Abstract
In wild aquatic birds and poultry around the world, influenza A viruses carrying 15 antigenic subtypes of hemagglutinin (HA) and 9 antigenic subtypes of neuraminidase (NA) have been described. Here we describe a previously unidentified antigenic subtype of HA (H16), detected in viruses circulating in black-headed gulls in Sweden. In agreement with established criteria for the definition of antigenic subtypes, hemagglutination inhibition assays and immunodiffusion assays failed to detect specific reactivity between H16 and the previously described subtypes H1 to H15. Genetically, H16 HA was found to be distantly related to H13 HA, a subtype also detected exclusively in shorebirds, and the amino acid composition of the putative receptor-binding site of H13 and H16 HAs was found to be distinct from that in HA subtypes circulating in ducks and geese. The H16 viruses contained NA genes that were similar to those of other Eurasian shorebirds but genetically distinct from N3 genes detected in other birds and geographical locations. The European gull viruses were further distinguishable from other influenza A viruses based on their PB2, NP, and NS genes. Gaining information on the full spectrum of avian influenza A viruses and creating reagents for their detection and identification will remain an important task for influenza surveillance, outbreak control, and animal and public health. We propose that sequence analyses of HA and NA genes of influenza A viruses be used for the rapid identification of existing and novel HA and NA subtypes.
Influenza virus types A, B, and C belong to the family of Orthomyxoviridae and have many biological properties in common (4). A key difference between them is their host range: whereas influenza viruses of types B and C are predominantly human pathogens that have sporadically been isolated from seals and pigs, respectively (11, 24), influenza A viruses have been isolated from many animal species, including humans, pigs, horses, mink, marine mammals, and a wide range of domestic and wild birds (23, 33). Wild birds, predominantly ducks, geese, and shorebirds, form the reservoir of influenza A viruses in nature. Avian influenza viruses preferentially infect cells lining the intestinal tract of birds and are excreted in high concentrations in their feces. While avian influenza viruses are generally nonpathogenic in wild birds, they sometimes cause significant morbidity and mortality upon transmission to other species, including domestic birds and mammals (23, 33).
Influenza A viruses are classified on the basis of the antigenic properties of the hemagglutinin (HA) and neuraminidase (NA) glycoproteins expressed on the surface of virus particles. To date, influenza A viruses representing 15 HA and 9 NA subtypes have been detected in wild birds and poultry throughout the world (14, 22, 26, 35). These antigenic subtypes are distinguished by double immunodiffusion assays with hyperimmune animal sera because such tests revealed antigenic relationships among influenza A virus isolates which were not apparent with other methods (34, 35). Disadvantages of immunodiffusion assays for this purpose are that the development of monospecific reagents (antisera and antigens) can be very time-consuming, that the outcome is heavily dependent on the quality of the reagents used, and that the assay provides qualitative rather than quantitative information.
The influenza A virus HA protein is encoded by viral gene segment 4 and is initially synthesized as a single polypeptide precursor (HA0). The mature HA forms homotrimers, and each monomer is generated upon cleavage of HA0 into HA1 and HA2 subunits by trypsin-like or furin-like proteases. The HA of influenza A virus mediates early steps of the viral replication cycle, receptor binding and membrane fusion. The cellular receptors are sialic acids of cell surface glycoproteins and glycolipids (reviewed in reference 28).
During surveillance studies of influenza A virus in wild birds in Northern Europe (7, 32), we isolated influenza A viruses from black-headed gulls (Larus ridibundus) in Sweden which could not be classified by using antisera raised against the 15 known HA subtypes. Genetic analyses of the HA genes and antigenic analyses with hemagglutination inhibition (HI) assays provided strong support for the classification of a novel HA subtype. This was subsequently confirmed by double immunodiffusion assays with hyperimmune rabbit antisera. We thus propose that the HA of these black-headed gull viruses represent a new HA subtype, H16, which is genetically distantly related to HA of subtype H13. Moreover, we suggest that, with the rapidly increasing number of HA nucleotide sequences available from public databases, novel HA subtypes can be defined on the basis of their amino acid sequences.
MATERIALS AND METHODS
Virus detection.
Black-headed gulls (L. ridibundus) were caught by using funnel traps at Ottenby Bird Observatory in Öland, Sweden, in August 1999. Additional black-headed gulls were caught by hand on their breeding grounds at various locations in Sweden and The Netherlands. Cloacal swabs were collected with cotton swabs and stored in transport medium at −70°C. Transport medium consisted of Hanks balanced salt solution supplemented with 10% glycerol, 0.5% lactalbumin, 200 U of penicillin/ml, 200 μg of streptomycin/ml, 100 U of polymyxin B sulfate/ml, 250 μg of gentamicin/ml, and 50 U of nystatin/ml (all from ICN, Zoetermeer, The Netherlands). RNA isolation, reverse transcription (RT)-PCR, and dot blot detection were performed as described elsewhere (6). In brief, RNA was isolated by using a high-pure RNA isolation kit (Roche Molecular Biochemicals) and viral sequences were amplified in a one-step RT-PCR with primers M52C (5′-CTT CTA ACC GAG GTC GAA ACG-3′) and M253R (5′-AGG GCA TTT TGG ACA AAG/T CGT CTA-3′) targeting the matrix gene. RT-PCR products were transferred to dot blots and visualized with enhanced chemiluminescence detection reagents and exposure to hyperfilm (Amersham Pharmacia Biotech Benelux, Roosendaal, The Netherlands) upon hybridization with the biotinylated oligonucleotide Bio-M93C (5′-CCG TCA GGC CCC CTC AAA GCC GA-3′).
Virus isolation and characterization.
For RT-PCR-positive samples, 200 μl of the original specimens was inoculated into the allantoic cavities of 11-day-old embryonated chicken eggs, and hemagglutination titers in allantoic fluids were determined with turkey erythrocytes by standard procedures (25). Virus isolates were characterized by hemagglutination (27) and neuraminidase (1) inhibition assays with subtype-specific hyperimmune rabbit antisera raised against HA and NA preparations of virus isolates representing all influenza A virus subtypes and by nucleotide sequence analysis (see below).
Nucleotide sequence analyses and phylogenetic trees.
RT-PCR specific for the conserved noncoding regions of avian influenza viruses was described by others (16). PCR products were purified by using the QIAquick gel extraction kit (QIAGEN, Leusden, The Netherlands) and sequenced directly or upon cloning in pCR2.1 (Invitrogen, Groningen, The Netherlands). Sequencing was performed with the Dyenamic ET terminator cycle sequencing kit (Amersham Pharmacia Biotech) and a 373 genetic analyzer (Applied Biosystems, Nieuwerkerk aan den IJssel, The Netherlands). Primer sequences are available upon request. Nucleotide sequences are available from GenBank (accession no. AY684874 to AY684913).
