Abstract
Adeno-associated virus (AAV) is a small single-stranded DNA virus that requires the presence of a helper virus, such as adenovirus or herpes virus, to efficiently replicate its genome. AAV DNA is replicated by a rolling-hairpin mechanism (Ward, 2006), and during replication several DNA intermediates can be detected. This detailed protocol describes how to analyze the AAV DNA intermediates formed during AAV replication using a modified Hirt extract (Hirt, 1967) procedure and Southern blotting (Southern, 1975).
Keywords: Adeno-associated virus, AAV, DNA replication, Replicative intermediates, Southern blot
Background
AAV DNA replication is carried out by a rolling hairpin mechanism in cells co-infected by AAV and helper viruses such as adenovirus or herpes virus (Ward, 2006). The AAV DNA consists of a 4.7 kb linear DNA molecule with inverted terminal repeats (ITRs) that fold back to form T-shaped hairpin structures. The 3’ end hairpin serves as a primer for the replication of the AAV DNA. These hairpin structures are regenerated by the AAV Rep protein, allowing further rounds of replication (Im and Muzyczka, 1990). Both + and - strands of the AAV DNA are packaged and are infectious ( Rose et al., 1969 ). When replicating AAV DNA is analyzed, several replicative intermediates can be detected ( Straus et al., 1976 ). The most abundant replicative intermediate is a linear monomeric duplex molecule, formed by one + and one - strand of the AAV DNA, which is thought to be the immediate precursor of progeny single-stranded molecules that will be packaged in pre-formed capsids ( Straus et al., 1976 ). Dimeric replicative intermediates are also common, and the AAV replication model is compatible with even larger replicative intermediates. The study of AAV replication benefitted from the discovery that AAV plasmids are infectious–the AAV DNA can be fully rescued from a plasmid (in the presence of helper virus) and its replication mimics that of the native virus ( Samulski et al., 1982 ). The method detailed here allows the investigation of the DNA intermediates formed during DNA replication initiated from an AAV plasmid, and was used to compare different mutants of the AAV Rep protein for their ability to support AAV replication. The same method can be used to study other aspects of the AAV life cycle that can affect DNA replication of this virus, such as the effect of helper virus proteins or other factors that restrict/enhance AAV replication.
Materials and Reagents
-
Transfection of 293T cells and wild type (wt) AAV production
100 mm dishes (Corning, catalog number: 430293)
293T cells (ATCC, catalog number: CRL-11268)
pAV2 plasmid ( Laughlin et al., 1983 , available from ATCC, catalog number: 37216)
Linear polyethylenimine (PEI), MW 25,000, at 1 mg/ml and pH 7.0 (Polysciences, catalog number: 23966-1)
wt adenovirus serotype 5 (Graham and Prevec, 1991, available from ATCC, catalog number: VR-1516)
Dulbecco modified Eagle’s medium (DMEM) (Thermo Fisher Scientific, catalog number: 31966021)
Foetal bovine serum (FBS) (Thermo Fisher Scientific, catalog number: 10270106)
1x Dulbecco’s phosphate-buffered saline (DPBS) (Thermo Fisher Scientific, GibcoTM, catalog number: 14190094)
-
Extraction of low molecular weight DNA
15 ml conical tube (Corning, catalog number: 430791)
1.5 ml tube
-
Hirt lysis buffer (see Recipes)
Sodium dodecyl sulfate (SDS) (Thermo Fisher Scientific, AmbionTM, catalog number: AM9822)
Tris (pH 7.5)
Ethylenediaminetetraacetic acid (EDTA) (Sigma-Aldrich, catalog number: E9884)
Proteinase K solution (20 mg/ml) (Thermo Fisher Scientific, InvitrogenTM, catalog number: 25530049)
Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: S3014)
Phenol-chloroform-isoamyl alcohol 25:24:1 (Sigma-Aldrich, catalog number: 77617)
