximal rates of dislocation require an intact UPR. The diminished capacity of XBP-1−/− cells to degrade endogenous H-2Kb, also in the absence of US11, would suggest a defect in the degradation machinery of type I transmembrane glycoproteins rather than in a US11-specific degradation pathway.
FIG. 1.
Class I MHC heavy chains are more stable in XBP-1−/− cells. (A) wt and XBP-1−/− MEF cells were pulse-labeled with [35S]methionine for 20 min and chased at 37°C up to 80 min. Cells were lysed in 1% SDS, and the lysate was then diluted to 0.07% SDS with NP-40 lysis mix followed by immunoprecipitation with anti-mouse H-2K serum (P8 antibody). The immunoprecipitates were analyzed by SDS-PAGE (12%). The background band is marked with an asterisk. (B) wt MEFs were transfected with US11 encoding pcDNA3, pulse-labeled with [35S]methionine for 20 min, and chased at 37°C up to 80 min in the presence or absence of the proteasome inhibitor ZL3VS. Cells were lysed in 1% SDS, and the lysate was then diluted to 0.07% SDS with NP-40 lysis mix followed by immunoprecipitation with anti-mouse H-2K serum (P8 antibody). The immunoprecipitates were analyzed by SDS-PAGE (12%). The background band is marked with an asterisk. (C) MEFs were transfected as described in Materials and Methods with an HLA-A2 encoding pcDNA3 with or without US11 encoding pcDNA3. Cells were pulse-labeled with [35S]methionine for 20 min and chased up to 80 min at 37°C in the presence of the proteasome inhibitor ZL3VS. Cells were lysed in 1% SDS, and the lysate was then diluted to 0.07% SDS with NP-40 lysis mix followed by immunoprecipitation with anti-class I heavy chain serum (αHC). US11 was sequentially immunoprecipitated from the zero time chase point (lower panel). Immunoprecipitates were analyzed by SDS-PAGE (12%). (D) Gels were quantified by phosphoimager.
US11-mediated degradation of HC is not affected by modulation of UPR in U373 cells.
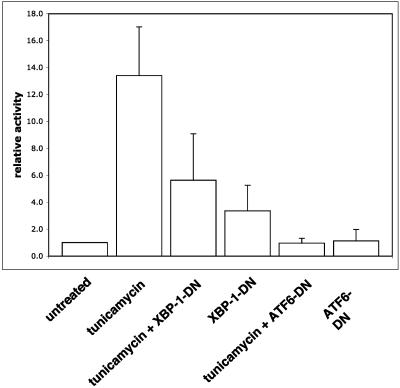
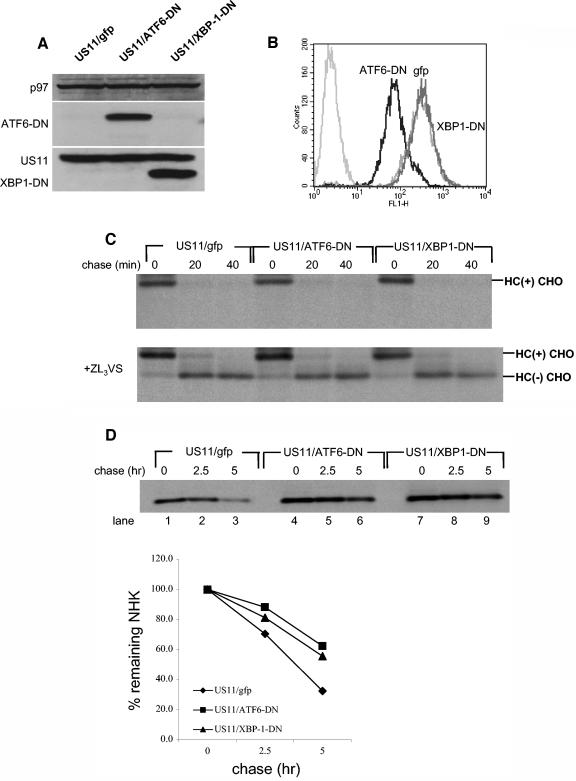
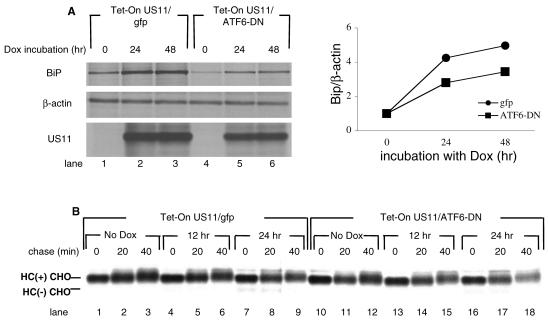
We used a dominant negative ATF6 construct (30) and a dominant negative XBP-1 construct (13) to further explore the influence of the UPR on the degradation of class I HC. To verify the dominant negative effect of the constructs used in U373 cells, we performed a dual-luciferase assay, using a luciferase reporter whose expression is under the control of a UPR responsive element (UPRE). Treatment with tunicamycin strongly activates the UPR, as seen by the induction in luciferase activity (Fig. 2). Both XBP-1-DN and ATF6-DN significantly attenuated the induction of the UPRE-luciferase construct by tunicamycin treatment. ATF6-DN was more potent than XBP-1-DN, reducing activity of the reporter to background levels (Fig. 2). However, when the dominant negative constructs were transfected into U373 cells without tunicamycin stimulation, we saw a moderate induction of the reporter by XBP-1-DN but not by ATF6-DN (Fig. 2). This effect is likely due to the fact that the XBP-1-DN construct maintains its DNA binding site. At high levels of expression, it can dimerize with endogenous transcription factors and direct them to the UPRE reporter. On the other hand, ATF6-DN harbors a mutation that prevents its DNA binding. Therefore, even at high levels, ATF6-DN cannot induce transcription. Regardless, we used both constructs in the remainder of our experiments. Due to their low transfection efficiency (20 to 30%), U373 cells were not suitable for measurement of the effect of the dominant negative constructs on US11-mediated degradation of HC by transient transfection. Therefore, we made cell lines that stably express ATF6-DN and XBP-1-DN. ATF6-DN and XBP-1-DN were cloned into the pMig murine stem cell virus retroviral construct, which