Abstract
Cytomegalovirus (CMV) elicits a potent T-cell response in humans that appears to protect the host from virus-associated disease. Despite facing strong host defense mechanisms, CMV remains as a lifelong infection that may reactivate and cause life-threatening disease in immunocompromised individuals. This persistence is probably assisted by expression of immune subversion proteins of the virus encoded by genes belonging to the US gene family. These proteins modulate major histocompatibility complex expression in infected cells and bias in vitro experiments toward the detection of only certain specificities. We have combined the use of recombinant CMV, lacking the US2 to US11 region genes, and cytoplasmic gamma interferon staining to define a more accurate assessment of CMV-specific responses in vivo. Recombinant CMV stimulation reveals a CD8 response much larger than that of parental virus in all donors tested. In some cases, this represented up to 10-fold increases in the number of cells detected. Responses were directed mainly against pp65, IE-1, and pp50 in the majority of donors. In addition, previously unreported IE-2-specific T-cell responses could be detected in a minority of cases. Furthermore, we observed a less marked increase in the response to mutant CMV by CD4 T cells in some donors. This suggests that a much broader T-cell response to CMV exists in vivo than is revealed by restimulation with wild-type virus and adds to the evidence that the efficacy of immune evasion strategies may not be as absolute as previously believed.
Human cytomegalovirus (CMV) causes major complications in immunosuppressed hosts, such as stem cell transplant recipients, human immunodeficiency virus-infected individuals, and congenitally infected newborns (31, 51). In the immunocompetent host, viral replication and associated disease is rare, implying the importance of protective host immune responses. Data from both murine models and immunosuppressed humans has highlighted the critical role of CD8+ cytotoxic T lymphocytes (CTL) (35, 40) and also more recently CD4+ helper T lymphocytes (33, 39, 48). T cells recognize viral antigens in the form of proteolytically processed peptides that are presented by major histocompatibility complex (MHC) molecules at the surfaces of infected cells.
In the mouse model, murine CMV (mCMV) infection appears to be controlled by CD8 T cells, with the immediate-early 1 (IE-1) antigen being a major target (36). In humans, the structural phosphoprotein pp65 (UL83) was identified as the major immunodominant T-cell antigen (29, 50), and recently, IE-1 has also been established as an immunodominant antigen (24). A number of MHC class I-restricted CMV-encoded peptides have been described (5, 6, 9). In the past few years, novel reagents, such as MHC-peptide tetramers and cytoplasmic gamma interferon (IFN-γ) staining have revealed frequencies of CMV-specific CD8 T cells up to several percent of the CD8 subset (9, 25). Despite the high frequencies of virus-specific CD8 T cells, CMV remains with the host for life as an asymptomatic persistent infection.
This relationship between CMV and its host is further complicated because the virus has acquired numerous strategies to evade immune detection. One such strategy involves the expression of a number of CMV-encoded gene products that interfere with the presentation of MHC-peptide complexes on the surfaces of infected cells. The best-described proteins are encoded by genes in the US2 to US11 region of the viral genome. These genes are expressed within hours after infection and can have several effects. The US3 gene product is expressed during the IE phase and causes intracellular retention of MHC class I complexes in infected fibroblasts (1, 20). US2 and US11 are expressed during the early (E) phase and direct the translocation of MHC class I to the cytosol and subsequent proteasomal degradation (21, 46, 49). Further interference can be attributed to the early-late-expressed US6 gene product, which appears to block TAP-mediated translocation of peptides into the endoplasmic reticulum (27). Very recently, a role for US10 in MHC class I interference has been described (7). Genes with similar immunosubversive properties operate in mCMV infection during the early phase of infection and are reviewed elsewhere (37). The overall effect is a down-regulation of surface MHC-viral-peptide expression, which serves as a mechanism for evading immune responses by rendering the virus immunologically silent. Under these circumstances, T-cell recognition may be limited to those peptides that are presented prior to the onset of US gene expression, i.e., proteins expressed during the immediate-early phase (such as IE-1) and structural proteins inside the virion (such as pp65). In addition, US2 and US3 have been shown to modulate MHC class II expression (11, 19, 44). However, despite these evasive tactics, humans and CMV have evolved to coexist with little evidence of disease.
Further complexity in the CMV-specific T-cell response is suggested by recent work on mCMV infection, which has demonstrated CD8 T-cell responses to a variety of CMV-encoded proteins across the viral replicative cycle in the face of immunosubversive early genes (13, 14, 15, 16). This has recently been corroborated by Elkington et al., who described broad T-cell responses against several novel CMV antigens in healthy virus carriers (6). Such findings imply that immune evasion strategies do not prevent the induction of a diverse T-cell response and that a number of responses are still unaccounted for in humans.
An area of interest in CMV is to design a vaccine that will induce long-term protective immunity. It would therefore be desirable to include multiple antigenic specificities across many HLA types. A more accurate knowledge of the global T-cell response is a prerequisite for such endeavors. The drawback of using tetramers or peptides is that only a minority of the 160 CMV antigens will be covered. Jones and Sun have described a genetically modified CMV (named RV798) lacking the US2 to US11 region that does not down-regulate MHC class I on infected cells (21). This may allow more successful antigen presentation to T cells. We sought to establish whether recombinant CMV preparations could serve as tools for more accurately defining CMV-specific responses in vivo. Using a number of laboratory CMV strains (AD169, Towne, and Toledo) and recombinant CMV viruses, we have compared ex vivo T-cell responses by IFN-γ staining and characterized their target antigens after expansion in vitro.
MATERIALS AND METHODS
Cell lines.
Primary chicken embryo fibroblasts (CEF) were produced from eggs obtained from a specific-pathogen-free colony (Institute for Animal Health, Compton, United Kingdom). Baby hamster kidney (BHK-21) cells and human fetal fibroblasts (HFF) were obtained from the European Collection of Animal Cell Cultures. CEF, BHK-21 cells, and HFF were all maintained in Dulbecco's modified Eagle's Medium (DMEM) supplemented with 2 mM glutamine, 100 IU of penicillin/ml, 100 μg of streptomycin/ml, and 10% fetal calf serum (FCS). Epstein-Barr virus (EBV)-transformed B lymphoblastoid cell lines (LCLs) were also generated in vitro for the donors studied, using the B95.8 strain of EBV. LCLs were maintained in RPMI supplemented with 10% FCS, 2 mM l-glutamine, 100 IU of penicillin/ml, and 100 μg of streptomycin/ml.
CMV propagation.
The following CMV strains were used in this study: AD169 (a kind gift from G. Wilkinson, Cardiff, United Kingdom), Towne and Toledo (kind gifts from E. Mocarski, Stanford, Calif.), and the recombinant CMV strains RV35, RV47, and RV798 (kind gifts from Thomas Jones, Wyett Research Institute) (described in reference 21). They were passaged onto HFF by infecting the cells at a multiplicity of infection (MOI) of 0.1 and growing them in DMEM as described above with the inclusion of 4.5 μg of glucose/ml in the medium. The supernatants were harvested at weekly intervals until 5 days after 100% cytopathic effect was observed, and virus pellets were isolated by ultracentrifugation of the clarified supernatants. CMV stocks were titered by plaque assay on HFF.
Primary human fibroblasts were established from skin biopsies for nine CMV-seropositive and three CMV-seronegative donors and grown in DMEM. For cytotoxicity assays, the fibroblasts were grown to 80% confluence in 75-cm2 flasks and infected at an MOI of 5:1 at various time points prior to the assay. MHC expression on virus-infected fibroblasts was determined using the anti-HLA-ABC (W6/32) and anti-HLA-DP/DQ/DR (CR3/43) monoclonal antibodies (both from Dako, Ely, United Kingdom).
Generation of recombinant MVA.
