Publisher's Note: There is an Inside Blood Commentary on this article in this issue.
Key Points
Single-cell gene expression analysis reveals CML stem cell heterogeneity and changes imposed by TKI therapy.
A subpopulation with primitive, quiescent signature and increased survival to therapy can be high-purity captured as CD45RA−cKIT−CD26+.
Abstract
Understanding leukemia heterogeneity is critical for the development of curative treatments as the failure to eliminate therapy-persistent leukemic stem cells (LSCs) may result in disease relapse. Here we have combined high-throughput immunophenotypic screens with large-scale single-cell gene expression analysis to define the heterogeneity within the LSC population in chronic phase chronic myeloid leukemia (CML) patients at diagnosis and following conventional tyrosine kinase inhibitor (TKI) treatment. Our results reveal substantial heterogeneity within the putative LSC population in CML at diagnosis and demonstrate differences in response to subsequent TKI treatment between distinct subpopulations. Importantly, LSC subpopulations with myeloid and proliferative molecular signatures are proportionally reduced at a higher extent in response to TKI therapy compared with subfractions displaying primitive and quiescent signatures. Additionally, cell surface expression of the CML stem cell markers CD25, CD26, and IL1RAP is high in all subpopulations at diagnosis but downregulated and unevenly distributed across subpopulations in response to TKI treatment. The most TKI-insensitive cells of the LSC compartment can be captured within the CD45RA− fraction and further defined as positive for CD26 in combination with an aberrant lack of cKIT expression. Together, our results expose a considerable heterogeneity of the CML stem cell population and propose a Lin−CD34+CD38−/lowCD45RA−cKIT−CD26+ population as a potential therapeutic target for improved therapy response.
Introduction
A groundbreaking example of molecular therapy of malignant disease is the development of tyrosine kinase inhibitors (TKIs) that specifically target the breakpoint cluster region (BCR)–Abelson (ABL), the result of the [9;22] translocation in chronic myeloid leukemia (CML).1-4 Although TKI treatment of CML is effective, a fraction of cells with leukemia-initiating capacity appear insensitive to TKIs, causing relapse upon TKI cessation even in patients with undetectable BCR-ABL levels.5 It is believed that this TKI insensitivity is a result of heterogeneity within the CML leukemic stem cell (LSC) compartment where primitive, quiescent subpopulations are inherently insensitive to TKIs and not dependent on BCR-ABL for survival.6-9 Thus, development of improved therapy for CML needs to be targeted at residual LSCs that persist under TKI therapy. However, LSCs are considered to be phenotypically similar to healthy hematopoietic stem cells (HSCs) and enriched in the Lin−CD34+CD38−/low stem cell compartment of the bone marrow (BM),10,11 herein referred to as “stem cell population” or “LSC population.” Several advances in defining CML LSCs have been made through the identification of aberrant expression of cell surface molecules such as CD33, CD123, IL1RAP, CD26, and CD25.12-16 Despite the potential of these markers to efficiently discriminate between leukemic and healthy cells within the stem cell population of CML patients, their specificity for different LSC subpopulations remains unknown. In addition, these previous efforts have focused on analysis of chronic phase (CP) CML at diagnosis, and their potential to capture persistent, TKI-insensitive cells has not been addressed.
Recent advances in single-cell gene expression analysis make possible the identification and characterization of molecularly distinct subpopulations and the subsequent delineation of heterogeneous hematopoietic cell fractions.17-23 In leukemia, single-cell methods additionally offer the opportunity to discriminate between leukemic and healthy cells, thereby allowing for specific characterization of the infrequent residual LSC population even months into treatment. Here we have dissected the heterogeneity of the CML LSC population both at diagnosis and following 3 months of TKI treatment. By combining and correlating large-scale single-cell gene expression analysis with cell surface marker screens, we reveal changes in the composition and the immunophenotype of the LSC compartment upon TKI treatment. In addition, we define a subpopulation with a quiescent, primitive molecular signature that shows increased relative survival to TKI therapy. This population is elusive to several previously suggested CML-specific LSC markers but can instead be high-purity prospectively isolated as a Lin−CD34+CD38−CD45RA−cKIT−CD26+ subfraction of putative CML LSCs.
Methods
Patient material
In total, 22 CP-CML patients and 5 age-matched healthy controls (normal BM [nBM]) were included in this study (supplemental Table 1, available on the Blood Web site). BM was aspirated from the posterior iliac crest after informed consent according to protocols approved by the regional research ethics committees of sites in Lund, Helsinki, Uppsala, Aarhus, and Stockholm. All samples were enriched for mononuclear or CD34+ cells and cryopreserved prior to analysis.
Flow cytometry
Mononuclear cells (MNCs) were isolated using Lymphoprep kits (Axis Shield), and CD34+ cells were enriched using magnetic microbeads (Miltenyi). Cells were stained with antibodies against lineage-specific markers not reported to be expressed on LSCs, together with antibodies listed in supplemental Table 2. Fluorescence-activated cell sorting (FACS)/analysis was performed using a FACSARIAII/III or LSRFORTESSA (BD Biosciences).
For antibody screens, MNCs were divided on 96-well plates containing commercially available screening panels according to the manufacturer’s protocol (BioLegend) and analyzed using the high-throughput sampler of FACSCANTOII (BD Biosciences). Data analysis was performed using FlowJo software (Tree Star).
Single-cell gene expression analysis
Single-cells (Lin−CD34+CD38−/low) were sorted into 4 µL lysis buffer.24 Preamplification was performed using Taqman-primers and Taq/SSIII reaction mix (Invitrogen). Linearity control and negative controls were included in each plate. Preamplification was performed according to a published polymerase chain reaction (PCR) protocol24 with an extended 50°C cycle. Complementary DNA was added to 96.96 Dynamic Array Chips (Fluidigm) with individual TaqMan-assays (supplemental Table 3) and quantitative PCR (qPCR)–analyzed on BioMarkHD (Fluidigm).
Data analysis
qPCR data were analyzed in the BiomarkHD analysis software (quality threshold 0.60, automatic global Ct threshold, and linear derivative baseline correction). After excluding controls, data were preprocessed in SCExV25 where Ct values were inverted, normalized to median expression of each cell, and z-score normalized. Unsupervised clustering analysis was performed using random forest clustering (RFC)26 and principal component analysis (PCA), and correlations were measured using the Spearman rank method. The raw data are available at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=yjevqoaylbafbqd&acc=GSE84507.