Nucleotide and amino acid sequences were aligned by using Clustal W running under Bioedit, version 5.0.9 (12). DNA maximum-likelihood trees were generated with Phylip, version 3.6 (5), with 100 bootstraps and 3 jumbles. The consensus tree was used as the user tree in Dnaml to recalculate branch lengths, and trees were rerooted at the midpoint. To circumvent computational limitations, we generated a tree representing all NA sequences available from public databases by using the unweighted pair group method with arithmetic mean (UPGMA) clustering method of the Neighbor program of Phylip. Amino acid sequence alignments were bootstrapped 100 times, and distance matrices were generated by using Kimura's distance. The consensus of 100 UPGMA trees was calculated, and this consensus tree was used for recalculation of branch lengths with the Fitch program of Phylip (5).
For Dnaml trees, we used sequences representing Eurasian avian influenza A viruses when available. The accession numbers that were used are as follows: for NA, AF523393, AJ416628, AY180837, AY207530, AY207552, AY340077, AY684892 to AY684895, L06574, M11445, and M24740; for HA, AF091313, AJ427297, AY083840, AY338460, D90304, D90307, L43916, M21647, M25283, M26089 to M26091, and M35997; for internal genes, see reference 8. The full list of accession numbers for all full-length NA (n = 649), HA0 (n = 541), HA1 (n = 1,959), and HA2 (n = 608) sequences available from public databases in June 2004, used for Fig. 2 and 5a, is available on request.
FIG. 2.
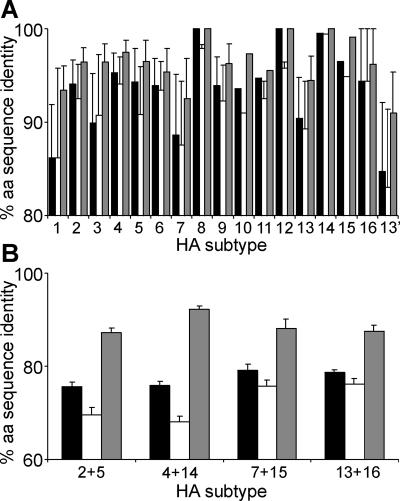
Amino acid sequence identity between pairs of HA sequences. The average percent amino acid (aa) sequence identities and standard deviations are shown for all pairs of HA sequences within a subtype (A) or for all pairs of HA sequences from two closely related subtypes (B). Black, white, and gray bars represent HA0, HA1, and HA2 sequences, respectively. HA subtype 13′ represents the combined group of H13 and H16 HA sequences. The percentages and standard deviations were calculated by using all HA sequences available from public databases upon sequence alignment per subtype (A) or per two subtypes (B) and subsequent calculation of pairwise Hamming distances (13).
FIG. 5.
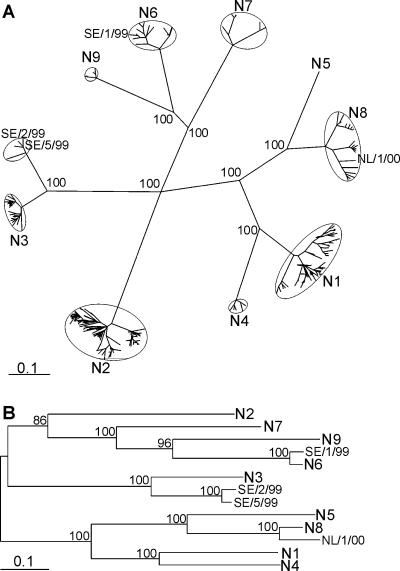
Phylogeny of NA genes from Swedish and Dutch black-headed gull influenza A viruses. A tree was generated with all 649 full-length NA amino acid sequences available from public databases (A). Aligned sequences were bootstrapped 100 times, and the Kimura amino acid distance matrices were converted to trees by using a UPGMA clustering algorithm. Very similar trees were obtained when distance matrices were generated by using Hamming distances or the Jones-Taylor-Thornton model (5, 13). The consensus tree was used for recalculation of the branch lengths with the Fitch program of Phylip. Representative nucleotide sequences, when available from Eurasian avian origin, were used to generate a DNA maximum-likelihood tree (B). This tree was built by using 100 bootstraps and 3 jumbles, after which branch lengths were recalculated for the consensus tree. NL/1/00, SE/1/99, SE/2/99, and SE/5/99 represent the NA sequences of black-headed gull viruses of subtypes H13N8, H13N6, H16N3, and H16N3 from The Netherlands and Sweden, respectively. Scale bars roughly represent 10% of changes between close relatives. Small numbers in trees represent bootstrap values.
DNA immunization of rabbits.
The HA gene segments of A/Black-headed Gull/Sweden/5/99 and A/Gull/Maryland/704/77 were cloned in expression plasmid pcDNA3. Both constructs were verified by sequencing. Rabbits were immunized with 100 μg of plasmid DNA mixed with 100 μl of Lipofectin as previously described for chickens (31). After 4 immunizations at weeks 0, 3, 6, and 9, the HI antibody titers were detectable but low (∼12). After two more immunizations at weeks 12 and 19 with 500 μg of plasmid DNA, the HI antibody titers were ∼96 and the animals were euthanized at week 20 for serum collection.
Reference sera.
Hyperimmunized rabbit antisera were generated against the following influenza reference strains: A/Puerto Rico/8/34 (H1N1), A/Fort Monmouth/1/47 (H1N1), A/Swine/Shope/1/56 (H1N1), A/Duck/Alberta/35/76 (H1N1), A/Singapore/1/57 (H2N2), A/Hong Kong/1/68 (H3N2), A/Equine/Miami/1/63 (H3N8), A/Duck/Ukraine/1/63 (H3N8), A/Duck/Czechoslovakia/1/56 (H4N6), A/Tern/South Africa/61 (H5N3), A/Duck/Hong Kong/205/77 (H5N3), A/Turkey/Massachusetts/65 (H6N2), A/Shearwater/Australia/1/72 (H6N5), A/Equine/Prague/1/54 (H7N7), A/Seal/Massachusetts/1/80 (H7N7), A/Turkey/Ontario/6118/68 (H8N4), A/Turkey/Wisconsin/1/66 (H9N2), A/Chicken/Germany/N/49 (H10N7), A/Duck/England/1/56 (H11N6), A/Duck/Memphis/546/74 (H11N9), A/Duck/Alberta/60/76 (H12N5), A/Gull/Maryland/704/77 (H13N6), A/Gull/Gurjev/263/82 (H14N5), A/Duck/Australia/341/83 (H15N8), and A/shearwater/West Australia/2576/79 (H15N9). Reference strains were propagated in embryonated chicken eggs, and afterwards, the allantoic fluid was cleared by centrifugation for 10 min at 1,000 × g and filtration through a 0.45-μm-pore-size filter. The virus was pelleted by centrifugation for 1.5 h at 150.000 × g at 4°C and resuspended in phosphate-buffered saline (PBS). Virus was treated overnight at 4°C with 1% Triton X-100, loaded on a layer of 25% sucrose in PBS and centrifuged for 1.5 h at 250.000 × g at 4°C. The top layer containing HA and NA proteins was inspected for purity and quantity on 12.5% sodium dodecyl sulfate-polyacrylamide gels stained with Coomassie brilliant blue. Animals were immunized with approximately 500 μg of HA/NA proteins in 2.4 ml of a Specol water-in-oil emulsion (2) at 1-month intervals with 0.4 ml of antigen intramuscularly and 2 ml of antigen subcutaneously. After 3 immunizations, HI antibody titers against the homologous viruses were determined, and if necessary, the rabbits were immunized once more.