Sodium acetate (CH3COONa) (Sigma-Aldrich, catalog number: 71183)
2-propanol (EMD Millipore, catalog number: 109634)
Ethanol (EMD Millipore, catalog number: 100983)
ddH2O (Thermo Fisher Scientific, AmbionTM, catalog number: AM9937)
RNaseA (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: EN0531)
-
Southern blot assay using Rep and Amp probes
WhatmanTM paper (GE Healthcare, catalog number: 3030-917)
Amersham Hybond-N+ membrane (GE Healthcare, catalog number: RPN203B)
Hybridisation tubes (Chemglass Life Sciences, catalog number: CG-1140-05)
Quick Spin Columns for radiolabeled DNA purification (Roche Diagnostics, catalog number: 11273973001)
DpnI (New England Biolabs, catalog number: R0176S)
UltraPureTM agarose (Thermo Fisher Scientific, InvitrogenTM, catalog number: 16500500)
-
TAE (Tris-acetate-EDTA) buffer (see Recipes)
Tris (Roche Diagnostics, catalog number: 10708976001)
Glacial acetic acid (Sigma-Aldrich, catalog number: ARK2183)
Ethylenediaminetetraacetic acid (EDTA) (Sigma-Aldrich, catalog number: E9884)
1 kb DNA ladder (New England Biolabs, catalog number: N3232)
-
20x saline sodium citrate (SSC) solution (see Recipes)
Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: S3014)
Sodium citrate (Sigma-Aldrich, catalog number: W302600)
-
Denaturing solution (see Recipes)
Sodium chloride (NaCl)
Sodium hydroxide (NaOH) (Sigma-Aldrich, catalog number: 71687)
-
Neutralising solution (see Recipes)
Tris (pH 7.4)
Sodium chloride (NaCl)
-
Rep probe primers
fw: AACTGGACCAATGAGAACTTTCC; rv: AAAAAGTCTTTGACTTCCTGCTT
-
Amp probe primers
fw: AATCAGTGAGGCACCTATCTCAGC; rv: AACTCGGTCGCCGCATACACTATT
GoTaq® Colorless Master Mix PCR Kit (Promega, catalog number: M7132)
QIAquick Gel Extraction Kit (QIAGEN, catalog number: 28704)
dCTPs, [α-32P]- 6,000 Ci/mmol 20 mCi/ml, 250 µCi (PerkinElmer, catalog number: BLU013Z250UC)
Prime-It RmT Random Primer Labeling Kit (Agilent Technologies, catalog number: 300392)
-
Nylon Wash solution (see Recipes)
Sodium phosphate dibasic (Na2HPO4) (Sigma-Aldrich, catalog number: S3264)
Ethylenediaminetetraacetic acid (EDTA)
Sodium dodecyl sulfate (SDS) (Sigma-Aldrich, catalog number: L3771)
Equipment
Pipette
CO2 incubator (Thermo Fisher Scientific, Thermo ScientificTM, model: HeraCell 240)
Hemocytometer
Refrigerated benchtop microcentrifuge (Thermo Fisher Scientific, Thermo ScientificTM, model: HeraeusTM FrescoTM 17)
Refrigerated centrifuge (Thermo Fisher Scientific, Thermo ScientificTM, model: HeraeusTM MultifugeTM X3R)
Microbiological safety cabinet (Medical Air Technology, model: BioMAT2 Class II)
NanoDrop spectrophotometer (Thermo Fisher Scientific, Thermo ScientificTM, model: NanoDrop 1000 or NanoDrop 2000)
Gel electrophoresis system (Thermo Fisher Scientific, Thermo ScientificTM, model: OwlTM A1)
Platform shaker (Heidolph Instruments, model: Polymax 1040)
Glass plate
Glass tray
Ultraviolet crosslinker (UVP, model: CL-1000)
Hybridisation oven (UVP, model: HB-1000)
Perspex screen (Thermo Fisher Scientific, Thermo ScientificTM, model: NalgeneTM Acrylic Benchtop Beta Radiation Shield, catalog number: 67002418)
Typhoon Molecular Dynamics phosphor/fluorescence imager (GE Healthcare, model: Trio Variable Mode Imager)
PCR thermal cycler (Eppendorf, model: Mastercycler®)
Heat block (Cole-Parmer, Stuart, catalog number: SBH200DC)
Storage Phosphor screen (GE Healthcare, catalog number: 28-9564-75)
Exposure cassette (GE Healthcare, catalog number: 63-0035-44)
Software
ImageQuant analysis software (GE Healthcare)
Procedure
-
Transfection of 293T cells and AAV production
Seed 4.3 x 106 293T cells in a 10 cm dish in 10 ml of growth medium (DMEM + 10% FBS) for transfection next day. Include an additional plate to determine cell number at the time of infection with adenovirus. Cells are maintained for 24 h at 37 °C and 5% CO2.