Results
Antibody screens identify CML LSC-specific cell surface markers
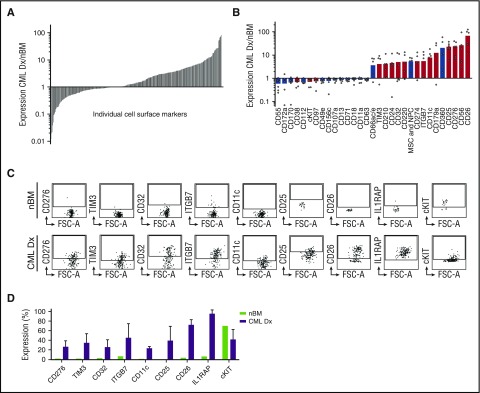
To identify cell surface proteins that are aberrantly expressed on CML LSCs, we performed antibody screens on CP-CML patient BM obtained at diagnosis. MNCs from each sample were antibody-stained for the stem cell population and divided into 332 individual wells, each containing a unique antibody against a specific surface marker. Subsequent flow cytometry analysis revealed a large number of differentially expressed markers in the LSC population as compared with the normal HSCs in 9 individual screens (CML at diagnosis, n = 6; nBM, n = 3) (Figure 1A). To identify markers with the potential to define subpopulations within the LSC population, we selected candidates that divided the LSCs (expressed on 10% to 90% of cells) in at least 3 CML patients but were either absent (<10%) or expressed on a large majority (>90%) in the stem cell population of healthy controls (Figure 1B). These included previously described LSC-specific cell surface molecules in CML (CD2515 and CD2613) and in acute myeloid leukemia (TIM327 and CD3228) together with a range of putative candidate CML LSC markers. To validate our screen, we individually analyzed 15 of the candidates together with the previously published CML LSC marker IL1RAP on additional diagnostic patients and an nBM control. Among these 16 markers, 8 positive (CD11c, CD25, CD26, CD32, CD276, IL1RAP, ITGB7, and TIM3) and 1 negative (cKIT) displayed an aberrant expression in validation experiments (Figure 1C-D).
Figure 1.
High-throughput antibody screens identify novel CML LSC-specific cell surface markers. (A) Overview of expression for 332 cell surface markers analyzed in the screen. The figure depicts the average cell surface marker expression in the Lin−CD34+CD38−/low LSC population of 6 CML patients at diagnosis (CML Dx) relative to the expression within the same population of 3 healthy controls. Each column represents an individual marker ordered from low to high relative expression compared with the controls. (B) Identification of candidate markers that aberrantly divided the CML LSC population (ie, <10% or >90% expressed in nBM HSCs, but >10% or <90% expressed on CML LSCs). The bars represent relative expression in LSCs compared with HSCs, and the dots denote the relative expression of single patients. The red color indicates the markers that were selected for validation experiments. (C) Data from 9 markers that showed consistent aberrant expression in validation experiments using additional CML Dx samples. The plots show representative FACS data for each marker as well as pooled data (D) from 3 different CML Dx’s compared with a new healthy nBM control. Gates are set based on background expression of isotype control antibodies. The error bars show standard deviation.
Single-cell molecular profiling reveals heterogeneity within the CML LSC compartment
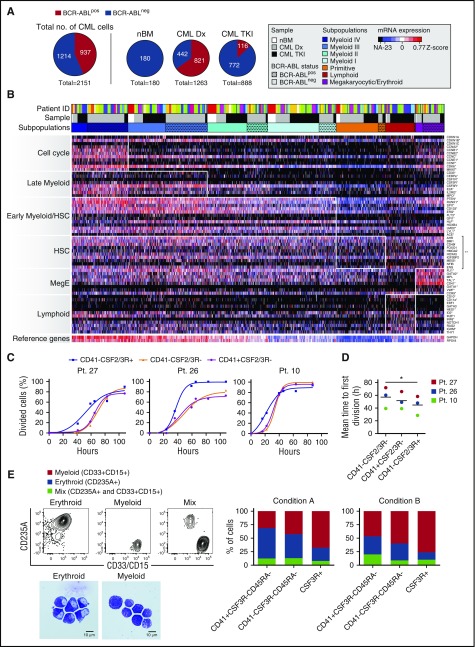
To define the heterogeneity within the LSC population, we next performed large-scale single-cell gene expression analysis on Lin−CD34+CD38−/low cells from CP-CML patients at diagnosis (n = 13) and 1 to 3 months following TKI treatment given that Ph+ cells could be detected by conventional BM cytogenetics (n = 10). Ninety-six to 192 single cells from each patient and from nBM were sorted and qPCR-analyzed against a panel of gene-specific primers (supplemental Table 3) selected based on literature and published gene expression analyses of hematopoietic and leukemic cells.29 These included housekeeping genes, established stem cell regulators, cell cycle molecules, lineage markers, and candidate cell surface molecules identified in the antibody screen together with the established CML LSC markers CD26 and IL1RAP. To discriminate between leukemic and normal cells, 2 primer assays targeting BCR-ABL were included. By analyzing a combination of cell lines expressing a variety of BCR-ABL transcripts and the LSC population of a CML patient with known frequency of Ph+ cells,30 we confirmed a high specificity and sensitivity of the BCR-ABL primers at the single-cell level and a close to absolute correlation between the proportion of Ph+ cells and the fraction of BCR-ABL–expressing primary LSCs (supplemental Figure 1). Additionally, we performed a number of control experiments to confirm that any effects on the data from pretreatment of cells (eg, cryopreservation and FACS), messenger RNA (mRNA) preamplification, and technical noise is minimal and significantly lower than observed cell-to-cell heterogeneity (supplemental Figure 2).