Double immunodiffusion assays.
Double immunodiffusion assays were performed essentially as described previously (17). Approximately 5-mm-thick 1% agarose gels, containing 2% polyethylene glycol in PBS, were used in wells which were punched with a Pasteur pipette approximately 1 cm apart. Antisera and antigens were loaded in these wells, and afterwards, the gels were placed in a humidified chamber for 48 h at room temperature. The gel was washed twice with PBS for 24 h and once with water for 4 h. Gels were covered with filter paper and dried overnight. Dried gels were stained for 10 min in 0.5% (wt/vol) Coomassie brilliant blue, 40% (vol/vol) ethanol, and 10% (vol/vol) glacial acetic acid and destained in 15% (vol/vol) ethanol and 5% (vol/vol) glacial acetic acid.
Sera were concentrated 8 times by precipitation with ammonium sulfate and 10 times by filtration through Mircocon centrifugal filter devices with a 100,000-Da molecular mass cutoff (Millipore Corporation, Bedford, Mass.). We used 20 μl of concentrated polyclonal antiserum obtained from rabbits hyperimmunized by DNA vaccination (see above). As antigens, we used 20 μl of whole-virus lysates, prepared by pelleting virus from the allantoic fluid of infected chicken eggs and resuspending it in 2.5% of the original volume of PBS with 1% Triton X-100.
RESULTS
Influenza A virus in black-headed gulls in Europe.
Within the framework of our ongoing influenza A virus surveillance studies in wild birds (7, 32), we obtained 10 samples from black-headed gulls in 1999 that were screened for influenza A virus by RT-PCR specific for the matrix gene. These samples were obtained from juvenile birds that were less than 1 year old and were sampled between August 10 and 14 at the Ottenby Bird Observatory in Öland, Sweden. Cloacal swabs obtained from six of these birds, 8-224, 8-251, 8-291, 8-292, 8-298, and 8-510, were positive for influenza A virus by RT-PCR and agarose gel electrophoresis or dot blot hybridization (data not shown). We next attempted to isolate influenza A virus from the RT-PCR-positive samples in 11-day-old embryonated chicken eggs. From samples 8-224, 8-291, 8-292, and 8-298, influenza A virus isolates were obtained upon the first inoculation of eggs, as determined by the presence of hemagglutinating activity in the allantoic fluid. We were unable to detect hemagglutinating activity in the allantoic fluid of eggs inoculated with samples 8-251 and 8-510, but after blind passage of the allantoic fluid in embryonated eggs, an isolate was obtained from sample 8-251. Further attempts to isolate virus from sample 8-510 were not successful. The virus isolates were named A/Black-headed Gull/Sweden/1/99 (8-251), A/Black-headed Gull/Sweden/2/99 (8-298), A/Black-headed Gull/Sweden/3/99 (8-224), A/Black-headed Gull/Sweden/4/99 (8-291), and A/Black-headed Gull/Sweden/5/99 (8-292).
Between 17 June and 6 July 2000, we performed the same testing of 90 cloacal swab samples obtained from juvenile black-headed gulls in Pieterburen, The Netherlands. Of these, three were positive by RT-PCR but only one was positive upon repeated attempts to isolate a virus (sample 17-73, A/Black-headed Gull/The Netherlands/1/00). Additional black-headed gull samples (5 from The Netherlands, December 1998, and 351 from Venan, Hoenborop, Kirokskar, Malmö, Kalmar, Umeå, and Lund, Sweden in June 2000) were all negative by RT-PCR (data not shown).
HI assays with influenza A viruses isolated from black-headed gulls.
The six influenza A virus isolates obtained from black-headed gulls in Sweden and The Netherlands were used in HI assays with hyperimmunized rabbit antisera representing all HA subtypes. Virus isolates A/Black-headed Gull/The Netherlands/1/00 and A/Black-headed Gull/Sweden/1/99 reacted specifically with the rabbit antiserum raised against A/Gull/Maryland/704/77 (H13N6, homologous HI antibody titer of 2,560), giving rise to HI antibody titers of 640 and 2,560 (Table 1), indicating that these viruses contained a HA of subtype H13. The other four virus isolates did not react specifically with the H13 antiserum (Table 1) or any of the antisera in the H1 to H15 reference panel (data not shown).
TABLE 1.
Antigenic characterization of Northern-European gull influenza A viruses by HI assays with antisera raised in rabbits and ferretsa
| Virus | Subtype | Titer for virus (subtype) with sera from:
|
||||
|---|---|---|---|---|---|---|
| Hyperimmunized rabbits
|
Postinfection ferrets
|
|||||
| A/Duck/Memphis/546/76 (H11) | A/Gull/Maryland/704/77 (H13) | A/BHG/Sweden/2/99 (H16) | A/BHG/Sweden/2/99 (H16) | A/BHG/Sweden/5/99 (H16) | ||
| A/Duck/Memphis/546/76 | H11 | 3,840 | <10 | <10 | <10 | <10 |
| A/Gull/Maryland/704/77 | H13 | <10 | 2,560 | 40 | <10 | <10 |
| A/BHG/The Netherlands/1/00 | H13 | <10 | 640 | NT | NT | NT |
| A/BHG/Sweden/1/99 | H13 | <10 | 2,560 | <10 | <10 | <10 |
| A/BHG/Sweden/2/99 | H16 | <10 | <10 | 2,560 | 320 | 20 |
| A/BHG/Sweden/3/99 | H16 | <10 | <10 | 7,680 | 320 | 20 |
| A/BHG/Sweden/4/99 | H16 | <10 | <10 | 2,560 | 640 | 80 |
| A/BHG/Sweden/5/99 | H16 | <10 | <10 | 3,840 | 80 | 480 |
Homologous HI titers are in boldface type and are underlined. NT, not tested; BHG, black-headed gull.
We generated a new hyperimmunized rabbit antiserum raised against one of the unidentifiable virus isolates, A/Black-headed Gull/Sweden/2/99. To this end, virus was propagated in embryonated eggs and purified from the allantoic fluid. The virus was treated with Triton X-100, and the HA and NA proteins were separated from viral ribonucleoprotein complexes by centrifugation through 25% sucrose. Rabbits were immunized three times with these partially purified HA and NA protein preparations. Using the hyperimmune rabbit antisera raised against A/Black-headed Gull/Sweden/2/99, we obtained a homologous HI titer of 2,560 and titers of 2,560 to 7,680 against A/Black-headed Gull/Sweden/3/99, A/Black-headed Gull/Sweden/4/99, and A/Black-headed Gull/Sweden/5/99. This antiserum did not react with the H13 viruses A/Black-headed Gull/Sweden/1/99 and A/Black-headed Gull/The Netherlands/1/00 and reacted only marginally with A/Gull/Maryland/704/77 (Table 1). Low, nonspecific, HI antibody titers of 20 to 80 are occasionally observed between antisera and antigens representing distinct HA subtypes (data not shown).
