Abstract
G protein-coupled receptors (GPCRs) promote cytoplasmic signalling by activating heterotrimeric G proteins in response to extracellular stimuli such as light, hormones and nucleosides. Structure determination of GPCR–G protein complexes is central to understanding the precise mechanism of signal transduction. However, these complexes are challenging targets for structural studies due to their conformationally dynamic and inherently transient nature. We recently developed an engineered G protein, mini-Gs, which addressed these problems and allowed the formation of a stable GPCR–G protein complex. Mini-Gs facilitated the structure determination of the human adenosine A2A receptor (A2AR) in its G protein-bound conformation at 3.4 Å resolution. Here, we describe a step by step protocol for the expression and purification of A2AR, and crystallisation of the A2AR–mini-Gs complex.
Keywords: Adenosine A2A receptor , A2AR , Active state, GPCR, G protein-coupled receptor, Mini G protein, Mini-Gs, G protein complex
Background
We recently developed an engineered minimal G protein, mini-Gs (Carpenter and Tate, 2016), which facilitated the structure determination of the human adenosine A2A receptor (A2AR) in its active state ( Carpenter et al., 2016 ). Mini-Gs stabilises the active conformation of A2AR sufficiently to allow crystallization of the complex by vapour diffusion in the detergent octylthioglucoside. Here, we describe a detailed protocol for the expression and purification of A2AR, which is adapted from a previously described method developed in our laboratory ( Lebon et al., 2011a and 2011b; Tate and Lebon, 2015). We also describe a step by step procedure for the preparation and crystallisation of the A2AR–mini-Gs complex, earlier described in Carpenter et al. (2016). Expression and purification of mini-Gs is described in a companion manuscript (Carpenter and Tate, 2017).
Materials and Reagents
Serological pipette
Pipette tips (STARLAB INTERNATION)
Plastic spatula
50 ml tubes (SARSTEDT, catalog number: 62.547.254)
15 ml tubes (SARSTEDT, catalog number: 62.554.002)
5 ml tubes (Eppendorf, catalog number: 0030119401)
1.5 ml tubes (SARSTEDT, catalog number: 72.690.001)
0.5 ml tubes (SARSTEDT, catalog number: 72.699)
Steritop 0.22 μm filter unit (EMD Millipore, catalog number: SCGPT01RE)
Amicon Ultra-15 concentrator 50 kDa cut-off (EMD Millipore, catalog number: UFC905024)
Plastic column (e.g., empty PD-10 column) (GE Healthcare, catalog number: 17043501)
PD-10 desalting column (GE Healthcare, catalog number: 17085101)
Amicon Ultra-4 concentrator 50 kDa cut-off (EMD Millipore, catalog number: UFC805024)
MRC 96-well 2-drop crystallization plates (Molecular Dimensions, catalog number: MD11-00-100)
Ni2+-NTA Superflow 5 ml prepacked column (QIAGEN, catalog number: 30760)
Superdex 200 GL 10/300 gel filtration column (GE Healthcare, catalog number: 17517501)
Trichoplusia ni (T. ni) insect cell line (Expression Systems, catalog number: 94-002F)
pBacPAK8 plasmid (Takara Bio, Clontech, catalog number: PT1262-5)
flashBAC ULTRA DNA (Oxford Expression Technologies, catalog number: 100300)
ESF921 insect cell media (Expression Systems, catalog number: 96-001-01)
Fetal bovine serum (Sigma-Aldrich, catalog number: F9665)
cOmplete, EDTA-free protease inhibitor tablets (Roche Diagnostics, catalog number: 11873580001)
PMSF (Sigma-Aldrich, catalog number: P7626)
Liquid nitrogen
NECA (Sigma-Aldrich, catalog number: E2387)
Imidazole (Sigma-Aldrich, catalog number: 56748)
Sodium chloride (NaCl) (Fisher Scientific, catalog number: 10598630)
n-Decyl β-maltoside (DM) detergent (Anatrace, catalog number: D322)
TEV protease (produced in-house)
Ni2+-NTA agarose (QIAGEN, catalog number: 30210)
Magnesium chloride (MgCl2) (Fisher Scientific, catalog number: BP214-500)
Apyrase (Sigma-Aldrich, catalog number: A6535)
Sodium acetate (Fisher Scientific, catalog number: 10794761)
MemGoldTM crystallisation screen (Molecular Dimensions, catalog number: MD1-39)
-
PEG 2000 (Sigma-Aldrich, catalog number: 81221)
Note: This product has been discontinued.
