Abstract
A rationally-designed library of ternary siRNA polyplexes was developed and screened for gene silencing efficacy in vitro and in vivo with the goal of overcoming both cell-level and systemic delivery barriers. [2-(dimethylamino)ethyl methacrylate] (DMAEMA) was homopolymerized or colpolymerized (50 mol% each) with butyl methacrylate (BMA) from a reversible addition – fragmentation chain transfer (RAFT) chain transfer agent, with and without pre-conjugation to polyethylene glycol (PEG). Both single block polymers were tested as core-forming units, and both PEGylated, diblock polymers were screened as corona-forming units. Ternary siRNA polyplexes were assembled with varied amounts and ratios of core-forming polymers to PEGylated corona-forming polymers. The impact of polymer composition/ratio, hydrophobe (BMA) placement, and surface PEGylation density was correlated to important outcomes such as polyplex size, stability, pH-dependent membrane disruptive activity, biocompatibility, and gene silencing efficiency. The lead formulation, DB4-PDB12, was optimally PEGylated not only to ensure colloidal stability (no change in size by DLS between 0 and 24 hr) and neutral surface charge (0.139 mV) but also to maintain higher cell uptake (>90% positive cells) than the most densely PEGylated particles. The DB4-PDB12 polyplexes also incorporated BMA in both the polyplex core- and corona-forming polymers, resulting in robust endosomolysis and in vitro siRNA silencing (~85% protein level knockdown) of the model gene luciferase across multiple cell types. Further, the DB4-PDB12 polyplexes exhibited greater stability, increased blood circulation time, reduced renal clearance, increased tumor biodistribution, and greater silencing of luciferase compared to our previously-optimized, binary parent formulation following intravenous (i.v.) delivery. This polyplex library approach enabled concomitant optimization of the composition and ratio of core- and corona-forming polymers (indirectly tuning PEGylation density) and identification of a ternary nanomedicine optimized to overcome important siRNA delivery barriers in vitro and in vivo.
Keywords: Gene delivery, Nanomedicine, RNA interference, Pharmacokinetics, Cancer
Graphical Abstract
Introduction
RNA interference (RNAi) was initially observed by Mello and Fire et al. almost twenty years ago[1], yet cellular and systemic delivery barriers have continued to limit the clinical application of siRNA.[2] Naked siRNAs do not readily enter cells, have no inherent mechanism for endosome escape, and are rapidly cleared through filtration in the kidneys after systemic administration.[3, 4] Thus, the use of siRNA as a safe and efficacious therapeutic is contingent upon its effective delivery to the desired tissue, cell type, and sub-cellular compartment of interest. To date, a variety of methodologies have been developed to overcome the challenge of siRNA delivery, including covalent modifications[5, 6], antibody-protamine fusion[7], liposomal encapsulation[8], and nanoparticle formulations of cationic lipids or polymers.[9–11]
Packaging of siRNA into cationic polymer- or lipid-based nanoparticles is one of the most investigated approaches.[12, 13] By this approach, siRNA is packaged onto nanocarriers with an excess of cationic charge which serves to drive cellular uptake through interaction with the anionic cellular membrane.[14] Moreover, surface PEGylation has been widely employed as a strategy to neutralize siRNA nanocarriers to reduce opsonization and increase stealth from the mononuclear phagocyte system (MPS) following systemic administration.[15–17] However, polyplexes formulated from diblock polymers comprising PEG and a purely cationic polymer block suffer from a lack of stability in vivo.[11, 18–20] This class of polyplexes, formed solely through electrostatic interactions, is disassembled at the glomerular basement membrane (GBM) and cleared primarily through the kidneys, resulting in modest increases of circulation time over naked siRNA.[21, 22] In previous work, we endeavored to improve the performance of cationic polyplexes through incorporation of hydrophobicity into the core of the polyplex to create siRNA nanocarriers stabilized by a combination of both electrostatic and hydrophobic interactions.[23, 24] This approach produced polyplexes with improved stability against destabilization by polyanions such as the herapan sulfates found in the GBM, longer in vivo circulation times, enhanced intracellular delivery of siRNA due to pH-dependent membrane disruptive function tuned to the endolysosomal environment, and improved in vivo bioactivity in the liver, kidneys, and spleen.
Efforts to develop effective siRNA transfection reagents through combinatorial approaches have yielded potent reagents that rival viral constructs. For example, the laboratories of Green, Langer, Anderson, et al. have developed large libraries of poly(β-amino esters) (PBAEs) and lipid or lipid-like chemistries through combinatorial methods with great success. Green et al. initially developed a library of cationic PBAEs in which the lead reagents were able to achieve pDNA transfection comparable to adenoviruses[25] and have more recently developed libraries which yielded PBAE derivatives highly effective at delivering siRNA and pDNA to glioblastomas.[26, 27] Anderson, Langer, and others have used combinatorial synthesis methods to develop large libraries of cationic lipid and lipid-like nanoparticles, reporting the most potent in vivo siRNA knockdown in multiple animal models to date.[9, 28–31] Recently, Siegwart and coworkers utilized combinatorial synthesis to build new chemical classes of biodegradable dendrimer and polymer constructs that were incorporated as the cationic component of lipid nanoparticles.[32, 33] This combinatorial approach allowed for the rapid screening and narrowing of a large chemical space which produced lead compounds highly effective for both siRNA and miRNA delivery in vivo.
Library-based screens, such as those highlighted above, used high throughput synthesis methods to screen different compositions of cationic lipids[9, 29, 30, 34, 35] or polymers[25, 26, 31–33, 36–38]. This approach has proven powerful in elucidation of siRNA carrier structure-function relationships, especially for endpoints focused on in vitro activity and/or in vivo liver gene silencing. In these studies, the authors did not utilize a PEGylation component[25, 26, 36, 37] or used a simple amphiphile (i.e., (mPEG2000-carbamoyl)-1,2-di-O-tetradecyl-sn-glyceride (PEG-DMG) lipid) to sterically stabilize the surface of the resultant nanoparticles[9, 29, 30, 32–35, 38]. Here, we sought to create a library for simultaneous investigation of the composition (cationic versus balanced hydrophobic/cationic) and the relative quantity of both core- and (PEG-containing) corona-forming polyplex components. Moreover, we chose to focus our analysis on polyplex characteristics that are important for overcoming systemic barriers (stability for long circulation and reduced renal clearance) in addition to cell-level barriers (uptake/endosomal escape) that drive in vitro activity; the outcome that most previous screens have focused on. This multiparametric approach[29] expands upon our previous findings showing the importance of hydrophobe incorporation into the core of PEG-stabilized cationic polyplexes[23, 24] and provides a systematic study of structure-function relationships of this class of new class of ternary, PEGylated siRNA polyplexes.
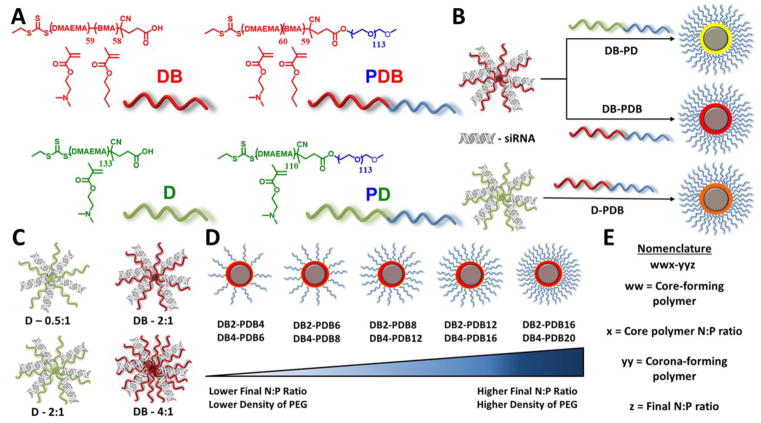
In this study, poly(2-(dimethylamino)ethyl methacrylate) (pDMAEMA, D), poly[(2-(dimethylamino)ethyl methacrylate)-co-(butyl methacrylate)] (p(DMAEMA-co-BMA), DB), poly[(ethylene glycol)-b-(2-(dimethylamino)ethyl methacrylate)] (PEG-b-p(DMAEMA), PD), and poly[(ethylene glycol)-b-[(butyl methacrylate)-co-(2-(dimethylamino)ethyl methacrylate)]] (PEG-b-p(BMA-co-DMAEMA), PDB) were synthesized via RAFT polymerization (Figure S1). Ternary siRNA polyplexes (si-NPs) were formed at varying N+:P− ratios (ratio of polymer amines:nucleic acid phosphates) and varying ratios of the core- (D/DB) to corona-forming polymers (PD/PDB) for three classes of formulations: DB core/PD corona [DB-PD], DB core/PDB corona [DB-PDB], and D core/PDB corona [D-PDB] to produce a library of precisely defined polyplexes with a range of physicochemical properties (Figure 1). This strategy affords surface charge neutral, siRNA core-loaded si-NPs stabilized by electrostatic and hydrophobic interactions with the ability to rapidly tune the polyplex core-corona composition and degree of surface PEGylation. Through this combinatorial ternary si-NP approach, we were able to systematically study important polyplex characteristics such as surface PEGylation density, size, stability, endosomolysis, biocompatibility, cell uptake, and target gene silencing.
Figure 1.
Ternary si-NP synthesis and structural characterization. (A) The four RAFT-synthesized polymers utilized for siRNA packaging and delivery. (B) Three sub-classes of ternary si-NPs were formed with a balance of hydrophobic BMA monomer copolymerized with cationic DMAEMA in either the core (DB-PD), corona (D-PDB), or both (DB-PDB). For formulation of ternary si-NPs, (C) core-forming, binary pre-NPs were made at varying N:P ratios (D: 0.5:1, 2:1; DB: 2:1, 4:1). (D) Ternary si-NPs were then formed by adding a PEGylated, corona-forming polymer to the binary complexes. The relative amount of the second polymer added dictates the degree of surface PEGylation. (E) Nomenclature used to identify ternary si-NPs.
Materials and Methods
Subhead 1: Polymer Synthesis and Characterization
1.1 Materials
All chemicals were purchased from Sigma-Aldrich (St. Louis, MO, USA) unless otherwise specified. DMAEMA and BMA monomers were passed twice through a basic alumina gravity column prior to use in order to remove inhibitors. 2,2-Azobis(2-methylpropionitrile) (AIBN) was recrystallized twice from methanol. All cell culture reagents were purchased through Fischer Scientific unless otherwise specified. Cell culture media and reagents, including Dulbecco’s modified eagle medium (DMEM), fetal bovine serum (FBS), PBS (−/−), PBS (+/+), Pen/Strep, and gentamycin were purchased through Life Technologies (Grand Island, NY, USA). For DLS experiments, dsDNA was used as a model for siRNA. DLS measurements confirmed that si-NPs formed with dsDNA and siRNA are the same size (Supplementary Figure S1). For all fluorescent measurements, fluorophore-labeled dsDNA was used a model of siRNA. A list of oligonucleotides is provided in the supplement (Table S1).