contains an IRES-GFP element. U373 cells stably expressing US11 were infected with the retroviruses and GFP-positive cells were obtained by sorting. Expression of the dominant negative proteins was confirmed by Western blotting (Fig. 3A). GFP expression in ATF6-DN-expressing cells was significantly lower than in cell lines expressing either an empty vector or XBP-1-DN (Fig. 3B). ATF6-DN, which is highly expressed, may be toxic, perhaps due to its greater potency as a dominant negative effector. We compared the rate of US11-mediated degradation of HC in the three cell lines. Cells were pulse-labeled for 10 min and chased in 20 min intervals. Under these conditions, a complete conversion of the glycosylated class I MHC precursor to the deglycosylated intermediate is expected to be seen at the 20-min chase point. Kinetics of decay of HC were superimposable for the three cell lines (Fig. 3C); hence, XBP-1-DN and ATF6-DN were without effect on dislocation of class I HC in U373 cells.
FIG. 2.
Expression of XBP-1 and ATF6 dominant negatives attenuates tunicamycin-induced UPR. U373 cells were transfected as described in Materials and Methods with plasmids encoding firefly luciferase under the UPRE promoter and Renilla luciferase under thymidine kinase promoter. Plasmids encoding XBP-1-DN or ATF6-DN were cotransfected as indicated. Twenty-four hours after transfection, 1 μg of tunicamycin/ml was added for overnight treatment. Cells were harvested the next day and the dual-luciferase assay was performed. The results are averages from four independent experiments ± standard deviations.
FIG. 3.
US11-mediated degradation of HC is not affected by modulation of UPR in U373 cells. (A) Total cell lysates of U373 cells stably expressing XBP-1-DN-IRES-GFP, ATF6-DN-IRES-GFP, or an empty IRES-GFP construct were resolved by SDS-PAGE, and protein expression was examined by Western blotting. p97 was used as a loading control. (B) GFP levels were measured by flow cytometry. Untransfected U373 cells were measured as background. (C) U373 cell lines were pulse-labeled with [35S]methionine for 10 min and chased up to 40 min in the absence (upper panel) or presence (lower panel) of the proteasome inhibitor ZL3VS. Cells were lysed in 1% SDS, and the lysate was then diluted to 0.07% SDS with NP-40 lysis mix, followed by immunoprecipitation with αHC and analyzed by SDS-PAGE (12%). (D) U373 cell lines were transfected with pcDNA3 encoding the α1-AT NHK. Forty-eight hours after transfection, cells were pulse-labeled with [35S]methionine for 20 min and chased up to 5 h. Cells were lysed in NP-40 lysis buffer followed by immunoprecipitation with anti- α1-AT, and the immunoprecipitates were analyzed by SDS-PAGE (12%). Autoradiograms were quantified by phosphoimager.
The lack of sensitivity of US11-mediated degradation of HC to modulation of the UPR by the dominant negative constructs may be due to insufficient expression levels of the dominant negatives or it could be that US11-mediated degradation is simply not sensitive to UPR modulation. To distinguish between those two possibilities, we transiently transfected the same cells with α1-AT NHK, the degradation of which utilizes the Ire1/XBP-1 pathway (33). Stability of α1-AT NHK was measured by pulse-chase analysis, which was followed by immunoprecipitation and SDS-PAGE. α1-AT NHK was more stable in cells expressing either XBP-1-DN or ATF6-DN than in untransfected controls (Fig. 3D), suggesting that the dominant-negative proteins indeed attenuate the UPR and thereby affect the stability of α1-AT NHK, consistent with the published data (13, 33). We conclude that US11-mediated degradation of HC is less sensitive to modulation of the UPR than is degradation of α1-AT NHK.
Expression of US11, but not US2, triggers UPR.