In total, 2151 single LSCs (CML Dx, n = 1263; TKI-treated CML, n = 888; nBM, n = 180) were included in the gene expression analysis after quality control (Figure 2A). BCR-ABL was expressed in 65% of the single-cells at diagnosis but reduced to 13% during TKI treatment (Figure 2A). Importantly, no BCR-ABL signal was detected in nBM cells, further excluding the possibility for false-positive identification of leukemic cells (Figure 2A). To define the heterogeneity within the data set we performed unsupervised RFC analysis to classify each cell based on their expression of the entire gene panel. This approach allowed for identification of 7 subpopulations with distinct molecular signatures reflecting identity and state (Figure 2B). Importantly, cells from several donors were represented in each subpopulation, excluding patient-specific skewing of the data (Figure 2B). Four of the subpopulations could be distinguished based on their increasing expression of genes involved in myeloid commitment (Myeloid I-IV). Myeloid I cells displayed a signature characterized by expression of genes involved in early myeloid commitment (eg, SPI1, ERG, RUNX1, HLF). Myeloid II and III populations were defined by their successive activation of a later myeloid program (eg, IL2RG, CSF2RA, CSF3R), associated with expression of various cell cycle genes (CDK6, CCNB1, CCNE1). The Myeloid IV subpopulation additionally expressed genes related to myeloid commitment (MPO, E2A, CD33) and displayed robust expression of a large cell cycle program including the proliferation marker KI67. We could additionally distinguish 2 subpopulations based on their expression of lymphoid-related genes (eg, CCR9, CD10, CD11a, IRF8) or a megakaryocytic/erythroid program (eg, CD41, GATA1, VWF, TAL1) and thus referred to them as Lymphoid and Meg/E, respectively. Finally, we identified a population with a quiescent molecular program accompanied with low expression of lineage molecules. This population displayed a molecular signature dominated by expression of genes from the HSC-related panel (eg, BMI1, FOXO1, HMGA2, HOXA5) and was thus termed Primitive. Although lacking expression of a majority of the lymphoid genes, we detected some expression of early myeloid and Meg/E genes in this subpopulation. However, these mainly included molecules also identified as important for the most primitive cells (ie, HLF, ERG, GATA2, MPL, and TAL1) (Figure 2B).
Figure 2.
Single-cell molecular analysis delineates the heterogeneity of the CML LSC population. (A) Overview of donor cell distribution and BCR-ABL expression in Lin−CD34+CD38−/low from CML patients analyzed for single-cell gene expression at diagnosis (CML Dx) (n = 13) and 1 or 3 months following TKI treatment (TKI) (n = 10) compared with nBM. (B) Heat map of normalized gene expression for 71 genes in 2331 Lin−CD34+CD38− single cells from all donors. Red indicates high expression; blue indicates low expression. Genes are listed to the right and ordered in groups based on which cell state or cell type they characterize, shown to the left. Each row in the heat map corresponds to a specific gene; each column corresponds to a particular single cell. RFC identified 7 subgroups of cells (Myeloid I-IV, Primitive, Lymphoid, and Megakaryocytic/Erythroid [Meg/E]), based on their expression of the predefined gene groups (indicated by white boxes in the heat map). The single asterisk indicates significant upregulated gene expression (P < .001) comparing the cells in the white boxes to all other cells in the heat map. The double asterisk indicates significant upregulated expression comparing the genes in the white box compared with all other genes within the Primitive subpopulation (P < 1 × 10−31). The row closest to the heat map represents the color coding for each subpopulation where BCR-ABLpos cells are indicated with dots. The color coding of the second row (sample) indicates donor status (nBM, white; CML Dx, gray; CML patient during TKI treatment [CML TKI], black), and the third row shows the donor distribution (“Patient ID”), where each color represents a donor. (C) Cumulative plots of the time to first division of single CD41+CSF2/3R−, CD41−CSF2/3R−, and CSF2/3R+ LSCs enriching for subpopulations expressing myeloid, primitive, and Meg/E signatures, respectively. Each plot represents an individual CML patient at diagnosis. Dead cells are excluded. (D) Mean time to first division in the 3 patients analyzed (*P = .002). (E) Lineage distribution of clones derived from single LSCs subdivided by the indicated markers to enrich for subpopulations expressing myeloid, primitive, and Meg/E signatures. The single cells were cultured in stem cell factor, thrombopoietin, interleukin-3 (IL-3), and erythropoietin (condition A) or stem cell factor, thrombopoietin, IL-3, erythropoietin, IL-6, and granulocyte colony-stimulating factor (condition B). The clones were scored by FACS analysis as erythroid (CD235a+), myeloid (CD15+/CD33+), or mixed (CD235a+ and CD15+/CD33+). Morphology analysis confirmed that the CD235a+ and CD15+/CD33+ cells represented erythroid and myeloid cells, respectively. The columns represent pooled data for lineage distribution for 230 single-cell clones from 3 CML Dx patients (condition A) and 164 clones from 2 CML Dx patients (condition B).
To assess whether the molecular signatures that defined the subgroups also reflected functional properties, we performed single-cell in vitro proliferation and differentiation experiments on LSCs expressing cell surface markers that were differentially expressed in the gene expression analysis. As expected from the gene expression data, LSCs expressing CSF2R or CSF3R on the surface (enriching for the Myeloid II, III, and IV population) rapidly entered proliferation and almost exclusively differentiated into cells expressing mature myeloid markers (CD15/CD33) following 2 weeks of culture (Figure 2C-E). In contrast, CD41+ LSCs (enriched for the Meg/E LSCs) differentiated more into cells expressing the erythroid marker CD235a. Moreover, cells that lacked expression of both markers (enriched for Myeloid I and Primitive) exhibited a delayed entry into cycle and multilineage differentiation capacity in our assay. Using morphology analysis, we could confirm that the CD235a+ and CD15+/CD33+ cells represented erythroid and myeloid cells, respectively (Figure 2E). Together, these data suggest that the observed molecular heterogeneity within the LSC population reflects functional differences in cell cycle status and lineage potential.
The most TKI-insensitive cells express a primitive, quiescent molecular program
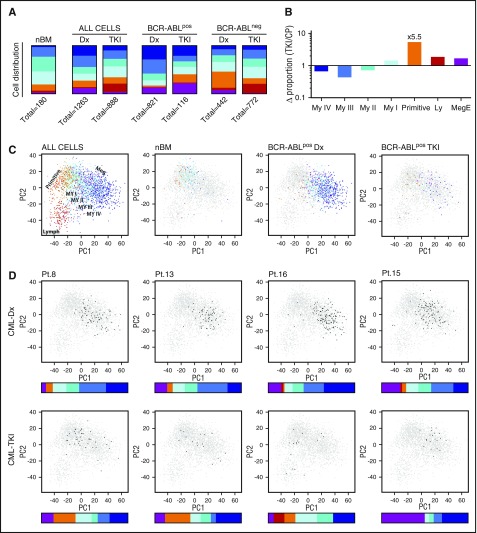
Based on the defined heterogeneity of the LSC population, we explored how BCR-ABL translocation and subsequent TKI treatment affects the composition of the stem cell pool. nBM contained primarily subpopulations with nonproliferating, primitive, or early myeloid molecular signatures (Primitive, Myeloid I and II) accounting for 71% of the total HSC compartment (Figure 3A). In contrast, the LSCs from diagnostic CML patients consisted predominantly of subpopulations with more mature proliferative, myeloid, or Meg/E molecular signatures, suggesting that BCR-ABL drives HSCs toward myeloid or Meg/E differentiation and expansion. Discrimination between leukemic and healthy cells in the analysis further strengthened this observation. Although the BCR-ABLpos diagnostic LSC pool consisted to 85% of cells belonging to subpopulations with late myeloid or Meg/E molecular signatures, the residual BCR-ABLneg HSC population resided to a large extent in the Primitive and Lymphoid subpopulations (Figure 3A).