1.2 Synthesis of 4-cyano-4-(ethylsulfanylthiocarbonyl)sulfanylpentanoic acid (ECT) and poly(ethylene glycol)-ECT (PEG-ECT) macro-Chain Transfer Agent (macro-CTA)
ECT, the RAFT CTA, was synthesized according to a previously reported procedure[39]. The terminal carboxylic acid of ECT was then conjugated to PEG[23]. Briefly, methoxy-PEG (2 mmol, 10 g, Mn = 5000 Da), ECT (4 mmol, 1.045 g), and Dimethylaminopyridine (DMAP, 0.08 mmol, 10 mg) were dissolved in dry DCM (50 mL), and dicyclohexylcarbodiimide (DCC, 4 mmol, 0.82 g) was added while stirring. The reaction mixture was stirred for 48 h at room temperature (RT). Precipitated cyclohexyl urea was removed by filtration (0.2 μM pore size). The DCM layer was concentrated and precipitated into diethyl ether twice. The precipitated PEG-ECT was washed thrice with diethyl ether and dried under vacuum. 1H NMR (400 MHz Sprectrometer, Bruker, CDCl3) showed 94% ECT conjugation to PEG[23].
1.3 Polymer Synthesis
RAFT controlled polymerization was used to synthesize four polymers, either from ECT or the PEG-ECT macro-CTA. P(DMAEMA-co-BMA) (DB) and pDMAEMA (D) were synthesized from ECT (Figure S1). In both cases, the target degree of polymerization was 150, reaction volume was 3 mL (Dioxane), degassing was done for 30 min by nitrogen purge, and polymerizations proceeded at 70°C for 20 h using AIBN as an initiator at 5:1 (CTA:AIBN) molar ratio. Reactions were stopped by removing the flask from heat and opening the reaction to air. The reaction mixtures were transferred to dialysis tubing and dialyzed one day against methanol and subsequently one day against diH2O to remove unreacted monomers prior to lyophilization. PEG-b-pDMAEMA (PD) and PEG-b-p(BMA-co-DMAEMA) (PDB) were synthesized from the PEG-ECT macro-CTA (Figure S1). The target degrees of polymerization were both 150. Reaction volumes were 3 mL (Dioxane), and degassing was done for 30 min by nitrogen purge. The polymerizations proceeded at 70 °C for 24 h, and AIBN was used as an initiator at 5:1 (macro-CTA:AIBN) molar ratio. Reactions were stopped by removing the flask from heat and opening the reaction to air. The resulting diblock copolymers were precipitated in a cold solution of pentane:diethyl ether (90:10). The isolated polymers were dried, re-dissolved in ethanol, dialyzed one day against diH2O and lyophilized to yield the final product.
1.4 Polymer Characterization
Polymers were characterized for composition and molecular weight (Mn) by 1H NMR (400 MHz Sprectrometer, Brüker) in either D2O (D) or CDCl3 (DB, PD, and PDB). Absolute molecular weight and polydispersity (PI) were further determined by gel permeation chromatography (GPC) using DMF + 0.1 M LiBr as the mobile phase with inline Agilent refractive index and Wyatt miniDAWN TREOS light scattering detectors. Serial dilutions (10 mg/ml – 0.25 mg/ml) in DMF were measured on a digital refractometer to determine dn/dc values which were used for calculating absolute molecular weight on GPC.
Subhead 2: Formulation and physicochemical characterization of ternary si-NPs
2.1 Assembly of ternary si-NPs and size, zeta potential, and stability characterization
Three combinations of polymers were utilized for forming ternary si-NPs: DB-PD, DB-PDB, and D-PDB. All polymers were dissolved in pH 4.0 citric acid buffer (10 mM). The dsDNA was pre-condensed with the binary, core-forming polymer at each specified N+:P− ratio for 15 min at 0.5 mg/ml polymer concentration. Next, differing amounts of ternary, corona-forming polymer (3.33 mg/mL, 10 mM citric acid buffer at pH 4.0) were added in order to give the appropriate final N+:P− ratio, and these solutions were incubated at room temperature for an additional 30 min. Polymer amounts needed to yield final N+:P− ratios were determined according to the following two equations for binary (1) and ternary (2) si-NPs:
| (1) |
| (2) |
where nmol Pol is the nmol of binary core-forming polymer, nmol NA is the nmol of nucleic acid, N:P is the ratio of amines to phosphates, RU DMAEMA is the number of repeating units of DMAEMA within the polymer backbone, nmol Pol2 is the nmol of ternary corona-forming polymer, nmol Pol1 is the nmol of binary core-forming polymer, RU DMAEMA1 is the number of repeating units of DMAEMA within the binary core-forming polymer backbone, and RU DMAEMA2 is the number of repeating units of DMAEMA within the ternary corona-forming polymer.
After formulation of both the core- and corona-forming polymers with the siRNA at pH 4, a 5-fold excess of pH 8.0 phosphate buffer (10 mM) was added to the samples before filtering through 0.45 μm pore syringe filters. The hydrodynamic diameter (Dh) and zeta potential (ζ) of the resulting ternary si-NPs were then measured at 0 and 24 h using dynamic light scattering (DLS) (Malvern Zetasizer Nano ZS, Malvern, UK). The naming scheme used for ternary si-NP formulations is as follows: [Binary Polymer](Binary N+:P−)-[Ternary Polymer](Ternary N+:P−). For example, the lead ternary si-NP which contains a DB core formulated at 4:1 N+:P− and PDB corona formulated to a final N+:P− of 12:1 is referred to as DB4-PDB12.
Subhead 3: In vitro assessment of ternary si-NPs
3.1 Characterization of pH-dependent membrane disruption by the hemolysis assay
The hemolysis assay was used to assess all ternary si-NP formulations for cytocompatibility and the potential to escape the endolysosomal pathway. Red blood cells (RBCs) were obtained from anonymous donors and isolated by a well-established protocol[40]. After isolation, RBCs were incubated with varying concentrations (5, 15, and 30 μg/mL total polymer concentration) of each ternary si-NP formulation at four pH’s representative of extracellular and endolysosomal ranges (7.4, 6.8, 6.2, 5.6). After 1 h of incubation, intact RBCs and cellular debris were centrifuged out, and supernatants were removed. The supernatants were measured for absorbance at 451 nm (hemoglobin absorbance) and percent hemolysis was determined relative to 1% Triton-X100 detergent.
3.2 Cell Culture
Human triple negative breast cancer cells (MDA-MB-231) were cultured in DMEM supplemented with 10% FBS and 50 μg/mL gentamicin. Mouse embryonic fibroblasts (NIH3T3), and mouse mesenchymal stem cells (MSCs) were cultured in DMEM supplemented with 10% FBS and 1% Pen/Strep. MDA-MB-231, NIH3T3, and MSC cells were transduced with lentivirus encoding firefly luciferase, Green Fluorescent Protein (GFP), and Blasticidin resistance, enabling the generation of stable luciferase expressing-MDA-MB-231 (L231), NIH3T3 (L3T3), and MSC (LMSC) cell lines.
3.3 Cytocompatability of ternary si-NPs
Cytocompatability of all ternary si-NP formulations was evaluated by adding scrambled siRNA containing ternary si-NPs to L231 and L3T3 cells and measuring relative cell number based on cellular luminescence. L231 and L3T3 cells were seeded to 96-well black-walled plates at a density of 5,000 cells/well and allowed to adhere overnight. Ternary si-NP formulations were then added to each well in full serum media (DMEM, 10% FBS, 50 μg/mL gentamicin/1% Pen/Strep) at a final siRNA concentration of 100 nM and incubated for 24 h. After 24 h, media was replaced with luciferin containing media (150 μg/ml) and luminescence signal was collected on a Lumina III IVIS system (Xenogen Corporation, Alameda, CA, USA). Cell viability was then calculated as the ratio of luminescence of siScrambled siRNA treated cells to non-treated cells.
3.4 Cell uptake of ternary si-NPs
Cell uptake was evaluated in both MDA-MB-231 and NIH3T3 cell types by flow cytometry. MDA-MB-231 and NIH3T3 cells were seeded into 24-well plates at a density of 30,000 cells/well and allowed to adhere overnight. Alexa488-labeled model siRNA was loaded into ternary si-NPs which were added to each well to give a final nucleic acid concentration of 100 nM. These treatments were incubated with cells in full serum media (DMEM, 10% FBS, 50 μg/mL gentamicin/1% Pen/Strep) for 24 h. After 24 h, media with treatments was removed, and cells were washed with PBS (−/−), trypsinized (0.25%), transferred to microcentrifuge tubes, and centrifuged at 420g for 7 min to yield a cell pellet. Pellets were re-suspended in 0.4 mL PBS(−/−) with 0.04% trypan blue to quench extracellular fluorescence, and intracellular si-NP delivery was measured by FACS (FACSCalibur, BD Biosciences, Franklin Lakes, NJ, USA) at excitation wavelength of 488 nm and emission wavelength of 519 nm.
3.5 In vitro siRNA silencing by ternary si-NPs
The gene silencing profile of ternary si-NP formulations was screened within L3T3s, L231s, and LMSCs. Cells were initially seeded in black-walled, 96-well plates at a density of 2,000 cells/well and allowed to adhere overnight. Cells were then treated for 24 h in full-serum media (DMEM, 10% FBS, 50 μg/mL gentamicin/1% Pen/Strep) with all ternary si-NP formulations prepared as described above and containing an anti-luciferase or scrambled siRNA sequence (100 nM). After 24 h, treatment media was replaced by luciferin containing media (150 μg/ml), and cellular luminescence was measured using an IVIS Lumina III imaging system (Xenogen Corporation, Alameda, CA, USA). The cells were then incubated for an additional 24 h after luciferin containing media was replaced by low-serum media (DMEM, 1% FBS, 50 μg/mL gentamicin/1% Pen/Strep), which was used to minimize cellular overgrowth. Cellular luminescence was re-measured at 48 h and normalized to treatment with analogous formulations containing a scrambled control siRNA sequence in all cases.