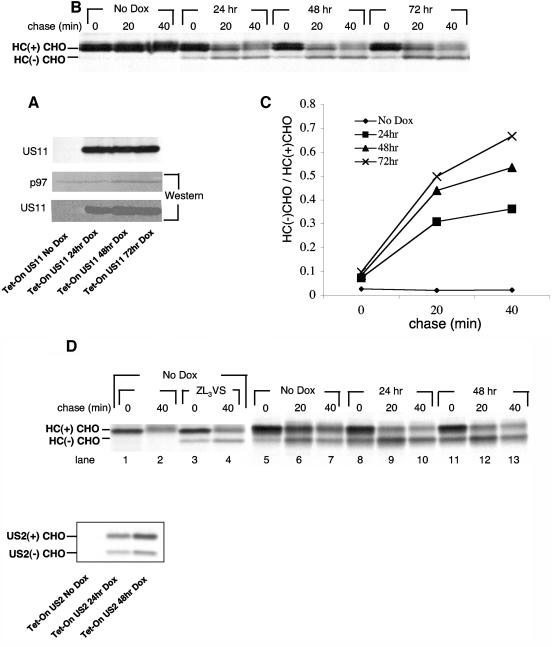
If an active UPR indeed facilitates the degradation of class I HC, as inferred from results obtained with the XBP-1−/− cells, what induces the UPR when cells express US11? We considered the possibility that the expression of US11 by itself is sufficient to induce a UPR. We generated a tetracycline-inducible (Tet-On) expression system for US11 and control proteins in U373 cells. We monitored the rate of dislocation of class I HC in the Tet-On US11 cells following induction with DOX. In parallel, we compared the levels of US11 expression by immunoblotting. US11 expression reaches maximum levels after 48 h of DOX treatment (Fig. 4A), but dislocation of HC further increases from 48 to 72 h after treatment with DOX as deduced from the relative increase in the dislocation intermediate (Fig. 4B, quantification in Fig. 4C). We suggest that somehow adaptation to expression of US11 is responsible for the enhancement of the dislocation reaction, even at comparable levels of US11 expression. Such a mechanism might involve the UPR. Of note, steady-state levels of US11 in the Tet-On cells were approximately 80% of the levels seen with the U373 transfectants that express US11 constitutively. Despite this level of US11, a considerable portion of the HC was spared from dislocation.
FIG. 4.
MHC class I HC dislocation rate increases after US11 reaches steady state. (A) Tet-On US11 cells were incubated in the presence of DOX (1 μg/ml) for the indicated times. Cells were then harvested and divided into two portions. One-half of the samples were lysed, and US11 protein expression was examined by Western blotting. p97 was used as a loading control. (B) The second half of the samples were pulse-labeled with [35S]methionine for 10 min and chased for 40 min in the presence of the proteasome inhibitor ZL3VS. Cells were lysed in 1% SDS, and the lysate was then diluted to 0.07% SDS with NP-40 lysis mix followed by immunoprecipitation with αHC and analysis by SDS-12% PAGE. US11 was sequentially immunoprecipitated from the zero time point and similarly analyzed (A, upper panel). (C) Autoradiograms were quantified as mentioned above, and the deglycosylated HC to glycosylated HC ratio was then calculated. (D) Left, Tet-On US2 cells were incubated in the absence of DOX. Cells were pulse-labeled with [35S]methionine for 10 min and chased for 40 min in the presence or absence of the proteasome inhibitor ZL3VS. HC was immunoprecipitated as described for panel B. Immunoprecipitates were analyzed by SDS-PAGE (12%). Right, Tet-On US2 cells were incubated in the presence of DOX (1 μg/ml) for the indicated times. Cells were pulse-labeled with [35S]methionine for 10 min and chased for 40 min in the presence of the proteasome inhibitor ZL3VS. HC was immunoprecipitated as described for panel B. Lower panel, US2 was immunoprecipitated sequentially from the zero time chase point (right panel, lanes 5, 8, and 11). Immunoprecipitates were analyzed by SDS-PAGE (12%).
We also established a Tet-On expression system of US2. We analyzed several single cell clones and observed that every single clone showed a significant rate of dislocation, even in the absence of added DOX, as indicated by the appearance of the deglycosylated intermediate when cells were chased in the presence of ZL3VS (Fig. 4D, left panel, lane 2 versus lane 4). This finding suggests that unlike the situation for US11, the US2 promoter is leaky in the Tet-On cells. When US2 expression was measured in a time-course experiment following DOX addition, as performed for US11 (Fig. 4A), very little if any US2 was observed in the absence of DOX (Fig. 4D, lower panel, left lane), even though we observed robust dislocation of class I HC (Fig. 4D, right panel, lanes 5 to 7). These results suggest that US2 efficiently dislocates class I HC even when present in minute quantities. Since US2 itself undergoes rapid degradation (15), it is unlikely that accumulation of US2 in the ER is required for optimal activity.
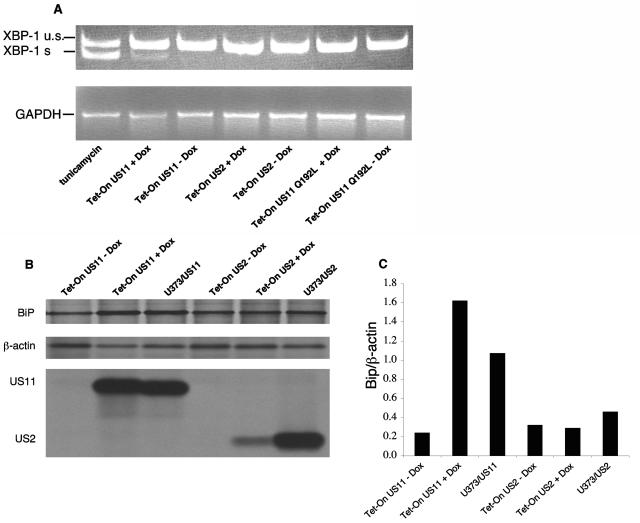
Next, we tested whether US11 expression also provokes a UPR. As an indication for successful induction of the UPR, we measured the splicing of XBP-1 by RT-PCR and the upregulation of the ER chaperone Bip. At first, we compared the effects of US11 induction to that of US2. US2 is an HCMV-encoded glycoprotein capable of catalyzing class I HC dislocation (26, 31), but it does so by a mechanism distinct from US11 (15). Induction of US11 expression resulted in splicing of XBP-1, as seen by the appearance of the expected 119-bp product, although clearly less than that induced by tunicamycin treatment (Fig. 5A). Neither induction of US2 nor of the inactive US11 Q192L mutant resulted in XBP-1 splicing. We compared the rate of Bip synthesis after induction of US11 or US2 in the Tet-On cells, and compared it also to the rate of Bip synthesis in cells that express US11 or US2 constitutively. For comparison, β-actin was immunoprecipitated as a control from carefully calibrated amounts of input radiolabeled lysate. The rate of synthesis of Bip was then estimated by calculating the Bip β-actin ratio. Induction of US11 significantly elevated the rate of synthesis of Bip (three- to fivefold), and this rate was maintained in cells that express US11 constitutively. Conversely, Bip synthesis is not affected by expression of US2, either inducibly or constitutively (Fig. 5B). Since US11 or US2 show comparable dislocation of HC, even though US2 induction was less efficient than US11 induction in the Tet-On system (Fig. 4 and 5), the induction of the UPR may be a trait more typical of US11, as opposed to being simply a response to ongoing dislocation of HC.
