Abstract
Molecular inversion probes (MIPs) in combination with massively parallel DNA sequencing represent a versatile, yet economical tool for targeted sequencing of genomic DNA. Several thousand genomic targets can be selectively captured using long oligonucleotides containing unique targeting arms and universal linkers. The ability to append sequencing adaptors and sample-specific barcodes allows large-scale pooling and subsequent high-throughput sequencing at relatively low cost per sample. Here, we describe a “wet bench” protocol detailing the capture and subsequent sequencing of >2000 genomic targets from 192 samples, representative of a single lane on the Illumina HiSeq 2000 platform.
Keywords: Molecular inversion probes, Massively parallel sequencing, Real-time PCR, Exonuclease cleanup and gel electrophoresis
1 Introduction
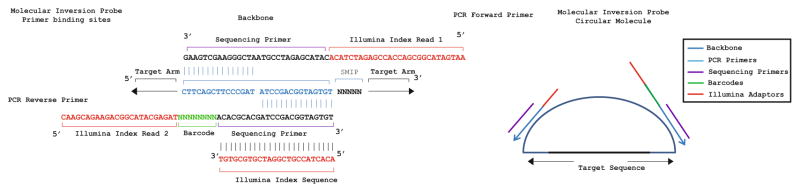
The ability to selectively enrich thousands of genomic DNA targets and sequence them in parallel has tremendously impacted the way genomes can be interrogated on a large scale [1]. Molecular inversion probes (MIPs) represent one such approach based on target circularization of single-stranded oligonucleotides consisting of a common DNA backbone flanked by target-specific sequences [2] (Fig. 1). Following hybridization of site-specific targeting arms, non-strand displacing DNA polymerase and deoxynucleotides facilitate extension (gap-closure) between targeting arms and the intervening sequence. The addition of DNA ligase completes the covalently closed circular molecule and exonuclease treatment removes linear DNA that failed to form a closed circle. PCR using universal primers complementary to the MIP backbone completes the DNA capture reaction and the library is, in principle, ready for DNA sequencing [3, 4].
Fig. 1.
Text-based primer map for molecular inversion probes. Sequence overlaps are annotated against the MIP backbone (blue ). Positions of the forward and reverse PCR primers are annotated in black with Illumina index primers annotated in red. Forward and reverse sequencing primers (purple) overlap the MIP backbone and the MIP PCR primers. The 8 bp sample-specific barcode is annotated in (green) and the small molecular tag in (grey)
The success of any targeted enrichment approach is directly impacted by the performance of the DNA capture reaction. The MIP protocol has proven to be adaptable and the integration of recent technical advances has led to notable improvements in MIP performance [4–7]. MIPs demonstrate consistent capture uniformity (~98 % of captured targets), capture specificity (>99 % target overlap), and multiplex scalability (thousands of capture targets) [4, 5]. Improvements to MIP-design tools also allow in silico predictions of assay success leading to increases in capture efficiency [6]. In addition, the use of single-molecule tagging, by adding random unique barcode tags to each molecule (termed smMIPs), has also facilitated the quantitation of individual capture events, allowing for highly sensitive variant calling and precise quantitation of somatic or mosaic events [7]. The simplicity of the workflow procedure, low sample input requirements, and cost-effectiveness of the MIP protocol have proven advantageous for the detection of rare and de-novo mutations in large disease cohorts [5, 8]. This protocol therefore describes in detail a method for large-scale resequencing of several thousand genomic targets using MIPs.
2 Materials
Prepare all dilutions using nuclease-free water. All enzymes and mastermixes should be stored at −20 °C unless otherwise noted. All reagents should be thawed and prepared on ice unless otherwise noted. All waste disposal regulations must be followed when disposing of hazardous materials.
-
Components for MIP pooling and phosphorylation.
70mer oligonucleotides synthesized at the 25 nanomole (nM) scale and hydrated to 100 micromole (μM) in 1× TE Buffer, pH 8.0. Store at −20 °C.
T4 DNA Ligase Reaction Buffer with 10 mM ATP (New England Biolabs). Store at −20 °C.
T4 Polynucleotide Kinase. Store at −20 °C.
ABgene 8-Flat-Cap Strip Tubes.