Recombinant modified vaccinia virus Ankara (MVA) expressing either pp65, pp150, IE-1, IE-2, glycoprotein B (gB), or pp50 was used to determine the antigen specificities of recombinant CMV-specific T cells. Briefly, cDNA was made from RNA extracted from CMV-infected human fibroblasts. The pp65 gene was amplified from a pp65 expression plasmid kindly provided by Andreas Moosmann (Munich, Germany). For pp65 (UL83) PCR, the primers were 5′-AGTTCAGGATCCATGGAGTCGCGCGGTCG and 3′-AGTCGTGGGCCCGCTAGCTCAACCTCGGTGCTTTTTGG. The IE-1 (UL123) primers were 5′AGTCTGGGATCCATGGAGTCCTCTGCCAAGAGA and 3′AGTCTTGCTAGCCCCGGGTTACTGGTCAGCCTTGCTTC. The IE-2 (UL122) primers were 5′-AGTTCAAGATCTATGCTGCCCCTCATCAAACAG and 3′-AGTCTTGCTAGCCCCGGGTTACTGAGACTTGTTCCTCAGG. The pp50 (UL44) primers were 5′-AGTTCAGGATCCATGGATCGCAAGACGCGCCT and 3′-AGTCGTGGGCCCGCTAGCCTAGCCGCACTTTTGCTTCTTG. The glycoprotein B (gB-UL55) primers were 5′-AGTCTGGGATCATGGAATCCAGGATCTGGT and 3′-AGTCGTGGGCCCGCTAGCTCAGACGTTCTCTTCTTCGT. The pp150 (UL32) primers were 5′-AGTTCAAGATCTATGAGTTTGCAGTTTATCGGTCTAC and 3′-AGTCGTGCGGCCGCGCTAGCCTATTCCTCCGTGTTCTTAATCTTC. The PCR products for pp65, pp50, and gB (BamHI and NheI), IE-1 (BamHI and SmaI), IE-2 (BglII and SmaI), and pp150 (BglII and NheI) were digested with restriction enzymes and ligated with a modified version of the vaccinia virus shuttle vector pSC11. Restriction digestion and DNA sequencing with gene-specific primers confirmed all plasmid constructs.
Recombinant MVAs were generated by transfecting the pSC11 plasmid into 106 CEF. These CEF were previously infected with wild-type MVA at an MOI of 0.1 in 25-cm2 tissue culture flasks. Recombinant MVA plaques were then selected by β-galactosidase screening. After six rounds of plaque purification, the viruses were expanded by infecting serially greater numbers of BHK-21 cells and were harvested after visible cytopathic effect occurred. Virus stocks were resuspended in phosphate-buffered saline and then subjected to three freeze-thaw cycles and sonication. Cell debris was removed by low-speed centrifugation prior to storage of the virus in aliquots at −80°C. Virus titers were determined by plaque assay on CEF and/or BHK-21 cells. Typical titers were 5 × 107 to 5 × 108 PFU/ml. A control MVA was also generated, which incorporated empty PSC11 plasmid sequences.
Protein expression of MVA recombinants was evaluated by Western blotting, IFN-γ staining of virus-infected peripheral blood mononuclear cells (PBMC), and cytotoxicity assays using specific T-cell clones or T-cell lines. Target cells used for MVA infection in cytotoxicity assays were autologous LCLs.
Cytoplasmic IFN-γ staining.
Human fibroblasts (2 × 105) were infected with AD169, Towne, Toledo, RV35, RV47, or RV798 viruse at an MOI of 5:1 for 36 h and either pretreated with IFN-γ or left untreated. The monolayers were then coincubated with freshly isolated PBMC for at least 6 h at 37°C and 5% CO2. As a positive control, PBMC were stimulated with staphylococcal enterotoxin B. After the first 2 h of coincubation, brefeldin A (Sigma Aldrich, Poole, United Kingdom) was added to the cells at a final concentration of 10 μg/ml. The PBMC were then removed from the fibroblast monolayer and washed prior to being surface stained with phycoerythrin-labeled anti-CD4 (Dako) and tricolor-labeled anti-CD8 monoclonal antibodies (Caltag, San Francisco, Calif.). After being washed again, the cells were fixed and permeabilized and then stained for IFN-γ using a fluorescein isothiocyanate-labeled anti-IFN-γ monoclonal antibody (BD Biosciences, Oxford, United Kingdom), followed by a final wash and then cytofluorometric analysis. This was carried out on a Coulter XL flow cytometer, and later examination was assisted by using Win MDI version 2.8 software (available at the Scripps Research Institute website [http://facs.scripps.edu/software.html]). Lymphocytes were gated by side scatter and forward scatter, and IFN-γ-producing cells were counted for both CD4 and CD8 T-cell subsets. All statistical analyses were performed using the Mann-Whitney test with Graphpad Prism software.
MHC-peptide tetramer staining.
Tetramers were synthesized as described elsewhere (2) with some minor modifications. The tetramers used incorporated the following peptides: HLA-B8-restricted IE-1 epitopes (QIKVRVDMV [QIK] and ELKRKMIYM [ELK]) and HLA-A1-restricted pp50 epitope (VTEHDTLLY [VTE]); for pp65 epitopes, HLA-A1 restricted (YSEHPTFTSQY [YSE]), HLA-A*0201 restricted (NLVPMVATV) (NLV), and HLA-B*0702 restricted (RPHERNGFTVL and TPRVTGGGAM). Staining experiments were performed at 37°C for 20 min, followed by washing and staining for surface CD8 on ice for 20 min.
Expansion of recombinant CMV-specific CTL and specificity analysis.
Virus-stimulated T cells were removed from fibroblast monolayers after 6 h and then plated out and incubated at 37°C in RPMI supplemented with 10% FCS, 1% human serum, 2 mM l-glutamine, and penicillin-streptomycin in the presence of interleukin 7 (IL-7) (Peprotech, London, United Kingdom). After 7 days, the cells were fed with fresh medium and 50 U of IL-2 (Chiron, Emeryville, United Kingdom)/ml. The cultures were then restimulated by incubation with virus-infected fibroblasts for 6 h, and thereafter, the medium was changed twice weekly. For specificity analysis, EBV-transformed LCLs were used as target cells. LCLs were infected with MVA viruses at an MOI of 2:1 overnight at 37°C and then loaded with 51Cr (Amersham, Little Chalfont, United Kingdom) for 2 h, washed, and then plated out at 2,500 cells per well. T cells were added to the targets at various effector/target ratios and incubated at 37°C for 5 h before the supernatants were harvested and tested for 51Cr release. All results were calculated with the following formula: (test release − spontaneous release)/(maximal release − spontaneous release) × 100. MHC-peptide tetramer complexes were used to track peptide-specific T-cell expansion in virus-stimulated T-cell lines.
RESULTS
Effects of US gene deletion on surface MHC expression.
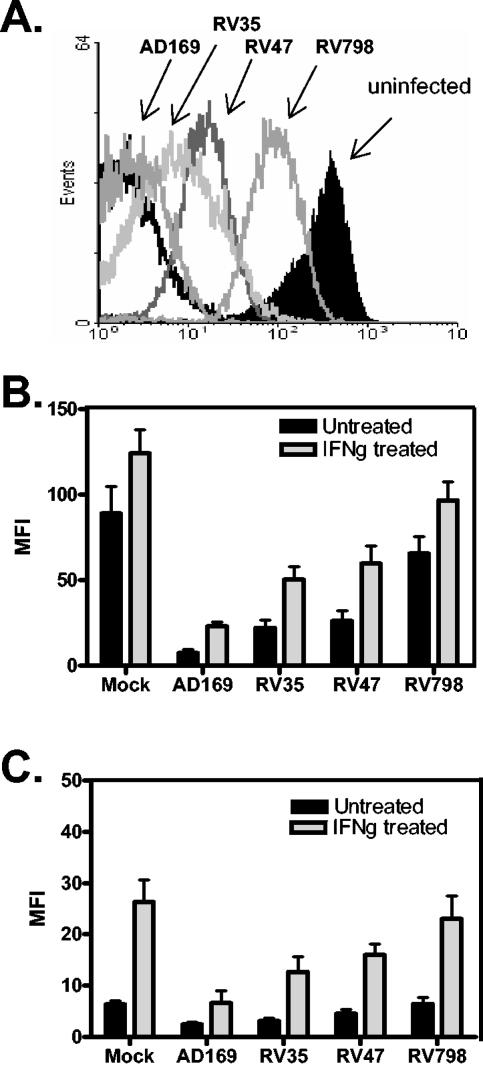
We first aimed to establish the effects of recombinant CMV infection on surface MHC levels in human fibroblasts. Infection experiments were carried out in three donors with a range of HLA types. Fibroblasts were infected with either the parental AD169 CMV or one of three mutant viruses, RV35 (lacking US6 to US11), RV47 (lacking US2 and US3), and RV798 (lacking US2 to US11). The results show that after overnight infection with AD169 virus, there is almost 2 orders of magnitude down-regulation of MHC class I compared with uninfected human fibroblasts (Fig. 1A). In comparison, fibroblasts infected with recombinant CMVs, RV35 (US6 to US11 mutant) and RV47 (US2 and US3 mutant), displayed up to 1 order of magnitude reduced MHC class I. RV798 (US2 to US11 mutant) had the least effect, with approximately threefold reduction in one case (Fig. 1A).