Figure 3.
The effect of BCR-ABL translocation and subsequent TKI treatment on the heterogeneity of the CML LSC population. (A) Distribution of BCR-ABLpos and/or BCR-ABLneg cells within the 7 Lin−CD34+CD38−/low subpopulations in nBM, CP-CML at diagnosis (CML Dx), and following TKI treatment (CML TKI). (B) Fold change in the proportion of BCR-ABLpos cells from patients where analysis was performed both at diagnosis (Dx) and following TKI treatment (TKI). (C) PCA plots of the single-cell gene expression data displaying how Lin−CD34+CD38−/low heterogeneity is changed in CP-CML at diagnosis (Dx) compared with healthy nBM, and how cells with more myeloid or Meg/E molecular signatures are eradicated during treatment (TKI). Each dot represents a single cell that has been color coded according to the RFC. The legend on top of the plots (ALL CELLS, nBM, BCR-ABLpos-Dx, BCR-ABLpos-TKI) indicates cells that are colored; other cells are depicted as gray. (D) PCA plots and distribution of BCR-ABLpos cells according to RFC (colored bar) for 4 individual patients. Black dots represent cells from the specific patient at the indicated point of analysis (diagnosis [Dx], following TKI treatment [TKI]); the rest of the data set is depicted in gray.
In the TKI-treated CML samples, the LSC heterogeneity resembled healthy hematopoiesis with the exception of a proportional increase in the subpopulation expressing a lymphoid gene program (Figure 3A). However, this Lymphoid population almost exclusively consisted of BCR-ABLneg cells. Among BCR-ABLpos cells, TKI treatment had shifted the heterogeneity toward the Primitive, Myeloid I, and Meg/E subpopulations as compared with at diagnosis (Figure 3A). To measure the difference in TKI sensitivity between the different subpopulations, we compared the proportion of each BCR-ABLpos fraction in TKI samples with the diagnostic samples of 7 patients that had detectable levels of BCR-ABLpos LSCs at both time points. Interestingly, BCR-ABL inhibition by TKIs had the strongest suppressing effect on subpopulations with late myeloid molecular signatures, whereas the Myeloid I, Primitive, Lymphoid, and Meg/E stem cell subpopulations were proportionally increased by treatment. Strikingly, the strongest effect observed was the 5.5-fold proportional increase in the Primitive subpopulation (Figure 3B).
To alternatively visualize our single-cell data, we performed PCA analysis, which, compared with RFC, is a lower-dimensional method. In accordance with the RFC-generated data, the single-cells clustered into a distinct subgroup expressing a lymphoid molecular signature, a cluster with a primitive signature, and subgroups with an increasingly myeloid or Meg/E program (Figure 3C). PCA analysis showed that BCR-ABLpos Lin−CD34+CD38−/low cells at diagnosis to a large extent clustered away from the subpopulations with a more primitive signature, which mainly make up the nBM HSC population. However, following TKI treatment, the majority of persisting BCR-ABLpos cells instead accumulated near the primitive part of the PCA plot, supporting the RFC data. The low number of BCR-ABLpos cells present in each patient after TKI treatment does not allow for precise conclusions regarding patient-to-patient heterogeneity. However, all 4 patients with >10 BCR-ABLpos LSCs following treatment displayed a robust reduction of the cells with late myeloid signature that dominated the stem cell population at diagnosis. In 3 out of 4 patients, TKI treatment pushed the heterogeneity toward the Primitive subpopulation, while the Meg/E subpopulation was proportionally increased in the fourth (Figure 3D). Together these data suggest that the most persistent subpopulation within the CML LSC fraction during TKI treatment expresses a primitive, quiescent molecular program.
CML-specific cell surface markers are unequally distributed across LSC subpopulations
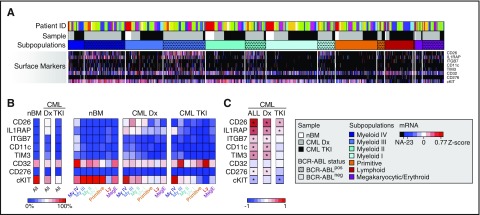
We next set out to map cell surface markers to the identified subpopulations. In accordance with our FACS-screen analysis, mRNA expression of the majority of positive CML-specific markers was absent in nBM Lin−CD34+CD38−/low cells, whereas the candidate negative marker for CML cKIT was expressed in nearly all normal cells of the HSC population (Figure 4A-B).
Figure 4.
Cell surface marker expression in CML LSCs changes during TKI treatment. (A) Cell surface marker expression extracted from the RFC heat map of single-cell gene expression (red indicates high expression, blue low expression, and black no expression). The rows represent mRNA expression for each marker, and the columns each single cell. The white boxes highlight BCR-ABLpos cells. (B) Heat map depicting the proportion of cells expressing mRNA for the indicated cell surface markers in the Lin−CD34+CD38−/low fraction of nBM samples, CML patients at diagnosis (CML Dx, n = 13), and of CML patients during TKI therapy (CML TKI, n = 10), as well as in each respective subpopulation (red represents high proportion, and blue low proportion). (C) Heat map of Spearman rank correlation coefficients for coexpression between BCR-ABL and indicated cell surface markers in all, diagnostic (Dx), or TKI-treated (TKI) CML-LSC samples, respectively (red indicates high correlation, and blue anticorrelation; *P < .05 according to Student t test).
We detected a sharp increase in the proportion of cells expressing IL1RAP and CD26 mRNA, as well as a small increase in TIM3pos cells in the diagnostic BCR-ABLpos LSC population (Figure 4A-B). In contrast, cKIT mRNA was expressed in a substantially lower proportion compared with the nBM HSC compartment (Figure 4B). CD11c and ITGB7 showed low expression levels, whereas CD32 was highly expressed in both nBM and CML cells. We next compared mRNA expression to the heterogeneity of BCR-ABLpos cells and observed aberrant IL1RAP, CD26, and TIM3 mRNA expression on a larger number of cells of the subpopulations with myeloid molecular signature compared with the other subfractions, while cKIT downregulation was strongest in the Primitive subpopulation (Figure 4B).