Costar* Microcentrifuge Tubes 1.7 mL; Color: Natural (holds 1.5 mL).
Nuclease-free water or equivalent. Store at room temperature.
-
Components for targeted capture.
Ampligase 10× Reaction Buffer (Epicentre). Store at −20 °C.
Ampligase® DNA Ligase (Epicentre). Store at −20 °C.
Hemo Klentaq (New England Biolabs). Store at −20 °C.
10 mM Deoxynucleotide (dNTP) set. Product should be diluted fresh for each capture reaction 1:40 (0.25 mM). Store at −20 °C.
Nuclease-free water or equivalent. Store at room temperature.
Eppendorf skirted 96-well plates or equivalent (clear).
Thermo Scientific* ABgene* Adhesive PCR Film or equivalent.
-
Components for exonuclease treatment.
Exonuclease I (E. coli). Store at −20 °C.
Exonuclease III (E. coli). Store at −20 °C.
-
Components for PCR.
iProof High Fidelity Master Mix (Bio-Rad). Store at −20 °C.
SYBR® Green I nucleic acid gel stain (Invitrogen). Store at −20 °C. Keep away from light.
Oligonucleotides. Synthesized at the 25 nM scale and hydrated to 100 μM in 1× TE Buffer, pH 8.0. Store at -20 °C.
Low−Profile 0.2 mL 8-Tube Strips Without Caps.
Optical Flat 8-Cap Strips.
-
Clean-up protocol components.
Agencourt AMPure XP beads. Store at 4 °C.
Ethanol (100 %). Store at room temperature.
DynaMag™-2 magnet (ThermoFisher Scientific).
Buffer EB (Qiagen). Store at room temperature.
-
Agarose gel electrophoresis components.
E-Gel® EX Gel, 2 % (Invitrogen).
E-Gel® Low Range Quantitative DNA Ladder (Invitrogen). Store at 4 °C.
-
Sequencing components.
Qubit dsDNA High Sensitivity Assay Kit. Store at room temperature.
0.5 mL tubes (for Qubit) or equivalent.
Illumina MiSeq Reagent Kit (300 cycles PE). Store at −20 °C.
3 Methods
3.1 Oligonucleotide Pooling and Phosphorylation
Design MIPs using an existing pipeline [6] (see Notes 1 and 2).
Pool oligonucleotides at equimolar concentrations by plate, by combining 5 μL of each MIP (100 μM/μL) into a single 1.5 mL tube. Each individual 1.5 mL tube will represent a combined sum of 96 MIPs for a total volume of 480 μL (see Note 3).
Take 9.6 μL of each individual MIP pool (0.1 μL multiplied by the number of MIPs in each plate) and combine these into a single tube to generate a MIP megapool.
Phosphorylate the MIP megapool by combining 25 μL of the MIP megapool, 3 μL of 10× T4 DNA Ligase Reaction Buffer, 1 μL of T4 Polynucleotide Kinase (10 U), and 1 μL of nuclease- free water in a total reaction volume of 30 μL. Using a thermo-cycler, incubate the reaction mix at 37 °C for 45 min with a final denaturation step of 65 °C for 20 min. Store un- phosphorylated MIPs at -20 °C for future use.
3.2 Targeted MIP Capture
-
1
Calculate the volume of the MIP megapool required in the capture reaction based on the ratio of desired MIP copies to DNA copies. This example will assume a megapool of 2000 MIPs captured using 100 ng of total genomic DNA, for a total ratio of 800 MIP copies to 1 DNA copy.
-
2
Calculate the expected number of MIP copies required given an input of 100 ng of genomic DNA, e.g., 800 × 33,000 haploid genome copies=2.64×107 MIP copies required.
-
3
Transform the number of MIP copies to picomoles (pmol) using Avogadro’s number (6.02 × 1023), e.g., (2.64 × 107/6.02×1023) (1×1012)=4.38×10−5 pmol.
Calculate the picomole per μL concentration of the MIP megapool:
e.g., 0.1 μL×100 μM/2000 MIPs = 0.005 μM (0.005×25 μL)/30 μL=0.004 pmol/μL
-
4
Calculate the volume of 1× MIP megapool required in the capture reaction (see Note 4): e.g., 4.38×10-5 pmol/0.004 pmol/μL = 0.011 μL per capture reaction.