FIG. 1.
Human fibroblasts were grown in monolayers at 37°C and infected with AD169 virus or one of the recombinant CMV viruses: RV35 (US6 to US11 deleted), RV47 (US2 and US3 deleted), or RV798 (US2 to US11 deleted). After overnight infection, the fibroblasts were trypsinized, washed, stained for surface MHC class I and class II using monoclonal antibodies, and analyzed by flow cytometry. (A) Representative histogram showing mean fluorescence intensity (MFI) of fibroblasts under each set of conditions. The black-line overlay represents the isotype control. (B) MHC class I and (C) MHC class II down-regulation in CMV-infected human fibroblasts. The fibroblasts were treated with IFN-γ 24 h before infection. The data shown in panels B and C were pooled from three different donor-derived fibroblast lines. The error bars represent one standard deviation from the mean.
The results of several experiments, conducted either with or without pretreatment of cells with IFN-γ to enhance the baseline levels of MHC expression in uninfected cells, are summarized in Fig. 1B and C. Both MHC class I and II levels were increased when uninfected fibroblasts were pretreated with IFN-γ. Indeed, for MHC class II expression, IFN-γ treatment was obligatory, as baseline levels were extremely low. After infection with AD169 virus, down-regulation of both MHC class I and class II was observed, although it was substantially less than with untreated cells. With recombinant CMVs, IFN-γ treatment virtually overcame the down-regulation observed, with RV798 again being the least inhibitory.
Improved T-cell recognition of RV798-infected fibroblasts by pp65- and IE-1-specific T-cell clones.
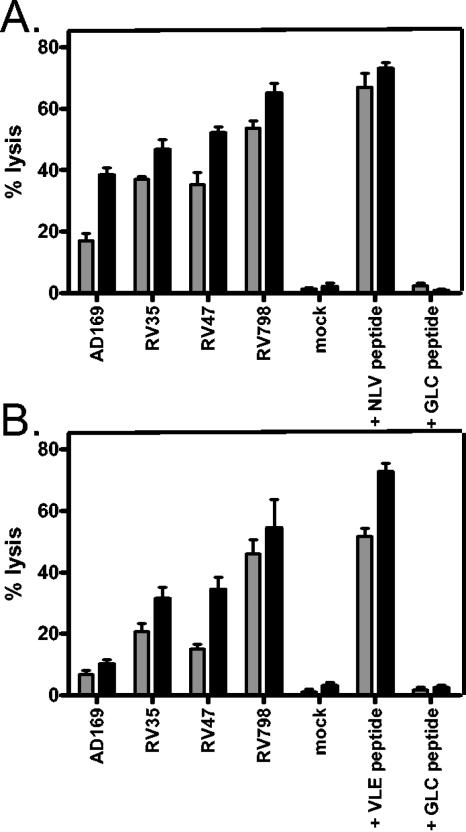
Having established that fibroblasts infected with recombinant CMV had higher levels of MHC than AD169-infected cells, we then proceeded to test their sensitivity to T-cell recognition using pp65- and IE-1-specific CD8 T-cell clones as effectors in 5-h cytotoxicity assays. pp65-specific clones had been generated previously and were directed toward the HLA-A*0201-restricted NLVPMVATV peptide. IE-1-specific clones were directed against the HLA-A*0201-restricted VLEETSVML peptide. The results indicated that enhanced lysis could be achieved when clones were incubated with targets incubated with recombinant CMV viruses, with the greatest enhancement in lysis observed with RV798 (Fig. 2). Most striking was the restoration of very good IE-1-specific target cell recognition, which was not demonstrable using AD169 virus.
FIG. 2.
Improved recognition by pp65- and IE-1-specific T-cell clones. CD8 T-cell clones were tested against CMV-infected autologous fibroblasts in 5-h chromium release assays. Target cells were pretreated with IFN-γ and then infected at an MOI of 5:1 overnight prior to being labeled and incubated with effectors at 37°C. Mock-infected cells were used as controls. The effector/target ratios used were 5:1 (grey bars) and 10:1 (black bars). (A) As positive controls for pp65-specific clones, target cells were pulsed with the cognate pp65 peptide NLVPMVATV. (B) For IE1-specific clones, target cells were pulsed with the cognate IE-1 peptide VLEETSVML. Irrelevant peptide (EBV-encoded HLA-A*0201-restricted GLCTLVAML)-loaded fibroblasts were used as a further control. The error bars represent one standard deviations from the mean.
Screening for ex vivo total CMV-specific responses.
We therefore used recombinant CMV-infected IFN-γ-treated fibroblasts as antigen-presenting cells in ex vivo assays of CMV-specific T-cell responses in immune donor PBMC. After the coincubation of PBMC with virus-infected fibroblasts, responder cells were stained for surface CD4 and CD8 and then, after permeabilization, for cytoplasmic IFN-γ. The initial experiments required optimization of the method. For maximal responses, fibroblasts needed to be pretreated with IFN-γ and then infected with recombinant virus strains for 36 h before they were used in the assay, whereas maximal AD169-specific responses were reached by 16 h postinfection (data not shown). Thereafter, 6- to 12-h coincubation of PBMC with infected fibroblasts was necessary to detect IFN-γ responses.
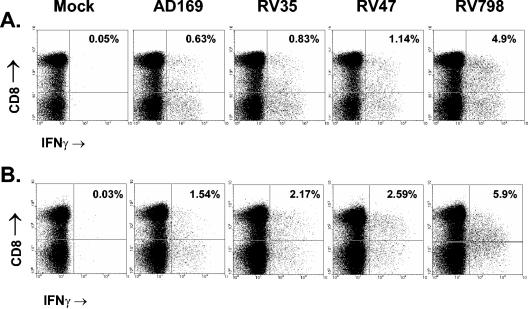
Nine CMV-seropositive donors were tested with this assay using a number of different CMV strains. In all cases, RV798 virus-infected stimulators consistently induced stronger responses than unmodified AD169 (or Towne and Toledo) virus-infected cells. Representative plots are shown in Fig. 3, where the RV798 response is significantly higher than that detected using AD169. Furthermore, there was also an increase with RV35 and RV47, although it was less pronounced, which probably reflects cooperative interference with antigen presentation.
FIG. 3.
Elevated frequencies of CMV-specific T cells using recombinant CMV-infected fibroblasts. PBMC were incubated with virus-infected autologous fibroblasts for 12 h and then removed and stained with monoclonal antibodies for surface CD4 and CD8 receptors. The cells were then fixed and permeabilized, followed by staining for intracellular IFN-γ. Flow cytometric analysis was carried out, with all plots shown gated on lymphocytes by forward scatter and side scatter. Mock-infected cells were used as negative controls, and staphylococcal enterotoxin B was used to stimulate T cells as a positive control (not shown) for IFN-γ production. CD8 T-cell responses of donor 8 (A) and donor 9 (B) against mock-infected and virus-infected fibroblasts are shown.
