Abstract
Next-generation DNA sequencing has revealed the complete genome sequences of numerous organisms, establishing a fundamental and growing understanding of genetic variation and phenotypic diversity. Engineering at the gene, network and whole-genome scale aims to introduce targeted genetic changes both to explore emergent phenotypes and to introduce new functionalities. Expansion of these approaches into massively parallel platforms establishes the ability to generate targeted genome modifications, elucidating causal links between genotype and phenotype, as well as the ability to design and reprogramme organisms. In this Review, we explore techniques and applications in genome engineering, outlining key advances and defining challenges.
Canadian author and Nobel Laureate Saul Bellow observed that “a writer is a reader moved to emulation” (REF. 1). Biological ‘reading’ and ‘writing’ are founded on pivotal discoveries regarding the structure, interpretation and manipulation of DNA, and have spurred a new biotechnological era in which the analysis and synthesis of whole genomes is now possible. Fruitful scientific investigations in the 1960s and 1970s led to the discovery of two ultimately interconnected techniques. Chain-termination sequencing developed by Frederick Sanger and Walter Gilbert provided an increasingly facile approach for reading the contents of genetic material2. Around the same time, investigations into bacterial resistance to bacteriophage infection led to the discovery of restriction endonucleases and DNA ligases3. Soon after, these natural mechanisms were used in recombinant DNA technologies to assemble DNA fragments controllably and reproducibly in the first examples of ‘copy-and-paste’ functionality.
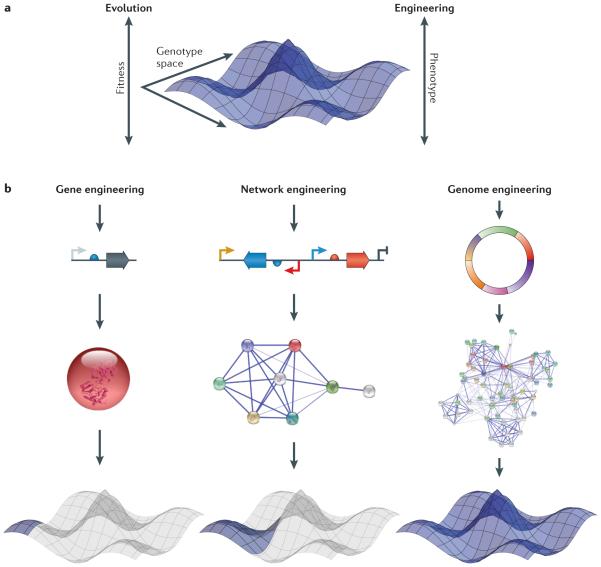
Early large-scale genome modification techniques (for example, transposon mutagenesis4) sought to affect phenotypic traits involving complex components but were limited by their untargeted nature, as much of the diversified population contained mutations that were unrelated to the cellular process of interest. Thus, studying numerous targeted mutations across genomes has been largely intractable. Genome engineering utilizes rationality and precision in genome design and mutagenesis to enable rapid and controlled exploration of a phenotype landscape (FIG. 1a). The goals of this process are varied and include elucidating combinatorial genetic variants that contribute to organism behaviour, producing expanded phenotype topologies via new construction components such as non-natural biochemical building blocks and increasing evolutionary robustness5 (FIG. 1b).
Figure 1. Genes, networks and genomes by design.
a | Both Darwinian evolution and rational genome engineering rely on the introduction of mutations to traverse a genotype space and explore the resultant changes in cellular phenotype. The high-dimensional nature of genotype landscapes presents a key challenge in resolving causal genotype–phenotype relationships and engineering organisms with prescribed behaviours. Genome engineering seeks to leverage existing knowledge, modern DNA manipulation techniques and evolution to search through complex genotype landscapes more efficiently. b | The genome can first be thought of as a physical collection of its constituent DNA elements. These constituent parts can be conceptualized as nodes in a network in which edges represent functional connections (for example, genes involved in the same cellular process). Networks can range in size from small collections of genes in biosynthetic clusters to the genome-scale network of all genes and cellular processes. The space of all possible genotypes can be visualized as a phenotypic landscape where each genotype is associated with a particular level of performance with respect to various phenotypic traits. Manipulation of the genome can alter the connections in cellular networks or add new nodes. These new networks can represent a new location in the phenotype landscape or alter the phenotype landscape itself. Changes at the gene and small-network level enable exploration of broadened phenotype landscapes, whereas large-scale genome modification can yield disruptive and novel fitness landscapes.
Laboratory studies of global evolution lack control over mutational processes, which limits the specific genotype landscapes explored over the course of a single experiment6. Furthermore, evidence suggests that evolution is driven by the cumulative effect of multiple, moderately beneficial mutations rather than individual events that become fixed across a population7. Coupling these observations elucidates unique challenges for genome engineering efforts: accessing multiple mutations within a single generation and enabling researchers to bypass adaptive valleys observed in non-engineered evolutionary processes.
Different conceptual approaches exist to traverse phenotype landscapes8. For example, de novo DNA synthesis and assembly techniques leverage extensive in silico design phases to anticipate and build a limited number of episomal constructs for characterization. Alternatively, as the effects of large-scale genomic changes on fitness are often unpredictable, many technologies seek to explore a large combinatorial space of genomic alterations. Rather than direct phenotyping, these larger chromosomal libraries necessitate sorting or selections in which beneficial elements of genotypic diversity are propagated via their phenotypes9. Uniquely, genome engineering approaches can be utilized to sample a targeted, combinatorial genetic landscape in order to elucidate causal links between genotype and phenotype.
The microbial biome is a rich, largely untapped resource10 that has a central role in ecological homeostasis11, and promises new medicinal compounds, routes to natural bioremediation and platforms for metabolic engineering. Owing to their relatively small genome sizes, well-described and malleable mechanisms of DNA repair, and fast reproduction rate, microbial genomes are favourable targets for genetic engineering. Genome sequencing and concurrent computational advances have established a knowledge base that now necessitates nucleotide resolution for functional genetic testing and reprogramming. A mutable, controllable genome is required to fully interrogate and manipulate the phenotype landscape. This Review is chiefly concerned with the rationale, technologies and applications of genome design. When coupled with next-generation sequencing (NGS), the iterative genome engineering process — design, build, test, evolve and learn — has become a powerful pipeline for the exploration of emergent and novel phenotypes.
Gene-scale engineering
Although many cellular properties are multifactorial and emerge from systems-level interactions encoded across many genomic loci, gene-scale engineering targets single genes contributing to a desired phenotype. Conceptually, we divide gene engineering efforts into two functions: the identification of a gene that is capable of eliciting a desired phenotype; and the targeted modification of a host genome. Directed evolution of single genes is a mature platform where a large, diverse library of sequences produced in vitro by methods such as DNA shuffling12 and PCR mutagenesis13 are subsequently screened or selected for function. An inherent limitation of directed evolution strategies is that the very large sequence libraries may contain gene variants that are capable of driving the desired phenotype at very low frequencies. One solution to this challenge is to reduce the complexity of the sequence space by first designing sequences to test in silico14 and then synthesizing them de novo.
These techniques create static, large libraries of DNA sequences that require researcher participation for further diversification and multiple rounds of selection. Solving the challenge of continuous evolution promises to enable autonomous diversity production. One such approach was identified in the field of biomolecule evolution in the form of phage-assisted continuous evolution15. By linking the expression of phage protein III (pIII) to the activity of their target gene, Liu and colleagues15 formulated a selection to engineer and modify bimolecular interactions of interest. Through suppressed proofreading16 and increased error-prone lesion bypass17, the authors suggest that the entirety of a random single- and double-mutant genotype landscape may be explored over a single evolutionary generation. Although the original platform was limited to selection by affinity (that is, positive selection), a subsequent study identified a dominant negative mutant of pIII that could be used to drive a negative selection, enabling evolution of T7 RNA polymerase enzymes with altered substrate specificities18.
One of the unifying factors of these gene design and selection strategies is that diversified libraries are typically cloned to plasmids or phage. In contrast to chromosomal targets, episomal constructs have downsides, including biosynthetic burden, poor persistence over time19, highly variable expression20 and an associated decrease in organismal fitness20,21. Genome engineering of the host chromosome (or chromosomes) circumvents pleiotropic plasmid effects while also ensuring the maintenance of physiological gene copy numbers.
Nuclease-based genome editing
Nuclease-based genome editing approaches enable gene disruption via non-homologous end-joining or targeted editing using homologous templates (FIG. 2a). Both zinc-finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs) are composed of an engineered DNA recognition domain fused to the FokI endonuclease catalytic domain22–24. Engineered ZFNs are assembled by linking individual modular domains with empirically determined target specificities23. TALENs utilize a similarly modular approach, but each domain contains repeat-variable di-residues that determine specificity in a highly reproducible manner23. The key difference between these targeting methods is that each zinc-finger module is thought to associate with a three-base sequence, whereas TALENs have single-base resolution. Both ZFNs and TALENs are areas of fruitful, ongoing investigation24,25.
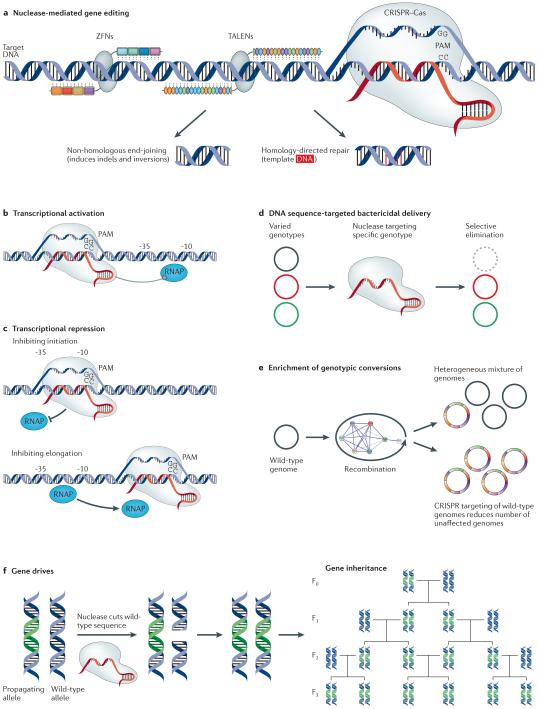
Figure 2. Nuclease-mediated genome engineering.
a | Nuclease-based gene editing using zinc-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs) or CRISPR–Cas targeting induces double-stranded breaks that are repaired in a template-dependent (homology-directed repair) or -independent (non-homologous end-joining) manner. b | Modified CRISPR–Cas systems have been used as transcriptional activators by stabilizing RNA polymerase (RNAP) binding at specified sites. c | Transcriptional repressors that use modified CRISPR–Cas to block transcriptional initiation or elongation have also been developed. d | Targeting CRISPR–Cas cleavage to sequences that are unique to a specific genome results in the selective removal of that genome from a bacterial population. e | In the context of recombination-mediated genome engineering, targeting CRISPR–Cas cleavage to the wild-type sequence has been used to remove unaltered cells and enrich for mutations. f | Gene drives using CRISPR–Cas systems cut homologous chromosomes and promote their own incorporation via homology-directed repair. Placement of constructs near genes of interest can facilitate their spread throughout a population. PAM, protospacer adjacent motif.