-
5
Prepare a 15 μL capture reaction on ice by combining 2.5 μL of Ampligase 10× Reaction Buffer, 0.0032 μL of 0.006 mM dNTP mix, 0.32 μL Klentaq (10 U/μL), 0.01 μL of Ampligase (100 U/μL), 0.0105 μL of MIP megapool, and 12.16 μL of nuclease-free water. The total volume of DH2O can be scaled depending on your DNA concentration requirements and the volumes are based on processing 192 samples (see below).
-
6
Plate 10 μL of DNA into a 96-well plate format (10 μL at 10 ng/μL). A range of 100–200 ng total DNA can be used in the final capture reaction.
-
7
Add 15 μL of capture reaction to each individual DNA sample.
-
8
Seal with adhesive PCR film (see Note 5).
-
9
Using a thermocycler, incubate the reaction mix at 95 °C for 10 min and 60 °C for 22 h. Remove plates and immediately place on chilling blocks (see Note 6).
-
10
Exonuclease treatment.
Immediately following capture, prepare a Exonuclease clean-up master mix containing 0.5 μL of Exonuclease I, 0.5 μL of Exonuclease III, 0.2 μL Ampligase 10× Reaction Buffer and 0.8 μL nuclease-free water per sample.
Add 2.0 μL of Exonuclease clean-up mix to each 25 μL capture reaction (see Note 7).
Using a thermocycler, incubate the reaction at 37 °C for 45 min and 95 °C for 2 min. Cool reaction plates to 4 °C (see Note 5).
Samples may be stored at 4 °C for a short term until PCR, or -20 °C for longer periods.
-
11
Real-Time PCR.
Prepare a RT-PCR master mix by combining 12.5 μL of 2 × iProof High Fidelity Master Mix, 0.125 μL of 100 μM universal MIP barcode forward primer, 0.125 μL of 100× SYBR® Green I nucleic acid gel stain and 6.125 μL of nuclease-free water.
Add 18.75 μL of RT-PCR master mix to each well.
Add 1.25 μL of 10 μM individual barcode primers and 5 μL of exonuclease-treated MIP capture reaction to each individual well (seeNote 8).
Using an RT-PCR thermocycler, amplify the reaction until the reaction begins to plateau under the following conditions: 98 °C for 30 s, followed by 20–25 cycles of 98 °C for 10 s, 60 °C for 30 s, and 72 °C for 30 s (see Note 9).
-
12
Standard PCR.
Prepare a PCR master mix by combining 12.5 μL of 2 × iProof High Fidelity Master Mix, 0.125 μL of 100 μM universal MIP barcode forward primer, and 6.25 μL of nuclease-free water.
Add 18.75 μL of RT-PCR master mix to each well.
Add 1.25 μL of 10 μM individual barcode primers and 5 μL of exonuclease-treated MIP capture reaction into each individual well.
Using a PCR thermocycler, amplify the reaction under the following conditions: 98 °C for 30 s, followed by 20–25 (established in step 11 of the real-time PCR protocol) cycles of 98 °C for 10 s, 60 °C for 30 s, and 72 °C for 30 s with a final extension time of 72 °C for 2 min and 4 °C forever.
-
13
Product pooling, clean-up, and gel electrophoresis.
For each plate of DNA samples pool 5 μL of each PCR reaction into a 1.5 mL tube (5 μL ×96=480 μL) (see Note 3).
Determine the correct ratio of beads to pooled MIP library by using a bead titration (see Note 10).
Add 0.9 μL of Agencourt AMPure XP beads per 1 μL of pooled PCR reaction, e.g., (432 μL per 480 μL of pooled PCR reaction). Vortex the tube thoroughly and pulse spin down to remove the beads from within the cap (see Note 11).
Incubate the sample pool with the beads for 10 min at room temperature.
Place the tube on the DynaMag™-2 magnet, lift the cap and allow the beads to adhere to the side of the tube nearest the magnet for 5 min.
Slowly remove the supernatant using a pipette without disturbing the bead pellet. If the bead pellet is disturbed, pipette them back into the tube and wait a further 1–3 min for the beads to re-bind.
Wash the bead pellet by adding 1 mL of 70 % ethanol to fully immerse the beads while the tube is still attached to the magnet. Do not disturb the bead pellet and incubate for 30 s.