-
PEG 2000 MME (Sigma-Aldrich, catalog number: 81321)
Note: This product has been discontinued.
PEG 400 (Hampton Research, catalog number: HR2-603)
Glycerol (VWR, catalog number: 24388.320)
HEPES (Sigma-Aldrich, catalog number: H3375)
EDTA
Absolute ethanol (VWR, catalog number: 20821.330)
DMSO (Sigma-Aldrich, catalog number: D2650)
n-Octyl-β-D-thioglucoside (OTG) detergent (Glycon, catalog number: D20014)
Precision Plus SDS-PAGE molecular weight standards (Bio-Rad Laboratories, catalog number: 161-0373)
4-20% Tris-glycine SDS-PAGE gels (Fisher Scientific, catalog number: EC60255BOX)
Buffer A (see Recipes)
PMSF stock solution (see Recipes)
NECA stock solution (see Recipes)
DM stock solution (see Recipes)
Buffer B (see Recipes)
Buffer C (see Recipes)
Buffer D (see Recipes)
Buffer E (see Recipes)
OTG stock solution (see Recipes)
Apyrase stock solution (see Recipes)
Buffer F (see Recipes)
Equipment
Pipettes (STARLAB INTERNATION)
Optimum GrowthTM 5 L flask (Thompson Instrument Company, catalog number: 931116)
Shaker incubator (Infors, model: Multitron Standard)
High speed centrifuge (e.g., Beckman Coulter, model: Avanti J-26XP, catalog number: 393124)
Type 45 Ti (Ti45) ultracentrifuge rotor (Beckman Coulter, catalog number: 339160)
Type 45 Ti (Ti45) ultracentrifuge bottle assembly (Beckman Coulter, catalog number: 355622)
Water bath (Julabo)
ULTRA-TURRAX T25 homogeniser (IKA, model: ULTRA-TURRAX T25 Homogeniser)
Magnetic stirring bar
Type 70 Ti (Ti70) ultracentrifuge rotor (Beckman Coulter, catalog number: 337922)
Type 70 Ti (Ti70) ultracentrifuge bottle assembly (Beckman Coulter, catalog number: 355618)
Peristaltic pump (e.g., GE Healthcare, model: Pump P-1, catalog number: 18-1110-91)
Refrigerated benchtop centrifuge (e.g., Eppendorf, catalog number: 5430 R)
Roller mixer (IKA, model: ROLLER 6 digital, catalog number: 0004011000)
TLA55 benchtop ultracentrifuge rotor (Beckman Coulter, catalog number: 366725)
Optima L100 XP preparative ultracentrifuge (Beckman Coulter, model: OptimaTM L-100 XP)
Optima MAX benchtop ultracentrifuge (Beckman Coulter, model: OptimaTM MAX)
Refrigerated microcentrifuge (e.g., Eppendorf, catalog number: 5418 R)
Rotor capable of spinning 1 L bottles (e.g., JLA-8.1000) (Beckman Coulter, catalog number: 363688)
ÄKTA Purifier chromatography system (GE Healthcare, model: ÄKTA Purifier)
Mosquito® Crystal protein crystallisation robot (TTP Labtech, model: mosquito® Crystal)
Software
UNICORN (GE Healthcare)
Graphical software (e.g., Prism 7) (GraphPad)
Procedure
-
Expression of A2AR in insect cells
Human A2AR, which was truncated to remove the flexible C-terminus (see Note 1) and contained the N154A mutation to remove a potential N-linked glycosylation site, was cloned into the transfer vector pBacPAK8, and baculoviruses were prepared using the flashBAC ULTRA system, following the manufacturer’s instructions. The A2AR construct contained a C-terminal 10x histidine tag to facilitate purification (see Note 2) and a TEV protease cleavage site to allow removal of the histidine tag (Figure 1).
Dilute T. ni cells (grown in ESF921 serum-free media) to 2,730 ml in a 5 L optimum growth flask at a density of 1 x 106 cells/ml, and incubate overnight at 27 °C, shaking at 124 rpm.
Once the density reaches 3.3 x 106 cells/ml, add 150 ml of fetal bovine serum (final concentration of 5% v/v) and 120 ml of passage three A2AR baculovirus (final concentration of 4% v/v).