The emergence of CRISPR–Cas24 is rapidly changing the field of microbial genome editing (FIG. 2b,c). CRISPR–Cas9 is a bacterial adaptive immunity system that genomically stores foreign DNA in response to infection26. In type II CRISPR–Cas, arrays of foreign DNA are transcribed and processed into CRISPR–Cas9 RNAs (crRNAs), annealed to trans-activating crRNA (tracr-RNA) and complexed with the Cas9 endonuclease. This complex can then target foreign genetic material for cleavage22. It was later shown that the crRNA and tracr-RNA can be engineered into a single construct (known as guide RNA (gRNA)) that is sufficient for targeted nuclease activity27. The endonuclease complex cleaves a 23 bp target sequence comprising 20 bases of guide sequence and the required protospacer adjacent motif (PAM)24,28. Unlike target specificities for ZFNs and TALENs, target specificity for CRISPR–Cas is derived from the gRNA sequence rather than protein modules, and CRISPR–Cas9 systems are easily modified for, in principle, any genomic target proximal to a PAM. Numerous improvements have already been described to address off-target cleavage events29, including the use of paired nicking CRISPR–Cas9 (REF. 30) or FokI31. CRISPR–Cas9 has been shown to enable direct editing in many settings22,24, including human cells32–34. Furthermore, applications in transcriptional regulation35,36 (FIG. 2b,c), targeted bactericidal activity37,38 (FIG. 2d,e) and gene drives39,40 (FIG. 2f) have emerged rapidly. Here, we focus on two distinct genome engineering challenges: combinatorial genome editing and small chromosomal library insertion.
The natural function of CRISPR–Cas9 in adaptive bacterial immunity suggested that multiple encoded guide sequences might be tiled into a single array for multiplexed editing of a single genome. In their pioneering study, Cong et al.34 showed deletion of two targets in eukaryotic cells with a single array. Furthermore, a study in Saccharomyces cerevisiae reported 43% and 19% efficiencies for disrupting loss-of-function mutations when targeting two and three sites, respectively41. Although earlier examples of multiplexed editing used individual gRNA for each target site, this approach has several draw-backs, including transfection of multiple, rapidly diluted plasmids, and variable copy numbers42. One study used a single lentiviral vector expressing Cas and up to four gRNAs to convert four loci in three out of ten cells42. More recently, CRISPR–Cas9 complexes were used to transcriptionally activate up to ten sites in a controlled manner43, simultaneously showing that pooling of gRNAs led to decreased efficiencies. Although this result has not been recapitulated in the context of genome editing, these findings suggest that CRISPRs may have increased multiplexing capacity for gene disruption. However, organism-dependent upper limits on double-stranded breaks and nicks are currently unknown.
In addition to direct editing of arbitrary genomes, CRISPR–Cas9 may enable the use of chromosomal, rather than episomal, gene libraries. As discussed, plasmid libraries for gene-scale engineering have several limitations, whereas homologous recombination is too inefficient for DNA library insertion and requires numerous antibiotic markers. In a recent study, Ryan et al.41 reported that the use of error-prone PCR to create a library of gene mutants, followed by CRISPR–Cas9-mediated insertion into the S. cerevisiae genome, improved consumption of the industrially relevant compound disaccharide cellobiose. This report highlights the potential synergies between CRISPR–Cas9 gene editing and genome engineering in organisms with well-described mechanisms of homologous recombination. Given the rapid developments in CRISPR–Cas9 technologies, we anticipate numerous imminent advances that will improve upon mutation efficiency, expand the already broad range of substrate organisms and increase throughput as required to further develop these platforms for large-scale genome engineering. Furthermore, proof-of-concept multiplex CRISPR–Cas9 studies have largely addressed gene disruption, suggesting that efficient, high-throughput homology-directed repair with provided templates requires further investigation.
Recombineering for gene and network design
Whereas nuclease-based engineering technologies leverage DNA-binding domains to confer target specificity, recombineering (recombination-mediated genetic engineering) relies on homologous recombination to recombine single-stranded DNA (ssDNA) or double-stranded DNA in the genome (FIG. 3a). In seminal work, it was shown that functional replacement of Escherichia coli RecBCD with the phage lambda Red operon (including the Beta, Exo and Gam genes) yielded a strain that was capable of recombining short, linear DNA fragments at a significantly improved rate44,45. As a result, homologous recombination was made accessible by the use of primers and facile amplification with PCR. It has been proposed that the oligonucleotides act as synthetic Okazaki fragments and are incorporated in the lagging strand of the replication fork44,45. Subsequent work by Costantino and Court44 improved the recombination rate at a single locus by >100-fold by removing E. coli mismatch repair.
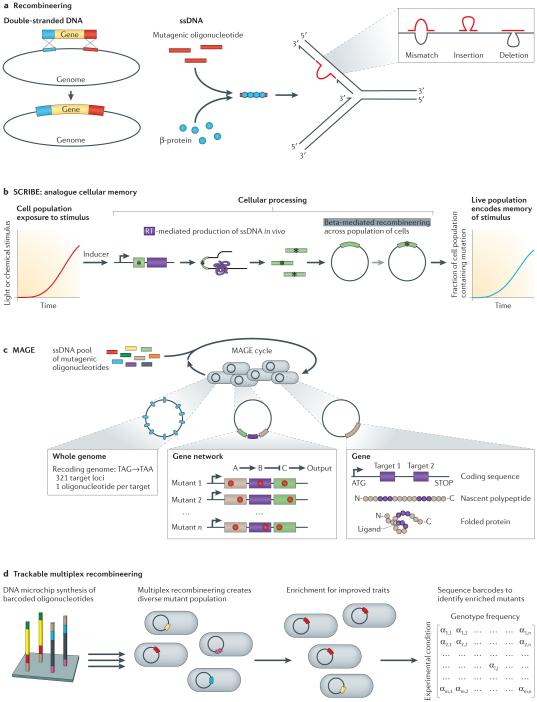
Figure 3. Gene and network-scale recombineering.
a|Recombineering (recombination-mediated genetic engineering) uses cellular homologous recombination enhanced by phage proteins to recombine single-stranded DNA (ssDNA) or double-stranded DNA into the genome. This enables site-specific introduction of mutations ranging in resolution from single base pair changes to the introduction of entire genes. b | The SCRIBE (Synthetic Cellular Recorders Integrating Biological Events) analogue memory system uses reverse transcriptase (purple) to produce ssDNA in vivo, which is then used in recombineering. SCRIBE is able to introduce heritable genetic changes (*) in response to a predetermined stimulus. These heritable changes act as a cellular memory of exposure to the stimulus. The fraction of cells containing this mutation serves as an approximation of the cumulative stimulus input function. c | Multiplex automated genome engineering (MAGE) uses the introduction and incorporation of synthetic ssDNA at the replication fork to introduce desired mutations. Mutations can be targeted to multiple residues in a single gene, multiple genes in a pathway, or across the genome. d | Trackable multiplex recombineering (TRMR) utilizes recombineering and high-throughput profiling to introduce mutations and to identify variants that provide a fitness benefit under different conditions. Each recombineering cassette contains a selectable marker, targeting homology domain, and a sequence to increase or decrease gene expression. Sequencing of genetic barcodes associated with each mutation reveals the resultant enrichment of mutations that confer a fitness benefit.
Recombineering with the lambda Red genes and homologues has been extended from E. coli to numerous organisms46–48, and has been combined with CRISPR–Cas9 and gRNA to enable 65% mutation efficiency in E. coli 49. By utilizing a dual-RNA–Cas9-directed cleavage to kill unmated cells, the authors showed that direct selection against non-edited cells is a powerful approach for efficient recovery of mutated genomes. Although this method has a high probability of yielding a desired genotype, it yields relatively few surviving clones, limiting potential library size. Notably, the same study observed nearly 100% CRISPR–Cas9 efficiencies in the highly recombinogenic Streptococcus pneumoniae, again suggesting41 that the host recombination machinery may prove to be a crucial adjuvant for CRISPR–Cas9 in microorganisms in which recombineering is not possible.
Recently, recombineering was utilized to create an analogue memory system in vivo50. The SCRIBE (Synthetic Cellular Recorders Integrating Biological Events) approach uses an encoded reverse transcriptase to produce a hybrid RNA–ssDNA molecule from an arbitrary provided sequence (FIG. 3b). When utilized alongside the Beta protein, this approach drove recombination at approximately 10−4 events per generation. By linking the expression of these components to inputs such as small-molecule inducers or optogenetic control, the authors describe a cumulative effect function in which the proportion of the population containing a mutation is proportional to the total cellular input signal, regardless of distribution over time. We anticipate that the key innovation presented in the SCRIBE study — that is, the use of reverse transcriptase to drive in vivo oligonucleotide production coupled with lambda Red recombination — will provide myriad downstream applications in genome engineering. For example, error-prone RNA polymerases51 or reverse transcriptases52 may enable the production of diversified oligonucleotides for autonomous in vivo evolution. This study also suggests that multi-site continuous recombination is possible, but will require concomitant increases in efficiency.