Dilute 17.5 µg of pAV2 plasmid in 500 µl of DMEM (no serum added, SF DMEM).
Dilute 94 µl of PEI in 500 µl of SF DMEM and gently mix. The transfection reagents should be at room temperature (RT) before mixing. Incubate 5 min at RT.
Combine the PEI and DNA and gently mix by pipetting. Incubate at RT for 20 min.
Add the mixture to the medium in which the cells are growing. Pipet carefully to avoid detaching the cells.
4 h after transfection, determine the cell number (for example using a hemocytometer) in the extra plate.
Infect the cells using 5 pfu/cell of adenovirus. Include a condition without adenovirus as a negative control for AAV replication.
-
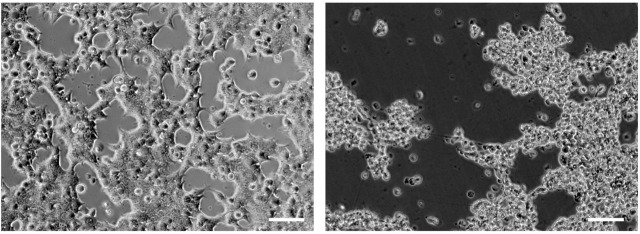
40 to 50 h post transfection a cytopathogenic effect is expected. Cells will round up and eventually detach (Figure 1).
Note: The amount of adenovirus necessary to fully support AAV replication and to cause a cytopathogenic effect at 40 to 50 h post transfection can vary depending on the health of the cell line used and its passage number. This may need to be optimized before starting this protocol: infect cells with increasing amounts of adenovirus and monitor the appearance of a cytopathogenic effect (Figure 1).
-
Extraction of low molecular weight DNA
Collect cells and supernatant in a 15 ml conical tube. Cells should be loosely adherent and can be detached by pipetting. Wash the plate with 2 ml 1x DPBS and add to the 15 ml tube.
Centrifuge for 5 min at 500 × g at RT.
Discard the supernatant, and resuspend the cell pellet in 1 ml DPBS. Transfer 250 µl to a new 1.5 ml tube. The remainder of the cells can be harvested for additional analyses, e.g., gene and protein expression.
Centrifuge for 5 min at 500 × g at RT in a benchtop centrifuge and discard supernatant.
-
Add 500 µl of Hirt lysis buffer and mix by flicking or inverting the tube.
Note: The lysate will be very viscous and mixing by pipetting will create bubbles.
Incubate for 10 min at RT.
Add 1.25 µl of proteinase K (50 µg/ml final concentration) and incubate for 1 h at 37 °C.
Add 120 µl of 5 M NaCl (1 M final concentration). Mix gently by flicking/inverting the tube.
Incubate overnight at 4 °C. This step precipitates the high molecular weight (chromosomal) DNA.
Centrifuge for 1 h at maximum speed (17,000 × g) at 4 °C in a refrigerated table-top centrifuge.
-
Carefully transfer the supernatant (approximately 500 µl) to a new 1.5 ml tube.
Note: If the supernatant cannot be separated sufficiently from the viscous pellet, a longer centrifugation may be required.
Under a fume hood, add 1 volume (500 µl) of phenol/chloroform/isoamyl alcohol and mix by inverting the tube until homogeneous.
Centrifuge for 10 min at 17,000 × g at 4 °C.
Carefully transfer the upper phase to a new 1.5 ml tube (approximately 450 µl).
Add 50 µl 3 M sodium acetate (0.3 M final concentration) and mix.
Incubate overnight at -20 °C.
Centrifuge for 20 min at 17,000 × g at 4 °C.
Carefully remove and discard the supernatant, then wash the DNA pellet with 100 µl of 70% ethanol.
Centrifuge for 10 min at 17,000 × g at 4 °C.
Remove supernatant and air-dry the DNA pellet.