FIG. 5.
Expression of US11 triggers the UPR. (A) Tet-On cells were incubated for 48 h in the presence or absence of DOX (1 μg/ml). Total RNA was extracted, and XBP-1 splicing was analyzed by RT-PCR. PCR products were analyzed by 11% PAGE and visualized by ethidium bromide staining. (B) Tet-On cells and U373 cells that express US2 or US11 constitutively were pulse-labeled with [35S]methionine for 20 min. Cells were lysed in 1% SDS, and the lysate was then diluted to 0.07% SDS with NP-40 lysis mix. Samples were divided into three parts. One portion was immunoprecipitated with αKDEL monoclonal antibodies, the second portion was immunoprecipitated with anti-β-actin, and the third portion was immunoprecipitated with anti-US11 or anti-US2 antibodies. Loading was normalized to equal the number of trichloroacetic acid (TCA)-precipitable counts. (C) Autoradiograms were quantitated as mentioned above, and the ratio of Bip to β-actin was calculated.
The membrane-spanning domain of US11 mediates the induction of UPR.
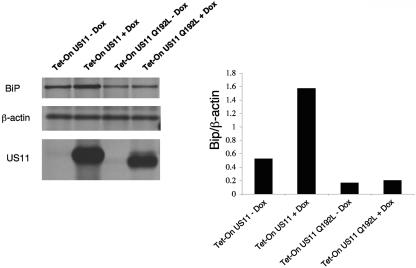
US11 induces the UPR either by its mere presence in the ER or through a mechanism that involves interaction with the dislocation machinery. To distinguish between these two mechanisms, we took advantage of the US11 Q192L mutant. This single amino acid mutation is located within the transmembrane domain of US11 and abolishes entirely its ability to dislocate class I MHC molecules (16). The phenotype of this mutant is explained by the specific interaction with the transmembrane protein Derlin-1, an essential factor in the US11 dislocation machinery (15). Unlike wt US11, the Q192L mutant of US11 failed to induce splicing of XBP-1 (Fig. 5A) and did not induce Bip synthesis (Fig. 6). We conclude that the induction of the UPR by US11 is not a response of the ER to the overexpression of US11 per se, but rather somehow depends on the interaction of US11 with Derlin-1 within the lipid bilayer of the ER.
FIG. 6.
The membrane-spanning domain of US11 mediates the induction of UPR. Tet-On cells for US11 or US11 Q192L mutant were incubated for 48 h in the presence or absence of DOX (1 μg/ml). Cells were pulse-labeled with [35S]methionine for 20 min. Cells were lysed in 1% SDS, and the lysate was then diluted to 0.07% SDS with NP-40 lysis buffer. Samples were divided into three parts. One portion was immunoprecipitated with αKDEL monoclonal antibodies, the second portion was immunoprecipitated with anti-β-actin, and the third portion was immunoprecipitated with anti-US11. Loading was normalized to equal the number of TCA-precipitable counts. (C) Autoradiograms were quantitated as mentioned above, and the ratio of Bip to β-actin was calculated.
Interference with the UPR attenuates the induction of Bip and decreases the dislocation of HC soon after turning on US11 expression:
To investigate whether the induction of the UPR by US11 plays a role in the dislocation of HC, we stably expressed ATF6-DN in the Tet-On US11 cells, using the same retroviral construct as used above. An empty IRES-GFP construct was used as the negative control. Cells were pulse-labeled for 20 min, and Bip synthesis was assayed as before. In the absence of US11, Bip synthesis was attenuated by the presence of ATF6-DN (Fig. 7A, lane 4 versus lane 1). As expected, the expression of US11 in the presence of DOX induced the synthesis of Bip (Fig. 7A, lanes 2 to 3 versus lane 1). In the presence of ATF6-DN, however, US11 was less effective at inducing Bip (Fig. 7A, lanes 5 and 6 versus lane 4). Although the induction of Bip synthesis was seen in both cell lines, the magnitude of Bip synthesis in response to US11 expression was reduced in the presence of ATF6-DN.
FIG. 7.