Network design technologies
Chromosomal network engineering
Recombineering technologies laid the groundwork for parameter optimization and the ability to target multiple genomic loci for oligonucleotide-mediated allelic replacement (FIG. 3c). Inspired by advances in ssDNA recombineering, multiplex automated genome engineering (MAGE) uses lambda Red recombination and synthetic phosphorothioate-protected oligonucleotides to introduce targeted mutations across the genome53,54, enabling user-defined indels or mismatches with efficiencies of ≤30%. Although MAGE was developed with the goal of whole-genome redesign and recoding of E. coli (as discussed below), stochastic incorporation of oligonucleotides across multiple genetic loci enabled the rapid creation of combinatorial genetic diversity within a cell population and facilitated its immediate use for pathway diversification. In a proof-of-concept demonstration of targeted, multi-site genome diversification, MAGE was used to optimize the 1-deoxy-d-xylulose-5-phosphate (DXP) pathway for lycopene production. The ribosome-binding sites of 20 enzymes in the DXP pathway were targeted using oligonucleotides containing partial degeneracy biased towards Shine–Dalgarno consensus sequences, enabling the identification of optimal cellular production from the diversified population. Additional nonsense mutations were targeted to four enzymes in competing pathways in order to maximize flux through the DXP pathway. The combination of these efforts yielded strains that were capable of up to a fivefold increase in lycopene production and, through comparison of mutations identified by sequencing, the authors elucidated the causal genetic basis for improved phenotypes. Subsequent work has shown that selections can dramatically increase observed allelic-replacement frequencies55,56 and have been used in the context of promoter engineering57. DNA microarrays can be a facile oligonucleotide source for lambda Red genome engineering58, as shown in a proof-of-concept study that created a library of ~13,000 130 nt ssDNA oligonucleotides (BOX 1). These experiments highlight the importance of creating combinatorial genetic diversity and subsequent characterization when optimizing network phenotypes.
Box 1. Large-scale DNA synthesis and assembly.
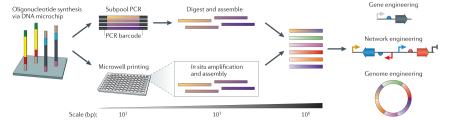
Advances in Sanger sequencing and next-generation sequencing (NGS) have enabled reading of genomic texts, whereas de novo DNA synthesis serves as the author’s ink. Solid-phase phosphoramidite chemistries that developed in the 1980s are the workhorses of column-based oligonucleotide synthesis methods, enabling the production of short single-stranded DNA (ssDNA) with moderate parallelization (≤384 oligonucleotides)19. New, large-scale DNA microchip-based synthesis methods look to provide a scale that parallels NGS more closely129–131 (FIG. 6).
DNA sourced from microarrays permits high-density synthesis of ~105 customized oligonucleotides129,132. Microarray synthesis yields approximately picomolar quantities, requiring PCR amplification of select pools of oligonucleotides. Two complementary approaches, barcoding with pool amplification133 and microwell assembly134 coupled with error correction129,131, have enabled the use of microarray-based oligonucleotide pools129,135 as a low-cost source for large numbers of oligonucleotides. Gene synthesis, the overlap assembly of short oligonucleotides (~50–200 nt) into larger fragments, relies on an array of approaches, including PCR136, circular amplification137 and enzymatic assembly138,139 (see the figure).
As shown in the figure, DNA microchip oligonucleotide synthesis can be coupled with either subpool PCR or microwell printing for the construction of intermediate-length constructs, which can then be assembled in vitro or in vivo for use in gene, network and genome engineering.
Building on a seminal study on microarray-based synthesis of translational genes131 and subsequent improvements in oligonucleotide quality, one study by Goodman et al.81 used a library of >14,000 microarray-synthesized combinations of promoters, ribosome-binding sites and amino-terminal codons to elucidate roles of codon bias in bacterial protein expression, showing that large synthetic DNA libraries can provide a platform for testing hypotheses about fundamental biological questions. De novo synthesis has become a viable and increasingly facile tool for the production of libraries of rational designer genes.
As efforts turn to bridging these advanced DNA synthesis capabilities with de novo synthesis of whole genomes79, the ‘design, build and test’ engineering design cycle will require significant optimization. Synthesis affords complete control over new genome architectures, but informing the design process remains a challenge. Over the past 5 years, microarray-based synthesis and associated assembly technologies have increased quality and reduced the cost of large-scale gene synthesis, establishing preliminary progress on future multiplexed synthesis and assembly of refactored pathways69. Efforts to scale towards synthesis of multiple genomes will require improved assembly technologies performed in vitro and in vivo with the aid of automation. Striking advances in rapid and facile phenotypic characterization will be essential for the testing of designer genomes.
The selection of MAGE targets requires previous knowledge of the biological processes under investigation. Trackable multiplex recombineering (TRMR) utilizes barcoded synthetic DNA cassettes to introduce expression-modulating mutations across the genome, enabling the identification of fitness-improving genes under previously uncharacterized biological or environmental conditions59 (FIG. 3d). Each cassette is designed to contain a selectable marker, homology domains that target it to an individual gene in E. coli, and a sequence designed either to increase or to decrease gene expression. With this system, Gill and colleagues60 were able to generate a mutant population in which more than 95% of E. coli genes had modified expression. The authors illustrated the utility of TRMR by exposing the mutant population to various environmental conditions and measuring the differential abundance of mutants before and after the population exposure. The abundance of each mutant was determined by sequencing the barcodes present in the two populations, subsequently enabling identification of genes that improved fitness. In one experiment, TRMR revealed several mutations that enabled cells to better tolerate lignocellulosic hydrolysate, including mutations affecting vitamin metabolic processes and in the gene encoding adenylate cyclase60. This broad, genome-scale approach can help to establish novel connections between genes and cellular fitness.
Further advances in multiplexed recombineering are still required, as the platform has only been explored in a select number of hosts with variable efficiency61,62, and ongoing researcher input is required to continue cycling cells through the experimental pipeline. Recent work has enabled oligonucleotide-based genome engineering alongside active mismatch repair machinery through the use of modified nucleotides, among other approaches63. Future combinations of these multiplexed recombineering approaches and continuous in vivo oligonucleotide production via reverse transcriptases promise to dramatically increase the efficiencies and range of genomic alterations. Bacterial artificial chromosomes are known to tolerate large synthetic constructs64 and may be capable of driving the production of >103 oligonucleotides for continuous and autonomous multi-site genome modifications in vivo. Oligonucleotide-mediated engineering is a facile, inexpensive, and powerful approach for large-scale, in vivo genome manipulation.
Episomal network engineering
In many cases, it is desirable to import functionality from unculturable, slow-growing, or otherwise challenging organisms into a more facile host such as E. coli. One such example, the 32 kb 6-deoxyerythronolide B synthase gene cluster, provided an early proof of concept for the de novo synthesis of whole biological networks65 (FIG. 4a). Genes in polyketide synthase (PKS) clusters are kilobases in size, and alteration of individual modules can lead to the biosynthesis of novel polyketides. To facilitate this exchange, a subsequent study introduced design elements to the PKS cluster in the form of unique restriction sites, enabling the assembly of 154 combinations of PKS clusters66. These studies reflect synergies between bioprospecting (the systematic search for medically or industrially relevant pathways and small molecules), massively parallel genome sequencing and de novo DNA synthesis, enabling the importing and optimization of frequently minimally expressed pathways in genetically tractable hosts.
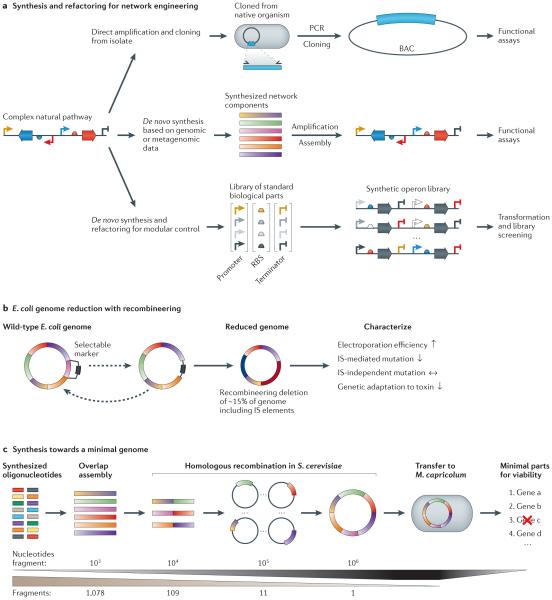
Figure 4. Refactored pathways and minimal genomes.
a | There are numerous approaches to the transfer of gene clusters. Biosynthetic clusters can be cloned from their native organism and transferred to another organism for heterologous expression. For organisms that are recalcitrant to laboratory culture techniques, gene sequences of interest can be identified from genomic or metagenomic databases and subsequently synthesized and introduced into a production organism. The native regulatory elements of a heterologous pathway may not be optimal for expression in the host organism. Refactoring approaches seek to rebuild the pathways using well-characterized modular regulatory elements and removing all native regulation. b | Recombineering (recombination-mediated genetic engineering) was used to delete up to 15% of the wild-type Escherichia coli K12 genome, including insertion sequence (IS) elements. This led to a more stable and streamlined genetic architecture that manifested itself in altered cellular phenotypes. c | The de novo synthesis of the Mycoplasma mycoides genome utilized synthetic DNA oligonucleotides assembled first in vitro and then in Saccharomyces cerevisiae. Following completion of the ~1 Mb genome, the whole construct was transplanted to Mycoplasma capricolum, yielding a viable organism. BAC, bacterial artificial chromosome; RBS, ribosome-binding site.
One appealing method to disentangle and control the components of a complex genetic circuit is to begin by refactoring the interwoven molecular parts into modular, defined units67,68. This engineering goal is intended to facilitate inter-species transfer and, simultaneously, to enable exogenous control over the system69. In a pioneering study by Endy and colleagues68, nearly 30% of the T7 bacteriophage genome was replaced with engineered DNA, resulting in viable phage that was simultaneously simpler to model and manipulate. In recent work, Voigt and colleagues67 used large-scale synthesis to address a long-standing challenge in understanding and optimizing the nitrogen fixation (nif) gene cluster in Klebsiella oxytoca for use in a facile host67,69. Using a refactored nif cluster, the authors designed a combinatoric library of 3 promoters, 12 ribosome binding sites, 7 spacers and 6 genes that underwent codon optimization across a total of 84 constructs. They then introduced the constructs into a K. oxytoca strain deficient for the six genes and screened for function, recovering a number of well-performing clusters. They repeated this process iteratively across different subsets of the nif cluster as well as on the entire 16-gene pathway. This study was able to close the design loop by performing functional assays, RNA sequencing, collecting growth information and feeding these data back into the cluster design. Although transfer to E. coli led to modest functionality, the computational (‘design’), synthesis (‘build’), and experimental (‘test’) pipelines in this study suggest a broad range of applications for heterologous pathway redesign, characterization and optimization. This work will benefit from future applications of recombineering and network mutagenesis technologies53,54,70 that introduce further genetic diversity to the synthesized pathways, effectively using de novo synthesis as a scaffold for in vivo evolution.