Incubate for 65-72 h at 27 °C, shaking at 124 rpm.
Harvest the cells by centrifugation at 5,000 × g for 10 min at 4 °C.
Resuspend the cell pellet to a final volume of 180 ml in buffer A, add 4 protease inhibitor tablets and PMSF to give a final concentration of 1 mM. Flash freeze in liquid nitrogen and store at -80 °C.
-
Insect cell membrane preparation
A crude membrane preparation is performed, which retains all insoluble intracellular components, whilst removing soluble proteins. This type of membrane preparation helps to limit the loss of membrane fragments and maximise the recovery of A2AR. This protocol describes the membrane preparation from three liters of insect cell culture, which is sufficient for one A2AR purification. However, if necessary, two batches (six liters of culture in total) can be processed simultaneously using a single Ti45 ultracentrifuge rotor.
Thaw the cell pellet from 3 L of insect cell culture in a water bath at room temperature.
Add fresh PMSF to give a final concentration of 1 mM, transfer the solution to three Ti45 tubes and centrifuge at 158,000 × g (45,000 rpm) in a Ti45 rotor for 2 h at 4 °C. Make sure the tubes are filled correctly (to the neck), top up with buffer A if necessary. (see Note 3)
Carefully remove the supernatant until level with the top of the pellet using a serological pipette (see Figure 2).
Add 25 ml of buffer A to each tube and break the pellet up into small pieces using plastic spatula.
Homogenise the pellet using an ULTRA-TURRAX T25 homogeniser operating at 10,000 rpm until no lumps remain. This should take approximately 30 sec per tube.
Top up the tubes with buffer A and centrifuge at 158,000 × g (45,000 rpm) in a Ti45 rotor for 2 h at 4 °C. (see Note 3)
Remove the supernatant until level with the top of the pellet using a serological pipette (see Figure 2).
Resuspend the pellet with the buffer that remains in the tubes using a plastic spatula followed by an ULTRA-TURRAX T25 homogeniser. Do not exceed a final volume of 160 ml. Aliquot the membrane suspension into 50 ml tubes, flash freeze in liquid nitrogen and store at -80 °C. Membranes can be stored under these conditions for up to three months.
-
Purification of A2AR
A2AR is purified in decylmaltoside (DM), in the presence of the agonist NECA, using a protocol adapted from a previously described method for the purification of a thermostabilised A2AR construct ( Lebon et al., 2011a and 2011b; Tate and Lebon, 2015). DM must be used if the complex is to be later exchanged into short chain detergents for vapour diffusion crystallisation. This protocol typically yields 3-6 mg of pure A2AR from 3 L of insect cell culture.
Thaw the membranes from 3 L of insect cell culture in a water bath at room temperature.
Add 4 protease inhibitor tablets, and NECA (100 μM), imidazole (10 mM), PMSF (1 mM), and NaCl (300 mM) to give the final concentrations indicated, stir slowly at room temperature for 30 min using a magnetic stirring bar to allow agonist binding.
Cool the membrane suspension to 4 °C in an ice bath. Solubilise the membranes by adding DM to give a final concentration of 2% (w/v) while stirring rapidly using a magnetic stirring bar, continue to stir slowly for 30 min at 4 °C.
Transfer the solution to eight Ti70 tubes and centrifuge at 265,000 × g (60,000 rpm) in a Ti70 rotor for 2 h at 4 °C.
Remove the supernatant and filter through a 0.2 μm Steritop filter unit. (see Note 4)
Load the filtrate (~150 ml) onto a 5 ml Ni2+-NTA Superflow column (pre-equilibrated with buffer B) at a flow rate of 2 ml/min at 4 °C, using a peristaltic pump. The filtrate should be kept on ice during loading.
Wash the column with 100 ml of buffer C at a flow rate of 5 ml/min at 4 °C.
Elute the column with 20 ml of buffer D at a flow rate of 2 ml/min at 4 °C, collecting a single 20 ml fraction.
Concentrate the eluate to 5 ml using an Amicon Ultra-15 concentrator (50 kDa molecular weight cut-off) in a refrigerated benchtop centrifuge at 4 °C. (see Note 5)
Exchange the concentrated protein into buffer E using two PD-10 desalting columns (load 2.5 ml onto each column). (see Note 6)
Add TEV protease to give a TEV:A2AR ratio of approximately 1:3 (w/w), incubate overnight on ice.