Interestingly, heterogeneous distribution of surface marker expression within the LSC population was even more pronounced during TKI treatment. Here, IL1RAP expression on BCR-ABLpos cells was found exclusively in subpopulations expressing late myeloid molecular signatures and in the Meg/E subpopulation (Figure 4B). Similarly, TIM3 expression was only detected in a low fraction of BCR-ABLpos cells within LSC subpopulations with late myeloid molecular signature. CD26 was also downregulated but still detected in the Myeloid I and III, Primitive, and Meg/E subfractions. In contrast, cKIT expression following TKI treatment was aberrantly absent on a majority of BCR-ABLpos cells of the subpopulations with more primitive and quiescent signatures as well as in the small Lymphoid subfraction of the LSC compartment (Figure 4B).
To investigate if mRNA expression of the cell surface markers additionally discriminated between BCR-ABLpos and BCR-ABLneg cells within LSC fractions, we performed a correlation analysis on the single-cell molecular data. Although expression of all positive markers except for CD32 significantly correlated with BCR-ABL expression at diagnosis, expression of the negative marker cKIT significantly correlated negatively to BCR-ABL expression. However, only cKIT, CD26, and IL1RAP expression significantly correlated with BCR-ABL expression following TKI treatment (Figure 4C). Together, these data demonstrate substantial heterogeneity in cell surface marker expression within the CML LSC population and reveal large differences in LSC immunophenotype following TKI treatment as compared with diagnosis.
The most TKI-insensitive LSC subpopulation can be defined as Lin−CD34+CD38−/lowCD45RA−cKIT−CD26+
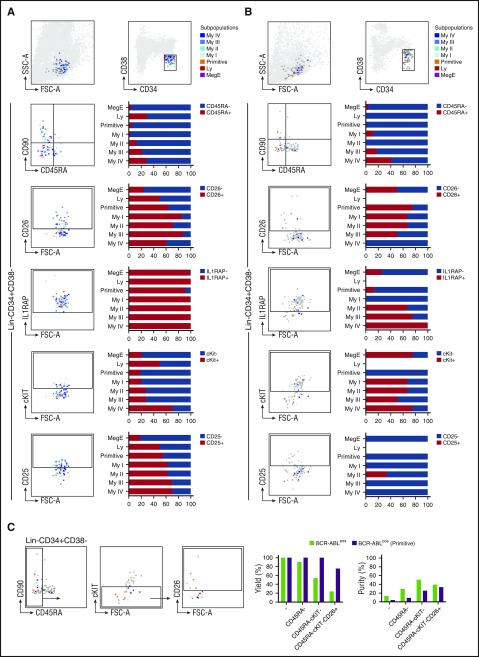
Based on the mRNA correlation data, single-cell molecular analysis from 3 patients was combined with index sorting for Lin, CD34, CD38, cKIT, CD26, and IL1RAP, as well as the established CML marker CD25 and the conventional HSC markers CD90 and CD45RA.31 This strategy enabled the sorting of single LSCs for molecular analysis and simultaneous acquisition of single-cell FACS data for all analyzed markers, thereby directly mapping the identified subpopulations to immunophenotype. All cells from subpopulations with more primitive molecular programs (Primitive and Myeloid I), as well as cells belonging to the Meg/E subpopulation, were CD45RA−, whereas the Lymphoid and later Myeloid (II-IV) subpopulations had variable CD45RA expression. In contrast, CD90 did not discriminate between different subpopulations within the LSC pool (Figure 5A). Moreover, in agreement with previous reports32 a majority, but not all, of the BCR-ABLpos cells expressed CD25 and/or CD26, while almost all leukemic cells were IL1RAP+ at diagnosis (Figure 5A). Corresponding to the mRNA analysis (Figure 4), the proportion of cells expressing cKIT on the cell surface was substantially downregulated in BCR-ABLpos cells of all LSC subpopulations, except for the large Myeloid IV population (Figure 5A).
Figure 5.
The most TKI-insensitive Primitive subpopulation can be defined as Lin−CD34+CD38−/lowCD45RA−cKIT−CD26+. Immunophenotypic characterization of sorted Lin−CD34+CD38−/low single cells at diagnosis (A) and at TKI treatment (B). The left panels depict representative FACS plots for cell surface expression of the indicated marker on individual BCR-ABLpos cells, color coded according to their subpopulation defined by the RFC of the single-cell gene expression data. The dark gray dots represent BCR-ABLneg cells, the lighter gray dots are unsorted but FACS recorded cells. The right panels show pooled data from 3 patients and the proportion of BCR-ABLpos cells in each subpopulations that express the indicated marker on their cell surface. (C) FACS analysis of Lin−CD34+CD38−/low with a combination of markers for prospective isolation of TKI-persistent CML LSCs. The FACS plots exemplify 1 representative CML patient following TKI treatment. The gates are set according to the isotype control antibody for CD26, IL1RAP, cKIT, and CD25, respectively. The bar charts depicts pooled data from 3 individual patients following 3 months of TKI treatment and display the relative yield and purity of total BCR-ABLpos cells (green) and of the subpopulation with Primitive molecular program (purple) that is achieved when using the Lin−CD34+CD38−/low protocol alone (-) or the indicated combination of markers.
To explore the potential for these markers to capture residual BCR-ABLpos cells during TKI treatment, we repeated this analysis on the same patients 3 months into imatinib or bosutinib therapy. As observed in the mRNA analysis (Figure 4), cell surface marker expression of TKI-treated BCR-ABLpos LSCs was considerably different compared with diagnostic samples. At both time points, BCR-ABLpos cells overlapped within the Lin−CD34+CD38−/low LSC gate, were almost exclusively CD45RA−, and could not be separated based on CD90 expression. Importantly, CD25 expression could not be detected on BCR-ABLpos cells following TKI treatment, while IL1RAP was exclusively expressed on subpopulations with Myeloid, Meg/E, and proliferative molecular programs (Figure 5B). In contrast, most of the cells from subpopulations with primitive, quiescent signatures (65% of the Myeloid I and 75% of the Primitive) were captured by CD26. Remarkably, all cells from the most TKI-insensitive Primitive subpopulation were negative for cKIT (Figure 5B), supporting the mRNA data from the single-cell gene expression analysis.