Interference with the UPR attenuates the induction of Bip and decreases the dislocation of HC soon after turning on US11 expression. (A) Tet-On cells for US11 expressing either ATF6-DN or empty vector as the control were incubated for the indicated times in the presence or absence of DOX (1 μg/ml). Cells were pulse-labeled with [35S]methionine for 20 min. Cells were lysed in 1% SDS, and the lysate was then diluted to 0.07% SDS with NP-40 lysis buffer. Samples were divided into three parts. One portion was immunoprecipitated with αKDEL monoclonal antibodies, the second portion was immunoprecipitated with anti-β-actin, and the third portion was immunoprecipitated with anti-US11. Loading was normalized to equal the number of TCA-precipitable counts. Autoradiograms were quantitated as mentioned above, and the ratio of Bip to β-actin was calculated. (B) Cells were incubated for the indicated times with DOX. Cells were then harvested and pulse-labeled with [35S]methionine for 10 min and chased for 40 min in the presence of the proteasome inhibitor ZL3VS. Cells were lysed in 1% SDS, and the lysate was then diluted to 0.07% SDS with NP-40 lysis mix, followed by immunoprecipitation with αHC and analyzed by SDS-PAGE (12%).
Because cells that constitutively express US11 were insensitive to the presence of ATF6-DN or XBP-1-DN, we examined whether ATF6-DN or XBP-1-DN modulates the onset of dislocation after US11 expression is induced. However, soon after the addition of DOX, cells that express ATF6-DN show only a modest reduction, if any, in the levels of the deglycosylated intermediate compared with levels for controls (Fig. 7B, lanes 13 to 15 versus lanes 4 to 6). We repeated this experiment in cells expressing XBP-1-DN with similar results.
XBP-1 splicing coincides with the expression of US11 upon HCMV infection.
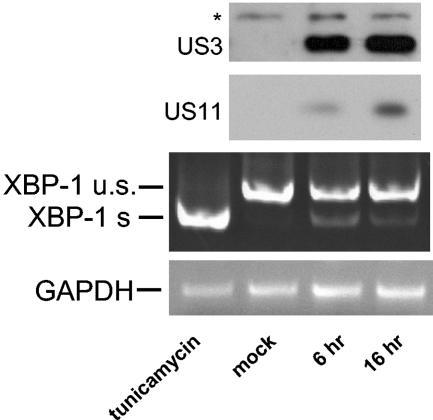
We examined whether induction of the UPR, as manifested by XBP-1 splicing, occurs in the context of HCMV infection. HFF cells were infected with AD169 HCMV at an MOI of 10. Six hours after virus removal (8 h after initial exposure to the virus), XBP-1 splicing was readily observed by RT-PCR (Fig. 8). US3 (IE gene) and US11 (E gene) expression were assessed by immunoblotting. US11 expression was detectable and increased with time. We conclude that XBP-1 splicing accompanies US11 expression in the context of HCMV infection.
FIG. 8.
UPR induction coincides with US11 expression upon HCMV infection. HFF cells were grown to 80% confluency and infected with HCMV at an MOI of 10. At the indicated times, cells were harvested. Ninety percent of the cells were used for RNA extraction and RT-PCR analysis of XBP-1 splicing as described for Fig. 5. The remainder of the samples were lysed in 1% SDS. Forty micrograms of total cell lysate was analyzed by Western blotting for the presence of US11 and US3.
DISCUSSION
To investigate the role of the UPR in the degradation of class I HC, a type I transmembrane glycoprotein, we initially measured the stability of endogenous HC in wt and XBP-1−/− MEFs. The rate of degradation of HC was slower in XBP-1−/− cells (Fig. 1A). Regardless of XBP-1 status, the presence of US11 did not further accelerate HC degradation (data not shown). In MEFs class I HC decayed rapidly, a process inhibited at reduced temperature and by inclusion of proteasome inhibitors (Fig. 1B and results not shown). These experiments suggest that for efficient HC degradation, whether mediated by US11 or caused by inefficient peptide loading onto class I HC, the IRE1/XBP-1 pathway is required in MEFs. To test this hypothesis in conditions where interactions of US11 and HC are optimal, HLA-A2 was coexpressed with US11 in MEFs. Again, degradation of HLA-A2 was faster in the wild-type cells than in XBP-1−/− cells (Fig. 1C), supporting the pattern seen for endogenous HC. In MEFs, the generation of properly spliced XBP-1 controls the expression of EDEM, a mannosidase I-like lectin that facilitates the degradation of several misfolded ER proteins (20, 22). In the absence of XBP-1, EDEM mRNA is not detected and is not induced by tunicamycin treatment (13). Since US11 accelerates the degradation of even nonglycosylated HC in tunicamycin-treated cells (31) and since US11 accelerates the degradation of HLA-A2 in XBP-1−/− MEFs, where EDEM expression is not seen, EDEM is most likely dispensable for US11-mediated degradation of class I MHC. We therefore conclude that either the absence of other targets of XBP-1 (13), or alterations in the levels of an assortment of ER proteins result in inhibition of HC degradation. Taken together, these findings indicate that it is clear that XBP-1 is not necessary for degradation of class I HC in MEFs, but an intact UPR pathway facilitates degradation of class I HC when misfolded or when degradation is assisted by US11.
The pathway for degradation of misfolded ER proteins in yeast cells, namely dislocation from the ER to the cytosol, ubiquitination and proteasomal degradation, is saturable. Two alternative routes for degradation were described in conditions of overexpression of misfolded proteins, combined with genetic manipulation of the classical pathways for protein degradation. The excess of misfolded proteins may be directed to an ER-to-vacuole pathway (25). Alternatively, misfolded proteins that escape the ER travel to the Golgi apparatus, from which they are dislocated to the cytosol, ubiquitinated by Rsp5p, and disposed of by proteasomal degradation (9). Both of these alternative pathways are regulated by the UPR. These findings implicate the UPR in yeast as an element that controls protein degradation at several points along the secretory pathway.