We also raised postinfection antisera in ferrets against A/Black-headed Gull/Sweden/2/99 and A/Black-headed Gull/Sweden/5/99. Such antisera, collected two weeks after infection, are generally more specific than hyperimmune rabbit antisera. Neither of these ferret antisera reacted with the H13 isolates tested. When tested against the four unidentifiable gull viruses, these antisera displayed some antigenic heterogeneity; the antiserum raised against A/Black-headed Gull/Sweden/2/99 gave rise to higher HI antibody titers against A/Black-headed Gull/Sweden/2/99 (titer of 320), A/Black-headed Gull/Sweden/3/99 (titer of 320), and A/Black-headed Gull/Sweden/4/99 (titer of 640) compared to A/Black-headed Gull/Sweden/5/99 (titer of 80), whereas the ferret antiserum raised against A/Black-headed Gull/Sweden/5/99 gave the opposite results (homologous titer of 480 and heterologous titers of 20, 20, and 80, respectively).
Thus, whereas influenza viruses A/Black-headed Gull/Sweden/1/99 and A/Black-headed Gull/The Netherlands/1/00 were identifiable as H13 viruses by HI assays, the other four viruses isolated from black-headed gulls were not. The latter four viruses were antigenically related but displayed some antigenic heterogeneity with ferret antisera.
Sequence analyses and phylogeny.
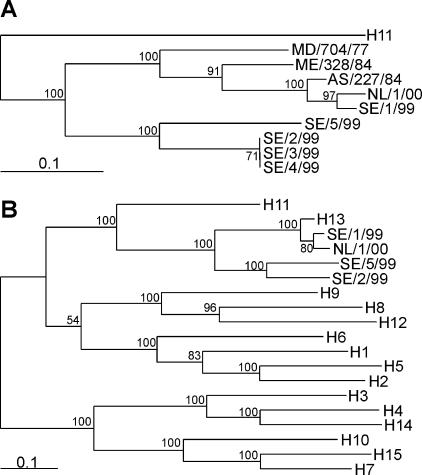
We next sequenced the HA open reading frame (ORF) for all six European black-headed gull viruses and compared them to sequences available from public databases. Upon analysis with the basic local alignment search tool (BLAST) available from GenBank, the HA ORF of A/Black-headed Gull/Sweden/1/99 and A/Black-headed Gull/The Netherlands/1/00 revealed high percentages of nucleotide and amino acid sequence identity to the HA of H13 influenza A viruses, whereas the four unidentifiable influenza A viruses revealed low homology at the nucleotide and amino acid levels, in agreement with the HI assay results (data not shown). We generated DNA maximum-likelihood trees for the HA ORFs of the six European black-headed gull viruses and those of all complete H13 HA ORFs available from public databases. This phylogenetic tree revealed that the HA ORFs of A/Black-headed Gull/Sweden/1/99 and A/Black-headed Gull/The Netherlands/1/00 were genetically most closely related to the HA ORF of another Eurasian gull virus, A/Gull/Astrakhan/227/84, and less related to the American H13 viruses A/Gull/Maryland/704/77 and A/Pilot Whale/Maine/328/84 (3). The four unidentifiable black-headed gull viruses from Sweden were found on a separate branch in this phylogenetic tree, distinct from the H13 viruses (Fig. 1a). The HA gene of A/Black-headed Gull/Sweden/5/99 was genetically distinct from the HA genes of the other three unidentifiable black-headed gull viruses, in agreement with the antigenic heterogeneity of these virus isolates in HI assays with postinfection ferret antisera. The HA genes of A/Black-headed Gull/Sweden/3/99 and A/Black-headed Gull/Sweden/4/99 were identical at the nucleotide level and differed by only one silent nucleotide substitution from the HA gene of A/Black-headed Gull/Sweden/2/99.
FIG. 1.
Phylogeny of HA genes from Swedish and Dutch black-headed gull influenza A viruses. DNA maximum-likelihood trees were generated by using the HA0 ORFs of prototype H13 influenza A viruses and all isolates described in this study (A) or representative sequences for all HA subtypes, when available from Eurasian avian origin (B). DNA maximum-likelihood trees were built by using 100 bootstraps and 3 jumbles, and branch lengths were recalculated for the resulting consensus tree. The scale bars roughly represent 10% of nucleotide changes between close relatives. MD, A/Gull/Maryland; ME, A/Pilot whale/Maine; AS, A/Gull/Astrakhan; NL, A/Black-headed Gull/The Netherlands; SE, A/Black-headed Gull/Sweden. Small numbers in trees represent bootstrap values.
To determine the significance of the branching of the black-headed gull HAs in the phylogenetic tree, we next constructed a new maximum-likelihood phylogenetic tree in which all HA subtypes were included by using HAs of Eurasian avian origin when available (Fig. 1b). This tree showed that the lengths of the branches between the HA genes of viruses of subtype H13 and those of the unidentifiable viruses (maximum-likelihood tree distances ranging from 0.39 to 0.42) were similar to the lengths of the branches between subtypes H2 and H5 (maximum-likelihood tree distance, 0.40), H7 and H15 (maximum-likelihood tree distance, 0.35) and H4 and H14 (ML tree distance 0.42), suggesting that the unidentified viruses represent a previously undescribed HA subtype, H16.
Identification of a novel HA subtype, H16, as evidenced by HA amino acid sequence comparisons.
We obtained all HA sequences available from public databases, irrespective of host species or geographical or temporal information and aligned these sequences per HA subtype. Alignments were made individually for HA0, HA1, and HA2, thereby including as many sequences representing an entire HA0, HA1, or HA2 domain as possible for analysis. Hamming amino acid distance matrices were generated for each alignment of sequences (13), and the average percent amino acid sequence identity and the standard deviation were calculated for each HA subtype (Fig. 2a). The average percent amino acid sequence identity for viruses characterized as HA subtypes H1 to H15 ranged from 86 to 100% for HA0 and HA1 and from 92 to 100% for HA2. The lowest average percent amino acid sequence identity was found for viruses within subtypes H1, H3, and H7. The relatively high diversification of these HA ORFs is likely due to the fact that many of these viruses are isolated frequently not only from avian hosts but also from nonavian hosts: H1 from humans and pigs; H3 from humans, pigs, and horses; and H7 from horses. When the H13 HA sequences were grouped separately from the putative H16 HA sequences, the average percent amino acid sequence identity between viruses within both subtypes was in the same range as for other avian HA subtypes (H13, 90, 89, and 95% for HA0, HA1, and HA2, respectively; putative H16, 94, 94, and 96% for HA0, HA1, and HA2, respectively). In contrast, when the H13 HA sequences were grouped together with the putative H16 sequences, the average percent amino acid sequence identities between the HA sequences were 85, 83, and 91% for HA0, HA1, and HA2, respectively. Thus, the combined group of H13 and H16 HA sequences would be more diverse than any known HA subtype identified to date, including those that diversified in nonavian hosts.