All of the results are summarized in Table 1. The highest response detected with AD169 stimulation was 2% of all CD8 T cells, and the mean response was 1.03%. Similar frequencies were detected using fibroblasts infected with Towne (0.99%), another high-passage strain of CMV, and with Toledo (0.95%), a low-passage strain.
TABLE 1.
Ex vivo CD8 T-cell responses against autologous fibroblasts infected with recombinant CMVs
| Virus | % IFN-γ staining of CD8 T cells for donor:
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CMV seropositive
|
CMV seronegative
|
||||||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | Mean | 10 | 11 | 12 | |
| AD169 | 1.7 | 0.7 | 0.33 | 0.36 | 0.07 | 1.9 | 2 | 0.63 | 1.54 | 1.03 | 0.03 | 0.18 | 0.09 |
| Towne | 1.53 | 0.76 | 0.24 | 0.31 | NT | 1.95 | 1.2 | 0.64 | 1.31 | 0.99 | NT | 0.12 | 0.11 |
| Toledo | 1.18 | 0.55 | NTb | 0.23 | NT | 1.45 | 1.4 | NT | 0.9 | 0.95 | NT | 0.02 | 0.11 |
| RV35 | 2.3 | 3.88 | 2.42 | 0.34 | 0.07 | 2.32 | 3.1 | 0.83 | 2.17 | 1.94 | 0.14 | 0.12 | 0.2 |
| RV47 | 2.55 | 7.3 | 2.85 | 1.57 | 0.19 | 2.8 | 3.62 | 1.14 | 2.59 | 2.85 | 0.07 | 0.13 | 0.16 |
| RV798 | 4.1 | 9.2 | 12.8 | 2.64 | 0.6 | 6.62 | 5.7 | 4.9 | 5.9 | 5.83 | 0.09 | 0.18 | 0.13 |
| Mock | 0.14 | 0.08 | 0.2 | 0.1 | 0.03 | 0.11 | 0.15 | 0.05 | 0.03 | 0.1 | 0.09 | 0.1 | 0.12 |
| Mismatcheda | 0.4 | 0.21 | 0.14 | 0.38 | NT | 0.2 | NT | 0.16 | 0.27 | 0.25 | 0.04 | 0.06 | 0.15 |
Mismatched fibroblasts were infected with RV798.
NT, not tested.
The mean response using RV798 virus was 5.83%, with the highest response at 12.8% of CD8 T cells. This increase in responding CD8 T cells between RV798 and AD169 was also statistically significant (P = 0.0008). The mean frequencies of responding CD8 T cells against RV35- and RV47-infected fibroblasts were 1.94 and 2.85%, respectively. The superiority of RV47 over RV35 suggests a greater role for US2 and US3 than for US6 to US11 in some donors. Three CMV-seronegative donors did not show detectable responses in this assay to any of the viruses tested.
For CD4 T cells, we also observed an increase in the number of cells producing IFN-γ, but not in all donors tested. Furthermore, the increase was less marked and was only evident with RV798 virus (Table 2). The highest response detected against AD169 virus was 3.2% of CD4 T cells, and the highest RV798-specific response was 4.43% of CD4 T cells. The mean responses were 1.24 and 1.55% of CD4 T cells for AD169 and RV798, respectively, but this did not represent a statistically significant difference (P = 0.6). Coincubation with mismatched virus-infected fibroblasts and supernatants from virus-infected fibroblasts could also stimulate good CD4 T-cell responses. This suggested that exogenous uptake and presentation of antigen by blood antigen-presenting cells, such as B cells, macrophages, or dendritic cells, were occurring. In fact, the response to mismatched AD169-infected fibroblasts was almost always equal to that against AD169-infected autologous fibroblasts (Table 2). However, these responses were usually 80 to 90% of those observed with RV798-infected autologous fibroblasts.
TABLE 2.
Ex vivo CD4 T-cell responses against autologous fibroblasts infected with recombinant CMVs
| Virus | % IFN-γ staining of total CD4 T cells for donor:
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CMV seropositive
|
CMV seronegative
|
||||||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | Mean | 10 | 11 | 12 | |
| AD169 | 1.9 | 1.3 | 3.2 | 0.34 | 0.15 | 0.59 | 1.24 | 0.97 | 1.5 | 1.24 | 0.1 | 0.07 | 0.04 |
| Towne | 1.63 | 0.9 | 2.87 | 0.32 | NT | 0.63 | 1.12 | 0.76 | 1.13 | 1.17 | NT | 0.1 | 0.05 |
| Toledo | 1.48 | 0.87 | NTb | 0.25 | NT | 0.52 | 0.9 | NT | 1.47 | 0.92 | NT | 0.06 | 0.02 |
| RV35 | 1.45 | 1.76 | 2.95 | 0.29 | 0.09 | 0.65 | 0.92 | 0.86 | 1.77 | 1.19 | 0.05 | 0.11 | 0.1 |
| RV47 | 1.9 | 1.64 | 3.5 | 0.3 | 0.16 | 0.81 | 0.96 | 1.18 | 1.26 | 1.3 | 0.1 | 0.09 | 0.11 |
| RV798 | 2.12 | 2.07 | 4.43 | 0.34 | 0.21 | 1.1 | 1.25 | 1.02 | 1.42 | 1.55 | 0.08 | 0.1 | 0.11 |
| Mock | 0.06 | 0.1 | 0.07 | 0.06 | 0.02 | 0.1 | 0.09 | 0.04 | 0.11 | 0.08 | 0.02 | 0.13 | 0.07 |
| Mismatcheda | 1.75 | 1.22 | 3.3 | 0.3 | NT | 0.64 | NT | 0.85 | 1.27 | 1.17 | 0.11 | 0.2 | 0.06 |
Mismatched fibroblasts were infected with AD169.
NT, not tested.
Definition of target antigens using polyclonal virus-specific effectors.
We then proceeded to identify the antigenic targets of recombinant CMV-specific T cells. For CD8 T cells, this was addressed by plating out responding PBMC after coincubation with virus-infected fibroblasts and propagation in vitro. In some cases, cultures were restimulated with virus-infected fibroblasts and maintained for a number of weeks. As expected, AD169-stimulated cultures showed good lysis of AD169-infected fibroblasts and RV798-stimulated cells recognized RV798-infected targets. However, AD169-stimulated cells were also reactive against RV798-infected targets, whereas RV798-stimulated cells showed poor recognition of AD169-infected targets (data not shown).
To screen these in vitro-restimulated effector populations, we generated recombinant MVAs expressing individual CMV target proteins, namely, IE-1 and IE-2 (both immediate-early proteins), pp50 (an early protein), and pp65, pp150, and gB (late proteins). EBV-transformed LCLs were infected separately with each one of the recombinant MVAs (expressing a CMV protein) or with control MVA.