Subhead 4: Comparative analysis of binary PDB and ternary DB4-PDB12 si-NPs
4.1 Cell uptake and endosomal escape of binary and ternary si-NPs by confocal microscopy
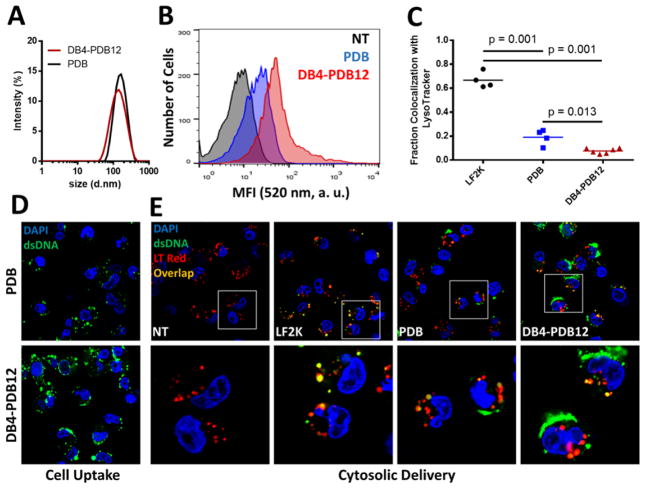
Cell uptake and endosomal escape of ternary si-NPs were imaged by confocal microscopy. MDA-MB-231 cells were seeded in 8-well chamber slides (Lab-Tek II Chambered Coverglass, Thermofisher) at a density of 20,000 cells/well and allowed to adhere overnight. Alexa488-labeled ternary si-NPs were added to each well to give a final nucleic acid concentration of 25 (LF2K) or 100 (PDB, DB4-PDB12) nM and incubated with cells in full serum media (DMEM, 10% FBS, 50 μg/mL gentamicin) for 24 h. After 24 h, treatment media was removed, cells were washed once with PBS (−/−) and imaged in PBS (−/−) containing DAPI nuclear stain. For visualizing endosomal escape after 2 h, cells were washed once with PBS (−/−) and incubated in PBS (−/−) containing DAPI nuclear stain and Lysotracker Red (75 nM) at 37 °C. Cells were imaged after 1 h incubation with Lysotracker Red. All images were processed using imageJ and colocalization was analyzed using Just Another Colocalization Plugin (JACoP) [41]. Using JACoP, Mander’s overlap coefficients were calculated to represent the fraction of green signal (Alexa488-dsDNA) overlapping with red signal (LysoTracker Red) to quantify colocalization (n ≥ 4 images).
4.2 Binary and ternary si-NP stability against heparin-saline disassembly
PDB and DB4-PDB12 si-NPs were loaded with Förster Resonance Energy Transfer (FRET, using Alexa Fluor 488 and Alexa Fluor 546) pair-labeled dsDNAs. Fluorescence intensity was measured using a microplate reader (Tecan Infinite F500, Männedorf, Switzerland) with an excitation wavelength of 488 ± 5 nm. Alexa Fluor 488 emission was collected at 519 ± 5 nm, and Alexa Fluor 546 emission was obtained at 573 ± 5 nm. FRET was calculated as a ratio of the fluorescent intensity as follows:
The stability of PDB and DB4-PDB12 si-NPs was measured in the presence of 2 to 100 U/mL of heparin sodium salt in DPBS [23]. The fluorescence emission at both wavelengths was measured over time following addition of heparin sodium salt. The EC50 of dissociation for PDB and DB4-PDB12 si-NPs was calculated according to the following equation:
4.3 Binary and ternary si-NP blood pharmacokinetics and biodistribution in vivo
Blood plasma pharmacokinetics
Blood was collected retro-orbitally at 5 min and 10 min post-injection, not exceeding two collections per animal. After 20 min, animals were sacrificed, and blood was immediately collected via the renal vein. Blood samples were centrifuged at 2000 × g for 5 min, and 5 μL of plasma was taken from the supernatant and diluted into 95 μL PBS (−/−). Fluorescence was quantified on an IVIS Lumina III imaging system (Xenogen Corporation, Alameda, CA, USA) at excitation wavelength of 620 ± 5 nm and emission wavelength of 670 ± 5 nm. A standard curve was generated by measuring the fluorescence of the initial fluorescent si-NP solution in PBS (−/−) over the range of 200% - 1.5% of the injected dose. The standard curve was utilized in order to calculate the percent of injected dose in each blood sample, and the calculated values were used to determine nucleic acid concentration in the plasma at each time point as well as area under the curve (AUC) values.
Biodistribution in athymic nude tumor-bearing mice
Athymic nude female mice (4–6 weeks old, Jackson Laboratory, Bar Harbor, ME, USA) were injected in each mammary fat pad with 1 × 106 L231 cells in DMEM:Matrigel (50:50). After 17 days, tumor-bearing mice were injected via the tail vein with 1 mg/kg (Cy5-dsDNA dose) of fluorescent si-NPs. After 20 min, animals were sacrificed and the organs of interest (heart, lungs, liver, spleen, kidneys, and tumors) were excised. The organs were fluorescently imaged and quantified on an IVIS Lumina III imaging system (Xenogen Corporation, Alameda, CA, USA) at excitation wavelength of 620 ± 5 nm and emission wavelength of 670 ± 5 nm.
Confocal imaging of tumor sections
Athymic nude female mice (4–6 weeks old, Jackson Laboratory, Bar Harbor, ME, USA) were injected in each mammary fat pad with 1 × 106 L231 cells in DMEM:Matrigel (50:50). After 17 days, tumor-bearing mice were injected via the tail vein with 1 mg/kg (Cy5-dsDNA dose) of fluorescent si-NPs. After 180 min, animals were sacrificed and tumors were excised. Tumor tissue was immediately submerged in optimal cutting temperature (OCT) compound and snap frozen. Cryo-sections were then cut at 5 μm thickness by the Vanderbilt Translational Pathology Core and imaged in PBS containing DAPI on a C1si confocal microscope system (Nikon Instruments, Melville, NY, USA) equipped with differential interference contrast transmitted light detector.
4.4 Target gene silencing by binary and ternary si-NPs in vivo
Athymic nude female mice (4–6 weeks old, Jackson Laboratory, Bar Harbor, ME, USA) were injected in each mammary fat pad with 1 × 106 L231 cells in DMEM:Matrigel (50:50). After 17 days, tumor-bearing mice were injected i.p. with luciferin substrate (150 mg/kg) and imaged for bioluminescence on an IVIS Lumina III imaging system (Xenogen Corporation, Alameda, CA, USA) 20 minutes post-injection. Next, the mice were injected via the tail vein with 1 mg/kg (siRNA dose) si-NPs containing either anti-luciferase siRNA, a scrambled control siRNA, or saline. Mice were imaged and dosed at days 17 and 18 and also imaged on days 19 and 20. Relative luminescence was determined by measuring the raw luminescent intensity of each tumor on each day and comparing to the initial signal at day 17.
Subhead 5: Statistics and Ethics Statement
5.1 Statistical methods
Treatment groups were compared using either two-tailed student’s t-test or one-way ANOVA test coupled with Tukey means comparison test, where a p-value < 0.05 was deemed representative of a significant difference between treatment groups. No outliers were removed from data. For all data, the arithmetic mean and standard error are shown.
5.2 Ethics Statement
The animal studies were conducted with adherence to the guidelines for the care and use of laboratory animals of the National Institutes of Health (NIH). Mice were fed a standard chow diet ad libitum and had free access to water. All protocols were approved by the Institutional Animal Care and Use Committee of Vanderbilt University and done in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Hemolysis assays were done on blood samples taken under a protocol approved by the Vanderbilt Institutional Review Board (IRB).
Results and Discussion
Subhead 1: Synthesis and multiparametric screen of ternary si-NPs
1.1 Polymer Synthesis and Characterization
Two core-forming polymers, pDMAEMA (D; Mn = 21 kDa and PDI = 1.02) and p(DMAEMA-co-BMA) (DB; Mn = 18 kDa and PDI = 1.05), and two corona-forming diblock polymers, PEG-b-pDMAEMA (PD; Mn = 22 kDa and PDI = 1.16) and PEG-b-p(BMA-co-DMAEMA) (PDB; Mn = 23 kDa and PDI = 1.03), were synthesized using RAFT polymerization to achieve polymers with low polydispersity and controlled monomer composition and molecular weight (Figure 1a, Table 1, and Supplemental Figures S2 and S3). RAFT polymerization is advantageous for synthesizing biomacromolecules of complex architectures with precision and low polydispersity.[42, 43] Moreover, the simple and single-step polymerizations used herein are scalable and yield polymers which can be rapidly purified.
Table 1.
Polymer degrees of polymerization (DP), composition (%BMA), number-average molecular weight (Mn), and polydispersity (PI) characterization.
| Polymer | DP (NMR) | Composition (%BMA, NMR) | Mn (Da, NMR) | PI (GPC) |
|---|---|---|---|---|
| D | 133 | 0 | 20,900 | 1.02 |
| DB | 117 | 49.6 | 17,500 | 1.05 |
| PD | 110 | 0 | 22,300 | 1.16 |
| PDB | 119 | 49.6 | 22,800 | 1.03 |
The well-defined D, DB, PD, and PDB polymers served as the base units of thirty ternary si-NPs formed by varying polymer to siRNA charge ratios (N+:P−) and the ratio of core:corona polymers within each ternary si-NP class (DB-PD, DB-PDB, and D-PDB) (Figure 1b–e). The D-PD combination[44] was not extensively pursued because of its lack of a hydrophobic component. Instead, we focused on “hydrophobized” ternary si-NPs which we hypothesized would outperform purely cationic ternary polyplexes based on our previous findings that balancing cationic and hydrophobic content of binary polyplexes increases stability, endosomolysis, and bioactivity.[23, 24] Importantly, a two-step mixing protocol was adapted from previous reports that show the initial complexing with a cationic component followed by “post-PEGylation” with a diblock, corona-forming polymer improves siRNA packaging over binary complexes with a covalently attached PEG block[44–46]. The three sub-classes are characterized primarily by the placement of the hydrophobic BMA monomer into either the core- (DB-PD), corona- (D-PDB), or both core- and corona-forming polymers (DB-PDB, Figure 1b). Within each sub-class, the amount of non-PEGylated, core-forming polymer was varied (Figure 1c) to form si-NP precursors (pre-NPs, Supplementary Figure S4). The amount of PEGylated, corona-forming polymer was also varied to generate different degrees of surface PEGylation and ratio of core:corona polymers within each sub-class (Figure 1d).