Remove the supernatant and repeat step (13 g).
Remove the supernatant completely from the tube, making sure that there is no ethanol left at the bottom of the tube without disturbing the bead pellet (see Note 12).
Allow the beads to dry for 5 min (see Note 13).
Remove the tube containing the beads from the magnet and add 100 μL of EB buffer; mix well by manually pipetting up and down at least ten times. Allow the beads to sit at room temperature for 1 min (see Note 14).
Transfer the tube back to the magnet and incubate for at least 1 min allowing the beads to separate from the EB buffer and adhere to the side of the tube.
Transfer the supernatant, which contains the cleaned MIP library, to a new 1.5 mL tube. Individual MIP libraries can be stored at 4 °C short term or −20 °C for longer periods.
Run the MIP library on a 2 % E-Gel® EX Gel by combining 2 μL of pooled MIP library with 18 μL of distilled water and loading 20 μL into the individual wells. Prepare a 100 bp DNA ladder by preparing a 1:1 ratio of E-Gel® Low Range Quantitative DNA Ladder with distilled water and load into the first or final wells in the gel (20 μL).
Run gel electrophoresis for 20 min using the E-Gel® EX Gel platform and confirm the presence of a 276 bp product (see Notes 15 and 16).
-
14
Massively parallel sequencing.
Quantitate and Pool MIP Libraries
-
(a)
Prepare individual pooled libraries for sequencing by normalizing each individual library against the concentration of the lowest library within the set pools.
-
(b)
Use the Qubit dsDNA High-Sensitivity assay kit to determine the concentration of each individually barcoded library [9, 10].
-
(c)
Combine each library at equal concentration and determine the final concentration of pooled MIP library as in step 14.b.
-
(d)
The size of the MIP megapool, the number of pooled samples, and the desired depth of coverage will determine the individual sequencing requirements. The following protocol uses the Illumina MiSeq platform to test and rebalance individual MIP libraries (see Note 17).
Denature and Dilute MIP libraries
-
(e)
Denature and dilute MIP libraries according to the Standard Normalization Methods described in the MiSeq Denature and Dilute Libraries Guide [11].
-
(f)
Prepare a fresh 0.2 N dilution of NaOH by combining 200 μL of stock 1 N NaOH and 800 μL of DH2O.
-
(g)
Dilute the MIP library to 2 nM; then add 5 μL of the library to 5 μL of 0.2 N NaOH.
-
(h)
Vortex the tube thoroughly and pulse spin down to remove the liquid from the lid. Incubate for 5 min at room temperature.
-
(i)
Prepare a 20 pmol denatured library by adding 990 μL of chilled HT1 Buffer to 10 μL of denatured MIP library.
-
(j)
Dilute the denatured 20 pmol MIP library according to desired MiSeq loading concentrations (6–20 pmol). 10 pmol is usually optimal for the majority of MIP libraries.
Loading the MiSeq Reagent Cartridge
-
(k)
Load the diluted MIP library (6–20 pmol) into the MiSeq reagent cartridge according to the MiSeq: Reagent Kit v3-Preparation Guide [12].
-
(l)
Prepare the forward, reverse, and index sequencing primers to a concentration of 10 μM and load into the MiSeq reagent cartridge, according to the MiSeq: Reagent Kit v3-Preparation Guide (see Note 18) [12].
-
(m)
Set up a sequencing run according to the MiSeq System User Guide [13].
-
15
Assessment of MIP performance.
Assess capture uniformity by plotting the depth of coverage for individually mapped MIPs.
Normalize read counts for each individual MIP by the total number of reads mapped.
Sort in descending order and plot the ranked uniformity of MIPs in Log10 scale.
Rebalance poor-performing MIPs by increasing the relative concentration of MIPs that are one order of magnitude lower in abundance (see Note 19) (Fig. 2).
Return to methods step 3.2 and set up the MIP capture using the rebalanced MIP megapool.
Fig. 2.