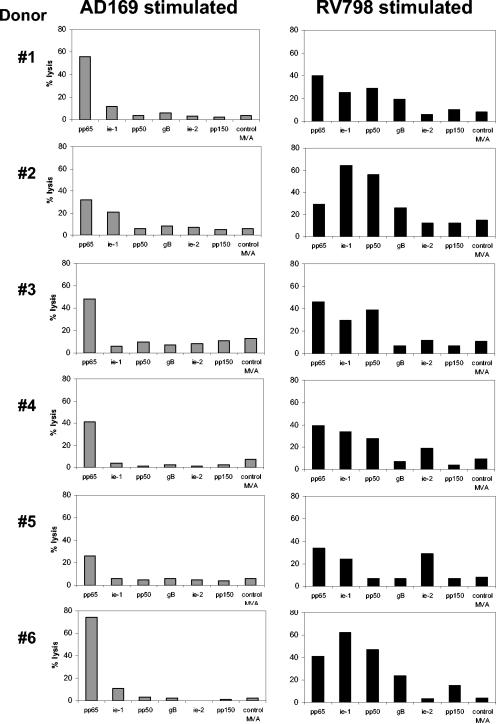
MVA screening assays showed that in all six CMV seropositive donors studied, pp65-specific lysis could be detected with either AD169- or RV798-stimulated bulk cultures (Fig. 4). Interestingly, using RV798, IE-1-specific responses were evident in all six donors, whereas only one of the six showed activity (at low levels) against IE-1 if stimulated with AD169. The early protein pp50 was revealed to be another major target, as five out of six donors showed good reactivity to this antigen, which was not observed in any of the donors when stimulated with AD169 (Fig. 4). In a few donors, low levels of gB, IE-2, and pp150-specific lysis were also observed with RV798-stimulated cells that again were not evident using the parental virus. In contrast, both AD169 and RV798 stimulation did not restimulate responses against any of the targets used in two CMV-seronegative donors (data not shown). This result confirmed that RV798 virus efficiently stimulates antigen-experienced T cells rather than naïve T cells and restimulates a much more representative range of CD8 T cells than AD169.
FIG. 4.
Recombinant CMV stimulation reveals broader target range of CMV-specific T cells in seropositive donors. Donor PBMC were stimulated with either AD169- or RV798-infected autologous fibroblasts and then expanded in vitro with the addition of recombinant IL-2 after 7 days. After 2 weeks, the cultures were restimulated and then, after another 7 days, tested for specificity by 5-h chromium release assays. The target cells were autologous LCLs infected overnight with MVAs at an MOI of 2:1. The effector/target ratios were 10:1 for the data shown.
Tetramer-guided tracking of T-cell expansion after recombinant CMV stimulation.
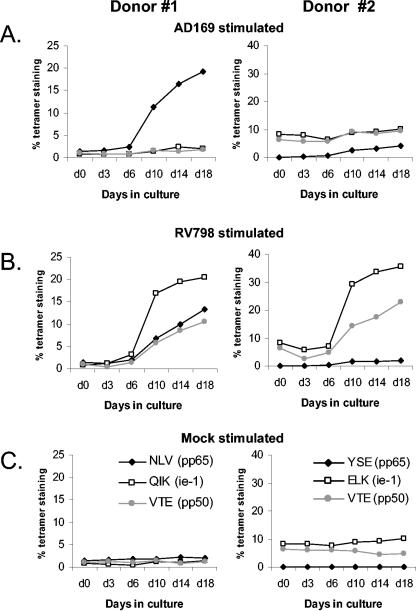
Two of the donors studied had simultaneously been characterized for circulating frequencies of CMV-specific CD8 T cells using MHC-peptide tetramers. Donors 1 and 2 had detectable responses against pp65, IE-1, and pp50. We wished to demonstrate directly that these specificities were restimulated using recombinant CMV. Virus-stimulated cells were cultured and harvested at given time points for analysis by tetramer staining (Fig. 5). For donor 1, AD169 clearly appeared to restimulate only the known pp65 (NLV)-specific CD8 T cells, with 10-fold expansions observed, whereas RV798 restimulated the pp65-specific response and also both IE-1 (QIK) and pp50-specific (VTE) responses. For donor 2, AD169 stimulated the low-frequency pp65-specific (YSE) response but did not affect the other responses despite the fact that in this donor the frequencies of IE-1 tetramer (ELK) and pp50 tetramer (VTE) binding cells were 9 and 6.5% of all CD8 T cells, respectively; in contrast, these dominant components were efficiently expanded by RV798 virus.
FIG. 5.
Tetramer-guided monitoring of T-cell expansion using recombinant CMV-infected target cells. PBMC were incubated with autologous fibroblasts infected as indicated. The PBMC were then expanded in vitro and stained with MHC-peptide tetramers at different time points to track the expansion of epitope-specific CD8 T cells. pp65-specific responses are shown by the black filled diamonds, IE-1-specific responses are shown by the empty squares, and pp50-specific responses are shown by the grey filled circles. For donor 1, we used the HLA-B8 QIK tetramer (for the IE-1 response), the HLA-A1 VTE tetramer (for the pp50 response), and the HLA-A2 NLV tetramer (for the pp65 response). For donor 2, we used the HLA-B8 ELK tetramer (for the IE-1 response), the HLA-A1 VTE tetramer (for the pp50 response), and the HLA-A1 YSE tetramer (for the pp65 response). Tetramer-staining populations were recorded as percentages of total CD8 T cells and plotted against time.
Detection of protein-specific CD8 T cells using recombinant MVA.
Having shown that CMV-infected autologous fibroblasts could be used as antigen-presenting cells in ex vivo assays of IFN-γ production from PBMC, we asked whether MVA-CMV gene recombinants could likewise be used to assay antigen-specific CD8 T-cell frequencies ex vivo. This proved to be the case, as shown in Fig. 6.
FIG. 6.
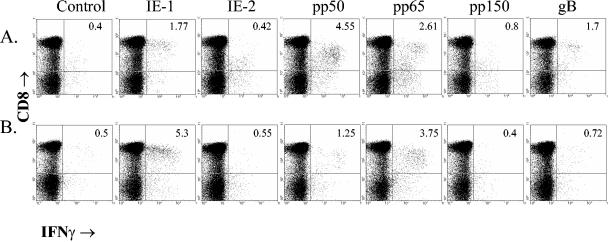
Protein-specific IFN-γ production by CD8 T cells using recombinant MVA. Autologous fibroblasts were infected with MVAs expressing one of six different CMV proteins or a control MVA at an MOI of 2:1 for 36 h. Fresh PBMC were added to the infected fibroblasts for 6 h, followed by staining for surface CD8 and cytoplasmic IFN-γ. Representative data for donors 6 (A) and 7 (B) are shown. All values indicate frequencies of CD8 T cells producing IFN-γ in response to the indicated antigen-expressing stimulator cell.
The results indicate that all seven donors tested had strong CD8 T-cell responses against at least two CMV proteins (Table 3). pp65 and IE-1 responses were the most common (seven of seven donors), followed by pp50 (six of seven donors), with frequencies of several percent of CD8 T cells for these three antigens. Responses were also common against gB but infrequent against IE-2 and pp150, and all were lower in magnitude. The total response against these six antigens was in some cases much lower than that measured against RV798-infected cells, in other cases equivalent, and in some cases slightly higher. The higher responses from MVA assays imply that some epitopes may be suboptimally presented in RV798-infected cells. Equivalent responses suggest that donor CMV responses are dominated by the six antigens expressed from MVAs in our panel. Lower responses imply that other CMV antigens (not in our panel) were significant targets for CD8 T cells in these donors.
TABLE 3.
Ex vivo CD8 T-cell responses against autologous fibroblasts infected with recombinant MVAs
| MVA expressing: | % IFN-γ staining of total CD8 T cells for donora:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| CMV seropositive
|
CMV seronegative
|
||||||||
| 1 | 2 | 3 | 4 | 6 | 7 | 9 | 10 | 12 | |
| IE-1 | 1.39 | 6.83 | 1.6 | 0.97 | 1.37 | 4.8 | 0.19 | 0 | 0 |
| IE-2 | 0 | 0.48 | 0 | 0.07 | 0 | 0.05 | 0 | 0 | 0.07 |
| pp50 | 0.79 | 3.33 | 1.41 | 0.27 | 4.15 | 0.75 | 0 | 0 | 0 |
| pp65 | 1.79 | 0.29 | 1.46 | 0.67 | 2.21 | 2.75 | 3.19 | 0.08 | 0.06 |
| pp150 | 0 | 0.03 | 0 | 0 | 0.4 | 0 | 0 | 0 | 0 |
| gB | 0.25 | 0.21 | 0.12 | 0 | 1.3 | 0.22 | 0.14 | 0.04 | 0.03 |
| Total | 4.22 | 11.2 | 4.59 | 1.98 | 9.45 | 8.57 | 3.55 | 0.18 | 0.16 |
| RV798 | 4.1 | 9.2 | 12.8 | 2.64 | 6.62 | 5.7 | 6.8 | 0.09 | 0.13 |
Total responses shown are the sum of protein-specific responses (after subtracting the background response (control MVA) from each recombinant MVA response).