1.2 Physicochemical characterization of ternary siRNA-loaded si-NPs
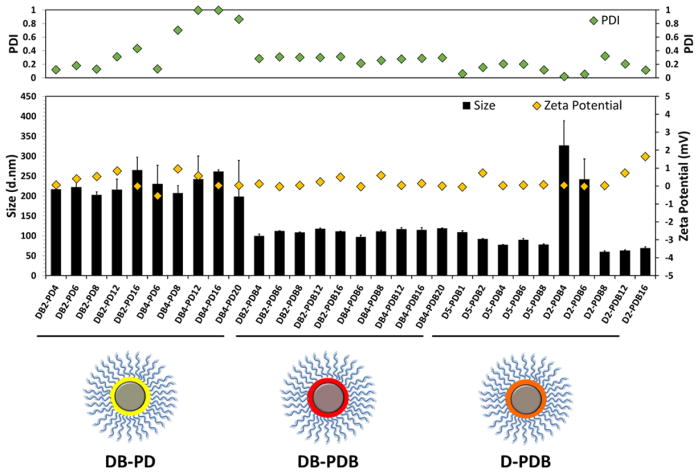
The physicochemical properties of si-NPs from each class were screened, including hydrodynamic diameter, polydispersity, and surface charge (zeta potential) at 0 h and 24 h post-fabrication. DB-PD, DB-PDB, and D-PDB ternary si-NPs each formed stable and compact si-NPs (~100–300 nm) (Figure 2, Supplemental Table S2). While D2-PDB4 and D2-PDB6 si-NPs significantly increased in size over 24 h, all other si-NPs retained their initial size. All si-NPs had approximately neutral surface charge which is ideal for systemic administration, with zeta potentials for each si-NP ranging from −1 to +2 mV. Ternary si-NPs utilizing the corona-forming polymer PDB (DB-PDB and D-PDB) displayed PDIs < 0.3, as compared to highly variable PDIs (0.1 – 1.0) seen in the PD-coated si-NPs (Figure 2, Supplemental Table S2). In addition, the non-PEGylated, pre-NPs (DB4) rapidly aggregate, forming large and extremely polydisperse structures (Supplementary Figure S5) at neutral pH (7.4). By contrast, PDB coating of DB4 to form ternary si-NPs gives nanoparticles in the same size range as the binary, parent PDB si-NPs (~120 nm; Supplementary Figure S5). Moreover, PDB polymer not loaded with nucleic acid exists as very small micelles at pH 7.4 (Supplementary Figure S5). In DLS spectra of DB4-PDB12 si-NPs, neither aggregates of the uncoated DB4 pre-NPs or small micelles of the empty PDB micelles appear, suggesting that PDB (when added to pre-NPs in a solution of pH 4) associates with the DB4 pre-NPs and successfully coats the pre-NPs, producing stabilized ternary nanoparticles. In sum, the initial physicochemical screening of this library of ternary si-NPs highlighted the promise of a subgroup of formulations (DB-PDB and D-PDB), which leveraged the PDB polymer as a corona-forming component. These two classes yielded stable si-NPs ~100 nm in diameter, with neutral surface charge and low polydispersity.
Figure 2.
Physicochemical characterization of ternary si-NP library including si-NP size, surface charge (zeta potential), and polydispersity (PDI). (Yellow: DB-PD, Red: DB-PDB, Orange: D-PDB)
1.3 Characterization of pH-dependent membrane disruption
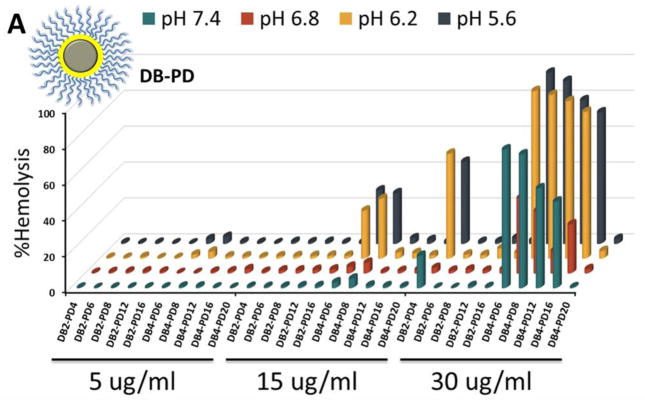
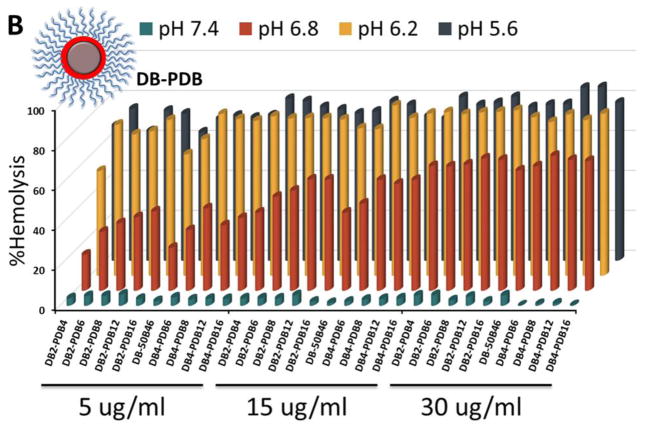
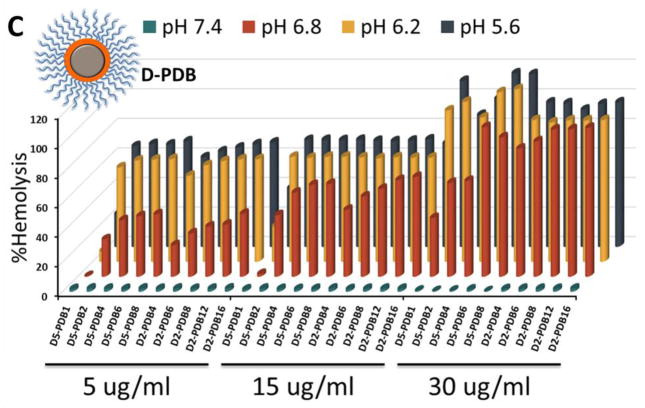
Entrapment in the endolysosomal pathway and subsequent degradation or trafficking out of the cell limit the effectiveness of many biologic drugs including siRNA.[47–49] Here, the pH-dependent hemolysis assay was used as a measure of active (non-proton sponge) endolysosomal escape capacity of the different ternary si-NP formulations[50]. The composition of the corona-forming polymer profoundly influenced si-NP hemolysis. PDB-containing si-NPs harbored “switch-like” endosomolytic behavior that is turned on at endosomal pH (6.8 and below) but not extracellular pH (7.4) (Figure 3). In contrast, PD-containing si-NPs showed weak hemolysis and were also hemolytic at pH 7.4, which is indicative of the potential for cytotoxicity (Figure 3a). For DB-PDB and D-PDB formulations, hemolysis increased proportionally with increasing amounts of the endosomolytic PDB, corona-forming polymer (Figure 3b, c). Therefore, ternary polyplexes which contained PDB as the corona-forming polymer exhibited optimal pH-dependent membrane disruptive behavior while PD-corona ternary polyplexes exhibited sporadic hemolysis and in some cases showed hemolysis at pH 7.4, which is generally correlated with cytotoxicity.
Figure 3.
Hemolysis profiles of (A) DB-PD, (B) DB-PDB, and (C) D-PDB ternary si-NPs.
1.4 In vitro cytocompatibility and cellular uptake of ternary si-NPs
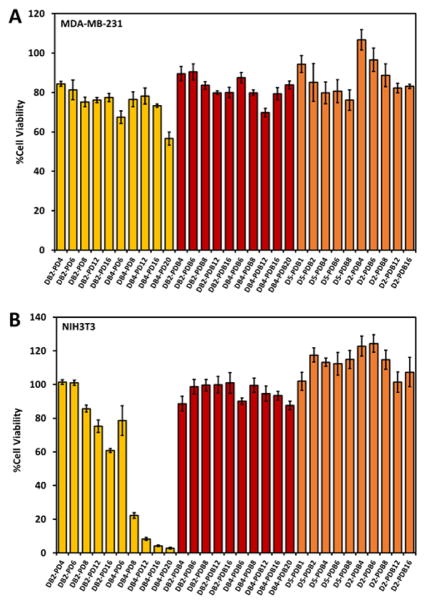
The relative cell uptake of all ternary si-NPs was initially screened in MDA-MB-231 (human triple negative breast cancer) and NIH3T3 (murine embryonic fibroblasts) cells, and cytocompatibility was assessed in MDA-MB-231 and NIH3T3 cells lentivirally-transduced to constitutively express luciferase (L231 and L3T3, respectively). Treatment of L231s for 24 h with all ternary si-NP formulations was well tolerated, although DB-PD si-NPs trended toward increased cytotoxicity as amount of PD polymer was increased (Figure 4a). Incubation of L3T3s for 24 h with DB-PD si-NPs loaded with scrambled siRNA exhibited increasing cytotoxicity proportional to the amount of polymer (Figure 4b). This is consistent with the observed hemolytic membrane disruption at pH 7.4 for DB-PD polyplexes. DB-PDB or D-PDB si-NPs were not significantly toxic in L3T3s, consistent with observations that they did not cause membrane disruption at pH 7.4 in hemolysis assays (Figure 4a–b). Finally, DB2 and DB4 pre-NPs are toxic to the L3T3 cells (~40% and ~80% toxicity, respectively) until being coated by the PEGylated, corona-forming PDB polymer (Supplementary Figure S6). Upon coating, cytotoxicity of the DB pre-NPs is entirely mitigated, further suggesting their successful coating and surface presentation of the biocompatible PEG molecules (Supplementary Figure S6). These results underscore the potential cytotoxicity of hemolytic materials which are not well-stabilized (such as DB-PD) and agree with previous reports of cell-type dependent cytotoxicity of nanomaterials[38, 51, 52].
Figure 4.
Cytocompatibility of DB-PD, DB-PDB, and D-PDB ternary si-NPs in (A) MDA-MB-231 and (B) NIH3T3 cells.
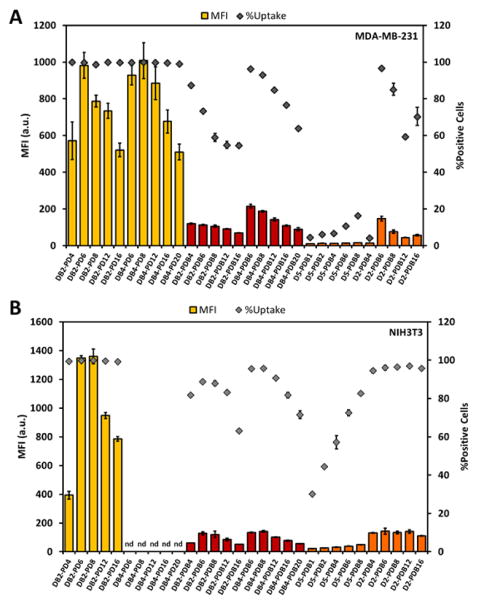
Next, ternary Alexa488-labeled si-NPs were used to track cell uptake of si-NPs by MDA-MB-231 and NIH3T3 cells. Cellular uptake was dependent upon the chemistry of the cationic block of the corona-forming ternary polymer as well as the ratio of the PEGylated, ternary and the core-forming, binary cationic polymers. Although cells treated with DB-PD ternary si-NPs exhibited nearly 10-fold higher mean fluorescence intensity than DB-PDB and D-PDB si-NPs (Figure 5), the total fraction of the cellular population positive for uptake of fluorescent nucleic acid cargo approached 100% for all ternary si-NPs tested. Further, cells treated with the D-PDB and DB-PDB ternary si-NP formulations displayed 5-fold higher mean fluorescence intensity than cells treated with the parental PDB binary si-NPs (Supplementary Figure S7). Decreasing cellular si-NP uptake was observed with increasing degree of PEGylation in both cell types. After reaching an ~ 8:1 final N+:P− ratio, each subsequent addition of more corona-forming PEGylated polymer generally decreased cell uptake (Figure 5). Importantly, being able to control the amount of corona-forming polymer and thereby the density of PEGylation affords the ability to choose an optimal PEGylation state which provides colloidal stability and also a high level of cell uptake.
Figure 5.