This same conclusion was reached when minimum rather than average percent amino acid sequence identity was calculated (data not shown). The minimum percent amino acid sequence identities between any pair of sequences in the H13-H16 combined subtype were 78, 74, and 86%, respectively, for HA0, HA1, and HA2. The minimum percent amino acid sequence identity was lower only for some pairs of HA0 and HA1 sequences of isolates of subtype H1 that have diversified in multiple hosts (75%).
We next compared the average percent amino acid sequence identity and standard deviation between strains of the most closely related HA subtypes: H2 versus H5, H4 versus H14, H7 versus H15, and H13 versus putative H16 (Fig. 2b). The average percent amino acid sequence identities between HA ORFs of isolates belonging to the H13 and putative H16 subtypes were 79, 76, and 88% for HA0, HA1, and HA2, respectively. These percentages were in the same range as those observed for other related HA subtypes (HA0, 76 to 79%; HA1, 68 to 76%; HA2, 87 to 92%).
Thus, the phylogenetic trees and analyses of HA amino acid sequences representing all previously described HA subtypes provide strong evidence for the classification of the HAs of A/Black-headed Gull/Sweden/2/99, A/Black-headed Gull/Sweden/3/99, A/Black-headed Gull/Sweden/4/99, and A/Black-headed Gull/Sweden/5/99 as a novel subtype, H16.
Identification of a novel HA subtype, H16, by classical criteria.
The classical way to identify a novel HA subtype is by using a double immunodiffusion assay (17, 26, 35). In such an assay, antibodies and antigens are loaded in separate wells in a gel-based matrix, after which diffusion gradients will form. When the diffusion gradients of antigens and antibodies cross, a line of immunoprecipitation may form to indicate the specific reaction between the antibodies and antigens. Because precipitations due to influenza virus antigens other than HA are undesired for HA typing, monospecific antisera or highly purified HA protein preparations (or both) are required. We generated hyperimmune rabbit antisera by DNA vaccination with cloned versions of the HA genes of influenza viruses A/Black-headed Gull/Sweden/5/99 and A/Gull/Maryland/704/77. The antisera were concentrated 80-fold by a combination of precipitation with ammonium sulfate and microfiltration because of the low titers of specific antibodies in the original antisera as determined in HI assays (HI antibody titers of 96). As antigens, we used whole-virus lysates prepared after 40-fold concentration of virus from the allantoic fluid of infected embryonated chicken eggs.
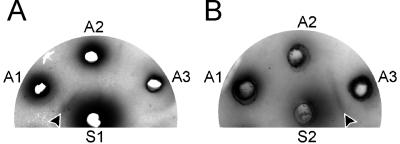
The antiserum raised against the H13 HA of A/Gull/Maryland/704/77 (Fig. 3a, serum S1) gave rise to antigen-antibody complexes when tested against virus lysate A/Gull/Maryland/704/77 (antigen A1) but not with virus lysates A/Shoveler/The Netherlands/18/99 (H11, antigen A2) and A/Black-headed Gull/Sweden/5/99 (putative H16, antigen A3). Conversely, the antiserum raised against the putative H16 HA of A/Black-headed Gull/Sweden/5/99 (Fig. 3b, serum S2) gave rise to antigen-antibody complexes when tested against virus lysate A/Black-headed Gull/Sweden/5/99 (antigen A3) but not with virus lysates A/Gull/Maryland/704/77 (antigen A1) and A/Shoveler/The Netherlands/18/99 (antigen A2). Thus, the HA of A/Black-headed Gull/Sweden/5/99 represents a previously unidentified HA subtype, H16, according to established criteria (35).
FIG. 3.
Double radial immunodiffusion assay with hyperimmune rabbit antisera raised against HA of gull influenza A viruses. Concentrated hyperimmune antisera raised against H13 HA of A/Gull/Maryland/704/77 (A) or HA of A/Black-headed Gull/Sweden/5/99 (B) were loaded in wells S1 and S2, respectively. Lysates prepared from concentrated virus stocks of A/Gull/Maryland/704/77 (H13), A/Shoveler/The Netherlands/18/99 (H11), and A/Black-headed Gull/Sweden/5/99 were loaded in antigen wells A1, A2, and A3, respectively. Precipitates were allowed to form for 48 h, after which the gels were dried and stained with Coomassie brilliant blue. Specific precipitates were observed only between wells A1 (H13, A/Gull/Maryland/704/77) and S1 (anti-H13, A/Gull/Maryland/704/77) and wells A3 (H16, A/Black-headed Gull/Sweden/5/99) and S2 (anti-H16, A/Black-headed Gull/Sweden/5/99), as indicated by arrowheads.
Predicted amino acid sequence of H16 HA.
In Fig. 4, the predicted amino acid sequences of the HA0 ORFs of the two antigenic variants of H16, obtained from A/Black-headed Gull/Sweden/2/99 (564 aa) and A/Black-headed Gull/Sweden/5/99 (566 aa), are shown. The predicted HA0 amino acid sequences are 88.3% identical, and those of the HA1 and HA2 subunits are 88.6 and 92.3% identical, respectively. The HA ORFs of A/Black-headed Gull/Sweden/2/99 and A/Black-headed Gull/Sweden/5/99 contain 9 and 8 potential N-linked glycosylation sites (N-X-T/S), respectively. With the exception of potential N-linked glycosylation site number 4 (Fig. 4), all of these sites are in common with H13 HA (3, 15). The protease cleavage site in H16 HA does not contain multiple basic amino acid residues (SIVER*GLFG, SVGER*GLFG) and is somewhat different from the cleavage site in H13 HA (AISNR*GLFG). Recently, differences in the receptor-binding site between HAs originating from Laridae (gulls) and Anatidae (ducks and geese) were described for amino acid positions 154, 191, 215, 222, 223, 227 to 229, and 231 (36). With the exceptions of amino acid positions 154 and 223, the following amino acid residues in the receptor-binding site of H16 HA were in agreement with these observations: 191Ala, 215Leu, 222Gly, 227Arg, 228Ser, 229Trp, and 231Lys (Fig. 4).
FIG. 4.
Alignment of predicted amino acid sequences of HA subtype H16. An alignment of representative H16 amino acid sequences (2/99, A/Black-headed Gull/Sweden/2/99; 5/99, A/Black-headed Gull Sweden/5/99) is shown with periods representing identical amino acid residues and dashes representing gaps. Potential N-linked glycosylation sites (N-X-T/S) are boxed and numbered. Asterisks above the sequences indicate amino acid positions at which differences were observed between HA genes from Laridae and Anatidae (positions 191, 215, 222, 227, 228, 229, and 231 in H3 HA) (36). The predicted signal peptide and the HA1 and HA2 domains are indicated.