Capture uniformity for 2196 MIPs “pre” (blue ) and “post” (red ) rebalancing. MIPs that perform poorly (one order of magnitude lower in abundance) are rebalanced at a ratio of 50:1 (bad vs. good MIPs) and a substantial number of MIPs are rescued (green) upon rebalancing
Acknowledgments
We thank Bradley P. Coe for his critical review of the manuscript and Tonia Brown for assistance with the manuscript preparation. We thank Brian J. O’Roak, Beth Martin, Evan A. Boyle, and Joseph B. Hiatt for their overall contributions to developing the MIP protocol. S.C. is supported by a National Health and Medical Research Council (NHMRC) CJ Martin Biomedical Fellowship (#1073726). H.A.S. is supported, in part, by the NHGRI Interdisciplinary Training in Genome Science Grant (T32HG00035). E.E.E. is an investigator of the Howard Hughes Medical Institute. J.S. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Download the MIPgen design and analysis suite of tools from GitHub (https://github.com/shendurelab/MIPGEN). Use MIPgen to design MIPs across your regions of interest. Note that there are several other dependencies for running this software (e.g., SAMtools, BWA, Tabix) successfully in your local environment.
MIPs can be customized to target moderate- and high- complexity DNA targets ranging from 120 to 250 base pairs in size. Low complexity and high GC regions of the genome perform poorly in this assay due primarily to the reliance of the method on PCR amplification and Illumina sequencing. Select your MIPs to be synthesized based on the SVR scores, logistic scores and failure flags (see the MIPGEN README file that accompanies this software package).
For ease of handling, use an 8-channel pipette to pool 5 μL of 100 μM MIPs from each well in the 96-well plate. Each tube in the 8-cap strip represents a combined sum of 12 wells or MIPs (5 μL ×12 wells=60 μL) which can be pooled together to generate a 96 MIP pool containing a volume of 480 μL (60 μL × 8 strip tubes).
If the volume of MIP megapool is too small for manual pipetting, dilute the MIP megapool to a lower concentration e.g., 1:1000, so a higher volume can be added. Dilutions should be made fresh for each capture reaction.
During this step be sure to create an air-tight seal using the adhesive PCR seal. Use a 10 °C lid offset for each step of the reaction. Failure to perform this step thoroughly will cause the DNA to evaporate during the capture reaction.
Capture incubation times may be reduced depending on input DNA concentration. Minimum working DNA stocks should not be less than 100 ng total for the MIP capture reaction.
Before adding the exonuclease treatment, cool down the capture plates using cold blocks and prepare the reaction mix on ice. Dispense the exonuclease clean-up mix in equal volumes across a set of 8-Flat-Cap Strip Tubes. Use an 8-channel pipette to dispense 2 μL of exonuclease reaction mix into each capture reaction.
RT-PCR is performed using a universal forward primer (MIP_ universal_forward: AATGATACGGCGACCACCGAGATC TACACATACGAGATCCGTAATCGGGAAGCTGAAG) and an individual reverse primer (MIP_barcode_reverse: CAAG CAGAAGACGGCATACGAGATNNNNNNNNACAC GCACGATCCGACGGTAGTGT) containing a unique 8mer barcode sequence, which is used for subsequent pooling and sequencing.
DNA samples extracted and stored under different conditions will reach plateau at different points during PCR cycling. It is recommended that RT-PCR be performed on a small number of samples representative of each particular sample set so that the correct number of cycles can be established. It is common for a percentage of samples to reach plateau at different cycle points. Select the cycle in which the majority of samples are still within log linear phase before plateau. Once completed, standard PCR may subsequently be performed using the correct number of cycles per sample set.
Small contaminants and undesired PCR products are removed during the bead clean-up. However, the ratio of beads to PCR product may vary depending on the size of the MIP library. Here, we use a concentration of 0.9× beads to clean up the pooled MIP library. To determine the quantity of beads to use, perform a bead titration by cleaning up control libraries with varied ratios of beads to MIP library (e.g., 0.8×–1.4× beads) and evaluating by agarose gel electrophoresis. As Agencourt AMPure XP beads preferentially bind to larger DNA fragments, the desired MIP PCR product (276 bp) can be saved while removing other nonspecific PCR products.
Allow 100 % Agencourt AMPure XP beads to come to room temperature before beginning clean-up. Vortex thoroughly to resuspend the beads into the buffer and dissolve the bead pellet at the bottom of the tube.
Tap the magnet gently to consolidate the ethanol at the bottom of the tube and use a p10 pipette tip to remove any residual ethanol.