We next explored the possibility to combine these markers into a protocol for prospective isolation of the most TKI-insensitive, Primitive subpopulation of CML LSCs (Figure 5C). Compared with the conventional Lin−CD34+CD38−/low marker combination, further separation of the TKI-treated LSC population based on CD45RA expression improved the purity of total BCR-ABLpos cells 2.5-fold without compromising the yield. Similarly, the BCR-ABLpos cells from the Primitive subpopulation represented only 3.36% of the total TKI-treated Lin−CD34+CD38−/low fraction but were enriched to a purity of 8.6% without loss of yield by exclusion of CD45RA+ cells. The entire BCR-ABLpos Primitive subpopulation was additionally captured within the cKIT− fraction, increasing purity to 25%. Finally, CD26 efficiently discriminated between BCR-ABLpos cells of the Primitive subpopulation and the remaining BCR-ABLpos cells, resulting in further enrichment of these cells to an ∼1 in 3 ratio.
Collectively, we have identified a CML LSC subpopulation with a primitive, quiescent molecular signature and increased TKI insensitivity. This subpopulation can be captured within the Lin−CD34+CD38−/lowCD45RA−cKIT−CD26+ fraction, at a 10-fold enrichment compared with conventional Lin−CD34+CD38−/low isolation protocols.
Discussion
Targeting therapy-persistent LSCs is a critical priority for improved treatment of CML.33,34 However, the LSC population is heterogeneous, consisting of a mixture of leukemic cells with differences in TKI sensitivity as well as residual healthy stem cells and progenitors.10,31,35 Single-cell gene expression analysis offers the possibility to identify distinct subpopulations within the stem cell population in CML while directly discriminating between leukemic and normal cells based on BCR-ABL expression. This is of particular importance in remission samples where only a minority of the stem cell population is leukemic. Here, we used qPCR technology for a preselected set of 95 genes, which we regard as a technically robust alternative36 (supplemental Figure 2) to genome-wide approaches that are likely confounded by technical noise. Analysis of >2000 Lin−CD34+CD38−/low single cells afforded the identification of 7 molecularly distinct subpopulations within the stem cell population. The HSC population from the nBM control was to a large extent made up by subpopulations expressing primitive or early myeloid molecular, quiescent signatures, likely reflecting the myeloid bias that has been reported in aged HSCs.37,38 However, in the diagnostic BCR-ABLpos LSC population, this heterogeneity was skewed toward subpopulations with cycling, late myeloid or Meg/E signatures in line with previous reports suggesting that BCR-ABL pushes HSCs into proliferation and differentiation.39 In contrast, identification of BCR-ABLpos cells within the subgroup that expressed a lymphoid molecular signature was infrequent. Still, we observed a proportional increase in the BCR-ABLneg Lymphoid subpopulation upon TKI treatment likely reflecting the previously reported lymphocyte expansion in response to TKIs.40
Analysis of TKI-treated patients clearly showed that the leukemic subpopulations with late myeloid programs were most affected by BCR-ABL inhibition. Interestingly, the Meg/E subpopulation was more TKI insensitive despite its proliferative signature, suggesting that TKI sensitivity is not dependent on cell cycle status per se. However, the most striking observation was the TKI-induced 5.5-fold relative expansion of the BCR-ABLpos subpopulation with a quiescent, primitive molecular profile.
In addition, we noted a fluctuation of reference gene expression levels between subpopulations that correlated with expression of the cell cycle program. Similar observations have been made in mice where the most primitive and quiescent HSC populations express low levels of total mRNA.41,42
A correlation between TKI insensitivity and primitiveness/quiescence has been suggested previously.6,7 Using our approach, we can now visualize primitive, quiescent, TKI-insensitive cells and correlate this distinct population with immunophenotype during TKI treatment. Using antibody screening, we could validate not only aberrant but also heterogeneous expression of several established CML-specific markers in the LSC compartment. In addition, we identified novel markers with the potential to specifically capture LSCs. However, the majority of these were substantially downregulated on BCR-ABLpos LSCs from TKI-treated patients, and only CD26, IL1RAP, and cKIT mRNA expression significantly correlated with BCR-ABL detection.
At diagnosis, CD25, CD26, and IL1RAP were frequently expressed also on the cell surface of BCR-ABLpos cells of all subpopulations within the stem cell fraction. In accordance with previous observations, IL1RAP marked the vast majority of CML LSCs, while not all BCR-ABLpos cells expressed CD26 and CD25.32 Of note, only subpopulations with a late myeloid or lymphoid molecular signature were CD45RA+ supporting previous reports describing that quiescent, diagnostic CML LSCs reside in the Lin−CD34+CD38−/lowCD45RA− population.43 The decreased cell surface expression of cKIT was clearly observed also in the index sorting of diagnostic samples. This downregulation was most pronounced in the more primitive populations.
Interestingly, TKI treatment resulted in a substantial change in immunophenotype of the residual leukemic cells of the stem cell pool. CD25 was absent on a large majority of TKI-treated BCR-ABLpos cells, and IL1RAP expression was considerably downregulated and almost exclusively detected on subpopulations expressing late myeloid signatures. These data indicate that the expression of these cell surface markers is directly or indirectly dependent on the activity of BCR-ABL, as has been suggested for IL1RAP in a TKI-treated CML cell line.12 Similarly, CD26 was downregulated on BCR-ABLpos cells upon TKI treatment but was in contrast to IL1RAP more frequently expressed on subpopulations with more primitive signatures. Furthermore, cKIT was to a large extent expressed on the subpopulations with late myeloid signatures but entirely absent on the subpopulation with a primitive molecular program. The observation that cKIT is downregulated in CML LSCs already at diagnosis and completely absent on the most persistent subpopulation after TKI treatment has potential implications for future CML therapy. Off-target effects on cKIT signaling have been suggested to affect the efficacy of TKIs given the tyrosine kinase activity of cKIT, where maximal effect is achieved when cKIT and BCR-ABL signaling are simultaneously blocked.44 It is possible that the constitutive activity of BCR-ABL at least in part can replace cKIT function in CML LSCs, resulting in cKIT independence and downregulation. Recent reports also demonstrate that the most quiescent HSCs in mouse are cKitlow.41,45 Our data suggest that TKI treatment, which inhibits both BCR-ABL and cKIT signaling,46 is most efficient on LSC subpopulations with a high frequency of cKIT+ cells but spares cKIT− quiescent, primitive subpopulations. Thus, clarification of the role for cKIT in inherent TKI insensitivity of LSCs represents an exciting topic for future investigations.
In conclusion, by directly comparing single-cell molecular signatures with therapy response and cell surface marker expression, we here reveal the heterogeneity of the CP-CML LSC population and identify a subpopulation with a primitive, quiescent molecular signature that persists during TKI treatment. These TKI-persistent cells can be high-purity captured within the CD45RA−cKIT−CD26+ LSC population, offering possibilities for characterization of therapy insensitivity in CML.