Sequence analyses of the NA genes of influenza A viruses isolated from European black-headed gulls.
We next amplified the NA gene segments of the black-headed gull influenza A viruses by RT-PCR with primers targeting the conserved noncoding regions and sequenced the obtained PCR products. Upon BLAST analysis, the NA gene of A/Black-headed Gull/The Netherlands/1/00 revealed closest resemblance to NA sequences of subtype N8, the NA gene of A/Black-headed Gull/Sweden/1/99 revealed closest resemblance to NA sequences of subtype N6, and the NA genes of the other four black-headed gull viruses revealed closest resemblances to NA sequences of subtype N3 (data not shown). A UPGMA phylogenetic tree, based on an amino acid sequence distance matrix in which all NA sequences available from public databases were included, was in agreement with these BLAST results (Fig. 5a). In this tree, each of the European black-headed gull viruses was found on a branch with previously identified gull viruses: A/Black-headed Gull/Sweden/1/99 (H13N6) with A/Gull/Maryland/704/77 (H13N6) and A/Gull/Astrakhan/227/84 (H13N6), A/Black-headed Gull/The Netherlands/1/00 (H13N8) with A/Herring Gull/Delaware/677/88 (H2N8), and the other black-headed gull viruses (H16N3) with A/Gull/Heuwiese/899-6/80 (H7N3). Whereas the NA genes of black-headed gull viruses of subtypes N6 and N8 are closely related to NA genes obtained from influenza A viruses isolated from other avian hosts (ducks and poultry), the N3 NA genes obtained from the H16 black-headed gull viruses form a distinct genetic lineage within the N3 NA cluster, together with the N3 genes of A/Tern/Astrakhan/775/83 (H13N3) and A/Gull/Heuwiese/899-6/80 (H7N3). Both the DNA maximum-likelihood tree (Fig. 5b) and the UPGMA tree based on the amino acid sequence distance matrix (Fig. 5a) revealed that the genetic distance between the European shorebird N3 sequences and the duck and poultry N3 sequences is smaller than the genetic distance between related NA subtypes such as N1 and N4, N6 and N9, and N5 and N8. In neuraminidase inhibition assays with hyperimmune rabbit antisera (1), the NA of the H16 black-headed gull viruses gave a weak positive reaction with an antiserum raised against A/Duck/Hongkong/207/77 (H5N3) (data not shown), indicating that they are antigenically related.
Sequence analyses of internal genes of influenza A viruses isolated from European black-headed gulls.
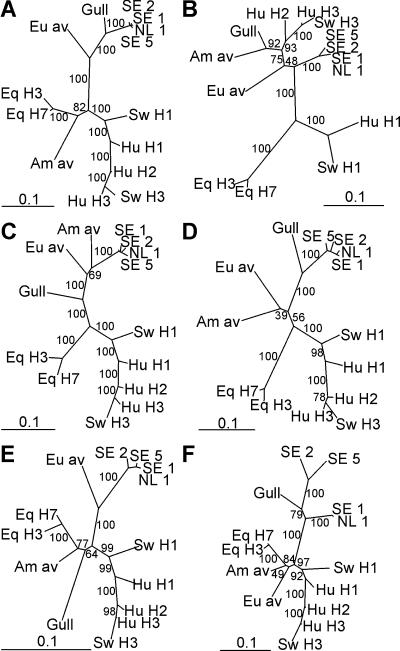
We sequenced the internal genes of the H13N6, H13N8, and representative H16N3 viruses (A/Black-headed Gull/Sweden/2/99 and A/Black-headed Gull/Sweden/5/99). These nucleotide sequences were analyzed in DNA maximum-likelihood phylogenetic trees in which sequences of prototypic influenza A virus isolates (8) from different hosts and geographical locations were included (Fig. 6). The PB2, NP, and NS genes of the black-headed gull viruses were closely related to those of previously characterized gull viruses, whereas the PB1 and MA genes were closer to sequences of Eurasian avian origin and the PA genes were closer to sequences of either American or Eurasian avian origin. GenBank BLAST analyses confirmed these results (data not shown). Thus, some of the internal genes of these European black-headed gull viruses are genetically distinct from those of the prototypic gull viruses, such as A/Gull/Maryland/704/77. The internal genes of the H13 and H16 European black-headed gull viruses are genetically closely related, although there was considerable heterogeneity between the NS genes of the H13 and H16 viruses (Fig. 6F).
FIG. 6.
Phylogenetic trees representing the internal genes of influenza A viruses. Trees were constructed based on a 2,262-nucleotide fragment of gene segment 1 (PB2) (A), 2,267 nucleotides of gene segment 2 (PB1) (B), 2,142 nucleotides of gene segment 3 (PA) (C), 1,486 nucleotides of gene segment 5 (NP) (D), 947 nucleotides of gene segment 7 (MA) (E), and 794 nucleotides of gene segment 8 (NS) (F). Sequences obtained from influenza viruses A/Black-headed Gull/The Netherlands/1/00 (H13N8) (NL 1), A/Black-headed Gull/Sweden/1/99 (H13N6) (SE 1), A/Black-headed Gull/Sweden/2/99 (H16N3) (SE 2), and A/Black-headed Gull/Sweden/5/99 (H16N3) (SE 5) were compared with those of reference strains available from GenBank, representing the known genetic lineages of influenza A virus (8). The DNA maximum-likelihood trees were built by using 100 bootstraps and 3 jumbles, and branch lengths were recalculated for the resulting consensus tree. Scale bars roughly represent 10% of nucleotide changes between close relatives. Small numbers in trees represent bootstrap values.
DISCUSSION
Here we identified a previously undescribed HA subtype of influenza A virus, H16, obtained from black-headed gulls in Sweden. Phylogenetic analyses of the HA nucleotide sequences and comparison of the predicted amino acid sequences of all HA genes available from public databases provided strong evidence for the classification of the HA of four of the Swedish black-headed gull viruses as H16. This classification was subsequently confirmed in a double radial immunodiffusion assay, according to classical criteria. The closest relative of the H16 HA gene was the HA gene of subtype H13.
HA of subtype H13 so far has been found exclusively in shorebirds, such as gulls, and in a pilot whale (potentially a spillover from shorebirds) but not in other avian species that are natural hosts of influenza A virus, such as ducks and geese (19). Since the HA of subtype H16 remained undetected until 1999, it is possible that the HA of subtype H16 is present only in a limited number of avian hosts. It is of interest that many amino acid residues, predicted to be in the receptor-binding site of HA, are in common in H13 and H16 gull influenza A viruses but distinct in HA of influenza A viruses isolated from ducks and geese (36). The receptor-binding site of HA could thus provide a molecular basis for the putative host preference of H13 and H16 influenza A viruses and could explain why these viruses have a specific ecological niche.