Cell uptake of DB-PD, DB-PDB, and D-PDB ternary si-NPs in (A) MDA-MB-231 and (B) NIH3T3 cells. (nd = no data)
1.5 siRNA silencing by ternary si-NPs in cultured mammalian cells
The panel of ternary si-NPs was initially evaluated for function based on in vitro target silencing of the model gene luciferase in L3T3 cells. After 24 h of treatment with ternary si-NPs loaded with anti-luciferase siRNA (siLuc), both DB-PD and DB-PDB classes of si-NPs significantly reduced protein-level expression of luciferase (Supplemental Figures S8 and S9). The DB-PDB group of ternary si-NPs (DB4-PDB6 – DB4-PDB20) achieved 65–85% reduction in luciferase activity at 48 h post-treatment (Supplemental Figures S8 and S9), which was greatest of all si-NP classes within our screen. Thus, despite their lower levels of cellular uptake, si-NPs from the DB-PDB are less toxic to cells at physiological pH, exhibit superior potential to escape endolysosomal compartments, and achieve more efficient target gene silencing in treated cells.
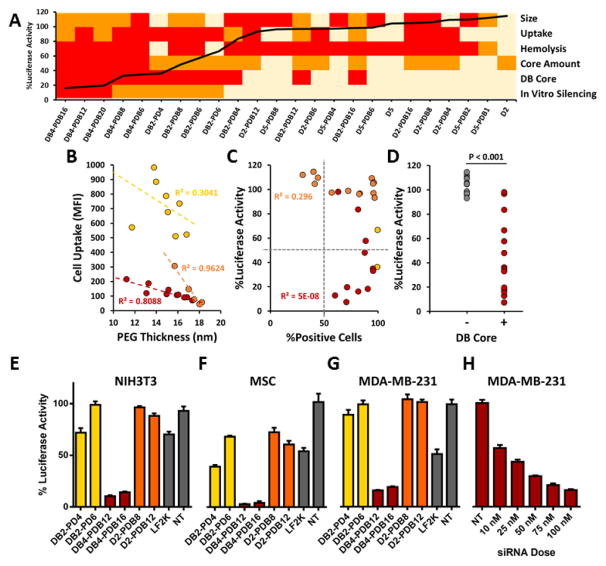
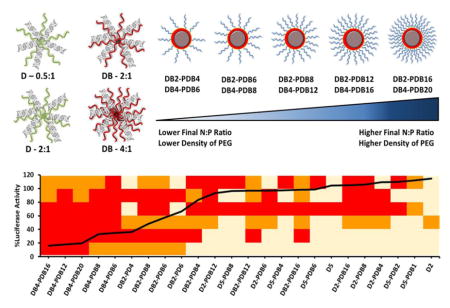
1.6 Multiparametric evaluation of combinatorial library of ternary si-NPs
The panel of siRNA formulations was developed based on the idea that si-NPs that concomitantly overcome multiple siRNA delivery barriers will perform best in in vivo models of disease. By integrating data regarding size, zeta potential, cytotoxicity, uptake, pH-dependent hemolysis, and target gene silencing for each si-NP formulation in our library, we were able to generate a heatmap to visualize the performance of each si-NP across a number of assays simultaneously[29] (Figure 6a). Importantly, ternary si-NPs that perform well across multiple screening assays achieve the greatest level of gene silencing in vitro (Figure 6a). The ternary si-NP surface PEG density (calculated as described in Supplemental Figure S10), cell uptake, and incorporation of the core-forming polymer DB were assessed more closely for correlation to knockdown activity. Within each class of ternary si-NPs, cell uptake was inversely proportional to the surface PEG density (Figure 6b). Although cell uptake was necessary for achieving knockdown, it was not sufficient, as many si-NPs with nearly 100% cellular uptake did not exhibit any knockdown (Figure 6c). Interestingly, the inclusion of the DB core was a key factor in achieving effective knockdown; nearly all DB-containing compounds produced target gene reduction, while all si-NPs without the DB core failed to knock down the target gene (Figure 6d).
Figure 6.
Multiparametric screen of ternary siRNA polyplexes reveals lead si-NP formulation DB4-PDB12. (A) si-NPs which are optimally tuned to overcome multiple siRNA delivery barriers such as size range, cell uptake, and endosomal escape achieve the highest target gene silencing in vitro (heat map parameters and thresholds are shown in Table S3; overlaid black line indicates level of residual luciferase activity for each si-NP formulation loaded with anti-luciferase siRNA). (B) Cell uptake trended inversely to si-NP surface PEG thickness (see PEG thickness calculations in Figure S8). (C) High cell uptake did not directly predict target gene silencing in vitro. (D) Incorporation of the hydrophobic and endosomolytic DB-core was the strongest predictor of gene silencing in vitro (p < 0.001). Lead si-NP formulations (DB4-PDB12 and DB4-PDB16; indicated in red) achieve > 80% gene silencing in (E) fibroblast (3T3s), (F) mesenchymal stem cells (MSCs), and (G) triple negative breast cancer (MDA-MB-231) cells. (H) The IC50 for DB4-PDB12 in MDA-MB-231 cells is 15.4 nM. (Yellow: DB-PD, Red: DB-PDB, Orange: D-PDB)
The model gene silencing results were confirmed using murine fibroblast (NIH3T3), murine mesenchymal stem cell (MSC), and human breast cancer (MDA-MB-231) cell lines, each with stable luciferase expression. Two lead candidates from each ternary si-NP class were screened for target (luciferase) gene silencing, revealing that cells treated with the DB-PDB class si-NPs achieved greater gene silencing (>80% protein level knockdown) as compared to analogous si-NPs loaded with scrambled siRNA or other classes of siLuc si-NPs (Figure 6e–g). Dose response analysis for MDA-MB-231 cells revealed an IC50 silencing value of 15.4 nM for DB4-PDB12 ternary si-NPs (Figure 6h).
A crux in the development of translatable siRNA therapies is the design of delivery systems which are stable in circulation and divert siRNA from clearance organs (liver, spleen, and kidneys) in order to consequently improve distribution to pathological sites. Effective nanocarriers will overcome systemic pharmacokinetic barriers, in addition to classical subcellular endolysosomal pathway barriers, in order to increase siRNA bioavailability within target cells. Previous combinatorial screens have yielded potent siRNA transfection reagents, but have focused primarily on in vitro and hepatic gene silencing[9, 25, 29, 30, 34, 53]. Alternatively, we envisioned the design of a combinatorial library with parallel aims of increasing in vitro potency (dictated largely by intracellular barriers) and preserving physicochemical characteristics that reduce clearance by organs such as the kidneys, liver, and spleen for better in vivo delivery to alternate sites (e.g., to tumors). With a set of comprehensive cell- and system-level barriers in mind, a multiparametric in vitro screening approach was conducted on this small library of rationally-designed nanocarriers. The initial physicochemical screening of this library of ternary si-NPs identified a subgroup (DB-PDB and D-PDB) of promising formulations for systemic administration, with appropriate size (~100 nm), zeta potential (~ 0 mV), and long-term colloidal stability in salts. Ternary si-NP cytotoxicity, cell uptake, and hemolysis assays subsequently revealed formulations (DB4-PDB6 – 20) which were ideally tuned for low toxicity and high potential to overcome siRNA delivery barriers. Importantly, these leading formulations achieved the highest gene silencing, and incorporation of the balanced cationic and hydrophobic DB core-component was identified as a crucial parameter for achieving potent RNAi in vitro. For further testing, we down-selected to the leading in vitro formulation, DB4-PDB12, and conducted analysis of biodistribution, pharmacokinetics, and pharmacodynamics compared to our previous gold standard formulation, PDB. New techniques are still desperately needed to enable high-throughput in vivo screening of the pharmacokinetics and biodistribution of siRNA polyplex libraries generated within this study and others. Toward this end, Dahlman et al. recently developed an approach based on barcoded nanoparticles which enable the simultaneous tracking of the biodistribution of up to 30 formulations within a single mouse[54]. In addition to in vivo screening, it is critical to continue developing high throughput, in vitro characterization approaches that can better identify candidate formulations that will optimally deliver siRNA into solid tumors.
Subhead 2: Comparative analysis of ternary and binary si-NPs in vitro and in vivo
2.1 DB4-PDB12 lead ternary si-NPs increase internalization and cytosolic dose of siRNA in vitro
The lead ternary si-NP, DB4-PDB12, was selected for further analysis in vitro and in vivo, benchmarking against the parental PDB binary si-NPs[23]. The DB4-PDB12 ternary si-NPs were 124.4 nm in diameter (Figure 7a) with approximately neutral surface charge (0.139 mV; Supplemental Table S2), had nearly 4-fold higher cell uptake than the parental PDB binary si-NPs, and decreased colocalization of nucleic acid cargo with endolysosomes (labeled with LysoTracker Red) by 8.7- and 2.5-fold compared to Lipofectamine 2000 (LF2K) and PDB si-NPs, respectively (Figure 7b–e and Supplemental Figures S11 and S12). Increased cytosolic delivery of nucleic acid cargo from DB4-PDB12 si-NPs through the endolysosomal pathway was also visualized by confocal microscopy of cells treated with fluorescently-tagged si-NPs and co-stained with Lysotracker Red (Figure 7e and Supplemental Figure S12). Thus, the ternary analog, DB4-PDB12, maintained the important physicochemical characteristics of PDB for systemic administration (size and surface charge) while increasing lipid bilayer translocation (cell uptake and endosomal escape) of the siRNA delivery system.
Figure 7.
Leading DB4-PDB12 ternary si-NPs have higher cytosolic delivery than the parent binary si-NPs. (A) DB4-PDB12 si-NPs are similar size (DB4-PDB12: 124.4 vs PDB: 129.5 nm) as PDB si-NPs. (B) DB4-PDB12 have increased cell uptake over the PDB parent si-NPs (60× mag). (C) DB4-PDB12 si-NPs exhibit significantly lower endolysosomal colocalization than PDB or Lipofectamine 2000 (LF2K; administered at the maximum tolerated dose of 25 nM) si-NPs (n ≥ 4 fields of view; 60× mag). (D) Increased DB4-PDB12 cell uptake visualized by confocal microscopy. (E) Increased cytosolic delivery of cargo by DB4-PDB12 si-NPs visualized by confocal microscopy in cells with acidic endolysosomal vesicles co-stained with Lysotracker Red (75 nM).