Although many of the discussed methods, from directed evolution to de novo pathway synthesis and refactoring primarily focus on the addition of gene function, recent developments in transcriptome engineering may provide the inverse functionality. Even though RNA interference (RNAi) is not utilized in bacteria, synthetic RNAs (sRNAs) composed of scaffold and target-binding sequences have been shown to be capable of strongly repressing gene expression in E. coli71–73. Similar efforts in S. cerevisiae have used engineered RNA regulators74,75 as well as RNAi76,77 to enable rapid strain prototyping in yeast. Taken together, these transcriptome modification technologies have shown great promise for facile, rapid and scalable engineering in microorganisms.
Design of reduced and chimeric genomes
The commoditization of DNA sequencing and synthesis and the emergence of large-scale assembly (BOX 1) and mutagenesis techniques have enabled genome-scale design and manipulation. One long-term area of investigation rewarded by these advances is the search for a minimal microbial genome64,78,79, which can then be used as an efficient foundation for next-generation applications. The first landmark in this effort was the transposon mutagenesis of Mycoplasma genitalium, which found that 55–73% of the 517 genes were essential80, a figure later revised to 79%81. In related work, Blattner and colleagues82 utilized recombineering in E. coli K12 to investigate phenotypic outcomes of reduced genome size5,6,82 (FIG. 4b). Rather than pursuing random mutagenesis, the authors used bioinformatics approaches to identify genome segments that were unique to K12 and then used recombineering to delete up to 15% of the genome, yielding a strain with increased transformation efficiencies. By specifically targeting insertion sequence (IS) elements and other mobile DNA elements that are frequently involved in recombination, the engineered strains displayed both increased genomic stability and the ability to maintain plasmids with deleterious genes.
The first whole-genome transplantation83, complete chemical synthesis of the M. genitalium chromosome64, and the synthesis followed by transplantation of the Mycoplasma mycoides genome79 marked important proof of concepts in genome engineering64,79 (FIG. 4c). In the case of the ~1 Mb M. mycoides genome, 11 ~100 kb assemblies were built from short DNA oligonucleotides using overlap assembly and then homologous recombination in S. cerevisiae. In parallel, constructs containing 1 of the 11 assemblies and supplemented with the remainder of the wild-type M. mycoides genome were used to evaluate the function of individual components. Although the first-stage intermediates were sequence-verified, one ~100 kb assembly contained a single point mutation in dnaA that precluded transplantation. Upon repair of this mutation and reassembly of the genome, the team was able to successfully transplant and sequence the M. mycoides genome in Mycoplasma capricolum.
This remarkable series of studies suggests that it may be possible to produce functional cells from pre-determined sequences. At the same time, because the de novo synthesis strategy relies on assembly outside the target cellular context, design errors or point mutations may dramatically alter the desired organismal phenotype. The cloning of a chimeric Synechocystis PCC6803 and Bacillus subtilis genome140 suggests that attention to design constraints (for example, a single ribosomal RNA species for each genome) or key culturing parameters may be required for genome construction efforts. Indeed, the synthesis of larger and more complex genomes may reveal otherwise masked epistatic relationships. The complete syntheses of Mycoplasma genomes and others84 have inspired genome engineering based on DNA synthesis and, when combined with other design principles such as refactoring, may deliver ‘plug-and-play’ production-optimized genomes.
Genomic recoding and orthogonal biology
Genetic code expansion
A central goal of genome-scale engineering is the ability to fundamentally redesign genomes to explore and uncover novel phenotypic landscapes. Questions regarding the redundancy, mutability, and structure of the genetic code are challenging to address6, and reconstruction of alternative genetic codes was, until recently, infeasible. Historically, genetic code expansion has the aim of increasing the biochemical diversity of translated proteins. Beyond the 20 standard amino acids, some species are able to utilize selenocysteine85 and pyrrolysine86, whereas further chemical space is accessible to many organisms via post-translational modifications (PTMs). Synthetic chemistry has enabled in vitro access to a wide variety of compounds, but these are not necessarily accessible to the replicating cell. Thus, with the vision of creating biological foundries and organisms with novel properties, considerable efforts have been extended towards the expansion of the cellular amino acid repertoire87.
Early studies recognized that chemically aminoacylated tRNA88 or amino acid auxotrophs89 could be used to drive utilization of a non-standard amino acid (nsAA). It was quickly recognized that in vivo genetically encoded translation using a nsAA would require the design of orthogonal translation systems (OTSs)90,91. An OTS is composed of a tRNA that does not interact with native aminoacyl-tRNA synthetases (aaRSs) and an aaRS that specifically and uniquely charges the tRNA with a nsAA. Furthermore, the orthogonal tRNA must then suppress a codon that does not otherwise encode a standard amino acid. The first such example in vivo utilized the tyrosyl-tRNA aaRS and tRNA from Methanococcus jannaschii to encode O-methyl-l-tyrosine in response to the amber stop codon TAG92. Although other methods including quadruplet-decoding ribosomes have found successful application93,94, amber suppression has become the most common method of genetic code expansion, partially because it is the least abundant codon across the genome.
Amber suppression has enabled the DNA template-controlled incorporation of diverse chemistries by >100 nsAAs, including tyrosyl-derivatives95 (see REF. 91 for an extensive list), and PTMs96,97. The multiple instances of TAG codons throughout the genome lead to uncontrolled proteome-wide nsAA incorporation, affecting cellular fitness. Furthermore, competition between the amber-suppressing tRNA and release factor 1 (RF1) limits the production of nsAA-containing polypeptides. For this reason, numerous efforts were made to alter both RF1 and RF2 (REFS 98,99) or to delete RF1 and supply essential TAG-terminating genes episomally100. These findings were extended by the discovery that the conversion of seven TAG codons to TAA in essential genes enabled deletion of RF1 and a subsequent increase in protein yields at the cost of cellular fitness97. One consequence of this process was the rapid evolution of glutamine amber suppressors, possibly relieving ribosomal stalling at more than 300 remaining TAG sites101.
Genomic recoding
The confluence of numerous technologies has enabled the first genome-wide replacement of a single codon and reassignment as an open coding channel101 (FIG. 5a). Recognizing that the genome of E. coli contains 321 TAG codons, MAGE53,54 was used to convert TAG codons to TAA in 32 separate strains102. By maintaining evolutionary context in vivo, each set of mutations could be evaluated for viability and phenotypic effects. As the E. coli genome was being edited, rather than transplanted79 or incorporated103,104, a new method for conjugative assembly genome engineering (CAGE)102,105 was developed. CAGE utilizes conjugation to hierarchically combine distinct genotypes from engineered E. coli into a single strain, enabling compilation of four partially recoded strains. In a subsequent study, it was shown that full genomic recoding enabled RF1 deletion and reassignment of the TAG codon to incorporate nsAA101. This process was without observable pleiotropic effects associated with genomic recoding (for example, the emergence of tRNA suppressors).
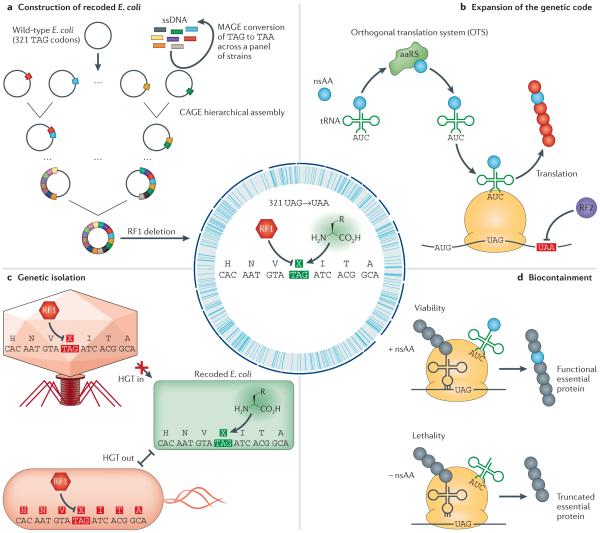
Figure 5. Genome recoding and new biological functions.
a | The genomically recoded organism was constructed by first using multiplex automated genome engineering (MAGE) to convert TAG stop codons to TAA in a panel of strains. Conjugative assembly genome engineering (CAGE) was then used for the hierarchical assembly of these partially recoded genomes into a fully recoded genome in which all 321 TAG stop codons are reassigned. Release factor 1 (RF1) was then deleted in order to establish TAG as an open codon for the incorporation of non-standard amino acids (nsAAs). b | The addition of an orthogonal translation system capable of charging a nsAA to a TAG-recognizing tRNA enables the genetic encoding of nsAA-incorporating proteins. c | The reassignment of TAG from a stop codon can render TAG-containing or -terminating genes non-functional in a different organism. This reduces the probability of horizontal gene transfer (HGT) between recoded and natural organisms. d | The incorporation of TAG codons in essential genes provides a method for the biocontainment of recoded organisms by linking its viability to the presence of an exogenous nsAA. aaRS, aminoacyl-tRNA synthetase.
The resultant genomically recoded organism (GRO) reveals a dramatically altered phenotypic landscape. As there are no TAG codons present in the genome, nsAA incorporation via amber suppression is exclusively directed by the DNA templates provided and uncontested by RF1, leading to high purity and yields of diverse proteins (FIG. 5b). Second, the recoded E. coli exhibits increased resistance to T7 bacteriophage, revealing that genome recoding may interfere with the function of genes transferred from viruses, between species and across ecosystems (FIG. 5c). Genes ending with TAG that are transferred into the recoded E. coli will not be translated properly, and TAG-containing genes transferred from this strain will result in premature truncation. These proof-of-concept horizontal gene transfer experiments suggest that further recoding101,106 will enable more stringent genetic isolation and multi-virus resistance.