Acknowledgments
The authors are grateful to all patients and healthy donors for their participation in this study. The authors would like to thank Ingbritt Åstrand-Grundström for expert advice on the morphology analysis, as well as Petter Uvesten for input on the bioinformatic analysis.
This work was supported by grants from the Swedish Cancer Society, the Ragnar Söderberg Foundation, the Knut and Alice Wallenberg Foundation, the Swedish Research Council, the Crafoord Foundation, Jaensson’s Foundation, a StemTherapy grant from the Swedish Research Council, a Pfizer investigator-initiated research grant, and regional research support from Region Skåne, Sweden. This work was performed together with the Nordic CML Study Group as a substudy to the Pfizer-sponsored bosutinib trial in first line chronic myelogenous leukemia treatment (BFORE).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: G.K. conceived, designed, and supervised the study; R.W., L.G., and M.N.E.S. conceived, performed, and analyzed the experiments; S.L. performed the majority of the bioinformatics analysis with assistance of M.N.E.S. and under the supervision of S.S.; T.R. provided critical assistance and advice regarding flow cytometry experiments; S.M., J.R., H.H.-H., L.S., J.S., and U.O.-S. provided patient material; S.M., J.R., J.S., and H.H.-H. were national investigators for the study; G.K. wrote the majority of the manuscript; and R.W., L.G., M.N.E.S., C.K., S.M., and J.R. discussed research and contributed to writing the manuscript with input from all authors.
Conflict-of-interest disclosure: S.M. and J.R. have received honoraria and/or research funding from Novartis, Pfizer, Bristol-Myers Squibb, and Ariad. The remaining authors declare no competing financial interests.
Correspondence: Göran Karlsson, Division of Molecular Hematology, BMC B12, SE-221 84 Lund, Sweden; e-mail: goran.karlsson@med.lu.se.
References
- 1.de Klein A, van Kessel AG, Grosveld G, et al. A cellular oncogene is translocated to the Philadelphia chromosome in chronic myelocytic leukaemia. Nature. 1982;300(5894):765-767. [DOI] [PubMed] [Google Scholar]
- 2.Schindler T, Bornmann W, Pellicena P, Miller WT, Clarkson B, Kuriyan J. Structural mechanism for STI-571 inhibition of Abelson tyrosine kinase. Science. 2000;289(5486):1938-1942. [DOI] [PubMed] [Google Scholar]
- 3.O’Brien SG, Guilhot F, Larson RA, et al. ; IRIS Investigators. Imatinib compared with interferon and low-dose cytarabine for newly diagnosed chronic-phase chronic myeloid leukemia. N Engl J Med. 2003;348(11):994-1004. [DOI] [PubMed] [Google Scholar]
- 4.Rowley JD. Letter: A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973;243(5405):290-293. [DOI] [PubMed] [Google Scholar]
- 5.Mahon FX, Réa D, Guilhot J, et al. ; Intergroupe Français des Leucémies Myéloïdes Chroniques. Discontinuation of imatinib in patients with chronic myeloid leukaemia who have maintained complete molecular remission for at least 2 years: the prospective, multicentre Stop Imatinib (STIM) trial. Lancet Oncol. 2010;11(11):1029-1035. [DOI] [PubMed] [Google Scholar]
- 6.Graham SM, Jørgensen HG, Allan E, et al. Primitive, quiescent, Philadelphia-positive stem cells from patients with chronic myeloid leukemia are insensitive to STI571 in vitro. Blood. 2002;99(1):319-325. [DOI] [PubMed] [Google Scholar]
- 7.Copland M, Hamilton A, Elrick LJ, et al. Dasatinib (BMS-354825) targets an earlier progenitor population than imatinib in primary CML but does not eliminate the quiescent fraction. Blood. 2006;107(11):4532-4539. [DOI] [PubMed] [Google Scholar]
- 8.Hamilton A, Helgason GV, Schemionek M, et al. Chronic myeloid leukemia stem cells are not dependent on Bcr-Abl kinase activity for their survival. Blood. 2012;119(6):1501-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corbin AS, Agarwal A, Loriaux M, Cortes J, Deininger MW, Druker BJ. Human chronic myeloid leukemia stem cells are insensitive to imatinib despite inhibition of BCR-ABL activity. J Clin Invest. 2011;121(1):396-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3(7):730-737. [DOI] [PubMed] [Google Scholar]
- 11.Eisterer W, Jiang X, Christ O, et al. Different subsets of primary chronic myeloid leukemia stem cells engraft immunodeficient mice and produce a model of the human disease. Leukemia. 2005;19(3):435-441. [DOI] [PubMed] [Google Scholar]
- 12.Järås M, Johnels P, Hansen N, et al. Isolation and killing of candidate chronic myeloid leukemia stem cells by antibody targeting of IL-1 receptor accessory protein. Proc Natl Acad Sci USA. 2010;107(37):16280-16285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herrmann H, Sadovnik I, Cerny-Reiterer S, et al. Dipeptidylpeptidase IV (CD26) defines leukemic stem cells (LSC) in chronic myeloid leukemia. Blood. 2014;123(25):3951-3962. [DOI] [PubMed] [Google Scholar]
- 14.Herrmann H, Cerny-Reiterer S, Gleixner KV, et al. CD34(+)/CD38(-) stem cells in chronic myeloid leukemia express Siglec-3 (CD33) and are responsive to the CD33-targeting drug gemtuzumab/ozogamicin. Haematologica. 2012;97(2):219-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kobayashi CI, Takubo K, Kobayashi H, et al. The IL-2/CD25 axis maintains distinct subsets of chronic myeloid leukemia-initiating cells. Blood. 2014;123(16):2540-2549. [DOI] [PubMed] [Google Scholar]
- 16.Nievergall E, Ramshaw HS, Yong AS, et al. Monoclonal antibody targeting of IL-3 receptor α with CSL362 effectively depletes CML progenitor and stem cells. Blood. 2014;123(8):1218-1228. [DOI] [PubMed] [Google Scholar]
- 17.Guo G, Luc S, Marco E, et al. Mapping cellular hierarchy by single-cell analysis of the cell surface repertoire. Cell Stem Cell. 2013;13(4):492-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karlsson G, Rörby E, Pina C, et al. The tetraspanin CD9 affords high-purity capture of all murine hematopoietic stem cells. Cell Reports. 2013;4(4):642-648. [DOI] [PubMed] [Google Scholar]