Unlike yeast, mammalian cells developed different mechanisms to cope with an excess of misfolded proteins in the ER. An early response, through the activation of PERK, attenuates translation and immediately alleviates the load of newly synthesized proteins that enter the secretory pathway (8). Should stress in the ER persist, activation of caspase 12 and downstream effectors may result in apoptosis (21, 24). Both attenuation of translation and control of apoptosis involve activation of NF-κB (10), suggesting that cell viability is carefully balanced by pro- and anti-apoptotic signals in response to ER stress.
We investigated whether attenuation of the UPR in cells that engage in US11- and US2-mediated degradation of class I MHC HC would improve the stability of HC. We used two constructs, ATF6-DN and XBP-1-DN, previously shown to have a strong dominant negative effect on the UPR (13, 30). By using a UPRE reporter assay, we confirmed that the two constructs indeed behave as dominant negatives in the U373 cell line (Fig. 2). Due to the low efficiency of transient transfection in U373 cells, we were obliged to establish cell lines stably expressing either ATF6-DN or XBP-1-DN. Overexpression of ATF6-DN apparently compromises cell viability (Fig. 3B) and limits the useful range over which ATF6-DN can be used to modulate the UPR. US11-mediated HC degradation was not affected by stably expressing either ATF6-DN or XBP-1-DN (Fig. 3C). As a functional readout for the manipulation of the UPR, we monitored the degradation of the NHK variant of α1-antitrypsin, reported to be sensitive to the IRE1/XBP-1 pathway. We saw that NHK degradation was slowed down by the expression of ATF6-DN or XBP-1-DN, confirming the dominant negative effect of the constructs. However, degradation of NHK was not blocked completely (Fig. 3D), as seen also in Ire1α−/− cells (33). These findings suggest that the UPR is attenuated, not blocked, by ATF6-DN or XBP-1-DN. We conclude that US11-mediated HC degradation is less sensitive to UPR modulation than is the degradation of α1-AT NHK. Different degradation pathways must operate for the two substrates.
If indeed optimal US11-mediated HC degradation relies on activation of the UPR, perhaps US11 itself might provide the necessary stimulus. We constructed a Tet-On system in U373 cells to measure changes diagnostic of induction of a UPR that occur when US11 expression is turned on. Initially, we carefully correlated the HC dislocation reaction to US11 expression. Interestingly, while US11 expression reached steady-state levels after 48 h of incubation in the presence of DOX, the rate of dislocation of HC did not reach a plateau until 72 h of DOX treatment (Fig. 4). For a given level of US11 expression, the cells could still modulate the rate of dislocation. This observation might reflect a process by which the cells gradually adapt the ER degradation machinery to the presence of US11, so as to optimize the rate of HC degradation. One such mechanism might be the UPR.
US2, on the other hand, induced HC dislocation even in the absence of DOX, conditions under which US2 expression is barely detectable (Fig. 4D). Unlike the US11 protein, of which a critical amount must accumulate in the ER to cause efficient HC dislocation, US2 mediates dislocation of HC even when present in low quantities. Since US2 itself is targeted for dislocation and degradation with a half-life of minutes, stress conditions in the ER are unlikely to assist US2-mediated degradation of HC.
We assessed the induction of a UPR as a consequence of US11 expression by measuring XBP-1 mRNA splicing and by examining the upregulation of ER chaperones. In contrast to the expression of US2, that of US11 induced a UPR (Fig. 5). Again, the difference between US11 and US2 might mirror the rather different half-lives of the viral proteins themselves. While US2 is a short-lived protein, US11 is more stable. Therefore, induction of ER stress seen for US11 and not for US2 might reflect the accumulation of US11 in the ER membrane, rather than a mechanism unique to US11. The high concentration of US11 in the ER might stress the ER folding machinery regardless of the function of the protein. It is unclear what exactly constitutes a high concentration of a misfolded protein, and obviously this value might well differ for different ER-resident proteins. To link the generation of US11 to induction of the UPR, we used the Q192L mutant of US11, which localizes to the ER like wild-type US11 and shows a similarly long half-life (16). The difference between the Q192L US11 and wild-type US11 protein is a single amino acid in the TM domain. This residue mediates interaction of US11 with Derlin-1, which is a constituent of the dislocation machinery. In the absence of US11-Derlin-1 interaction, class I heavy chains are more stable (16). The Q192L mutant did not induce the UPR (Fig. 6), and therefore the luminal domain of US11 is unlikely to account for the induction of the UPR. We induced UPR in cells stably expressing the US11 Q192L mutant by the expression of either spliced XBP-1, the 1-373 cytosolic fragment of wild-type ATF6, or by treatment with tunicamycin. None of these treatments restore HC dislocation (data not shown). Therefore, the interaction of the US11 TM domain with the dislocation machinery does not require the UPR. Supported by the finding that reduction in Derlin-1 levels induces the UPR (32), the determination of the mechanism by which US11-Derlin-1 interaction induces the UPR would be of considerable interest. Does Derlin-1 interact directly with the UPR machinery? Does the sequestration of Derlin-1 by wt US11 and not by the Q192L US11 mutant evoke the UPR indirectly, for example, by impairing the degradation of endogenous ER misfolded proteins? Finally, US11 might use a distinct, as yet undiscovered, UPR transducer that is sensitive to interactions within the ER membrane. The existence of additional transducers for the mammalian UPR is inferred from studies in XBP-1−/− cells that express a small interfering RNA construct for ATF6. Intriguingly, these cells still show an almost normal upregulation of Bip in response to tunicamycin (13).