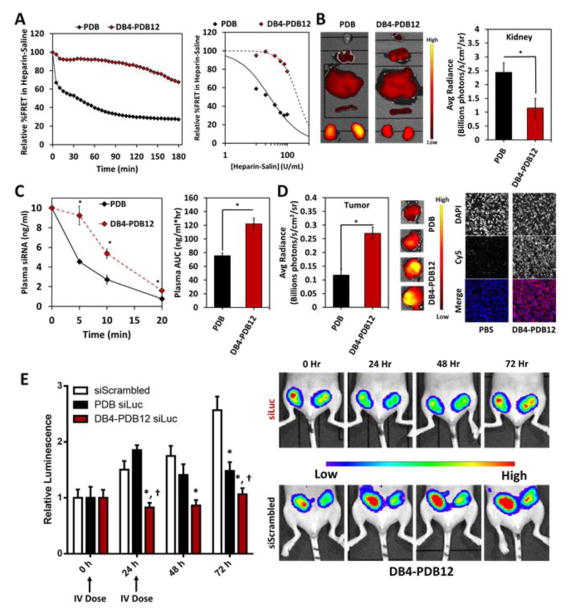
2.2 Pharmacokinetics, biodistribution, and bioactivity of binary PDB and ternary DB4-PDB12 si-NPs
To assess the stability of si-NPs in an assay that models the microenvironment of the kidney GBM, we measured binary PDB and ternary DB4-PDB12 si-NP stability in the presence of heparan sulfate. Using a FRET-based readout, DB4-PDB12 si-NPs were significantly more resistant to heparin-mediated disassembly over time when compared to PDB binary si-NPs (Figure 8a and Supplemental Figures S13 and S14). Consistent with these results, the fluorescently-labeled cargo of DB4-PDB12 ternary si-NPs exhibited lower concentration in the kidneys of athymic female tumor-bearing mice relative to PDB binary si-NPs following i.v. injection (Figure 8b and Supplementary Figure S15), suggesting that the ternary si-NPs have decreased susceptibility to GBM-triggered disassembly and renal clearance in vivo. In agreement with better stability within the circulation, DB4-PDB12 ternary si-NPs had a 1.6-fold greater area under the curve (AUC) value compared to PDB binary si-NPs (Figure 8c). Greater stability and bioavailability of DB4-PDB12 ternary si-NPs also correlated to a 2.6-fold increase in accumulation within orthotopic MDA-MB-231 breast tumor xenografts following i.v. injection relative to PDB binary si-NPs (Figure 8d and Supplementary Figures S15 and 16). The correlation between higher circulation time and greater tumor biodistribution is consistent with the principles of enhanced permeability and retention-(EPR−) based tumor accumulation[55, 56]. In total, DB4-PDB12 si-NPs showed increased blood plasma AUC, reduced kidney accumulation, and increased tumor accumulation when compared to our previously optimized PDB binary si-NP composition[23], all advantageous characteristics for effective systemic delivery of siRNA to solid tumors.
Figure 8.
Ternary DB4-PDB12 si-NPs improve pharmacokinetics and bioactivity of siRNA relative to binary PDB si-NPs after i.v. administration. (A) Ternary si-NPs have increased resilience to disassembly by polyanionic heparin in saline solution. (B) Biodistribution of si-NPs after i.v. administration (1 mg/kg Cy5-labeled dsDNA). Ternary si-NPs have significantly reduced concentration in the kidneys compared to parent PDB si-NPs in male CD-1 mice (n = 6; p < 0.001). (C) Ternary si-NPs persist longer in blood (n ≥ 3; p = 0.008) and (D) increase uptake into orthotopic breast tumors after i.v. administration relative to PDB binary si-NPs (n = 6; p = 0.03). (E) Treatment with 2 doses (0 and 24 h) at 1 mg/kg siRNA silences the model gene luciferase in luciferase-expressing orthotopic MDA-MB-231 xenografts and arrests growth in luminescence signal. DB4-PDB12 si-NPs enhance siRNA bioactivity over parent PDB binary si-NPs (n ≥ 5; p = 0.05).
The siRNA silencing efficacy of lead DB4-PDB12 ternary si-NPs was evaluated in vivo in orthotopic L231 (MDA-MB-231 triple negative breast cancer cells constitutively expressing luciferase) tumors. DB4-PDB12 ternary si-NPs or PDB binary si-NPs loaded with siLuc siRNAs were injected i.v in two doses, (days 17 and 18 after tumor inoculation), and tumor luciferase expression was assessed using intravital bioluminescence. As early as 24 h after treatment, luciferase was diminished by ~ 45% knockdown in tumors of mice treated with siLuc-loaded DB4-PDB12 si-NPs as compared to siScrambled control si-NPs (p = 0.01). Increased silencing by the DB4-PDB12 si-NPs was detected at 48 h and 72 h after treatment, with ~ 51% (p < 0.01) and ~ 59% (p < 0.01) luciferase attenuation, respectively (Figure 8e). Moreover, DB4-PDB12 ternary siLuc-NPs decreased luciferase signal significantly more than PDB binary siLuc-NPs at 24, 48, and 72 h (45% vs. −23%, 51% vs. 20%, and 59% vs. 42% reduction, respectively; Figure 8e) when compared head-to-head.
Recent studies have identified the kidneys as the major route for in vivo clearance of i.v.-injected siRNA polyplexes assembled by electrostatic interactions[21, 22]. In these studies, anionic proteoglycans such as heparan sulfates of the GBM were identified as culprits for polyplex disassembly and clearance through the kidneys/urine. Therefore, it is functionally significant that the DB4-PDB12 ternary si-NPs resist heparin-mediated disassembly, reduce clearance through the kidneys, and consequently achieve higher AUCs after i.v. administration when benchmarked against PDB binary si-NPs which previously were optimized to outperform PD (100% cationic) si-NPs[23]. Importantly, the optimized DB4-PDB12 formulation only accumulated ~50% within the kidneys (Supplementary Figure S14b), whereas siRNA polyplexes reported in the literature typically show > 75% renal accumulation[11, 19, 20, 28, 46, 57–59], severely attenuating the amount of injected dose available for delivery to non-renal target organs. The increased AUC of DB4-PDB12 ternary si-NPs also correlated to higher siRNA delivery to orthotopic breast tumors in our studies. Although recent studies highlight the variability of the EPR effect[60, 61], Clark et al. recently observed EPR-based tumor accumulation of CRLX101 in gastrointestinal adeno- or squamous cell carcinomas in human patients while the nanomedicine was absent in surrounding, non-neoplastic tissue[62]. Generally, it is accepted that EPR-based accumulation of nanoparticles within tumors occurs when particles fall within the appropriate size range and correlates with circulation AUC as observed within the current study[55, 56]. In addition to passive targeting by the EPR effect, active targeting through receptor-ligand interactions (e.g., transferrin, folic acid, RGD, HER2-ScFv) is a popular approach for increasing tumor retention after i.v. administration of si-NPs, but active targeting is inherently limited if the nanocarrier is rapidly cleared. For this reason, we focused on initially improving the pharmacokinetics of our ternary si-NPs through modulation of the si-NP core chemistry, and it is anticipated that inclusion of active targeting will further improve our lead si-NPs in the future.
Conclusions
Many siRNA transfection reagents have been primarily optimized to achieve gene silencing in vitro, and therefore, utilize design principles (e.g., extreme cationic charge for cell internalization[14]) which do not translate well to in vivo settings, especially for delivery to non-hepatic targets. Due to the shortcomings of strictly cationic polyplexes (such as colloidal instability and short persistence in circulation in vivo), we combinatorially incorporated hydrophobicity into the core, corona, and both core and corona of a rationally-designed library of 30 distinct ternary architecture si-NPs to stabilize them through both electrostatic and van der Waals forces. Within each class, the amount of core-forming polymer, corona-forming polymer, and the ratio of the two were varied in order to systematically study the effects of each component as well as investigate structure-function relationships. The full library was screened by a multiparametric strategy which facilitated identification of a lead candidate that is optimized to overcome both cell-level (e.g., lipid bilayer translocation) and systemic (e.g., stability in blood and rapid renal clearance) delivery barriers. Our lead candidate from this screen, DB4-PDB12, is small (~100 nm), stable in size, efficiently internalized by cells, potently endosomolytic, and effective at gene silencing in vitro. DB4-PDB12 si-NPs were benchmarked against our previous “gold-standard” PDB si-NPs[23] and exhibited superior cell internalization and cytosolic delivery of nucleic acid cargo, increased resilience to heparan sulfate-mediated disassembly, reduced renal clearance, increased blood circulation AUC, and improved target gene silencing within tumors after intravenous delivery. These results confirmed that a multiparametric screening approach identified new lead formulations within the PEGylated polyplex class of siRNA carriers. New insights were also gleaned regarding the value of ternary over binary formulations, tuning of PEG density, and comprehensive incorporation of balanced cationic and hydrophobic content within both core- and corona-forming polyplex components.
Supplementary Material
Acknowledgments
We are grateful to the Vanderbilt Institute for Nanoscale Science and Engineering (VINSE) for access to DLS for nanoparticle characterization.
Funding: We are grateful to the National Science Foundation for supporting the Graduate Research Fellowship Program (NSF#1445197). We are grateful to the National Institutes of Health for financial support (NIH R01 EB019409). We are grateful to the Department of Defense Congressionally Directed Medical Research program for financial support (DOD CDMRP OR130302). We are grateful to the Vanderbilt School of Engineering for institutional and financial support.