DISCUSSION
This study highlights the problem of detecting CMV-specific T cells by using stimulator cells infected with the conventional AD169 and Towne virus strains. In earlier reports of limiting-dilution experiments using AD169 or Towne virus as stimulation, there appeared to be a focusing of the response on pp65, with much lower frequencies of IE-1-specific T cells detected (29, 50). However, staining with MHC-peptide tetramers or measurement of peptide-induced IFN-γ production showed that pp65, and also IE-1, can be an immunodominant protein (24, 25). This discrepancy is probably explained by the concerted action of CMV-encoded genes present within AD169 and Towne strains that modulate MHC class I antigen presentation from the IE phase onward. Antigen presentation in CMV-infected cells is therefore limited either to proteins expressed very early, before immune evasion is fully operative, or to protein constituents of the incoming virus particle. The abundance of pp65, which is a major constituent of both virus particles and noninfectious particles known as dense bodies (22, 45), provides the rationale for the observed bias toward pp65 as the immunodominant antigen.
We have shown that higher frequencies of CMV-specific CD8 T cells can be detected with a mutant CMV that is deficient in the US2 to US11 region (RV798), leading in some cases to a 10-fold increase in the frequency of responding cells. Furthermore, this approach can give a global picture of CMV immunity and is not restricted to certain HLA types or particular CMV proteins, as is the case with other methods. Partial deletions of the US region had less profound effects on stimulatory function, although our data do suggest a more active role for US2 and US3, as opposed to US6 to US11, in restricting antigen presentation in this experimental system. The necessity to fully delete the US2 to US11 region to optimize epitope display clearly demonstrates that immune subversion of MHC-peptide presentation involves cooperative effects of the different US gene products. This lack of redundancy is in agreement with mCMV data using a number of deletion mutant viruses (23, 47). Interestingly, the RV798 virus did cause some MHC class I down-regulation, which was not described previously (21). Contamination of our RV798 virus stocks was ruled out after the absence of wild-type AD169 DNA was verified by PCR. Alternatively, this may have been a consequence of virus-mediated cytopathic effect over the extended infection period.
With regard to antigen specificity, it is clear from our results that pp65-specific responses can be restimulated with both parental and mutant viruses. However, we also show good IE-1 responses with RV798 virus, but not with AD169, in all donors tested. This is an interesting result, as it has been shown that pp65 can inhibit the presentation of IE-1 peptides (8) in CMV-infected fibroblasts. If this phenomenon were true, then we would not expect increased detection of IE-1-specific CTL using RV798 virus, which also contains pp65. This suggests that US protein-mediated inhibition is more important in blocking IE-1 presentation. The identification of pp50 as a third immunodominant antigen is in agreement with other work (6) and also intriguing. pp50 is expressed during the E phase of the human CMV replicative cycle, when US family-mediated immune subversion is operative. In addition, we have identified weaker responses against another immediate-early antigen, IE-2. These are also undetected in AD169-based experiments, again implying successful inhibition of IE-2 presentation by US proteins in this in vitro system.
Very recently, Manley et al. and Mutimer et al. published work with RV798 virus showing that IE-1, pp65, pp150, and gB are targets that often represent a minority of the T-cell clones generated in vitro after RV798 stimulation (28, 30). Furthermore, the authors showed that the majority of RV798-specific clones recognize as-yet-undefined IE- and/or E-phase antigens (28). Our study confirms this, since we have identified pp50 as an immunodominant E-phase antigen and IE-2 as a subdominant IE antigen. In fact, we have detected strong ex vivo responses (up to 7% of CD8 T cells) against the HLA-A1-restricted pp50 epitope recently identified by Elkington et al. (6), often in the absence of strong pp65 or IE-1 responses in PBMC of several donors (data not shown). These authors also described low-frequency ex vivo responses against other proteins, such as pp28, gH, UL16, and even US2 and US3. It is thus clear from our work and the recent literature that immune evasion proteins do not prevent the priming and long-term persistence of a diverse CD8 T-cell response against CMV. The critical questions are whether the various specificities are maintained by direct priming or cross-priming (26) of CMV-specific T cells and whether they have a protective role in vivo.
Cross-priming by dendritic cells processing exogenously acquired antigen may allow responses to be induced against a wide variety of CMV proteins, but many of these cells may be biologically ineffective, since their cognate epitope is not presented on CMV-infected stromal cells in vivo because of US2 to US11 expressed in the same cell; indeed, this might be seen as an elaborate virus tactic for tricking the immune system into sustaining an ineffective response. This was elegantly demonstrated in mCMV infection for an immunodominant Db-restricted E-phase-derived antigenic peptide (17). Despite the high-frequency response, these T cells could only protect mice against infection with mutant CMV lacking the “immunoevasin” m152 but not against infection with wild-type virus. In human CMV infection, this may also be the case for some of the responses detected.
The fact that equally high-magnitude immune responses can be detected against antigens expressed across the replicative cycle suggests that cross-priming plays a major part in response induction, given that in naturally infected cells immunosubversive genes are expressed from the immediate-early phase onward. pp65, however, may be a special case, because from in vitro work this protein, as a component of the virion, is actually available for presentation in infected cells prior to any viral-gene expression (41). However, it must be remembered that the virus preparations used for in vitro work are often composed of pp65-rich nonviral particles, as well as authentic virions, and this may not reflect the situation in vivo. Consequently, it is as likely that pp65 responses are also cross-primed; certainly exogenous uptake of pp65 and cross-presentation has been demonstrated in vitro (42).
In addition, we have demonstrated a few cases in which the CD4 T-cell response is enhanced by using RV798 virus. In contrast to the massive increase in CD8 T-cell response against RV798 virus, the effect on the CD4 response is more subtle, with increases of <2-fold. Furthermore, experiments with CMV-infected MHC-mismatched fibroblasts showed that the majority of the response was probably stimulated by uptake and presentation via antigen-presenting cells, such as B cells and dendritic cells, in the responding PBMC fraction. This is not an unexpected result, because exogenous uptake of antigen is the conventional pathway for MHC class II antigen processing and presentation. However, despite the increased CD4 responses observed in some donors by using RV798 virus, it is not clear whether this slight enhancement can be attributed to improved endogenous MHC class II presentation in fibroblasts.
Some caution must be exercised when conclusions are drawn from this type of study. Unlike laboratory CMV strains, which replicate efficiently only in fibroblasts, natural CMV infection involves a range of cell types, such as endothelium, hepatocytes, smooth muscle cells, epithelial cells, monocytes (18, 32, 43), and even dendritic cells, where it can also impair the antigen-presenting function (3, 34). However, it is unclear whether immune-evasive proteins are as effective in different cell types (12, 38). Furthermore, IFN-γ treatment appears to overcome the inhibitory effect of US proteins in vitro, which suggests that there may be ongoing competition between CMV immune evasion proteins and inflammatory cytokines in vivo (4). Interestingly, Gold and colleagues have shown that mCMV immune evasion genes are not necessary for virus persistence. The sizes and phenotypes of virus-specific CD8 T-cell responses were similar in mice infected with either wild-type virus or mutant virus (lacking mCMV immune evasion genes). Moreover, both the wild-type and mutant viruses could be reactivated from immunosuppressed mice (10).
Importantly, the overwhelming fact is that despite CMV immune subversion, humans have managed to survive and control infection without any obvious signs of disease. This suggests that at least some of the T-cell specificities against CMV detected by experiments in vitro are protective and keep the virus under control. It remains to be seen whether the existence of a pool of “irrelevant” T-cell specificities is detrimental to the host, possibly crowding out more biologically relevant responses for immune space.