Add 2 ml of Ni2+-NTA agarose resin (pre-equilibrated with buffer E), mix on a roller mixer for 30 min at 4 °C.
Pour the suspension onto 1 ml of Ni2+-NTA agarose resin (pre-equilibrated with buffer E) packed into a disposable plastic column and let it run through by gravity.
Collect the flow through and wash the column with 2 x 3 ml of buffer E.
Pool the wash with the flow through and concentrate to 0.2 ml using an Amicon Ultra-15 concentrator (50 kDa cut-off) in a refrigerated benchtop centrifuge at 4 °C.
Centrifuge the concentrated protein at 135,000 × g (55,000 rpm) in a TLA55 rotor using a benchtop ultracentrifuge for 10 min at 4 °C to remove aggregates.
Load the supernatant onto a Superdex 200 GL 10/300 gel filtration column, (pre-equilibrated with buffer E) at a flow rate of 0.4 ml/min at 4 °C, collecting 0.5 ml fractions.
Pool peak fractions (typically 3-4 ml) based on the gel filtration chromatogram. (see Note 7)
Concentrate the pooled fractions to 0.2 ml using an Amicon Ultra-4 concentrator (50 kDa cut-off) in a refrigerated benchtop centrifuge at 4 °C.
Determine the protein concentration using the amido black assay (Schaffner and Weissmann, 1973). (see Note 8)
If required, purified A2AR can be aliquoted, flash frozen in liquid nitrogen and stored at -80 °C. (see Note 9)
-
Crystallisation of the A2AR–mini-Gs complex
The A2AR–mini-Gs complex is first prepared in DM, once the complex is formed it is stable enough to be exchanged into octylthioglucoside (OTG) for vapour diffusion crystallisation trials. A detailed protocol for the expression and purification of mini-Gs is provided in a companion manuscript (Carpenter and Tate, 2017).
Aliquot 2 mg of A2AR into a microcentrifuge tube and add a 1.2-fold molar excess of mini-Gs construct 414 (1.8 mg). Adjust the volume to 0.2 ml with buffer E, add MgCl2 to give a final concentration of 1 mM and add 0.1 units of apyrase. Incubate for 3-16 h on ice. (see Note 10)
Start the detergent exchange (from DM to OTG) by diluting the complex 1:10 in buffer F, and re-concentrating to 0.2 ml using an Amicon Ultra-4 concentrator (50 kDa cut-off) in a refrigerated benchtop centrifuge at 4 °C.
Centrifuge the concentrated protein at 135,000 × g (55,000 rpm) in a TLA55 rotor using a benchtop ultracentrifuge for 10 min at 4 °C to remove aggregates.
Complete the detergent exchange by loading the supernatant onto a Superdex 200 GL 10/300 gel filtration column (pre-equilibrated with buffer F) at a flow rate of 0.5 ml/min at 4 °C, collecting 0.5 ml fractions.
Pool peak fractions (typically 2-3 ml) based on the gel filtration chromatogram (Figure 3).
Concentrate the pooled fractions to ≥ 20 mg/ml using an Amicon Ultra-4 concentrator (50 kDa cut-off) in a refrigerated benchtop centrifuge at 4 °C.
Centrifuge the concentrated protein at 55,000 rpm (135,000 × g) in a TLA55 rotor using a benchtop ultracentrifuge for 10 min at 4 °C to remove aggregates.
Transfer the supernatant to a new tube and discard the pellet.
Accurately determine the concentration of the complex using the amido black assay (Schaffner and Weissmann, 1973).
Dilute the complex to 20 mg/ml and immediately set up crystallisation trials in 96-well sitting drop plates, dispensing 120 nl of protein and 120 nl of precipitant solution using a Mosquito crystallisation robot situated in a cold room at 4 °C. (see Note 12)
The initial hit for the A2AR–mini-Gs414 complex came from the MemGoldTM screen (0.1 M sodium acetate pH 5.5, 8.8% [w/v] PEG 2000 MME). The two crystals that were used for structure determination were grown in either: 0.1 M sodium acetate pH 5.5, 10% (w/v) PEG 2000; or 0.1 M sodium acetate pH 5.7, 9.5% (w/v) PEG 2000 MME.