The long-term evolutionary outcomes of genetic code expansion in E. coli or other hosts are as yet unknown6. Despite deep conservation of the genetic code, there is evidence of TAG and TAG codon reassignments in bacteriophages and TAG reassignments in bacteria107. A recent study involving exposure of phage to 3-iodotyrosine-incorporating E. coli revealed that the phage mutated to preferentially incorporate the nsAA in the lysis-involved protein type II holin, yielding an improved evolutionary phenotype108. Similar experiments in E. coli exploring evolution in the context of non-natural biochemical building blocks109,110 may require the development of OTSs with improved functionality as current platforms are several orders of magnitude less efficient than native aaRS–tRNA pairs95,111–113, providing a potential counter-selection to widespread nsAA incorporation.
Recoding of other codons appears to be a plausible aim, as evidenced by a study that utilized DNA microchips to investigate removal of 13 rare codons from a panel of 42 highly expressed essential genes106. This report observed that synonymous codons used at a single locus within an essential gene yielded unpredictable variability, reinforcing the importance of a rapid design and test process in vivo. Furthermore, evolutionarily conserved codons may be recalcitrant to even synonymous mutations. Two more notable challenges await these efforts. First, even the rarest remaining codons have thousands of instances across the genome. Second, precise re-engineering of translational components will be required to discriminate between required natural codon function and reassigned codons. For example, converting a sense codon in E. coli into an open channel will require elimination of tRNA or altering the specificity of tRNA participating in wobble base-pairing interactions, and engineering of aaRSs that are capable of discriminating between natural and orthogonal tRNA.
Beyond modified inter-species barriers and the production of biomolecules with novel functions, the combination of genomic recoding and nsAA incorporation offers a novel mechanism to dynamically alter an organism’s fitness landscape. A pair of recent studies established that the recoded E. coli tolerates the insertion of nsAAs into essential genes in both functionally conserved residues (for example, the active site of a protein)110 and computationally redesigned protein cores109. Additional genome engineering could then render the organism dependent on nsAA supplementation for viability — a condition that could not be rescued with metabolic cross-feeding or mutation in cis (FIG. 5d). These results indicate that genetic code expansion can be used to create phenotype landscapes that are dependent on the presence of synthetic amino acids.
Design principles applied to synthetic yeast chromosomes
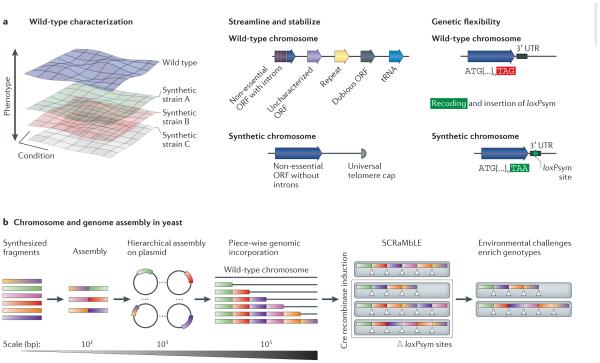
The synthesis of the S. cerevisiae chromosome IX right arm (IXR) blended the synthesis of ~9 × 104 bases with three design principles for a synthetic genome: first, an approximate wild-type phenotype; second, the removal of destabilizing elements; and third, the introduction of genetic flexibility104 (FIG. 6a). By removing retrotransposons, subtelomeric repeats, introns and relocating tRNA genes, the authors sought to increase genomic stability. To increase genetic flexibility, two key changes were made. Although gene order was maintained, symmetrical loxP (loxPsym) sites114 were added to the 3′ untranslated regions of non-essential genes, forming the SCRaMbLE toolkit. By activating a Cre recombinase, this system enables the large-scale induction of genomic inversions and deletions, producing a large pool of genetic diversity (FIG. 6b) across a population of living cells. This mutated library of cells can then be studied for diverse emergent phenotypes in an unbiased manner. By using group II introns to insert loxP sites in microorganisms, Ellington and colleagues115 showed that the same recombinase system can be used to create large chromosomal inversions, insertions and deletions.
Figure 6. Design principles and assembly: synthetic yeast genome project.
a | Design principles in the synthetic yeast genome project include: wild-type phenotypes, unburdened and stabilized chromosomes via the removal of numerous types of genetic elements, and increased genetic flexibility through recoding of all TAG codons to TAA and introduction of symmetrical loxP (loxPsym) sites after non-essential genes. b | Synthetic DNA assembled in vitro is incorporated into the genome in a piece-wise manner, permitting phenotype characterization of each hierarchical segment. Induction of Cre recombinase drives activity of loxPsym, leading to chromosomal rearrangements that eliminate and translocate segments, enabling rapid exploration of dramatically altered genotypes. ORF, open reading frame; UTR, untranslated region.
The second facet of genetic flexibility was the recoding of TAG stop codons to TAA in a manner analogous to the genomically recoded E. coli101,102. Alongside the synthetic IXR (synIXR), the authors designed a semisynthetic segment of the left arm of chromosome VI (semisynVIL). Of note, when incorporated into host strains, neither major fitness effects nor compensatory transcriptomic changes were observed. With the assistance of an undergraduate course116, a subsequent study carried this project to its logical conclusion with the complete synthesis of a 2 × 105 bp S. cerevisiae chromosome 3 (synIII)103. In contrast to prior work in Mycoplasma, in this first example of a synthetic eukaryotic chromosome, the conversion occurred in the target organism, rather than via an intermediate, and it remains to be seen whether this process can be replicated for the remainder of the genome. Three key distinctions of this approach are as follows: the introduction of new design features into the eukaryotic genome; an iterative synthesis and characterization approach capable of detecting deleterious mutations before final assembly; and parameters that enable future genome evolution via SCRaMbLE.
Perspectives
In his book History of Writing, Fischer suggests that “all modern society rests on writing’s plinth” (REF. 117). To date, reading capacity in the biomedical sciences has far outpaced writing capability, but recent developments reflect dramatic progress in this area. As described in this Review, numerous recent approaches for genome design and engineering have enabled a breadth of applications, including the production of biomolecules101, elucidating the complex genetic basis of phenotypes and strengthening the barriers between laboratory organisms and the environment101,109,110. Near-term technological goals have been identified, including improvements in multiplexing capability34 and target specificity for nuclease29 and oligonucleotide methods. Furthermore, de novo synthesis and refactoring of pathways69 will benefit from ongoing in silico improvements and integration with in vivo methods that permit combinatorial expansion of genetic landscapes53, rapid functional genetic testing and characterization of new genome designs. We anticipate that breakthroughs in these areas will lead to their adoption by the wider biological community.
We envision that the future of genome engineering sits at the interface between in vivo genome evolution and de novo synthesis. As phenotypes are fundamentally unpredictable, it is essential to explore a combinatorial space of genotypes, rather than a single, static design. The scale of phenotype exploration is a factor of two variables: ease of genotypic diversification; and power of selection. De novo synthesis alone is currently capable of assembling thousands of user-defined genes, hundreds of networks and single genomes for individual iteration, but in vivo evolution strategies produce billions of combinatorial variants per day54,57. The union of design, de novo synthesis and in vivo evolution will lead to the creation of large and diverse populations, highlighting that both computation and phenotype selections are key limitations.
The vast combinatorial complexity enabled by genome engineering technologies is too large to be explored through experiments alone. Computational approaches such as computational protein folding118 and flux balance analysis119 can be used to help to refine the space in which genome engineering is applied. Computational analyses of protein sequence or pathway conservation may help to narrow the scope of mutations to be created and evaluated120. Additional benefits can be obtained through the use of computational genome design tools and models that are iteratively refined through multiple rounds of experiments.
Screens and selections are commonly used to evaluate mutant populations and isolate enriched mutations. Screens such as colorimetric assays and fluorescence-activated cell sorting (FACS) provide researchers with a quick and easily measured readout for the biological property under optimization121. However, screening the large number of variants created in many genome-engineering workflows with limited-throughput screening assays can quickly become impractical.
Selections provide a more efficient method for identifying high-performing variants. Given the vast space of possible mutants, this efficiency is key to identifying rare, high-performing mutants, which might be missed in a screen15. However, selective pressures can often lead to enrichment of ‘cheaters’ that have evolved an alternative path to fitness that is decoupled from the biological activity being optimized. Rounds of positive and negative selection, together with incremental increases in the stringency of selection, have been shown to reduce the prevalence of such ‘cheaters’ and improve the enrichment of high-performing variants122. Effective selections are dependent on establishing a strong correlation between fitness and the biological property being optimized.
Alongside screens and selections, in vivo sensors can be used to enrich complex populations of cells for desirable genotypes or phenotypes. Many metabolite-specific markers have been identified and used to optimize production in various biochemical pathways122,123. However, the lack of in vivo sensors for all molecules of interests limits the application of this approach. Connection of protein domains that are capable of acting as biosensors to domains that are able to elicit a biological response offers promise as a method for linking biosensor recognition of a desired compound to a selectable physiological response. Biosensors that are able to link biochemical changes to a transcriptional response have been shown to be effective in metabolic engineering124. No universal solution exists, although recent work suggests that RNA and proteins provide viable sensors for many desirable biomolecules122,125,126.
Analogue, rather than digital, output signals offer a unique opportunity to increase the selection sensitivity for variables such as small-molecule and metabolite concentrations or gene expression levels. Colorimetric assays have already been utilized in varied genome engineering studies53,127, but the next step is to link analogue signalling to selections in order to reduce the time and effort spent on complex phenotype characterization69. We expect that the increasingly sophisticated circuits under development will contribute substantially to these aims128. The development of more flexible and powerful selections will fully enable rapid genome design manifesting organisms that are capable of sampling new evolutionary and fitness landscapes.
Acknowledgements
The authors thank M. Lajoie, the Isaacs laboratory and the anonymous reviewers for helpful comments on this paper. A.D.H. acknowledges support from US National Cancer Institute (NCI) award 1F30CA196191-01. P.M. is supported by the Raymond and Beverly Sackler Institute for Biological, Physical and Engineering Sciences. F.J.I. gratefully acknowledges support from the US Defense Advanced Research Projects Agency (N66001-12-C-4020, N66001-12-C-4211); US Department of Energy (152339.5055249.100); Gen9 Inc., DuPont Inc., and the Arnold and Mabel Beckman Foundation.
Glossary
- Directed evolution
A process that utilizes mutagenesis and a user-defined selective pressure to develop populations that are enriched for mutants exhibiting a desired function or behaviour.
- Nuclease-based genome editing
The use of engineered nucleases with a DNA-targeting mechanism (for example, a zinc-finger nuclease (ZFN), transcription activator-like effector nuclease (TALEN) or a clustered regularly interspaced short palindromic repeat (CRISPR)) for targeted mutagenesis in vivo.