Resuspend the pellet in 80 µl ddH2O.
Determine the concentration of DNA in the samples using a NanoDrop spectrophotometer. Typical DNA concentration is around 750 ng/µl.
-
Southern blot assay (gel transfer)
Digest approximately 1 µg of the extracted DNA with DpnI, for 2 h at 37 °C. DpnI specifically digests methylated DNA, e.g., DNA amplified in dam+ bacteria strains. This step ensures that the input plasmid DNA is digested while newly replicated AAV DNA will not be affected.
Separate the digested DNA on a 0.8% agarose gel overnight at 1 V/cm.
Remove the gel from the electrophoresis tank and incubate for 30 min in denaturing solution on a platform shaker at approximately 25 rpm.
Rinse the gel twice in ddH2O.
Incubate the gel for 30 min in neutralizing solution on a platform shaker at approximately 25 rpm.
Replace the neutralizing solution with fresh neutralizing solution and incubate for a further 30 min while shaking.
Rinse in ddH2O, and then proceed with the assembly of the transfer set-up as detailed in Figure 2.
Incubate overnight to transfer the DNA in the gel to the nylon membrane by the capillary method (Figure 2). To obtain the same sample orientation on the nylon membrane as on the gel, turn the gel upside down.
Disassemble the set-up and carefully remove the membrane.
Rinse the membrane in 2x SSC, and let it air-dry.
To fix the DNA to the membrane, UV cross-link the membrane with 120,000 µJ/cm2.
Pre-hybridize the membrane in 20 ml 0.75x nylon wash solution for at least 1 h at 65 °C in hybridization tubes.
-
Preparation of the Rep and Amp probes
-
The template to prepare the Rep and Amp specific probes is obtained by PCR on the pAV2 plasmid using the aforementioned primer pairs and the PCR conditions listed below. The PCR products are gel-purified and quantified using a NanoDrop spectrophotometer. The expected size of the PCR products is 338 base-pairs for the Rep PCR and 587 for the Amp PCR. 20 to 25 ng of purified PCR product DNA is used as DNA template to prepare the probes.
For both Rep and Amp PCRs, mix in a final volume of 25 µl:
GoTaq Colorless Master mix (2x) 12.5 µl
Fw Rep or Amp primer (10 µM) 1 µl
Rv Rep or Amp primer (10 µM) 1 µl
Template DNA 25 ng
ddH2O to 25 µl
Cycling conditions Rep PCR:
95 °C 2 sec
x 3095 °C 30 sec
60 °C 30 sec
72 °C 30 sec
72 °C 5 min
Cycling conditions Amp PCR:
95 °C 2 sec
x 3095 °C 30 sec
60 °C 30 sec
72 °C 40 sec
72 °C 5 min
Reconstitute the reaction mixture (provided as 0.2 ml tubes containing dehydrated random primers, dNTPs, buffer and cofactors) using ddH2O containing 20-25 ng of probe DNA to a final volume of 44 µl, as suggested in the Prime-It RmT Random Primer Labeling Kit. Heat at 95 °C for 5 min.
Add 1.5 µl of 32P-labelled dCTPs, and 3 µl of Magenta polymerase enzyme (tube with red cap in the labelling kit). Incubate for 8-10 min at 37 °C.
Add 2 µl of stop mix (tube with green cap in the labelling kit).
Separate labelled DNA (the probe) from unincorporated labelled nucleotides using the Quick Spin Columns for radiolabeled DNA purification.
Heat the probe at 95 °C for 5 min to denature the DNA.
Place tube on ice for 5 min then quickly spin to collect the probe at the bottom of the tube.
-
-
Southern blot assay (membrane hybridization)
Add the probe to the hybridization tube. Depending on the freshness of the radioactivity, one probe prepared as detailed above is sufficient to label 2-4 membranes.
Incubate overnight at 65 °C in a hybridization oven.
Wash membrane in 0.5x nylon wash solution for 20 min at 65 °C.
-
Wash membrane in 0.1x nylon wash solution for 20 min at 65 °C.
Note: If additional washing is required, repeat washing step using 0.01x nylon wash solution.
-
Expose the membrane to the Phosphoimager screen for at least 2 h.