Harvest the crystals in a cold room at 4 °C. Sequentially transfer the crystals between drops of mother liquor supplemented with increasing concentrations (10, 20, 30%) of the cryoprotectant PEG 400 (1 min in each solution), before flash freezing in liquid nitrogen. (see Note 13)
Figure 1. A2AR baculovirus expression construct.
A2AR was cloned into the baculovirus transfer vector pBacPAK8 using BamHI and NotI restriction sites (underlined), and baculoviruses were prepared using the flashBAC ULTRA system. The construct consists of residues 1-308 of human A2AR followed by a TEV protease cleavage site (highlighted in green) and C-terminal 10x histidine tag (highlighted in red). A2AR contains the N154A mutation (highlighted in yellow) to remove a potential N-linked glycosylation site. Start and stop codons are shown in bold.
Figure 2. Illustration of the membrane pellet after ultracentrifugation.
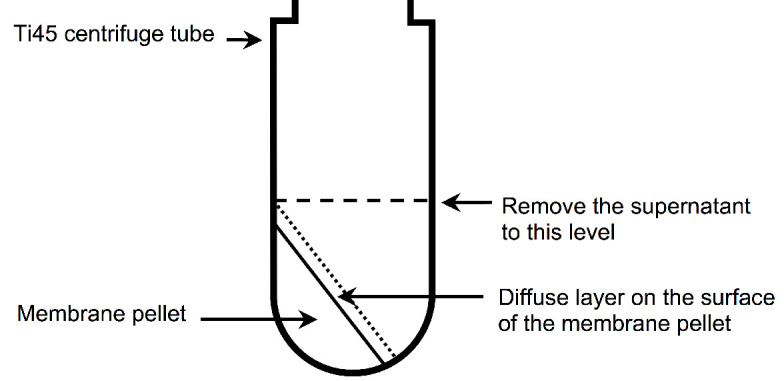
A diffuse layer, which contains a significant amount of A2AR, is observed on the surface of the membrane pellet after ultracentrifugation. It is important not to decant the supernatant, because this will remove the diffuse layer, resulting to a decreased yield of A2AR. Instead, the supernatant should be carefully aspirated using a serological pipette until level with the top of the membrane pellet (indicate by the dashed line).
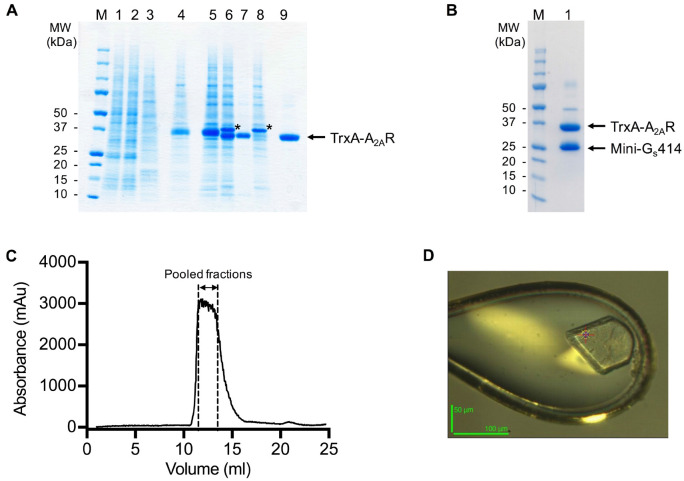
Figure 3. SDS-PAGE and gel filtration analysis of the A2AR purification.
A and B. SDS-PAGE analysis of the purification of a representative A2AR construct. Despite having molecular weights of 35 and 27 kDa respectively, A2AR and mini-Gs414 migrate at an identical rate on the SDS-PAGE gel, making visualisation of the complex difficult. Therefore, for demonstration purposes, we show SDS-PAGE gels of an A2AR construct that contains an N-terminal thioredoxin fusion (TrxA-A2AR), which increases its molecular weight to 46 kDa. TrxA-A2AR was purified in DM using an identical protocol to that described for A2AR, however, the TrxA-A2AR–mini-Gs complex, which was exchanged into OTG, did not yield crystals that diffracted to higher than 3.5 Å resolution. A. SDS-PAGE analysis of the TrxA-A2AR purification: (M) molecular weight markers; (1) Ni2+-NTA column loading material (1:10 dilution); (2) Ni2+-NTA column flow through (1:10 dilution); (3) Ni2+-NTA column wash; (4) Ni2+-NTA column eluate; (5) sample after concentration and buffer exchange; (6) sample after TEV digestion; (7) Ni2+-NTA negative purification flow through; (8) Ni2+-NTA negative purification eluate; (9) gel filtration pool. TEV protease is indicated by an asterisk. The samples should not be boiled prior to loading on the gel (see Note 11). B. SDS-PAGE analysis of the TrxA-A2AR–mini-Gs414 complex after detergent exchange into OTG: (M) molecular weight markers; (1) gel filtration pool. C. Preparative gel filtration chromatogram for the A2AR–mini-Gs414 complex in OTG, pooled fractions (typically 2-3 ml) are indicated by dashed lines. Note that the UV detector reaches saturation, which results in a truncated absorbance profile. D. A crystal of the A2AR–mini-Gs414 complex mounted in a loop on beamline ID23-2 at the European Synchrotron Radiation Facility (the scale bar is 50 x 100 μm).