- CRISPR–Cas
A bacterial adaptive immune system comprising clustered regularly interspaced short palindromic repeats (CRISPRs) and CRISPR-associated (Cas) endonucleases that has been repurposed for genome editing.
- Recombineering
(Recombination-mediated genetic engineering). A technique that utilizes homologous recombination to alter genetic loci using synthetic single-stranded DNA and double-stranded DNA.
- Bioprospecting
Searching and sourcing useful genes from other organisms to specifically confer a desired phenotype (for example, medically or industrially relevant pathways and small molecules).
- Refactoring
Reorganization of biological systems or pathways with the goal of improving the ease and predictability of future engineering efforts. This often entails the removal of native regulation, separation of overlapping genetic elements and the use of well-characterized regulatory elements.
- Codon optimization
A multifactorial process that utilizes synonymous codons to reflect the preferred codon frequency in order to optimize protein expression in a target species.
- Gene synthesis
De novo construction of user-defined double-stranded DNA.
- Genetic code expansion
Modification of the genetic code to enable site-directed incorporation of amino acids beyond the canonical 20 into proteins.
- Non-standard amino acid
(nsAA). An amino acid beyond the canonical 20 present in the genetic code. nsAAs generally fall into two classes: post-translational modifications (for example, phosphoserine); and synthetic amino acids that do not exist in nature.
- Orthogonal translation systems
(OTSs). Used in genetic code expansion to link incorporation of a non-standard amino acid (nsAA) to a target codon. These systems are composed of a tRNA that is selectively charged with a given nsAA by a cognate aminoacyl-tRNA synthetase (aaRS). Other possible components of OTSs include mutant elongation factor Tu and ribosomes designed to accommodate the given nsAA.
- Genomically recoded organism
(GRO). An organism in which codons have been reassigned to create an alternate genetic code. For example, an Escherichia coli strain in which all instances of the TAG stop codon have been mutated to TAA and release factor 1 (RF1) has been deleted, enabling the use of TAG as an open coding channel.
Footnotes
Competing interests statement
The authors declare no competing interests.
References
- 1.Johnson A. The Hidden Writer. Random House; 1997. [Google Scholar]
- 2.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc. Natl Acad. Sci. USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Loenen WA, Dryden DT, Raleigh EA, Wilson GG, Murray NE. Highlights of the DNA cutters: a short history of the restriction enzymes. Nucleic Acids Res. 2014;42:3–19. doi: 10.1093/nar/gkt990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simon R, Priefer U, Puhler AA. Broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat. Biotechnol. 1983;1:784–791. [Google Scholar]
- 5.Renda BA, Hammerling MJ, Barrick JE. Engineering reduced evolutionary potential for synthetic biology. Mol. Biosyst. 2014;10:1668–1678. doi: 10.1039/c3mb70606k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pal C, Papp B, Posfai G. The dawn of evolutionary genome engineering. Nat. Rev. Genet. 2014;15:504–512. doi: 10.1038/nrg3746. [DOI] [PubMed] [Google Scholar]
- 7.Desai MM, Fisher DS, Murray AW. The speed of evolution and maintenance of variation in asexual populations. Curr. Biol. 2007;17:385–394. doi: 10.1016/j.cub.2007.01.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Silver PA, Way JC, Arnold FH, Meyerowitz JT. Synthetic biology: engineering explored. Nature. 2014;509:166–167. doi: 10.1038/509166a. [DOI] [PubMed] [Google Scholar]
- 9.Neylon C. Chemical and biochemical strategies for the randomization of protein encoding DNA sequences: library construction methods for directed evolution. Nucleic Acids Res. 2004;32:1448–1459. doi: 10.1093/nar/gkh315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bunge J, Willis A, Walsh F. Estimating the number of species in microbial diversity studies. Annu. Rev. Stat. Appl. 2014;1:427–445. [Google Scholar]
- 11.Pace NR. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 12.Stemmer WP. Rapid evolution of a protein in vitro by DNA shuffling. Nature. 1994;370:389–391. doi: 10.1038/370389a0. [DOI] [PubMed] [Google Scholar]
- 13.Jackel C, Kast P, Hilvert D. Protein design by directed evolution. Annu. Rev. Biophys. 2008;37:153–173. doi: 10.1146/annurev.biophys.37.032807.125832. [DOI] [PubMed] [Google Scholar]
- 14.Das R, Baker D. Macromolecular modeling with Rosetta. Annu. Rev. Biochem. 2008;77:363–382. doi: 10.1146/annurev.biochem.77.062906.171838. [DOI] [PubMed] [Google Scholar]
- 15.Esvelt KM, Carlson JC, Liu DR. A system for the continuous directed evolution of biomolecules. Nature. 2011;472:499–503. doi: 10.1038/nature09929. This paper describes a mutagenesis and selection system that links a desired function to viral fitness and enables the continuous, directed evolution of proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fijalkowska IJ, Schaaper RM. Mutants in the Exo I motif of Escherichia coli dnaQ: defective proofreading and inviability due to error catastrophe. Proc. Natl Acad. Sci. USA. 1996;93:2856–2861. doi: 10.1073/pnas.93.7.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Opperman T, Murli S, Smith BT, Walker GC. A model for a umuDC-dependent prokaryotic DNA damage checkpoint. Proc. Natl Acad. Sci. USA. 1999;96:9218–9223. doi: 10.1073/pnas.96.16.9218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carlson JC, Badran AH, Guggiana-Nilo DA, Liu DR. Negative selection and stringency modulation in phage-assisted continuous evolution. Nat. Chem. Biol. 2014;10:216–222. doi: 10.1038/nchembio.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sorensen SJ, Bailey M, Hansen LH, Kroer N, Wuertz S. Studying plasmid horizontal transfer in situ: a critical review. Nat. Rev. Microbiol. 2005;3:700–710. doi: 10.1038/nrmicro1232. [DOI] [PubMed] [Google Scholar]
- 20.Bouma JE, Lenski RE. Evolution of a bacteria/ plasmid association. Nature. 1988;335:351–352. doi: 10.1038/335351a0. [DOI] [PubMed] [Google Scholar]
- 21.Betenbaugh MJ, Beaty C, Dhurjati P. Effects of plasmid amplification and recombinant gene expression on the growth kinetics of recombinant E. coli. Biotechnol. Bioengineer. 1989;33:1425–1436. doi: 10.1002/bit.260331110. [DOI] [PubMed] [Google Scholar]
- 22.Gersbach CA. Genome engineering: the next genomic revolution. Nat. Methods. 2014;11:1009–1011. doi: 10.1038/nmeth.3113. [DOI] [PubMed] [Google Scholar]
- 23.Gaj T, Gersbach CA, Barbas CF. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim H, Kim JS. A guide to genome engineering with programmable nucleases. Nat. Rev. Genet. 2014;15:321–334. doi: 10.1038/nrg3686. [DOI] [PubMed] [Google Scholar]
- 25.Miller JC, et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat. Biotechnol. 2007;25:778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- 26.Barrangou R, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 27.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. This seminal paper uncovers the Cas9 endonuclease guided by dual RNAs for site-specific DNA cleavage and presents the use of a single chimeric RNA for programming. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- 29.Fu Y, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ran FA, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsai SQ, et al. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat. Biotechnol. 2014;32:569–576. doi: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jinek M, et al. RNA-programmed genome editing in human cells. eLife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bikard D, et al. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 2013;41:7429–7437. doi: 10.1093/nar/gkt520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gilbert LA, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Citorik RJ, Mimee M, Lu TK. Sequence-specific antimicrobials using efficiently delivered RNA-guided nucleases. Nat. Biotechnol. 2014;32:1141–1145. doi: 10.1038/nbt.3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bikard D, et al. Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nat. Biotechnol. 2014;32:1146–1150. doi: 10.1038/nbt.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gantz VM, Bier E. Genome editing. The mutagenic chain reaction: a method for converting heterozygous to homozygous mutations. Science. 2015;348:442–444. doi: 10.1126/science.aaa5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.DiCarlo JE, Chavez A, Dietz SL, Esvelt KM, Church GM. RNA-guided gene drives efficiently reversibly bias inheritance wild yeast. BioRxiv. 2015 http://dx.doi.org/10.1101/013896.