- 19.Psaila B, Barkas N, Iskander D, et al. Single-cell profiling of human megakaryocyte-erythroid progenitors identifies distinct megakaryocyte and erythroid differentiation pathways. Genome Biol. 2016;17:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moore FE, Garcia EG, Lobbardi R, et al. Single-cell transcriptional analysis of normal, aberrant, and malignant hematopoiesis in zebrafish. J Exp Med. 2016;213(6):979-992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drissen R, Buza-Vidas N, Woll P, et al. Distinct myeloid progenitor-differentiation pathways identified through single-cell RNA sequencing. Nat Immunol. 2016;17(6):666-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Macaulay IC, Svensson V, Labalette C, et al. Single-cell RNA-sequencing reveals a continuous spectrum of differentiation in hematopoietic cells. Cell Reports. 2016;14(4):966-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paul F, Arkin Y, Giladi A, et al. Transcriptional heterogeneity and lineage commitment in myeloid progenitors [published correction appears in Cell. 2016;164(1-2):325]. Cell. 2015;163(7):1663-1677. [DOI] [PubMed] [Google Scholar]
- 24.Pina C, Fugazza C, Tipping AJ, et al. Inferring rules of lineage commitment in haematopoiesis. Nat Cell Biol. 2012;14(3):287-294. [DOI] [PubMed] [Google Scholar]
- 25.Lang S, Ugale A, Erlandsson E, Karlsson G, Bryder D, Soneji S. SCExV: a webtool for the analysis and visualisation of single cell qRT-PCR data. BMC Bioinformatics. 2015;16:320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi T, Seligson D, Belldegrun AS, Palotie A, Horvath S. Tumor classification by tissue microarray profiling: random forest clustering applied to renal cell carcinoma. Mod Pathol. 2005;18(4):547-557. [DOI] [PubMed] [Google Scholar]
- 27.Kikushige Y, Shima T, Takayanagi S, et al. TIM-3 is a promising target to selectively kill acute myeloid leukemia stem cells. Cell Stem Cell. 2010;7(6):708-717. [DOI] [PubMed] [Google Scholar]
- 28.Saito Y, Kitamura H, Hijikata A, et al. Identification of therapeutic targets for quiescent, chemotherapy-resistant human leukemia stem cells. Sci Transl Med. 2010;2(17):17ra9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bagger FO, Rapin N, Theilgaard-Mönch K, et al. HemaExplorer: a database of mRNA expression profiles in normal and malignant haematopoiesis. Nucleic Acids Res. 2013;41(D1):D1034-D1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mustjoki S, Richter J, Barbany G, et al. ; Nordic CML Study Group (NCMLSG). Impact of malignant stem cell burden on therapy outcome in newly diagnosed chronic myeloid leukemia patients. Leukemia. 2013;27(7):1520-1526. [DOI] [PubMed] [Google Scholar]
- 31.Majeti R, Park CY, Weissman IL. Identification of a hierarchy of multipotent hematopoietic progenitors in human cord blood. Cell Stem Cell. 2007;1(6):635-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Landberg N, Hansen N, Askmyr M, et al. IL1RAP expression as a measure of leukemic stem cell burden at diagnosis of chronic myeloid leukemia predicts therapy outcome. Leukemia. 2016;30(1):255-258. [DOI] [PubMed] [Google Scholar]
- 33.Abraham SA, Hopcroft LE, Carrick E, et al. Dual targeting of p53 and c-MYC selectively eliminates leukaemic stem cells. Nature. 2016;534(7607):341-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prost S, Relouzat F, Spentchian M, et al. Erosion of the chronic myeloid leukaemia stem cell pool by PPARγ agonists. Nature. 2015;525(7569):380-383. [DOI] [PubMed] [Google Scholar]
- 35.Holyoake T, Jiang X, Eaves C, Eaves A. Isolation of a highly quiescent subpopulation of primitive leukemic cells in chronic myeloid leukemia. Blood. 1999;94(6):2056-2064. [PubMed] [Google Scholar]
- 36.Guo G, Huss M, Tong GQ, et al. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev Cell. 2010;18(4):675-685. [DOI] [PubMed] [Google Scholar]
- 37.Rossi DJ, Bryder D, Zahn JM, et al. Cell intrinsic alterations underlie hematopoietic stem cell aging. Proc Natl Acad Sci USA. 2005;102(26):9194-9199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cho RH, Sieburg HB, Muller-Sieburg CE. A new mechanism for the aging of hematopoietic stem cells: aging changes the clonal composition of the stem cell compartment but not individual stem cells. Blood. 2008;111(12):5553-5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schemionek M, Elling C, Steidl U, et al. BCR-ABL enhances differentiation of long-term repopulating hematopoietic stem cells. Blood. 2010;115(16):3185-3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.El Missiry M, Adnan Awad S, Rajala HL, et al. Assessment of bone marrow lymphocytic status during tyrosine kinase inhibitor therapy and its relation to therapy response in chronic myeloid leukaemia. J Cancer Res Clin Oncol. 2016;142(5):1041-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Säwén P, Lang S, Mandal P, Rossi DJ, Soneji S, Bryder D. Mitotic history reveals distinct stem cell populations and their contributions to hematopoiesis. Cell Reports. 2016;14(12):2809-2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nestorowa S, Hamey FK, Pijuan Sala B, et al. A single-cell resolution map of mouse hematopoietic stem and progenitor cell differentiation. Blood. 2016;128(8):e20-e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holyoake TL, Jiang X, Jorgensen HG, et al. Primitive quiescent leukemic cells from patients with chronic myeloid leukemia spontaneously initiate factor-independent growth in vitro in association with up-regulation of expression of interleukin-3. Blood. 2001;97(3):720-728. [DOI] [PubMed] [Google Scholar]
- 44.Corbin AS, O’Hare T, Gu Z, et al. KIT signaling governs differential sensitivity of mature and primitive CML progenitors to tyrosine kinase inhibitors. Cancer Res. 2013;73(18):5775-5786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matsuoka Y, Sasaki Y, Nakatsuka R, et al. Low level of c-kit expression marks deeply quiescent murine hematopoietic stem cells. Stem Cells. 2011;29(11):1783-1791. [DOI] [PubMed] [Google Scholar]
- 46.Scheijen B, Griffin JD. Tyrosine kinase oncogenes in normal hematopoiesis and hematological disease. Oncogene. 2002;21(21):3314-3333. [DOI] [PubMed] [Google Scholar]