Data analysis
Chromatograms were visualised using UNICORN software and graphs were plotted using GraphPad Prism 7. Statistical analysis was not required for this work.
Notes
The A2AR construct described here contains a deletion to remove the flexible C-terminus, the length of this deletion was one area of optimisation used to improve the diffraction of the A2AR–mini-Gs crystals. Crystallisation trials of the A2AR–mini-Gs complex were performed using A2AR constructs that consisted of residues 1-308, 1-311 or 1-317. The best diffraction was observed using the construct consisting of residues 1-308 of A2AR. The A2AR–mini-Gs structure ( Carpenter et al., 2016 ) revealed that the C-terminal region of A2AR is involved in crystal contacts, which explains why A2AR constructs of different lengths affected the crystallisation of the complex.
A C-terminal 10x histidine tag and TEV protease cleavage site were utilised because this strategy has previously been used by our lab to purify and crystallise a thermostabilised A2AR construct ( Lebon et al., 2011a and 2011b; Lebon and Tate, 2015). A 10x histidine tag is preferred over a 6x histidine tag because it binds Ni2+-NTA resin with higher affinity and therefore allows higher imidazole concentrations (up to 80 mM) to be used during the wash step, which significantly improves the purity of the receptor.
It is important to fill the Ti45 ultracentrifuge tubes correctly (to the neck). Partially filled tubes can collapse during centrifugation causing damage to the cap assembly and rotor.
Filtration of the supernatant usually requires six Steritop filter units due to its high viscosity and the presence of particulate matter. However, this step greatly extends the life of the Ni2+-NTA column, which can be regenerated and reused up to 10 times.
It is important to use a refrigerated centrifuge when concentrating A2AR to maintain the temperature at a constant 4 °C. A non-refrigerated centrifuge located in a cold room will rapidly warm up during operation, which will result in the denaturation of the temperature-sensitive receptor.
Following buffer exchange, and during the overnight incubation with TEV protease, a white precipitate may be observed. The composition of this precipitate is unknown, but it does not contain a significant amount of A2AR and can be removed by centrifugation at 5,000 × g for 5 min at 4 °C prior to the Ni2+-NTA negative purification step.
A2AR usually elutes from the preparative gel filtration column as a broad asymmetric peak of 3-4 ml in volume. The reason for this is unknown, but it may be due to excess detergent, carried over from the solubilisation and purification, affecting the migration of A2AR through the gel filtration matrix. After pooling and concentrating the peak fractions from the preparative gel filtration run, a small analytical sample of A2AR (~100 μg) can be re-run on the same column. This sample should resolve as a sharp peak and is a better indicator of the quality of the purified receptor than the preparative gel filtration profile.
The amido black assay (Schaffner and Weissmann, 1973) is one of the most reliable ways to accurately determine the concentration of membrane proteins in the presence of detergent and ligand.
A2AR that has been frozen and thawed retains its ability to form a complex with mini-Gs and this complex can be crystallised with similar results to those observed when using freshly prepared receptor. Buffer E contains 10% glycerol, so additional cryoprotectants are not required.
The shortest incubation time tested was three hours, after which the A2AR–mini-Gs complex was fully formed. However, an overnight incubation does not negatively affect the stability of the complex and is often more convenient.
Membrane protein samples should never be boiled before loading on SDS-PAGE because this causes the protein to aggregate.
Crystallisation plates must be set up and incubated at 4 °C, even short-term exposure to elevated temperatures will result in denaturation of the complex.