The clinical interest in CMV is presently directed toward designing a vaccine that will elicit broad and lasting immunity. For CD8 T-cell responses, this would require that multiple antigens be included, with wide HLA coverage to benefit a larger population. Therefore, the search for novel antigenic targets is an area of great interest that will be the focus of many investigators. Here, we show the value of the RV798 virus and recombinant MVA-CMV gene constructs as tools applicable to all donors regardless of HLA type and which have the potential to reveal responses to any CMV antigen. Our data clearly identify IE-1 and pp50 as immunodominant antigens, in addition to pp65; future studies may add more proteins to this list.
Acknowledgments
This work was supported by program grant G9901249 from the Medical Research Council United Kingdom.
REFERENCES
- 1.Ahn, K., A. Angula, P. Ghazal, P. A. Peterson, Y. Yang, and K. Fruh. 1996. Human cytomegalovirus inhibits antigen presentation by a sequential multi-step process. Proc. Natl. Acad. Sci. USA 93:10990-10995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altman, J. D., P. A. Moss, P. J. R. Goulder, D. H. Barouch, M. G. McHeyzer-Williams, J. I. Bell, A. J. McMichael, and M. M. Davis. 1996. Phenotypic analysis of antigen-specific T lymphocytes. Science 274:94. (Erratum, 280:1821, 1998.) [DOI] [PubMed] [Google Scholar]
- 3.Andrews, D. M., C. D. Andoniou, F. Granucci, P. Ricciardi-Castagnoli, and M. A. Degli-Esposti. 2001. Infection of dendritic cells by murine cytomegalovirus induces functional paralysis. Nat. Immunol. 2:1077-1084. [DOI] [PubMed] [Google Scholar]
- 4.Benz, C., and H. Hengel. 2000. MHC class I subversive gene functions of cytomegalovirus and their regulation by interferons—an intricate balance. Virus Genes 21:39-47. [PubMed] [Google Scholar]
- 5.Diamond, D. J., J. York, J.-Y. Sun, C. L. Wright, and S. J. Forman. 1997. Development of a candidate HLA A*0201 restricted peptide-based vaccine against human cytomegalovirus infection. Blood 90:1751-1767. [PubMed] [Google Scholar]
- 6.Elkington, R., S. Walker, T. Crough, M. Menzies, J. Telham, M. Bharadwaj, and R. Khanna. 2003. Ex vivo profiling of CD8 T-cell responses to human cytomegalovirus reveals broad and multispecific reactivities in healthy virus carriers. J. Virol. 77:5226-5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Furman, M. H., N. Dey, D. Tortorella, and H. L. Ploegh. 2002. The human cytomegalovirus glycoprotein US10 gene product delays trafficking of major histocompatibility complex class I molecules. J. Virol. 76:11753-11756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gilbert, M. J., S. R. Riddell, B. Plachter, and P. D. Greenberg. 1996. Cytomegalovirus selectively blocks antigen processing and presentation of its immediate early gene product. Nature 383:720-722. [DOI] [PubMed] [Google Scholar]
- 9.Gillespie, G. M., M. R. Wills, V. Appay, C. O'Callaghan, M. Murphy, N. Smith, P. Sissons, S. Rowland-Jones, J. I. Bell, and P. A. Moss. 2000. Functional heterogeneity and high frequencies of cytomegalovirus-specific CD8+ T lymphocytes in healthy seropositive donors. J. Virol. 74:8140-8150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gold, M. C., M. W. Munks, M. Wagner, C. W. McMahon, A. Kelly, D. G. Kavanagh, M. K. Slifka, U. H. Kozinowski, S. P. Fling, D. H. Raulet, and A. B. Hill. 2004. Murine cytomegalovirus interference with antigen presentation has little effect on the size or the effector memory phenotype of the CD8 T cell response. J. Immunol. 172:6944-6953. [DOI] [PubMed] [Google Scholar]
- 11.Hegde, N. R., R. Tomazin, T. W. Wisner, C. Dunn, J. M. Boname, D. M. Lewinsohm, and D. C. Johnson. 2002. Inhibition of HLA-DR assembly, transport, and loading by human cytomegalovirus glycoprotein US3: a novel mechanism for evading major histocompatibility complex class II antigen presentation. J. Virol. 76:10929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hegde, N. R., and D. C. Johnson. 2003. Human cytomegalovirus US2 causes similar effects on both major histocompatibility complex class I and class II proteins in epithelial and glial cells. J. Virol. 77:9287-9294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holtappels, R., D. Thomas, J. Podlech, and M. J. Reddehase. 2002. Two antigenic peptides from genes m123 and m164 of murine cytomegalovirus quantitatively dominate CD8 T-cell memory in the H-2d haplotype. J. Virol. 76:151-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holtappels, R., J. Podlech, N. K. A. Grzimek, D. Thomas, M. F. Pahl-Seibert, and M. J. Reddehase. 2001. Experimental pre-emptive immunotherapy of murine cytomegalovirus disease with CD8 T-cell lines specific for ppM83 and ppM84, the two homologs of human cytomegalovirus tegument protein ppUL83 (pp65). J. Virol. 75:6584-6600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holtappels, R., D. Thomas, J. Podelch, G. Geginat, H. P. Steffens and M. J. Reddehase. 2000. The putative natural killer cell decoy early gene m04 (gp34) of murine cytomegalovirus encodes an antigenic peptide recognized by protective antiviral CD8 T cells. J. Virol. 74:1871-1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holtappels, R., N. K. A. Grzimek, C. O. Simon, D. Thomas, D. Dreis, and M. J. Reddehase. 2002. Processing and presentation of murine cytomegalovirus pORF m164-derived peptide in fibroblasts in the face of all immunosubversive early gene functions. J. Virol. 76:6044-6053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holtappels, R., J. Podlech, M. F. Pahl-Seibert, M. Julch, D. Thomas, C. O. Simon, M. Wagner, and M. J. Reddehase. 2004. Cytomegalovirus misleads its host by priming of CD8 T cells specific for an epitope not presented in infected tissues. J. Exp. Med. 199:131-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jarvis, M. A., and J. A. Nelson. 2002. Human cytomegalovirus persistence and latency in endothelial cells and macrophages. Curr. Opin. Microbiol. 5:403-407. [DOI] [PubMed] [Google Scholar]
- 19.Johnson, D. C., and N. R. Hegde. 2002. Inhibition of the MHC class II antigen presentation pathway by human cytomegalovirus. Curr. Top. Microbiol. Immunol. 269:101-115. [DOI] [PubMed] [Google Scholar]
- 20.Jones, T. R., E. J. Wiertz, L. Sun, K. N. Fish, J. A. Nelson, and H. L. Ploegh. 1996. Human cytomegalovirus US3 impairs transport and maturation of major histocompatibility complex class I heavy chains. Proc. Natl. Acad. Sci. USA 93:11327-11333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones, T. R., and L. Sun. 1997. Human cytomegalovirus US2 destabilizes major histocompatibility complex class I heavy chains. J. Virol. 71:2970-2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Irmiere, A., and W. Gibson. 1983. Isolation and characterization of a noninfectious virion-like particle released from cells infected with human strains of cytomegalovirus. Virology 130:118-133. [DOI] [PubMed] [Google Scholar]
- 23.Kavanagh, D. G., M. C. Gold, M. Wagner, U. Kozinowski, and A. B. Hill. 2001. The multiple immune evasion genes of murine cytomegalovirus are not redundant: m4 and m152 inhibit antigen presentation in a complementary and co-operative fashion. J. Exp. Med. 194:967-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kern, F., I. P. Surel, N. Faulhaber, C. Frommel, J. Scheinder-Mergener, C. Schonemann, P. Reinke, and H.-D. Volk. 1999. Target structures of the CD8 T-cell response to human cytomegalovirus: the 72-kDa major immediate early protein revisited. J. Virol. 73:8179-8184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khan, N., M. Cobbold, R. Keenan, and P. A. H. Moss. 2002. Comparative analysis of CD8 T cell responses against human cytomegalovirus proteins pp65 and immediate early 1 shows similarities in precursor frequency, oligoclonality and phenotype. J. Infect. Dis. 185:1025-1034. [DOI] [PubMed] [Google Scholar]
- 26.Kurts, C., W. R. Heath, F. R. Carbone, J. Allison, J. F. A. P. Miller, and H. Kosaka. 1997. Constitutive class-I restricted exogenous presentation of self antigens in vivo. J. Exp. Med. 184:923-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lehner, P. J., J. T. Kartlunen, G. W. Wilkinson, and P. Cresswell. 1997. The human cytomegalovirus US6 glycoprotein inhibits transporter associated with antigen processing-dependent peptide translocation. Proc. Natl. Acad. Sci. USA 94:6904-6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manley, T. J., L. Luy, T. R. Jones, M. Boeckh, H. Mutimer, and S. R. Riddell. 2004. Immune evasion proteins of human cytomegalovirus do not prevent a diverse CD8+ cytotoxic T cell response in natural infection. Blood 104:1075-1082. [DOI] [PubMed] [Google Scholar]
- 29.Mclaughlin-Taylor, E. H., H. Pande, S. Forman, B. Tanamachi, C. Li, J. Zaia, P. Greenberg, and S. R. Riddell. 1994. Identification of the major late cytomegalovirus matrix protein pp65 as a target antigen for CD8+ virus-specific cytotoxic T lymphocytes. J. Med. Virol. 43:103-110. [DOI] [PubMed] [Google Scholar]
- 30.Mutimer, H. P., Y. Akatsuka, T. Manley, E. L. Chuang, M. Boeckh, R. Harrington, T. R. Jones, and S. R. Riddell. 2003. Association between immune recovery uveitis and a diverse intraocular cytomegalovirus-specific cytotoxic T cell response. J. Infect. Dis. 186:701-705. [DOI] [PubMed] [Google Scholar]
- 31.Pass, R. F. Cytomegalovirus, p. 2675-2705. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams and Wilkins, Philadelphia, Pa.