- 41.Ryan OW, et al. Selection of chromosomal DNA libraries using a multiplex CRISPR system. eLife. 2014;3:e03703. doi: 10.7554/eLife.03703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kabadi AM, Ousterout DG, Hilton IB, Gersbach CA. Multiplex CRISPR/Cas9-based genome engineering from a single lentiviral vector. Nucleic Acids Res. 2014;42:e147. doi: 10.1093/nar/gku749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Konermann S, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Costantino N, Court DL. Enhanced levels of lambda Red-mediated recombinants in mismatch repair mutants. Proc. Natl Acad. Sci. USA. 2003;100:15748–15753. doi: 10.1073/pnas.2434959100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Court DL, Sawitzke JA, Thomason LC. Genetic engineering using homologous recombination. Annu. Rev. Genet. 2002;36:361–388. doi: 10.1146/annurev.genet.36.061102.093104. [DOI] [PubMed] [Google Scholar]
- 46.Binder S, Siedler S, Marienhagen J, Bott M, Eggeling L. Recombineering in Corynebacterium glutamicum combined with optical nanosensors: a general strategy for fast producer strain generation. Nucleic Acids Res. 2013;41:6360–6369. doi: 10.1093/nar/gkt312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y, et al. Bacillus subtilis genome editing using ssDNA with short homology regions. Nucleic Acids Res. 2012;40:e91. doi: 10.1093/nar/gks248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sawitzke JA, et al. Recombineering: in vivo genetic engineering in E. coli, S. enterica, and beyond. Methods Enzymol. 2007;421:171–199. doi: 10.1016/S0076-6879(06)21015-2. [DOI] [PubMed] [Google Scholar]
- 49.Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Farzadfard F, Lu TK. Genomically encoded analog memory with precise in vivo DNA writing in living cell populations. Science. 2014;346:1256272. doi: 10.1126/science.1256272. This work describes the generation of target mutations through the in vivo production of ssDNA and co-expression of recombineering machinery. The accumulation of these mutations in the population in response to various input signals acts as a genetic memory device. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brakmann S, Grzeszik S. An error-prone T7 RNA polymerase mutant generated by directed evolution. ChemBioChem. 2001;2:212–219. doi: 10.1002/1439-7633(20010302)2:3<212::AID-CBIC212>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 52.Bebenek K, Abbotts J, Wilson SH, Kunkel TA. Error-prone polymerization by HIV-1 reverse transcriptase. Contribution of template-primer misalignment, miscoding, and termination probability to mutational hot spots. J. Biol. Chem. 1993;268:10324–10334. [PubMed] [Google Scholar]
- 53.Wang HH, et al. Programming cells by multiplex genome engineering and accelerated evolution. Nature. 2009;460:894–898. doi: 10.1038/nature08187. This paper describes the use of efficient oligonucleotide-mediated recombineering to create targeted, multi-site diversity across the E. coli genome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gallagher RR, Li Z, Lewis AO, Isaacs FJ. Rapid editing and evolution of bacterial genomes using libraries of synthetic DNA. Nat. Protoc. 2014;9:2301–2316. doi: 10.1038/nprot.2014.082. [DOI] [PubMed] [Google Scholar]
- 55.Gregg CJ, et al. Rational optimization of tolC as a powerful dual selectable marker for genome engineering. Nucleic Acids Res. 2014;42:4779–4790. doi: 10.1093/nar/gkt1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Carr PA, et al. Enhanced multiplex genome engineering through co-operative oligonucleotide co-selection. Nucleic Acids Res. 2012;40:e132. doi: 10.1093/nar/gks455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang HH, et al. Genome-scale promoter engineering by coselection MAGE. Nat. Methods. 2012;9:591–593. doi: 10.1038/nmeth.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bonde MT, et al. Direct mutagenesis of thousands of genomic targets using microarray-derived oligonucleotides. ACS Synth. Biol. 2015;4:17–22. doi: 10.1021/sb5001565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mansell TJ, Warner JR, Gill RT. Trackable multiplex recombineering for gene-trait mapping in E. coli. Methods Mol. Biol. 2013;985:223–246. doi: 10.1007/978-1-62703-299-5_12. [DOI] [PubMed] [Google Scholar]
- 60.Warner JR, Reeder PJ, Karimpour-Fard A, Woodruff LB, Gill RT. Rapid profiling of a microbial genome using mixtures of barcoded oligonucleotides. Nat. Biotechnol. 2010;28:856–862. doi: 10.1038/nbt.1653. [DOI] [PubMed] [Google Scholar]
- 61.DiCarlo JE, et al. Yeast oligo-mediated genome engineering (YOGE) ACS Synth. Biol. 2013;2:741–749. doi: 10.1021/sb400117c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pines G, Freed EF, Winkler JD, Gill RT. Bacterial recombineering: genome engineering via phage-based homologous recombination. ACS Synth. Biol. 2015 doi: 10.1021/acssynbio.5b00009. http://dx.doi.org/10.1021/acssynbio.5b00009. [DOI] [PubMed]
- 63.Wang HH, Xu G, Vonner AJ, Church G. Modified bases enable high-efficiency oligonucleotide-mediated allelic replacement via mismatch repair evasion. Nucleic Acids Res. 2011;39:7336–7347. doi: 10.1093/nar/gkr183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gibson DG, et al. Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science. 2008;319:1215–1220. doi: 10.1126/science.1151721. [DOI] [PubMed] [Google Scholar]
- 65.Kodumal SJ, et al. Total synthesis of long DNA sequences: synthesis of a contiguous 32-kb polyketide synthase gene cluster. Proc. Natl Acad. Sci. USA. 2004;101:15573–15578. doi: 10.1073/pnas.0406911101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huang J, Koide A, Makabe K, Koide S. Design of protein function leaps by directed domain interface evolution. Proc. Natl Acad. Sci. USA. 2008;105:6578–6583. doi: 10.1073/pnas.0801097105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Temme K, Zhao D, Voigt CA. Refactoring the nitrogen fixation gene cluster from Klebsiella oxytoca. Proc. Natl Acad. Sci. USA. 2012;109:7085–7090. doi: 10.1073/pnas.1120788109. In this study, the essential genes in a heterologous biosynthetic pathway are identified, removed from their native context, and placed under well-characterized regulatory elements to facilitate more predictable pathway engineering. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chan LY, Kosuri S, Endy D. Refactoring bacteriophage T7. Mol. Syst. Biol. 2005;1 doi: 10.1038/msb4100025. 2005.0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Smanski MJ, et al. Functional optimization of gene clusters by combinatorial design and assembly. Nat. Biotechnol. 2014;32:1241–1249. doi: 10.1038/nbt.3063. [DOI] [PubMed] [Google Scholar]
- 70.Horwitz AA, et al. Efficient multiplexed integration of synergistic alleles and metabolic pathways in yeasts via CRISPR-Cas. Cell Syst. 2015 doi: 10.1016/j.cels.2015.02.001. http://dx.doi.org/10.1016/j. cels.2015.02.001. [DOI] [PubMed]
- 71.Isaacs FJ, et al. Engineered riboregulators enable post-transcriptional control of gene expression. Nat. Biotechnol. 2004;22:841–847. doi: 10.1038/nbt986. [DOI] [PubMed] [Google Scholar]
- 72.Isaacs FJ, Dwyer DJ, Collins JJ. RNA synthetic biology. Nat. Biotechnol. 2006;24:545–554. doi: 10.1038/nbt1208. [DOI] [PubMed] [Google Scholar]
- 73.Na D, et al. Metabolic engineering of Escherichia coli using synthetic small regulatory RNAs. Nat. Biotechnol. 2013;31:170–174. doi: 10.1038/nbt.2461. [DOI] [PubMed] [Google Scholar]
- 74.Bayer TS, Smolke CD. Programmable ligand-controlled riboregulators of eukaryotic gene expression. Nat. Biotechnol. 2005;23:337–343. doi: 10.1038/nbt1069. [DOI] [PubMed] [Google Scholar]
- 75.Qi LS, Arkin AP. A versatile framework for microbial engineering using synthetic non-coding RNAs. Nat. Rev. Microbiol. 2014;12:341–354. doi: 10.1038/nrmicro3244. [DOI] [PubMed] [Google Scholar]
- 76.Drinnenberg IA, et al. RNAi in budding yeast. Science. 2009;326:544–550. doi: 10.1126/science.1176945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Crook NC, Schmitz AC, Alper HS. Optimization of a yeast RNA interference system for controlling gene expression and enabling rapid metabolic engineering. ACS Synth. Biol. 2014;3:307–313. doi: 10.1021/sb4001432. [DOI] [PubMed] [Google Scholar]
- 78.Hutchison CA, et al. Global transposon mutagenesis and a minimal Mycoplasma genome. Science. 1999;286:2165–2169. doi: 10.1126/science.286.5447.2165. [DOI] [PubMed] [Google Scholar]
- 79.Gibson DG, et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science. 2010;329:52–56. doi: 10.1126/science.1190719. This work describes the creation of a synthetic genome from synthesized DNA fragments using a combination of in vitro assembly, propagation in E. coli and recombination cloning in S. cerevisiae. The resultant genome was successfully transplanted into a recipient cell to generate a cell run by the synthetic genome. [DOI] [PubMed] [Google Scholar]
- 80.Bornscheuer UT, Pohl M. Improved biocatalysts by directed evolution and rational protein design. Curr. Opin. Chem. Biol. 2001;5:137–143. doi: 10.1016/s1367-5931(00)00182-4. [DOI] [PubMed] [Google Scholar]
- 81.Goodman DB, Church GM, Kosuri S. Causes and effects of N-terminal codon bias in bacterial genes. Science. 2013;342:475–479. doi: 10.1126/science.1241934. [DOI] [PubMed] [Google Scholar]
- 82.Posfai G, et al. Emergent properties of reduced-genome Escherichia coli. Science. 2006;312:1044–1046. doi: 10.1126/science.1126439. This study uses recombination and selection methods to remove parts of the E. coli genome in a targeted and planned manner, and evaluates the resultant phenotypes of E. coli with reduced genomes. [DOI] [PubMed] [Google Scholar]
- 83.Lartigue C, et al. Genome transplantation in bacteria: changing one species to another. Science. 2007;317:632–638. doi: 10.1126/science.1144622. [DOI] [PubMed] [Google Scholar]
- 84.Fleishman SJ, et al. RosettaScripts: a scripting language interface to the Rosetta macromolecular modeling suite. PLoS ONE. 2011;6:e20161. doi: 10.1371/journal.pone.0020161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bock A, et al. Selenocysteine: the 21st amino acid. Mol. Microbiol. 1991;5:515–520. doi: 10.1111/j.1365-2958.1991.tb00722.x. [DOI] [PubMed] [Google Scholar]
- 86.Srinivasan G, James CM, Krzycki JA. Pyrrolysine encoded by UAG in Archaea: charging of a UAG-decoding specialized tRNA. Science. 2002;296:1459–1462. doi: 10.1126/science.1069588. [DOI] [PubMed] [Google Scholar]
- 87.Ambrogelly A, Palioura S, Soll D. Natural expansion of the genetic code. Nat. Chem. Biol. 2007;3:29–35. doi: 10.1038/nchembio847. [DOI] [PubMed] [Google Scholar]
- 88.Noren CJ, Anthony-Cahill SJ, Griffith MC, Schultz PG. A general method for site-specific incorporation of unnatural amino acids into proteins. Science. 1989;244:182–188. doi: 10.1126/science.2649980. [DOI] [PubMed] [Google Scholar]
- 89.van Hest JCM, Kiick KL, Tirrell DA. Efficient incorporation of unsaturated methionine analogues into proteins in vivo. J. Am. Chem. Soc. 2000;122:1282–1288. [Google Scholar]