Footnotes
Competing interests: The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Wang J, Lu Z, Wientjes MG, Au JS. Delivery of siRNA Therapeutics: Barriers and Carriers. AAPS J. 2010;12:492–503. doi: 10.1208/s12248-010-9210-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartlett DW, Davis ME. Effect of siRNA nuclease stability on the in vitro and in vivo kinetics of siRNA-mediated gene silencing. Biotechnology and Bioengineering. 2007;97:909–921. doi: 10.1002/bit.21285. [DOI] [PubMed] [Google Scholar]
- 4.Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Soutschek J, Akinc A, Bramlage B, Charisse K, Constien R, Donoghue M, Elbashir S, Geick A, Hadwiger P, Harborth J, John M, Kesavan V, Lavine G, Pandey RK, Racie T, Rajeev KG, Rohl I, Toudjarska I, Wang G, Wuschko S, Bumcrot D, Koteliansky V, Limmer S, Manoharan M, Vornlocher HP. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 6.Meade BR, Gogoi K, Hamil AS, Palm-Apergi C, van den Berg A, Hagopian JC, Springer AD, Eguchi A, Kacsinta AD, Dowdy CF, Presente A, Lonn P, Kaulich M, Yoshioka N, Gros E, Cui XS, Dowdy SF. Efficient delivery of RNAi prodrugs containing reversible charge-neutralizing phosphotriester backbone modifications. Nature biotechnology. 2014;32:1256–1261. doi: 10.1038/nbt.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Song E, Zhu P, Lee SK, Chowdhury D, Kussman S, Dykxhoorn DM, Feng Y, Palliser D, Weiner DB, Shankar P, Marasco WA, Lieberman J. Antibody mediated in vivo delivery of small interfering RNAs via cell-surface receptors. Nat Biotech. 2005;23:709–717. doi: 10.1038/nbt1101. [DOI] [PubMed] [Google Scholar]
- 8.Zimmermann TS, Lee ACH, Akinc A, Bramlage B, Bumcrot D, Fedoruk MN, Harborth J, Heyes JA, Jeffs LB, John M, Judge AD, Lam K, McClintock K, Nechev LV, Palmer LR, Racie T, Röhl I, Seiffert S, Shanmugam S, Sood V, Soutschek J, Toudjarska I, Wheat AJ, Yaworski E, Zedalis W, Koteliansky V, Manoharan M, Vornlocher HP, MacLachlan I. RNAi-mediated gene silencing in non-human primates. Nature. 2006;441:111–114. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]
- 9.Dong Y, Love KT, Dorkin JR, Sirirungruang S, Zhang Y, Chen D, Bogorad RL, Yin H, Chen Y, Vegas AJ, Alabi CA, Sahay G, Olejnik KT, Wang W, Schroeder A, Lytton-Jean AKR, Siegwart DJ, Akinc A, Barnes C, Barros SA, Carioto M, Fitzgerald K, Hettinger J, Kumar V, Novobrantseva TI, Qin J, Querbes W, Koteliansky V, Langer R, Anderson DG. Lipopeptide nanoparticles for potent and selective siRNA delivery in rodents and nonhuman primates. Proceedings of the National Academy of Sciences. 2014;111:3955–3960. doi: 10.1073/pnas.1322937111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rahman MA, Amin AR, Wang X, Zuckerman JE, Choi CH, Zhou B, Wang D, Nannapaneni S, Koenig L, Chen Z, Chen ZG, Yen Y, Davis ME, Shin DM. Systemic delivery of siRNA nanoparticles targeting RRM2 suppresses head and neck tumor growth. Journal of controlled release : official journal of the Controlled Release Society. 2012;159:384–392. doi: 10.1016/j.jconrel.2012.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christie RJ, Matsumoto Y, Miyata K, Nomoto T, Fukushima S, Osada K, Halnaut J, Pittella F, Kim HJ, Nishiyama N, Kataoka K. Targeted Polymeric Micelles for siRNA Treatment of Experimental Cancer by Intravenous Injection. ACS Nano. 2012;6:5174–5189. doi: 10.1021/nn300942b. [DOI] [PubMed] [Google Scholar]
- 12.Kanasty R, Dorkin JR, Vegas A, Anderson D. Delivery materials for siRNA therapeutics. Nat Mater. 2013;12:967–977. doi: 10.1038/nmat3765. [DOI] [PubMed] [Google Scholar]
- 13.Oh YK, Park TG. siRNA delivery systems for cancer treatment. Advanced Drug Delivery Reviews. 2009;61:850–862. doi: 10.1016/j.addr.2009.04.018. [DOI] [PubMed] [Google Scholar]
- 14.Mislick KA, Baldeschwieler JD. Evidence for the role of proteoglycans in cation-mediated gene transfer. Proceedings of the National Academy of Sciences. 1996;93:12349–12354. doi: 10.1073/pnas.93.22.12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Venkataraman S, Ong WL, Ong ZY, Joachim Loo SC, Rachel Ee PL, Yang YY. The role of PEG architecture and molecular weight in the gene transfection performance of PEGylated poly(dimethylaminoethyl methacrylate) based cationic polymers. Biomaterials. 2011;32:2369–2378. doi: 10.1016/j.biomaterials.2010.11.070. [DOI] [PubMed] [Google Scholar]
- 16.Mishra S, Webster P, Davis ME. PEGylation significantly affects cellular uptake and intracellular trafficking of non-viral gene delivery particles. European Journal of Cell Biology. 2004;83:97–111. doi: 10.1078/0171-9335-00363. [DOI] [PubMed] [Google Scholar]
- 17.Sato A, Choi SW, Hirai M, Yamayoshi A, Moriyama R, Yamano T, Takagi M, Kano A, Shimamoto A, Maruyama A. Polymer brush-stabilized polyplex for a siRNA carrier with long circulatory half-life. Journal of Controlled Release. 2007;122:209–216. doi: 10.1016/j.jconrel.2007.04.018. [DOI] [PubMed] [Google Scholar]
- 18.Verbaan FJ, Oussoren C, Snel CJ, Crommelin DJA, Hennink WE, Storm G. Steric stabilization of poly(2-(dimethylamino)ethyl methacrylate)-based polyplexes mediates prolonged circulation and tumor targeting in mice. The Journal of Gene Medicine. 2004;6:64–75. doi: 10.1002/jgm.475. [DOI] [PubMed] [Google Scholar]
- 19.Bartlett DW, Su H, Hildebrandt IJ, Weber WA, Davis ME. Impact of tumor-specific targeting on the biodistribution and efficacy of siRNA nanoparticles measured by multimodality in vivo imaging. Proceedings of the National Academy of Sciences. 2007;104:15549–15554. doi: 10.1073/pnas.0707461104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oe Y, Christie RJ, Naito M, Low SA, Fukushima S, Toh K, Miura Y, Matsumoto Y, Nishiyama N, Miyata K, Kataoka K. Actively-targeted polyion complex micelles stabilized by cholesterol and disulfide cross-linking for systemic delivery of siRNA to solid tumors. Biomaterials. 2014;35:7887–7895. doi: 10.1016/j.biomaterials.2014.05.041. [DOI] [PubMed] [Google Scholar]
- 21.Zuckerman JE, Choi CH, Han H, Davis ME. Polycation-siRNA nanoparticles can disassemble at the kidney glomerular basement membrane. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:3137–3142. doi: 10.1073/pnas.1200718109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Naeye B, Deschout H, Caveliers V, Descamps B, Braeckmans K, Vanhove C, Demeester J, Lahoutte T, De Smedt SC, Raemdonck K. In vivo disassembly of IV administered siRNA matrix nanoparticles at the renal filtration barrier. Biomaterials. 2013;34:2350–2358. doi: 10.1016/j.biomaterials.2012.11.058. [DOI] [PubMed] [Google Scholar]
- 23.Nelson CE, Kintzing JR, Hanna A, Shannon JM, Gupta MK, Duvall CL. Balancing Cationic and Hydrophobic Content of PEGylated siRNA Polyplexes Enhances Endosome Escape, Stability, Blood Circulation Time, and Bioactivity in Vivo. ACS Nano. 2013;7:8870–8880. doi: 10.1021/nn403325f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sarett SM, Werfel TA, Chandra I, Jackson MA, Kavanaugh TE, Hattaway ME, Giorgio TD, Duvall CL. Hydrophobic interactions between polymeric carrier and palmitic acid-conjugated siRNA improve PEGylated polyplex stability and enhance in vivo pharmacokinetics and tumor gene silencing. Biomaterials. 2016;97:122–132. doi: 10.1016/j.biomaterials.2016.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Green JJ, Zugates GT, Tedford NC, Huang YH, Griffith LG, Lauffenburger DA, Sawicki JA, Langer R, Anderson DG. Combinatorial Modification of Degradable Polymers Enables Transfection of Human Cells Comparable to Adenovirus. Advanced Materials. 2007;19:2836–2842. [Google Scholar]
- 26.Tzeng SY, Green JJ. Subtle Changes to Polymer Structure and Degradation Mechanism Enable Highly Effective Nanoparticles for siRNA and DNA Delivery to Human Brain Cancer. Advanced healthcare materials. 2013;2:468–480. doi: 10.1002/adhm.201200257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mangraviti A, Tzeng SY, Kozielski KL, Wang Y, Jin Y, Gullotti D, Pedone M, Buaron N, Liu A, Wilson DR, Hansen SK, Rodriguez FJ, Gao GD, DiMeco F, Brem H, Olivi A, Tyler B, Green JJ. Polymeric Nanoparticles for Nonviral Gene Therapy Extend Brain Tumor Survival in Vivo. ACS Nano. 2015;9:1236–1249. doi: 10.1021/nn504905q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dahlman JE, Barnes C, Khan OF, Thiriot A, Jhunjunwala S, Shaw TE, Xing Y, Sager HB, Sahay G, Speciner L, Bader A, Bogorad RL, Yin H, Racie T, Dong Y, Jiang S, Seedorf D, Dave A, Singh Sandhu K, Webber MJ, Novobrantseva T, Ruda VM, Lytton-JeanAbigail KR, Levins CG, Kalish B, Mudge DK, Perez M, Abezgauz L, Dutta P, Smith L, Charisse K, Kieran MW, Fitzgerald K, Nahrendorf M, Danino D, Tuder RM, von Andrian UH, Akinc A, Panigrahy D, Schroeder A, Koteliansky V, Langer R, Anderson DG. In vivo endothelial siRNA delivery using polymeric nanoparticles with low molecular weight. Nat Nano. 2014;9:648–655. doi: 10.1038/nnano.2014.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alabi CA, Love KT, Sahay G, Yin H, Luly KM, Langer R, Anderson DG. Multiparametric approach for the evaluation of lipid nanoparticles for siRNA delivery. Proceedings of the National Academy of Sciences. 2013;110:12881–12886. doi: 10.1073/pnas.1306529110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Akinc A, Zumbuehl A, Goldberg M, Leshchiner ES, Busini V, Hossain N, Bacallado SA, Nguyen DN, Fuller J, Alvarez R, Borodovsky A, Borland T, Constien R, de Fougerolles A, Dorkin JR, Narayanannair Jayaprakash K, Jayaraman M, John M, Koteliansky V, Manoharan M, Nechev L, Qin J, Racie T, Raitcheva D, Rajeev KG, Sah DWY, Soutschek J, Toudjarska I, Vornlocher H-P, Zimmermann TS, Langer R, Anderson DG. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nat Biotech. 2008;26:561–569. doi: 10.1038/nbt1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siegwart DJ, Whitehead KA, Nuhn L, Sahay G, Cheng H, Jiang S, Ma M, Lytton-Jean A, Vegas A, Fenton P, Levins CG, Love KT, Lee H, Cortez C, Collins SP, Li YF, Jang J, Querbes W, Zurenko C, Novobrantseva T, Langer R, Anderson DG. Combinatorial synthesis of chemically diverse core-shell nanoparticles for intracellular delivery. Proceedings of the National Academy of Sciences. 2011;108:12996–13001. doi: 10.1073/pnas.1106379108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hao J, Kos P, Zhou K, Miller JB, Xue L, Yan Y, Xiong H, Elkassih S, Siegwart DJ. Rapid Synthesis of a Lipocationic Polyester Library via Ring-Opening Polymerization of Functional Valerolactones for Efficacious siRNA Delivery. Journal of the American Chemical Society. 2015;137:9206–9209. doi: 10.1021/jacs.5b03429. [DOI] [PubMed] [Google Scholar]