- 32.Plachter, B., C. Singzer, and G. Jahn. 1996. Cell types involved in replication and distribution of human cytomegalovirus. Adv. Virus Res. 46:195-261. [DOI] [PubMed] [Google Scholar]
- 33.Polic, B., H. Hengel, A. Krmpotic, J. Trgovcich, I. Pavic, P. Lucin, S. Jonjic, and U. H. Kozinowski. 1998. Hierarchical and redundant lymphocyte subset control precludes cytomegalovirus replication during latent infection. J. Exp. Med. 188:1047-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raftery, M. J., M. Schwab, S. M. Elbert, Y. Samstag, H. Walczak, and G. Schonrich. 2001. Targeting the function of mature dendritic cells by human cytomegalovirus: a multilayered viral defense strategy. Immunity 15:997-1009. [DOI] [PubMed] [Google Scholar]
- 35.Reddehase, M. J. 1985. Interstitial murine cytomegalovirus pneumonia after irradiation: characterization of cells that limit viral replication during established infection of the lungs. J. Virol. 55:264-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reddehase, M. J., W. Mutter, K. Munch, H. J. Buhring, and U. H. Koszinowski. 1987. CD8-positive T lymphocytes specific for murine cytomegalovirus immediate-early antigens mediate protective immunity. J. Virol. 61:3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reddehase, M. J. 2002. Antigens and immunoevasins: opponents in cytomegalovirus immune surveillance. Nat. Immunol. Rev. 2:831-844. [DOI] [PubMed] [Google Scholar]
- 38.Rehm, A., A. Engelsberg, D. Tortorella, I. J. Korner, I. Lehmann, H. L. Ploegh, and U. E. Hopken. 2002. Human cytomegalovirus gene products US2 and US11 differ in their ability to attack major histocompatibility class I heavy chains in dendritic cells. J. Virol. 76:5043-5050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rentinaar, R. J., L. E. Gamadia, N. van Der Hoek, F. N. van Diepen, R. Boom, J. F. Weel, P. M. Wertheim-van Dillen, R. A. van Lier, and I. J. ten Berge. 2000. Development of virus-specific CD4+ T cells during primary CMV infection. J. Clin. Investig. 105:541-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reusser, P., S. R. Riddell, J. D. Meyers, and P. D. Greenberg. 1991. Cytotoxic T lymphocyte response to cytomegalovirus after human allogeneic bone marrow transplantation: pattern of recovery and correlation with cytomegalovirus infection and disease. Blood 78:1373. [PubMed] [Google Scholar]
- 41.Riddell, S. R., M. Rabin, A. Gaballe, W. Britt, and P. D. Greenberg. 1991. Class I MHC restricted cytotoxic T lymphocyte recognition of cells infected with cytomegalovirus does not require endogenous viral gene expression. J. Virol. 146:2795-2804. [PubMed] [Google Scholar]
- 42.Tabi, Z., M. Moutaftsi, and L. K. Borysiewics. 2001. Human cytomegalovirus pp65- and immediate early 1 antigen-specific HLA class I restricted cytotoxic T-cell responses induced by cross presentation of viral antigens. J. Immunol. 166:5685-5703. [DOI] [PubMed] [Google Scholar]
- 43.Taylor Weidemann, J., J. G. Sissons, L. K. Borysiewics, and J. H. Sinclair. 1991. Monocytes are the major site of persistence of human cytomegalovirus in peripheral blood mononuclear cells. J. Gen. Virol. 72:2059-2064. [DOI] [PubMed] [Google Scholar]
- 44.Tomazin, R., J. Boname, N. R. Hegde1, D. M. Lewinsohn, Y. Altschuler, T. R. Jones, P. Cresswell, J. A. Nelson, S. R. Riddell, and D. C. Johnson. 1999. Cytomegalovirus US2 destroys two components of the MHC class II pathway, preventing recognition by CD4+ T cells. Nat. Med. 5:1039-1045. [DOI] [PubMed] [Google Scholar]
- 45.Topilko, A., and S. Michelson. 1994. Morphological and cytochemical analysis of human cytomegalovirus inoculum: correlation of free particles in inoculum with counterparts in infected cells. Res. Virol. 145:65-73. [DOI] [PubMed] [Google Scholar]
- 46.Van der Wal, F. J., M. Kikkert, and E. Wiertz. 2002. The HCMV gene products US2 and US11 target MHC class I molecules for degradation in the cytosol. Curr. Top. Microbiol. Immunol. 269:37-55. [DOI] [PubMed] [Google Scholar]
- 47.Wagner, M., A. Gutermann, J. Podlech, M. J. Reddehase, and U. H. Kozinowski. 2002. Major histocompatibility complex class I allele-specific cooperative and competitive interactions between immune evasion proteins of cytomegalovirus. J. Exp. Med. 196:805-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walter, E. A., P. D. Greenberg, M. J. Gilbert, R. J. Finch, K. S. Watanabe, E. D. Thomas, and S. R. Riddell. 1995. Reconstitution of cellular immunity against cytomegalovirus in recipients of allogeneic bone marrow by transfer of T-cell clones from the donor. N. Engl. J. Med. 333:1038. [DOI] [PubMed] [Google Scholar]
- 49.Wiertz, E. J., T. R. Jones, L. Sun, M. Bogyo, H. J. Geuze, and H. L. Ploegh. 1996. The human cytomegalovirus US11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol. Cell 84:760-779. [DOI] [PubMed] [Google Scholar]
- 50.Wills, M. R., A. J. Carmichael, K. Mynard, X. Jin, M. J. Weekes, B. Plachter, and J. G. P. Sissons. 1996. The human cytotoxic T lymphocyte (CTL) response to cytomegalovirus is dominated by structural protein pp65: frequency, specificity, and T-cell receptor usage of pp65-specific CTLs. J. Virol. 70:7569-7579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zaia, J. A. 1990. Epidemiology and pathogenesis of cytomegalovirus disease. Semin. Hematol. 27:5-10. [PubMed] [Google Scholar]