With the exception of the NS genes, the internal genes of the H13 and H16 European black-headed gull viruses were genetically closely related (Fig. 6). The PB2, PB1, PA, NP, and MA genes were 97 to 99% identical at the nucleotide level, but for the NS gene, this was as low as 86%. It is unclear what the origin is of these distinct NS gene segments. In BLAST searches, the NS genes of all 4 European black-headed gull viruses revealed more nucleotide sequence identity to those of other gull viruses than to allele A and allele B avian NS genes. The fact that they still clustered within the gull lineage suggests that these NS genes have diversified in gulls or other shorebirds rather than that they were recently introduced in gulls from other avian hosts, as could be speculated for the NS genes of some American gull viruses (20, 30). The PB1, PA, and MA genes of the European black-headed gull viruses were genetically distinct from those of American gull viruses (18, 21, 33). In contrast, the PB2, NP, and NS genes of the European black-headed gull viruses were genetically most closely related to those of American gull viruses (9, 10, 20, 30, 33). Thus, the influenza A viruses isolated from gulls can be distinguished genetically from other avian influenza A viruses based on the PB2, NP, and NS genes but not necessarily based on PB1, PA, and MA.
The four influenza A viruses of subtype H16 contained NA genes that were closely related to NA of subtype N3. Genetically, these 4 NA genes clustered together with the NA genes from influenza A viruses isolated from Eurasian shorebirds (A/Tern/Astrakhan/775/83 [H13N3] and A/Gull/Heuwiese/899-6/80 [H7N3]). This Eurasian shorebird lineage of N3 genes is genetically distinct from the N3 genes detected in other wild birds and poultry around the world. The genetic distance between Eurasian shorebird N3 and other N3 sequences is somewhat smaller than between related NA subtypes (N1-N4, N5-N8, and N6-N9), and there is some reactivity in neuraminidase inhibition assays between antigens and antisera raised against the different N3 lineages. Thus, there is no strong argument to classify the Eurasian shorebird N3 genes as a separate NA subtype based on existing criteria.
Although the double radial immunodiffusion assay has proven to be valuable for the classification of antigenic subtypes of HA and NA of influenza A virus, the question arises whether it still provides the best classification today. The immunodiffusion assay was recommended for classification at a time when antigenic characterization was easier than nucleotide sequence analysis. A disadvantage of the immunodiffusion assay is that high-titer monospecific antisera or highly purified antigen preparations (or both) are required to prevent undesired immunoprecipitation reactions between additional influenza virus antigens and the antisera. The second disadvantage of such serological tests is that they provide quantitative rather than qualitative information because the serological assays (HI, neuraminidase inhibition, or immunodiffusion assays) depend on the amount of antigen and quality and quantity of antibodies used. Weak antisera may identify too many different subtypes and strong or less-specific antisera may identify too few subtypes, which may yield discrepant results in different laboratories.
Today, sequence analysis is a standard technique in most laboratories and may be a preferred method for classification. A wide variety of algorithms for clustering or inference of genetic relationships are available for analyses of nucleotide or amino acid sequences (29). Sequences can be grouped in existing or novel clusters based on preset criteria that could include the maximum distance between sequences within a cluster (subtype) and the minimum distance between sequences of different clusters (subtypes). Serological tests in a simple format, such as the HI and neuraminidase inhibition assays, would remain extremely useful as a high-throughput laboratory tool, and together with the sequence analysis of HA and NA genes, they could provide the most practical framework for assigning existing or newly discovered HA and NA subtypes to influenza A virus isolates.
With the current intensified surveillance for influenza A virus in wild and domestic animals, it is probable that further novel HA and NA subtypes will be detected. This could be either due to the emergence of new influenza A virus subtypes, perhaps in part as the result of genetic mixing or adaptation to new hosts, or due to the circulation of unknown influenza A virus subtypes in specific ecological niches that were previously not explored. Gaining information on the full spectrum of influenza A viruses circulating in our environment and developing reagents for the specific detection of these viruses will remain important tasks for influenza surveillance, outbreak control, and animal and public health.
Acknowledgments
We thank C. Eising and T. Groothuis for providing avian specimens, L. van der Kemp for technical assistance, J. C. de Jong for critically reading the manuscript, and R. G. Webster for providing influenza A virus reference strains.
This work was supported financially by the Dutch Ministry of Agriculture, the Framework V program “NovaFlu” from the European Union, the Health Research Council of Southeast Sweden, and the Medical Faculty of Umeå University. R.A.M.F. is a fellow of the Royal Dutch Academy of Arts and Sciences.
Footnotes
This is contribution no. 198 from Ottenby Bird Observatory.
REFERENCES
- 1.Aymard-Henry, M., M. T. Coleman, W. R. Dowdle, W. G. Laver, G. C. Schild, and R. G. Webster. 1973. Influenza neuraminidase and neuraminidase-inhibition test procedures. Bull. W. H. O. 48:199-202. [PMC free article] [PubMed] [Google Scholar]
- 2.Bokhout, B. A., C. van Gaalen, and P. J. van der Heijden. 1981. A selected water-in-oil emulsion: composition and usefulness as an immunological adjuvant. Vet. Immunol. Immunopathol. 2:491-500. [Google Scholar]
- 3.Chambers, T. M., S. Yamnikova, Y. Kawaoka, D. K. Lvov, and R. G. Webster. 1989. Antigenic and molecular characterization of subtype H13 hemagglutinin of influenza virus. Virology 172:180-188. [DOI] [PubMed] [Google Scholar]
- 4.Cox, N. J., F. Fuller, N. Kaverin, H. D. Klenk, R. A. Lamb, B. W. J. Mahy, J. McCauley, K. Nakamura, P. Palese, and R. Webster. 2000. Orthomyxoviridae, p. 585-597. In M. H. V. van Regenmortel, C. M. Fauquet, D. H. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy: seventh report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, Calif.