- 33.Zhou K, Nguyen LH, Miller JB, Yan Y, Kos P, Xiong H, Li L, Hao J, Minnig JT, Zhu H, Siegwart DJ. Modular degradable dendrimers enable small RNAs to extend survival in an aggressive liver cancer model. Proceedings of the National Academy of Sciences. 2016;113:520–525. doi: 10.1073/pnas.1520756113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Love KT, Mahon KP, Levins CG, Whitehead KA, Querbes W, Dorkin JR, Qin J, Cantley W, Qin LL, Racie T, Frank-Kamenetsky M, Yip KN, Alvarez R, Sah DWY, de Fougerolles A, Fitzgerald K, Koteliansky V, Akinc A, Langer R, Anderson DG. Lipid-like materials for low-dose, in vivo gene silencing. Proceedings of the National Academy of Sciences. 2010;107:1864–1869. doi: 10.1073/pnas.0910603106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whitehead KA, Dorkin JR, Vegas AJ, Chang PH, Veiseh O, Matthews J, Fenton OS, Zhang Y, Olejnik KT, Yesilyurt V, Chen D, Barros S, Klebanov B, Novobrantseva T, Langer R, Anderson DG. Degradable lipid nanoparticles with predictable in vivo siRNA delivery activity. Nature Communications. 2014;5:4277. doi: 10.1038/ncomms5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sunshine JC, Akanda MI, Li D, Kozielski KL, Green JJ. Effects of Base Polymer Hydrophobicity and End-Group Modification on Polymeric Gene Delivery. Biomacromolecules. 2011;12:3592–3600. doi: 10.1021/bm200807s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kozielski KL, Tzeng SY, Hurtado De Mendoza BA, Green JJ. Bioreducible Cationic Polymer-Based Nanoparticles for Efficient and Environmentally Triggered Cytoplasmic siRNA Delivery to Primary Human Brain Cancer Cells. ACS Nano. 2014;8:3232–3241. doi: 10.1021/nn500704t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yan Y, Liu L, Xiong H, Miller JB, Zhou K, Kos P, Huffman KE, Elkassih S, Norman JW, Carstens R, Kim J, Minna JD, Siegwart DJ. Functional polyesters enable selective siRNA delivery to lung cancer over matched normal cells. Proceedings of the National Academy of Sciences. 2016;113:E5702–E5710. doi: 10.1073/pnas.1606886113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Convertine AJ, Benoit DS, Duvall CL, Hoffman AS, Stayton PS. Development of a novel endosomolytic diblock copolymer for siRNA delivery. Journal of controlled release : official journal of the Controlled Release Society. 2009;133:221–229. doi: 10.1016/j.jconrel.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Evans BC, Nelson CE, Yu SS, Beavers KR, Kim AJ, Li H, Nelson HM, Giorgio TD, Duvall CL. Ex Vivo Red Blood Cell Hemolysis Assay for the Evaluation of pH-responsive Endosomolytic Agents for Cytosolic Delivery of Biomacromolecular Drugs. 2013:e50166. doi: 10.3791/50166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bolte S, CordeliÈRes FP. A guided tour into subcellular colocalization analysis in light microscopy. Journal of Microscopy. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- 42.Chiefari J, Chong YK, Ercole F, Krstina J, Jeffery J, Le TPT, Mayadunne RTA, Meijs GF, Moad CL, Moad G, Rizzardo E, Thang SH. Living Free-Radical Polymerization by Reversible Addition–Fragmentation Chain Transfer: The RAFT Process. Macromolecules. 1998;31:5559–5562. [Google Scholar]
- 43.Boyer C, Bulmus V, Davis TP, Ladmiral V, Liu J, Perrier S. Bioapplications of RAFT Polymerization. Chemical Reviews. 2009;109:5402–5436. doi: 10.1021/cr9001403. [DOI] [PubMed] [Google Scholar]
- 44.Kong WH, Sung DK, Shim YH, Bae KH, Dubois P, Park TG, Kim JH, Seo SW. Efficient intracellular siRNA delivery strategy through rapid and simple two steps mixing involving noncovalent post-PEGylation. J Control Release. 2009;138:141–147. doi: 10.1016/j.jconrel.2009.04.034. [DOI] [PubMed] [Google Scholar]
- 45.Guo S, Huang Y, Zhang W, Wang W, Wei T, Lin D, Xing J, Deng L, Du Q, Liang Z, Liang XJ, Dong A. Ternary complexes of amphiphilic polycaprolactone-graft-poly (N,N-dimethylaminoethyl methacrylate), DNA and polyglutamic acid-graft-poly(ethylene glycol) for gene delivery. Biomaterials. 2011;32:4283–4292. doi: 10.1016/j.biomaterials.2011.02.034. [DOI] [PubMed] [Google Scholar]
- 46.Huang Y, Lin D, Jiang Q, Zhang W, Guo S, Xiao P, Zheng S, Wang X, Chen H, Zhang HY, Deng L, Xing J, Du Q, Dong A, Liang Z. Binary and ternary complexes based on polycaprolactone-graft-poly (N, N-dimethylaminoethyl methacrylate) for targeted siRNA delivery. Biomaterials. 2012;33:4653–4664. doi: 10.1016/j.biomaterials.2012.02.052. [DOI] [PubMed] [Google Scholar]
- 47.Medina-Kauwe LK, Xie J, Hamm-Alvarez S. Intracellular trafficking of nonviral vectors. Gene therapy. 2005;12:1734–1751. doi: 10.1038/sj.gt.3302592. [DOI] [PubMed] [Google Scholar]
- 48.Gilleron J, Querbes W, Zeigerer A, Borodovsky A, Marsico G, Schubert U, Manygoats K, Seifert S, Andree C, Stoter M, Epstein-Barash H, Zhang L, Koteliansky V, Fitzgerald K, Fava E, Bickle M, Kalaidzidis Y, Akinc A, Maier M, Zerial M. Image-based analysis of lipid nanoparticle-mediated siRNA delivery, intracellular trafficking and endosomal escape. Nat Biotech. 2013;31:638–646. doi: 10.1038/nbt.2612. [DOI] [PubMed] [Google Scholar]
- 49.Li H, Nelson CE, Evans BC, Duvall CL. Delivery of Intracellular-Acting Biologics in Pro-Apoptotic Therapies. Current Pharmaceutical Design. 2011;17:293–319. doi: 10.2174/138161211795049642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Evans BC, Nelson CE, Yu SS, Beavers KR, Kim AJ, Li H, Nelson HM, Giorgio TD, Duvall CL. Ex vivo red blood cell hemolysis assay for the evaluation of pH-responsive endosomolytic agents for cytosolic delivery of biomacromolecular drugs. J Vis Exp. 2013:e50166. doi: 10.3791/50166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sohaebuddin SK, Thevenot PT, Baker D, Eaton JW, Tang L. Nanomaterial cytotoxicity is composition, size, and cell type dependent. Particle and Fibre Toxicology. 2010;7:1–17. doi: 10.1186/1743-8977-7-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xia T, Kovochich M, Liong M, Zink JI, Nel AE. Cationic Polystyrene Nanosphere Toxicity Depends on Cell-Specific Endocytic and Mitochondrial Injury Pathways. ACS Nano. 2008;2:85–96. doi: 10.1021/nn700256c. [DOI] [PubMed] [Google Scholar]
- 53.Frank-Kamenetsky M, Grefhorst A, Anderson NN, Racie TS, Bramlage B, Akinc A, Butler D, Charisse K, Dorkin R, Fan Y, Gamba-Vitalo C, Hadwiger P, Jayaraman M, John M, Jayaprakash KN, Maier M, Nechev L, Rajeev KG, Read T, Röhl I, Soutschek J, Tan P, Wong J, Wang G, Zimmermann T, de Fougerolles A, Vornlocher H-P, Langer R, Anderson DG, Manoharan M, Koteliansky V, Horton JD, Fitzgerald K. Therapeutic RNAi targeting PCSK9 acutely lowers plasma cholesterol in rodents and LDL cholesterol in nonhuman primates. Proceedings of the National Academy of Sciences. 2008 doi: 10.1073/pnas.0805434105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dahlman JE, Kauffman KJ, Xing Y, Shaw TE, Mir FF, Dlott CC, Langer R, Anderson DG, Wang ET. Barcoded nanoparticles for high throughput in vivo discovery of targeted therapeutics. Proceedings of the National Academy of Sciences. 2017 doi: 10.1073/pnas.1620874114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Torchilin V. Tumor delivery of macromolecular drugs based on the EPR effect. Adv Drug Deliv Rev. 2011;63:131–135. doi: 10.1016/j.addr.2010.03.011. [DOI] [PubMed] [Google Scholar]
- 56.Maeda H, Nakamura H, Fang J. The EPR effect for macromolecular drug delivery to solid tumors: Improvement of tumor uptake, lowering of systemic toxicity, and distinct tumor imaging in vivo. Adv Drug Deliv Rev. 2013;65:71–79. doi: 10.1016/j.addr.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 57.Gao S, Dagnaes-Hansen F, Nielsen EJB, Wengel J, Besenbacher F, Howard KA, Kjems J. The Effect of Chemical Modification and Nanoparticle Formulation on Stability and Biodistribution of siRNA in Mice. Mol Ther. 2009;17:1225–1233. doi: 10.1038/mt.2009.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Han S, Cheng Q, Wu Y, Zhou J, Long X, Wei T, Huang Y, Zheng S, Zhang J, Deng L, Wang X, Liang XJ, Cao H, Liang Z, Dong A. Effects of hydrophobic core components in amphiphilic PDMAEMA nanoparticles on siRNA delivery. Biomaterials. 2015;48:45–55. doi: 10.1016/j.biomaterials.2015.01.026. [DOI] [PubMed] [Google Scholar]
- 59.Merkel OM, Librizzi D, Pfestroff A, Schurrat T, Buyens K, Sanders NN, De Smedt SC, Béhé M, Kissel T. Stability of siRNA polyplexes from poly(ethylenimine) and poly(ethylenimine)-g-poly(ethylene glycol) under in vivo conditions: Effects on pharmacokinetics and biodistribution measured by Fluorescence Fluctuation Spectroscopy and Single Photon Emission Computed Tomography (SPECT) imaging. Journal of Controlled Release. 2009;138:148–159. doi: 10.1016/j.jconrel.2009.05.016. [DOI] [PubMed] [Google Scholar]
- 60.Hansen AE, Petersen AL, Henriksen JR, Boerresen B, Rasmussen P, Elema DR, Rosenschöld PMa, Kristensen AT, Kjær A, Andresen TL. Positron Emission Tomography Based Elucidation of the Enhanced Permeability and Retention Effect in Dogs with Cancer Using Copper-64 Liposomes. ACS Nano. 2015;9:6985–6995. doi: 10.1021/acsnano.5b01324. [DOI] [PubMed] [Google Scholar]
- 61.Miller MA, Gadde S, Pfirschke C, Engblom C, Sprachman MM, Kohler RH, Yang KS, Laughney AM, Wojtkiewicz G, Kamaly N, Bhonagiri S, Pittet MJ, Farokhzad OC, Weissleder R. Predicting therapeutic nanomedicine efficacy using a companion magnetic resonance imaging nanoparticle. Science Translational Medicine. 2015;7:314ra183–314ra183. doi: 10.1126/scitranslmed.aac6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Clark AJ, Wiley DT, Zuckerman JE, Webster P, Chao J, Lin J, Yen Y, Davis ME. CRLX101 nanoparticles localize in human tumors and not in adjacent, nonneoplastic tissue after intravenous dosing. Proceedings of the National Academy of Sciences. 2016 doi: 10.1073/pnas.1603018113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.