Exceeding 5 min drying time may result in a lower DNA yield.
Optimize the amount of elution buffer added to individual MIP libraries to achieve the desired concentration. Smaller MIP pools can typically be eluted in lower volumes.
The 276 bp MIP product is specifically based on capturing 162 bp of target sequence using targeting arm lengths of 40–45 bp and single-molecule tags of 5 bp.
A small amount of nonspecific product (150 bp) may still remain after bead clean-up; as long as the MIP library (276 bp) represents the predominant band, this should not impact further sequencing steps.
Paired-end 101 bp reads are sufficient to sequence individual MIP amplicons of 276 bp, capturing 162 bases with arm lengths of 40–45 bp, 5–8 bp single-molecule tags with enough overlap for read assembly. This can be modified according to the specifics of the individual sequencing library.
Sequencing is performed using forward primer: 5′ CATACGAGATCCGTAATCGGGAAGCTGAAG 3′, MIPseq reverse primer: 5′ ACACGCACGATCCGACGGTAGTGT 3′, and MIPseqindexprimer:5′ACACTACCGTCGGATCGTGCGT GT 3′.
Poor-performing MIPs can be recovered by “spiking” MIPs in increased relative concentrations, termed rebalancing. Separate MIPs that perform well at 1× concentration from MIPs that require rebalancing. Phosphorylate these MIP pools separately, then pool at a ratio 10:1, 50:1, and 100:1 (poor performers: good performers). MIPs that perform particularly poorly, for example, those that generate zero sequence reads, may not be recoverable and can affect the overall performance of the MIP pool. It is recommended to do a second test run of the rebalanced MIP pool before testing large sample numbers. Check for large proportions of off-target reads indicative of rare MIPs with high off-target capture. This can be avoided by checking output files from the MIPgen design files for MIPs that have over represented arm sequences.
Competing financial Interests
E.E.E. is on the scientific advisory board (SAB) of DNAnexus, Inc., and is a consultant for the Kunming University of Science and Technology (KUST) as part of the 1000 China Talent Program.
References
- 1.Mamanova L, Coffey AJ, Scott CE, et al. Target-enrichment strategies for next-generation sequencing. Nat Methods. 2010;7:111–118. doi: 10.1038/nmeth.1419. [DOI] [PubMed] [Google Scholar]
- 2.Hardenbol P, Baner J, Jain M, et al. Multiplexed genotyping with sequence-tagged molecular inversion probes. Nat Biotechnol. 2003;21:673–678. doi: 10.1038/nbt821. [DOI] [PubMed] [Google Scholar]
- 3.Porreca GJ, Zhang K, Li JB, et al. Multiplex amplification of large sets of human exons. Nat Methods. 2007;4:931–936. doi: 10.1038/nmeth1110. [DOI] [PubMed] [Google Scholar]
- 4.Turner EH, Lee C, Ng SB, et al. Massively parallel exon capture and library-free resequencing across 16 genomes. Nat Methods. 2009;6:315–316. doi: 10.1038/nmeth.f.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Roak BJ, Vives L, Fu W, et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science. 2012;338:1619–1622. doi: 10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyle EA, O’Roak BJ, Martin BK, et al. MIPgen: optimized modeling and design of molecular inversion probes for targeted resequencing. Bioinformatics. 2014;30:2670–2672. doi: 10.1093/bioinformatics/btu353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hiatt JB, Pritchard CC, Salipante SJ, et al. Single molecule molecular inversion probes for targeted, high-accuracy detection of low-frequency variation. Genome Res. 2013;23:843–854. doi: 10.1101/gr.147686.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O'Roak BJ, Stessman HA, Boyle EA, et al. Recurrent de novo mutations implicate novel genes underlying simplex autism risk. Nat Commun. 2014;5(5595):1–6. doi: 10.1038/ncomms6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qubit® Assays: Quick Reference Guide: Pub. no: MAN0010876
- 10.Qubit® 2.0 Fluorometer: MAN0003231
- 11.MiSeq: Denature and Dilute Libraries Guide: 15039740v1
- 12.MiSeq: Reagent Kit v3-Preparation Guide: Part#15044983
- 13.MiSeq System User Guide: part # 15027617