Note: The length of exposure necessary to obtain the best signal can vary depending on the strength of the signal.
Acquire images using the Typhoon phosphor imager. Image analysis can be performed using the ImageQuant analysis software (GE Healthcare).
Figure 1. AAV and adenovirus induced cytopathogenic effect.
Left panel: 24 h post-transfection cells are adherent and form a monolayer. Right panel: 55 h post-transfection the cytopathogenic is evident; cells have rounded up and are lifting from the plate. Scale bars = 50 µm.
Figure 2. Scheme of capillary transfer method for Southern blotting.
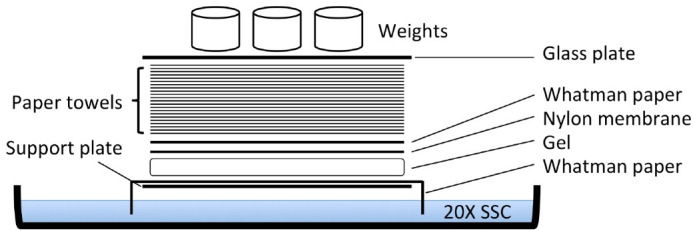
The support plate is a glass plate resting on the sides of the glass tray containing the 20x SSC. The paper towels layer is approximately 15 cm high. The weight used was approximately 600 g.
Data analysis
The gel images acquired following the procedure described above were analyzed using the ImageQuant analysis software (GE Healthcare). The level of AAV replication under different conditions is proportional to the intensity of the bands corresponding to viral replicative intermediates in DpnI-treated samples hybridized with the viral (Rep) probe (Figure 3). For wt AAV2 virus, monomeric and dimeric molecules of approximately 4.7 kb and 9.4 kb, respectively should be visible. Additional higher molecular weight intermediates can also be present. Membranes hybridized with the Amp probe allow for the assessment of efficiency of transfection and efficacy of DpnI digestion (Figure 3). The ImageQuant software supports band quantification by densitometry if a quantitative band comparison is required.
Figure 3. Analysis of AAV replication intermediates by Southern blot.
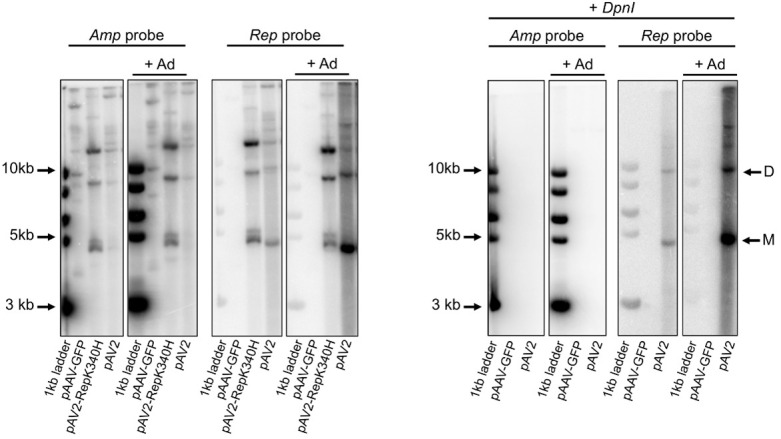
Left panel: pAAV-GFP (no Rep control), pAV2 (WT AAV plasmid) and pAV2-RepK340H (replication-deficient Rep mutant) were transfected in the presence (+ Ad) or absence of adenovirus. The Amp probe binds to all plasmids containing the ampicillin resistance gene. The Rep probe only binds to DNA containing the AAV Rep gene. Right panel: samples were treated with DpnI digestion to remove all input plasmid; only AAV DNA that is rescued from the plasmid and replicated is detected. Plasmids pAAV-GFP and pAV2 were transfected in the presence (+ Ad) or absence of adenovirus. Hybridisation with the Amp probe confirms that all input plasmid was digested by DpnI treatment. The Rep probe detects replicated AAV DNA. M: monomeric replicative form; D: dimeric replicative form; larger replicative intermediates are visible above. Adapted from Bardelli et al., 2016 .
Notes
Steps D3 to E5 should only be performed in areas designated for work with radioactivity. Please follow local directives.