Cryoprotection using 30% PEG 400 resulted in visible shrinkage of the crystals. The extent of this dehydration varied between crystals, meaning that datasets collected from different crystals were difficult to merge. Increasing the concentration of the cryoprotectant in a stepwise manner resulted in more uniform dehydration of the crystals, and allowed the collection of diffraction data that could be reliably merged.
PMSF solutions must be prepared in an anhydrous solvent (such as absolute ethanol), because even a small water content will cause rapid hydrolysis of the PMSF rendering it inactive. PMSF stock solutions prepared in absolute ethanol are stable and can be conveniently stored at 4 °C for several months. Note that the PMSF solution should be warmed to room temperature before opening to prevent condensation entering the solvent.
Recipes
-
Buffer A
20 mM HEPES, pH 7.5
1 mM EDTA
1 mM PMSF (add immediately before use)
-
PMSF stock solution
200 mM in absolute ethanol, store at 4 °C (see Note 14)
-
NECA stock solution
10 mM in DMSO, store at -20 °C
-
DM stock solution
20% w/v in Milli-Q water, store at -20 °C
-
Buffer B
20 mM HEPES, pH 7.5
300 mM NaCl
10 mM imidazole
100 μM NECA (add immediately before use)
0.15% (w/v) DM (add immediately before use)
Filter using a 0.22 μm Steritop or syringe filter
-
Buffer C
20 mM HEPES, pH 7.5
500 mM NaCl
80 mM imidazole
10% (v/v) glycerol
100 μM NECA (add immediately before use)
0.15 % (w/v) DM (add immediately before use)
Filter using a 0.22 μm Steritop or syringe filter
-
Buffer D
20 mM HEPES, pH 7.5
100 mM NaCl
300 mM imidazole
10% (v/v) glycerol
100 μM NECA (add immediately before use)
0.15% (w/v) DM (add immediately before use)
Filter using a 0.22 μm Steritop or syringe filter
-
Buffer E
10 mM HEPES, pH 7.5
100 mM NaCl
10% (v/v) glycerol
100 μM NECA (add immediately before use)
0.15% (w/v) DM (add immediately before use)
Filter using a 0.22 μm Steritop or syringe filter
-
OTG stock solution
15% w/v in Milli-Q water, store at -20 °C
-
Apyrase stock solution
1 unit/µl in Milli-Q water, store at -20 °C
-
Buffer F
10 mM HEPES, pH 7.5
100 mM NaCl
1 mM MgCl2
100 μM NECA (add immediately before use)
0.35% (w/v) OTG (add immediately before use)
Filter using a 0.22 μm Steritop or syringe filter
Acknowledgments
This work was funded by a grant from Heptares Therapeutics Ltd and core funding from the Medical Research Council [MRC U105197215]. Parts of this protocol was adapted from previously described methods ( Lebon et al., 2011a and 2011b; Tate and Lebon, 2015; Carpenter et al., 2016). We thank Rony Nehmé for comments on the manuscript.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1.Carpenter B., Nehmé R., Warne T., Leslie A. G. and Tate C. G.(2016). Structure of the adenosine A2A receptor bound to an engineered G protein . Nature 536(7614): 104-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carpenter B. and Tate C. G.(2016). Engineering a minimal G protein to facilitate crystallisation of G protein-coupled receptors in their active conformation. Protein Eng Des Sel 29(12): 583-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carpenter B. and Tate C. G.(2017). Expression and purification of mini G proteins from Escherichia coli . Bio-protocol 7(8): e2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lebon G., Bennett K., Jazayeri A. and Tate C. G.(2011). Thermostabilisation of an agonist-bound conformation of the human adenosine A2A receptor . J Mol Biol 409(3): 298-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lebon G., Warne T., Edwards P.C., Bennett K., Langmead C.J., Leslie A.G. and Tate C.G.(2011) Agonist-bound adenosine A2A receptor structures reveal common features of GPCR activation. Nature 474: 521-5. [DOI] [PMC free article] [PubMed]
- 6.Schaffner W. and Weissmann C.(1973). A rapid, sensitive, and specific method for the determination of protein in dilute solution. Anal Biochem 56(2): 502-514. [DOI] [PubMed] [Google Scholar]
- 7.Tate C. G. and Lebon G.(2015). Purification and crystallization of a thermostabilized agonist-bound conformation of the human adenosine A2A receptor . Methods Mol Biol 1335: 17-27. [DOI] [PubMed] [Google Scholar]