Next, we examined whether expression of the dominant negative constructs antagonizes the induction of the UPR induced by US11 and delays the onset of dislocation in the Tet-On US11 cells. Although the effect was modest at best, we consistently observed that at the early time points after US11 induction, when US11 levels in the cells are limited, dislocation of HC is reduced when the ATF6-DN is expressed (Fig. 7). At later (>24 h) time points, we no longer observed any difference between the two cell types (data not shown). We attribute the modest effect seen for the ATF-DN (or XBP-1-DN, not shown) to a combination of factors: (i) US11-mediated dislocation of HC is not critically dependent on the UPR; (ii) the expression of ATF-6-DN, which is limited by the viability of the cells, does not fully block the UPR; and (iii) HC dislocation in the Tet-On cells is only partial, even when US11 reaches its maximal level.
Finally, we tested whether UPR induction occurs in the context of an HCMV infection. Indeed, XBP-1 splicing was observed at a time point when expression of the early genes, such as US11, gets under way (Fig. 8). The UPR is probably induced by a combination of US11 itself and other HCMV glycoproteins, and thus its induction accompanies the HCMV immunoevasins expression. The relevance of this observation to immunodetection by class I-restricted T cells remains to be investigated. In addition, the UPR might play a greater role in cell types that express a less favorable repertoire of ER proteins for the dislocation reaction than that seen with U373 cells. We conclude that an active UPR is dispensable for dislocation of class I HC, yet is capable of tuning the early onset of US11-mediated class I degradation. US11 itself is sufficient to induce such a response.
Acknowledgments
B.T. is supported by a Dorot Foundation fellowship. A.-H.L. is supported by NIH Career Development Award 1P50CA100707, and N.N.I. is supported by an Irvington Institute Postdoctoral Fellowship Award. This study was supported by NIH grants AI32412 (L.H.G.) and 5R37-AI33456 (H.L.P.).
REFERENCES
- 1.Bonifacino, J. S., and A. M. Weissman. 1998. Ubiquitin and the control of protein fate in the secretory and endocytic pathways. Annu. Rev. Cell Dev. Biol. 14:19-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Casagrande, R., P. Stern, M. Diehn, C. Shamu, M. Osario, M. Zuniga, P. O. Brown, and H. Ploegh. 2000. Degradation of proteins from the ER of S. cerevisiae requires an intact unfolded protein response pathway. Mol. Cell. 5:729-735. [DOI] [PubMed] [Google Scholar]
- 3.Ellgaard, L., and A. Helenius. 2001. ER quality control: towards an understanding at the molecular level. Curr. Opin. Cell Biol. 13:431-437. [DOI] [PubMed] [Google Scholar]
- 4.Friedlander, R., E. Jarosch, J. Urban, C. Volkwein, and T. Sommer. 2000. A regulatory link between ER-associated protein degradation and the unfolded-protein response. Nat. Cell Biol. 2:379-384. [DOI] [PubMed] [Google Scholar]
- 5.Furman, M. H., J. Loureiro, H. L. Ploegh, and D. Tortorella. 2003. Ubiquitinylation of the cytosolic domain of a type I membrane protein is not required to initiate its dislocation from the endoplasmic reticulum. J. Biol. Chem. 278:34804-34811. [DOI] [PubMed] [Google Scholar]
- 6.Gewurz, B. E., R. Gaudet, D. Tortorella, E. W. Wang, H. L. Ploegh, and D. C. Wiley. 2001. Antigen presentation subverted: structure of the human cytomegalovirus protein US2 bound to the class I molecule HLA-A2. Proc. Natl. Acad. Sci. USA 98:6794-6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harding, H. P., M. Calfon, F. Urano, I. Novoa, and D. Ron. 2002. Transcriptional and translational control in the mammalian unfolded protein response. Annu. Rev. Cell Dev. Biol. 18:575-599. [DOI] [PubMed] [Google Scholar]
- 8.Harding, H. P., Y. Zhang, A. Bertolotti, H. Zeng, and D. Ron. 2000. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol. Cell 5:897-904. [DOI] [PubMed] [Google Scholar]
- 9.Haynes, C. M., S. Caldwell, and A. A. Cooper. 2002. An HRD/DER-independent ER quality control mechanism involves Rsp5p-dependent ubiquitination and ER-Golgi transport. J. Cell Biol. 158:91-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang, H. Y., S. A. Wek, B. C. McGrath, D. Scheuner, R. J. Kaufman, D. R. Cavener, and R. C. Wek. 2003. Phosphorylation of the alpha subunit of eukaryotic initiation factor 2 is required for activation of NF-κB in response to diverse cellular stresses. Mol. Cell. Biol. 23:5651-5663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kabani, M., S. S. Kelley, M. W. Morrow, D. L. Montgomery, R. Sivendran, M. D. Rose, L. M. Gierasch, and J. L. Brodsky. 2003. Dependence of endoplasmic reticulum-associated degradation on the peptide binding domain and concentration of BiP. Mol. Biol. Cell 14:3437-3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaufman, R. J. 1999. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 13:1211-1233. [DOI] [PubMed] [Google Scholar]