-
Note on controls: several controls should be used when running this assay.
+ AAV, - adenovirus: no replication control. In the absence of adenovirus, AAV replication is absent or very low.
- Rep, + adenovirus: no replication control. Instead of wt AAV, use a replication-deficient AAV mutant or recombinant AAV vector (for example pAAV-GFP). This will also control for the absence of wt AAV contamination in the adenovirus batch used in the experiment.
The Amp probe is used to assure that input DNA is digested.
No DpnI control: allows the comparison of input plasmid between different conditions, and controls for the binding of the Amp probe.
Recipes
-
Hirt lysis buffer
6 ml of 10% SDS
1 ml of 1 M Tris (pH 7.5) solution
2 ml of 500 mM EDTA (pH 8.0) solution
Bring the final volume to 100 ml with ddH2O
-
TAE (Tris-acetate-EDTA) buffer
Prepare 50x solution:
242 g Tris
57.1 ml glacial acetic acid
100 ml of 500 mM EDTA (pH 8.0) solution
Bring the final volume to 1 L ddH2O
Dilute 1:50 in ddH2O to make 1x TAE buffer
-
20x saline sodium citrate (SSC) solution
175.3 g NaCl
88.2 g sodium citrate
Bring the final volume to 1 L ddH2O
-
Denaturing solution
300 ml of 5 M NaCl solution
100 ml of 5 M NaOH solution
Bring to 1 L with ddH2O
-
Neutralising solution
500 ml of 1 M Tris (pH 7.4) solution
300 ml of 5 M NaCl solution
Bring to 1 L with ddH2O
-
Nylon wash solution (pH 7.2)
40.6 g Na2HPO4
18.65 g EDTA
500 g SDS
Bring the final volume to 3.58 L with ddH2O
Acknowledgments
This work was supported by the Pfizer Rare Diseases Consortium Award (to EH), NIH grant RO1-GM092854 (to CRE) and United Kingdom Medical Research Council grant 1001764 (to RML).
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1.Bardelli M., Zarate-Perez F., Agundez L., Linden R. M., Escalante C. R. and Henckaerts E.(2016). Identification of a functionally relevant Adeno-associated virus Rep68 oligomeric interface. J Virol 90(15): 6612-6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Graham F. L. and Prevec L.(1991). Manipulation of adenovirus vectors. Methods Mol Biol 7: 109-128. [DOI] [PubMed] [Google Scholar]
- 3.Hirt B.(1967). Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol 26(2): 365-369. [DOI] [PubMed] [Google Scholar]
- 4.Im D. S. and Muzyczka N.(1990). The AAV origin binding protein Rep68 is an ATP-dependent site-specific endonuclease with DNA helicase activity. Cell 61(3): 447-457. [DOI] [PubMed] [Google Scholar]
- 5.Laughlin C. A., Tratschin J. D., Coon H. and Carter B. J.(1983). Cloning of infectious adeno-associated virus genomes in bacterial plasmids. Gene 23(1): 65-73. [DOI] [PubMed] [Google Scholar]
- 6.Rose J. A., Berns K. I., Hoggan M. D. and Koczot F. J.(1969). Evidence for a single-stranded adenovirus-associated virus genome: formation of a DNA density hybrid on release of viral DNA. Proc Natl Acad Sci U S A 64(3): 863-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Samulski R. J., Berns K. I., Tan M. and Muzyczka N.(1982). Cloning of adeno-associated virus into pBR322: rescue of intact virus from the recombinant plasmid in human cells. Proc Natl Acad Sci U S A 79(6): 2077-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Southern E. M.(1975). Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol 98: 503-17. [DOI] [PubMed] [Google Scholar]
- 9.Straus S. E., Sebring E. D. and Rose J. A.(1976). Concatemers of alternating plus and minus strands are intermediates in adenovirus-associated virus DNA synthesis. Proc Natl Acad Sci U S A 73(3): 742-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ward P.(2006). Replication of adeno-associated virus DNA. In: Kerr, J. R., Cotmore, S. F., Bloom, M. E., Linden, R. M. and Parrish, C. R.(Eds.). Parvoviruses. Hodder Arnold pp: 189-211. [Google Scholar]