- 90.Chin JW. Expanding and reprogramming the genetic code of cells and animals. Annu. Rev. Biochem. 2014;83:379–408. doi: 10.1146/annurev-biochem-060713-035737. [DOI] [PubMed] [Google Scholar]
- 91.Liu CC, Schultz PG. Adding new chemistries to the genetic code. Annu. Rev. Biochem. 2010;79:413–444. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- 92.Wang L, Brock A, Herberich B, Schultz PG. Expanding the genetic code of Escherichia coli. Science. 2001;292:498–500. doi: 10.1126/science.1060077. This paper describes the development of an OTS that is capable of site-specifically incorporating a nsAA into proteins in E. coli. [DOI] [PubMed] [Google Scholar]
- 93.Neumann H, Wang K, Davis L, Garcia-Alai M, Chin JW. Encoding multiple unnatural amino acids via evolution of a quadruplet-decoding ribosome. Nature. 2010;464:441–444. doi: 10.1038/nature08817. [DOI] [PubMed] [Google Scholar]
- 94.Anderson JC, et al. An expanded genetic code with a functional quadruplet codon. Proc. Natl Acad. Sci. USA. 2004;101:7566–7571. doi: 10.1073/pnas.0401517101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.O’Donoghue P, Ling J, Wang YS, Soll D. Upgrading protein synthesis for synthetic biology. Nat. Chem. Biol. 2013;9:594–598. doi: 10.1038/nchembio.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Park HS, et al. Expanding the genetic code of Escherichia coli with phosphoserine. Science. 2011;333:1151–1154. doi: 10.1126/science.1207203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Heinemann IU, et al. Enhanced phosphoserine insertion during Escherichia coli protein synthesis via partial UAG codon reassignment and release factor 1 deletion. FEBS Lett. 2012;586:3716–3722. doi: 10.1016/j.febslet.2012.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Johnson DB, et al. RF1 knockout allows ribosomal incorporation of unnatural amino acids at multiple sites. Nat. Chem. Biol. 2011;7:779–786. doi: 10.1038/nchembio.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Johnson DB, et al. Release factor one is nonessential in Escherichia coli. ACS Chem. Biol. 2012;7:1337–1344. doi: 10.1021/cb300229q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mukai T, et al. Codon reassignment in the Escherichia coli genetic code. Nucleic Acids Res. 2010;38:8188–8195. doi: 10.1093/nar/gkq707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lajoie MJ, et al. Genomically recoded organisms expand biological functions. Science. 2013;342:357–360. doi: 10.1126/science.1241459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Isaacs FJ, et al. Precise manipulation of chromosomes in vivo enables genome-wide codon replacement. Science. 2011;333:348–353. doi: 10.1126/science.1205822. References 101 and 102 describe genome engineering technologies and the construction of a recoded strain of E. coli (that is, a GRO) in which all TAG codons have been converted to TAA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Annaluru N, et al. Total synthesis of a functional designer eukaryotic chromosome. Science. 2014;344:55–58. doi: 10.1126/science.1249252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dymond JS, et al. Synthetic chromosome arms function in yeast and generate phenotypic diversity by design. Nature. 2011;477:471–476. doi: 10.1038/nature10403. This study describes the design and construction of partially synthetic yeast chromosomes that incorporate multiple design features aimed at improving the ease of future evolution and engineering. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ma NJ, Moonan DW, Isaacs FJ. Precise manipulation of bacterial chromosomes by conjugative assembly genome engineering. Nat. Protoc. 2014;9:2285–2300. doi: 10.1038/nprot.2014.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lajoie MJ, et al. Probing the limits of genetic recoding in essential genes. Science. 2013;342:361–363. doi: 10.1126/science.1241460. [DOI] [PubMed] [Google Scholar]
- 107.Ivanova NN, et al. Stop codon reassignments in the wild. Science. 2014;344:909–913. doi: 10.1126/science.1250691. [DOI] [PubMed] [Google Scholar]
- 108.Hammerling MJ, et al. Bacteriophages use an expanded genetic code on evolutionary paths to higher fitness. Nat. Chem. Biol. 2014;10:178–180. doi: 10.1038/nchembio.1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mandell DJ, et al. Biocontainment of genetically modified organisms by synthetic protein design. Nature. 2015;518:55–60. doi: 10.1038/nature14121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rovner AJ, et al. Recoded organisms engineered to depend on synthetic amino acids. Nature. 2015;518:89–93. doi: 10.1038/nature14095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Umehara T, et al. N-acetyl lysyl-tRNA synthetases evolved by a CcdB-based selection possess N-acetyl lysine specificity in vitro and in vivo. FEBS Lett. 2012;586:729–733. doi: 10.1016/j.febslet.2012.01.029. [DOI] [PubMed] [Google Scholar]
- 112.Ling J, Reynolds N, Ibba M. Aminoacyl-tRNA synthesis and translational quality control. Annu. Rev. Microbiol. 2009;63:61–78. doi: 10.1146/annurev.micro.091208.073210. [DOI] [PubMed] [Google Scholar]
- 113.Tanrikulu IC, et al. Discovery of Escherichia coli methionyl-tRNA synthetase mutants for efficient labeling of proteins with azidonorleucine in vivo. Proc. Natl Acad. Sci. USA. 2009;106:15285–15290. doi: 10.1073/pnas.0905735106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hoess RH, Wierzbicki A, Abremski K. The role of the loxP spacer region in P1 site-specific recombination. Nucleic Acids Res. 1986;14:2287–2300. doi: 10.1093/nar/14.5.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Enyeart PJ, et al. Generalized bacterial genome editing using mobile group II introns and Cre-lox. Mol. Syst. Biol. 2013;9:685. doi: 10.1038/msb.2013.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Dymond JS, et al. Teaching synthetic biology, bioinformatics and engineering to undergraduates: the interdisciplinary Build-a-Genome course. Genetics. 2009;181:13–21. doi: 10.1534/genetics.108.096784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Fischer SR. History of Writing. Reaktion Books Ltd; 2001. [Google Scholar]
- 118.Dill KA, MacCallum JL. The protein-folding problem, 50 years on. Science. 2012;338:1042–1046. doi: 10.1126/science.1219021. [DOI] [PubMed] [Google Scholar]
- 119.Lewis NE, Nagarajan H, Palsson BO. Constraining the metabolic genotype–phenotype relationship using a phylogeny of in silico methods. Nat. Rev. Microbiol. 2012;10:291–305. doi: 10.1038/nrmicro2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Martin CH, Nielsen DR, Solomon KV, Prather KL. Synthetic metabolism: engineering biology at the protein and pathway scales. Chem. Biol. 2009;16:277–286. doi: 10.1016/j.chembiol.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 121.Mattanovich D, Borth N. Applications of cell sorting in biotechnology. Microb. Cell Fact. 2006;5:12. doi: 10.1186/1475-2859-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Raman S, Rogers JK, Taylor ND, Church GM. Evolution-guided optimization of biosynthetic pathways. Proc. Natl Acad. Sci. USA. 2014;111:17803–17808. doi: 10.1073/pnas.1409523111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zhang F, Carothers JM, Keasling JD. Design of a dynamic sensor-regulator system for production of chemicals and fuels derived from fatty acids. Nat. Biotechnol. 2012;30:354–359. doi: 10.1038/nbt.2149. [DOI] [PubMed] [Google Scholar]
- 124.Zhang F, Keasling J. Biosensors and their applications in microbial metabolic engineering. Trends Microbiol. 2011;19:323–329. doi: 10.1016/j.tim.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 125.Auslander S, et al. A general design strategy for protein-responsive riboswitches in mammalian cells. Nat. Methods. 2014;11:1154–1160. doi: 10.1038/nmeth.3136. [DOI] [PubMed] [Google Scholar]
- 126.Breaker RR. Prospects for riboswitch discovery and analysis. Mol. Cell. 2011;43:867–879. doi: 10.1016/j.molcel.2011.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mitchell LA, et al. Versatile genetic assembly system (VEGAS) to assemble pathways for expression in S. cerevisiae. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv466. http://dx.doi. org/10.1093/nar/gkv466. [DOI] [PMC free article] [PubMed]
- 128.Daniel R, Rubens JR, Sarpeshkar R, Lu TK. Synthetic analog computation in living cells. Nature. 2013;497:619–623. doi: 10.1038/nature12148. [DOI] [PubMed] [Google Scholar]
- 129.Kosuri S, Church GM. Large-scale de novo DNA synthesis: technologies and applications. Nat. Methods. 2014;11:499–507. doi: 10.1038/nmeth.2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Roy S, Caruthers M. Synthesis of DNA/RNA and their analogs via phosphoramidite and H-phosphonate chemistries. Molecules. 2013;18:14268–14284. doi: 10.3390/molecules181114268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Tian J, et al. Accurate multiplex gene synthesis from programmable DNA microchips. Nature. 2004;432:1050–1054. doi: 10.1038/nature03151. In this paper, array-based DNA synthesis, oligonucleotide selection, and assembly are described. These processes enable parallel gene synthesis. [DOI] [PubMed] [Google Scholar]
- 132.Cleary MA, et al. Production of complex nucleic acid libraries using highly parallel in situ oligonucleotide synthesis. Nat. Methods. 2004;1:241–248. doi: 10.1038/nmeth724. [DOI] [PubMed] [Google Scholar]
- 133.Kosuri S, et al. Scalable gene synthesis by selective amplification of DNA pools from high-fidelity microchips. Nat. Biotechnol. 2010;28:1295–1299. doi: 10.1038/nbt.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Quan J, et al. Parallel on-chip gene synthesis and application to optimization of protein expression. Nature Biotechnol. 2011;29:449–452. doi: 10.1038/nbt.1847. [DOI] [PubMed] [Google Scholar]
- 135.Heller MJ. DNA microarray technology: devices, systems, and applications. Annu. Rev. Biomed. Eng. 2002;4:129–153. doi: 10.1146/annurev.bioeng.4.020702.153438. [DOI] [PubMed] [Google Scholar]
- 136.Stemmer WP, Crameri A, Ha KD, Brennan TM, Heyneker HL. Single-step assembly of a gene and entire plasmid from large numbers of oligodeoxyribonucleotides. Gene. 1995;164:49–53. doi: 10.1016/0378-1119(95)00511-4. [DOI] [PubMed] [Google Scholar]
- 137.Bang D, Church GM. Gene synthesis by circular assembly amplification. Nat. Methods. 2008;5:37–39. doi: 10.1038/nmeth1136. [DOI] [PubMed] [Google Scholar]
- 138.Gibson DG. Synthesis of DNA fragments in yeast by one-step assembly of overlapping oligonucleotides. Nucleic Acids Res. 2009;37:6984–6990. doi: 10.1093/nar/gkp687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 140.Itaya M, Tsuge K, Koizumi M, Fujita K. Combining two genomes in one cell: stable cloning of the Synechocystis PCC6803 genome in the Bacillus subtilis 168 genome. Proc. Natl Acad. Sci. USA. 2005;102:15971–15976. doi: 10.1073/pnas.0503868102. [DOI] [PMC free article] [PubMed] [Google Scholar]