- 5.Felsenstein, J. 1989. PHYLIP-phylogeny inference package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 6.Fouchier, R. A., T. M. Bestebroer, S. Herfst, L. Van Der Kemp, G. F. Rimmelzwaan, and A. D. Osterhaus. 2000. Detection of influenza A viruses from different species by PCR amplification of conserved sequences in the matrix gene. J. Clin. Microbiol. 38:4096-4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fouchier, R. A., B. Olsen, T. M. Bestebroer, S. Herfst, L. van der Kemp, G. F. Rimmelzwaan, and A. D. Osterhaus. 2003. Influenza A virus surveillance in wild birds in Northern Europe in 1999 and 2000. Avian Dis. 47:857-860. [DOI] [PubMed] [Google Scholar]
- 8.Fouchier, R. A., P. M. Schneeberger, F. W. Rozendaal, J. M. Broekman, S. A. Kemink, V. Munster, T. Kuiken, G. F. Rimmelzwaan, M. Schutten, G. J. Van Doornum, G. Koch, A. Bosman, M. Koopmans, and A. D. Osterhaus. 2004. Avian influenza A virus (H7N7) associated with human conjunctivitis and a fatal case of acute respiratory distress syndrome. Proc. Natl. Acad. Sci. USA 101:1356-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gorman, O. T., W. J. Bean, Y. Kawaoka, I. Donatelli, Y. J. Guo, and R. G. Webster. 1991. Evolution of influenza A virus nucleoprotein genes: implications for the origins of H1N1 human and classical swine viruses. J. Virol. 65:3704-3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gorman, O. T., R. O. Donis, Y. Kawaoka, and R. G. Webster. 1990. Evolution of influenza A virus PB2 genes: implications for evolution of the ribonucleoprotein complex and origin of human influenza A virus. J. Virol. 64:4893-4902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo, Y. J., F. G. Jin, P. Wang, M. Wang, and J. M. Zhu. 1983. Isolation of influenza C virus from pigs and experimental infection of pigs with influenza C virus. J. Gen. Virol. 64:177-182. [DOI] [PubMed] [Google Scholar]
- 12.Hall, A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 13.Hamming, R. W. 1950. Error detecting and error correcting codes. Bell Syst. Tech. J. 9:147-160. [Google Scholar]
- 14.Hinshaw, V. S., G. M. Air, A. J. Gibbs, L. Graves, B. Prescott, and D. Karunakaran. 1982. Antigenic and genetic characterization of a novel hemagglutinin subtype of influenza A viruses from gulls. J. Virol. 42:865-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hinshaw, V. S., W. J. Bean, J. Geraci, P. Fiorelli, G. Early, and R. G. Webster. 1986. Characterization of two influenza A viruses from a pilot whale. J. Virol. 58:655-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoffmann, E., J. Stech, Y. Guan, R. G. Webster, and D. R. Perez. 2001. Universal primer set for the full-length amplification of all influenza A viruses. Arch. Virol. 146:2275-2289. [DOI] [PubMed] [Google Scholar]
- 17.Hornbeck, P. 2001. Double-immunodiffusion assay for detecting specific antibodies, p. 2.3.1-2.3.4. In J. E. Coligan, A. M. Kruisbeek, D. H. Margulies, E. M. Shevach, and W. Strober (ed.), Current protocols in immunology. John Wiley & Sons, New York, N.Y.
- 18.Ito, T., O. T. Gorman, Y. Kawaoka, W. J. Bean, and R. G. Webster. 1991. Evolutionary analysis of the influenza A virus M gene with comparison of the M1 and M2 proteins. J. Virol. 65:5491-5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawaoka, Y., T. M. Chambers, W. L. Sladen, and R. G. Webster. 1988. Is the gene pool of influenza viruses in shorebirds and gulls different from that in wild ducks? Virology 163:247-250. [DOI] [PubMed] [Google Scholar]
- 20.Kawaoka, Y., O. T. Gorman, T. Ito, K. Wells, R. O. Donis, M. R. Castrucci, I. Donatelli, and R. G. Webster. 1998. Influence of host species on the evolution of the nonstructural (NS) gene of influenza A viruses. Virus Res. 55:143-156. [DOI] [PubMed] [Google Scholar]
- 21.Kawaoka, Y., S. Krauss, and R. G. Webster. 1989. Avian-to-human transmission of the PB1 gene of influenza A viruses in the 1957 and 1968 pandemics. J. Virol. 63:4603-4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawaoka, Y., S. Yamnikova, T. M. Chambers, D. K. Lvov, and R. G. Webster. 1990. Molecular characterization of a new hemagglutinin, subtype H14, of influenza A virus. Virology 179:759-767. [DOI] [PubMed] [Google Scholar]
- 23.Murphy, B. R., and R. G. Webster. 1996. Orthomyxoviruses, p. 1397-1445. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed., vol. 1. Lippincott-Raven publishers, Philadelphia, Pa. [Google Scholar]
- 24.Osterhaus, A. D., G. F. Rimmelzwaan, B. E. Martina, T. M. Bestebroer, and R. A. Fouchier. 2000. Influenza B virus in seals. Science 288:1051-1053. [DOI] [PubMed] [Google Scholar]
- 25.Rimmelzwaan, G. F., M. Baars, E. C. Claas, and A. D. Osterhaus. 1998. Comparison of RNA hybridization, hemagglutination assay, titration of infectious virus and immunofluorescence as methods for monitoring influenza virus replication in vitro. J. Virol. Methods 74:57-66. [DOI] [PubMed] [Google Scholar]
- 26.Rohm, C., N. Zhou, J. Suss, J. Mackenzie, and R. G. Webster. 1996. Characterization of a novel influenza hemagglutinin, H15: criteria for determination of influenza A subtypes. Virology 217:508-516. [DOI] [PubMed] [Google Scholar]
- 27.Salk, J. E. 1944. Simplified procedure for titrating hemagglutinating capacity of influenza virus and the corresponding antibody. J. Immunol. 49:87-98. [Google Scholar]
- 28.Skehel, J. J., and D. C. Wiley. 2000. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 69:531-569. [DOI] [PubMed] [Google Scholar]
- 29.Sneath, P. H. A., and R. R. Sokal. 1973. Numerical taxonomy: the principles and practice of numerical classification. Freeman, San Francisco, Calif.
- 30.Suarez, D. L., and M. L. Perdue. 1998. Multiple alignment comparison of the non-structural genes of influenza A viruses. Virus Res. 54:59-69. [DOI] [PubMed] [Google Scholar]
- 31.Suarez, D. L., and S. Schultz-Cherry. 2000. The effect of eukaryotic expression vectors and adjuvants on DNA vaccines in chickens using an avian influenza model. Avian Dis. 44:861-868. [PubMed] [Google Scholar]
- 32.Wallensten, A., V. J. Munster, R. A. M. Fouchier, and B. Olsen. 2004. Avian influenza A virus in ducks migrating through Sweden, p. 771-772. In Y. Kawaoka (ed.), Options for the control of influenza V. Elsevier Science B.V., Amsterdam, The Netherlands.
- 33.Webster, R. G., W. J. Bean, O. T. Gorman, T. M. Chambers, and Y. Kawaoka. 1992. Evolution and ecology of influenza A viruses. Microbiol. Rev. 56:152-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.World Health Organization. 1971. A revised system of nomenclature for influenza viruses. Bull. W. H. O. 45:119-124. [PMC free article] [PubMed] [Google Scholar]
- 35.World Health Organization. 1980. A revision of the system of nomenclature for influenza viruses: a W.H.O. memorandum. Bull. W. H. O. 58:585-591. [PMC free article] [PubMed] [Google Scholar]
- 36.Yamnikova, S. S., A. S. Gambaryan, A. B. Tuzikov, N. V. Bovin, M. N. Matrosovich, I. T. Fedyakina, A. A. Grinev, V. M. Blinov, D. K. Lvov, D. L. Suarez, and D. E. Swayne. 2003. Differences between HA receptor-binding sites of avian influenza viruses isolated from Laridae and Anatidae. Avian Dis. 47:1164-1168. [DOI] [PubMed] [Google Scholar]