- 13.Lee, A. H., N. N. Iwakoshi, and L. H. Glimcher. 2003. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol. Cell. Biol. 23:7448-7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee, K., W. Tirasophon, X. Shen, M. Michalak, R. Prywes, T. Okada, H. Yoshida, K. Mori, and R. J. Kaufman. 2002. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 16:452-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lilley, B. N., and H. L. Ploegh. 2004. A membrane protein required for dislocation of misfolded proteins from the ER. Nature 429:834-840. [DOI] [PubMed] [Google Scholar]
- 16.Lilley, B. N., D. Tortorella, and H. L. Ploegh. 2003. Dislocation of a type I membrane protein requires interactions between membrane-spanning segments within the lipid bilayer. Mol. Biol. Cell 14:3690-3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ljunggren, H. G., N. J. Stam, C. Ohlen, J. J. Neefjes, P. Hoglund, M. T. Heemels, J. Bastin, T. N. Schumacher, A. Townsend, K. Karre, et al. 1990. Empty MHC class I molecules come out in the cold. Nature 346:476-480. [DOI] [PubMed] [Google Scholar]
- 18.Machold, R. P., E. J. Wiertz, T. R. Jones, and H. L. Ploegh. 1997. The HCMV gene products US11 and US2 differ in their ability to attack allelic forms of murine major histocompatibility complex (MHC) class I heavy chains. J. Exp. Med. 185:363-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Misaghi, S., Z. Y. Sun, P. Stern, R. Gaudet, G. Wagner, and H. Ploegh. 2004. Structural and functional analysis of human cytomegalovirus US3 protein. J. Virol. 78:413-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Molinari, M., V. Calanca, C. Galli, P. Lucca, and P. Paganetti. 2003. Role of EDEM in the release of misfolded glycoproteins from the calnexin cycle. Science 299:1397-1400. [DOI] [PubMed] [Google Scholar]
- 21.Nakagawa, T., H. Zhu, N. Morishima, E. Li, J. Xu, B. A. Yankner, and J. Yuan. 2000. Caspase-12 mediates endoplasmic-reticulum-specific apoptosis and cytotoxicity by amyloid-beta. Nature 403:98-103. [DOI] [PubMed] [Google Scholar]
- 22.Oda, Y., N. Hosokawa, I. Wada, and K. Nagata. 2003. EDEM as an acceptor of terminally misfolded glycoproteins released from calnexin. Science 299:1394-1397. [DOI] [PubMed] [Google Scholar]
- 23.Rehm, A., P. Stern, H. L. Ploegh, and D. Tortorella. 2001. Signal peptide cleavage of a type I membrane protein, HCMV US11, is dependent on its membrane anchor. EMBO J. 20:1573-1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rutkowski, D. T., and R. J. Kaufman. 2004. A trip to the ER: coping with stress. Trends Cell Biol. 14:20-28. [DOI] [PubMed] [Google Scholar]
- 25.Spear, E. D., and D. T. Ng. 2003. Stress tolerance of misfolded carboxypeptidase Y requires maintenance of protein trafficking and degradative pathways. Mol. Biol. Cell 14:2756-2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Story, C. M., M. H. Furman, and H. L. Ploegh. 1999. The cytosolic tail of class I MHC heavy chain is required for its dislocation by the human cytomegalovirus US2 and US11 gene products. Proc. Natl. Acad. Sci. USA 96:8516-8521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tirosh, B., M. H. Furman, D. Tortorella, and H. L. Ploegh. 2003. Protein unfolding is not a prerequisite for endoplasmic reticulum-to-cytosol dislocation. J. Biol. Chem. 278:6664-6672. [DOI] [PubMed] [Google Scholar]
- 28.Tortorella, D., C. M. Story, J. B. Huppa, E. J. Wiertz, T. R. Jones, I. Bacik, J. R. Bennink, J. W. Yewdell, and H. L. Ploegh. 1998. Dislocation of type I membrane proteins from the ER to the cytosol is sensitive to changes in redox potential. J. Cell Biol. 142:365-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Travers, K. J., C. K. Patil, L. Wodicka, D. J. Lockhart, J. S. Weissman, and P. Walter. 2000. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101:249-258. [DOI] [PubMed] [Google Scholar]
- 30.Wang, Y., J. Shen, N. Arenzana, W. Tirasophon, R. J. Kaufman, and R. Prywes. 2000. Activation of ATF6 and an ATF6 DNA binding site by the endoplasmic reticulum stress response. J. Biol. Chem. 275:27013-27020. [DOI] [PubMed] [Google Scholar]
- 31.Wiertz, E. J., T. R. Jones, L. Sun, M. Bogyo, H. J. Geuze, and H. L. Ploegh. 1996. The human cytomegalovirus US11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol. Cell 84:769-779. [DOI] [PubMed] [Google Scholar]
- 32.Ye, Y., Y. Shibata, C. Yun, D. Ron, and T. A. Rapoport. 2004. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature 429:841-847. [DOI] [PubMed] [Google Scholar]
- 33.Yoshida, H., T. Matsui, N. Hosokawa, R. J. Kaufman, K. Nagata, and K. Mori. 2003. A time-dependent phase shift in the mammalian unfolded protein response. Dev. Cell. 4:265-271. [DOI] [PubMed] [Google Scholar]
- 34.Yoshida, H., T. Matsui, A. Yamamoto, T. Okada, and K. Mori. 2001. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107:881-891. [DOI] [PubMed] [Google Scholar]
- 35.Zhou, M., and R. Schekman. 1999. The engagement of Sec61p in the ER dislocation process. Mol. Cell 4:925-934. [DOI] [PubMed] [Google Scholar]