Abstract
The papillomavirus capsid is a nonenveloped icosahedral shell formed by the viral major structural protein, L1. It is known that disulfide bonds between neighboring L1 molecules help to stabilize the capsid. However, the kinetics of inter-L1 disulfide bond formation during particle morphogenesis have not previously been examined. We have recently described a system for producing high-titer papillomavirus-based gene transfer vectors (also known as pseudoviruses) in mammalian cells. Here we show that papillomavirus capsids produced using this system undergo a maturation process in which the formation of inter-L1 disulfide bonds drives condensation and stabilization of the capsid. Fully mature capsids exhibit improved regularity and resistance to proteolytic digestion. Although capsid maturation for other virus types has been reported to occur in seconds or minutes, papillomavirus capsid maturation requires overnight incubation. Maturation of the capsids of human papillomavirus types 16 and 18 proceeds through an ordered accumulation of dimeric and trimeric L1 species, whereas the capsid of bovine papillomavirus type 1 matures into more extensively cross-linked forms. The presence of encapsidated DNA or the minor capsid protein, L2, did not have major effects on the kinetics or extent of capsid maturation. Immature capsids and capsids formed from L1 mutants with impaired disulfide bond formation are infectious but physically fragile. Consequently, capsid maturation is essential for efficient purification of papillomavirus-based gene transfer vectors. Despite their obvious morphological differences, mature and immature capsids are similarly neutralizable by various L1- and L2-specific antibodies.
Papillomaviruses, which are etiologically implicated in the development of warts, cervical cancer, and other epithelial tumors, replicate in the stratified squamous epithelium of the skin or mucous membranes (reviewed in reference 18). The papillomavirus life cycle is closely tied to the epithelial differentiation program, which takes many days to complete. The eventual release of the nonenveloped virion is thought to depend primarily on the normal cellular disintegration typically seen near the surface of squamous epithelia.
The complexity of mimicking stratified epithelial differentiation in culture has made it challenging to study the papillomaviral life cycle in vitro. As a result, much of what is known about papillomavirus morphogenesis has been learned by studying recombinant versions of the two viral structural proteins, L1 and L2. The major capsid protein, L1, can spontaneously self-assemble into 72-pentamer virus-like particles (VLPs) that closely resemble native papillomavirus virions (20). The minor capsid protein, L2, is important for papillomavirus infectivity (38), but its arrangement in the virion and its role in encapsidation of the 8-kb double-stranded circular viral genomic DNA remain unclear.
We have recently described a system for generating high-titer papillomavirus-based gene transfer vectors (also known as pseudoviruses) by coexpression of L1 and L2 in mammalian cells cotransfected with various <8-kb reporter plasmids (2). The production system represents a tractable in vitro model in which papillomavirus capsid morphogenesis can be related to infectivity. Using this system, we have found that papillomavirus capsids expressed in mammalian cells undergo a morphological maturation process following cell lysis. Although maturation is a typical feature of viral capsids, it is generally a rapid process coordinated by encapsidation of viral nucleic acids, ejection of scaffolding proteins, and/or proteolytic cleavage of virion components. In contrast to the maturation of most other viral capsids, maturation of the papillomavirus capsid takes many hours to complete and appears to be driven primarily by the formation of intermolecular disulfide bonds. Although immature papillomavirus capsids are infectious, they are physically fragile. Incorporation of a maturation step is therefore essential for production of purified papillomavirus-based vector stocks.
MATERIALS AND METHODS
Plasmids.
Nucleotide maps of the plasmids used in this work are available at the website http://ccr.cancer.gov/Staff/links.asp?profileid=5637. Expression plasmids carrying codon-modified papillomavirus structural genes have been previously reported (2, 22, 26, 37). In Fig. 3 through 6, below, constructs encoding bovine papillomavirus type 1 (BPV1) L1 and L2 (pADAP-L1P and pZ-L2P) (2, 37), human papillomavirus type 16 (HPV16) L1 and L2 (p16L1h and p16L2h) (22), or HPV18 L1 and L2 (peL1BHB and peL2BHB) (26) were used. In each case, plasmids encoding the structural genes were cotransfected with a 5.9-kb target plasmid, pfwB, expressing enhanced green fluorescent protein (GFP; Clontech) under control of the human elongation factor 1 alpha promoter. pfwB also encodes the woodchuck hepatitis virus posttranscriptional regulatory element (10) to augment GFP expression. The backbone of pfwB was generated by recombination of plasmids pUno and pVac (Invivogen) with pSU5697 (2). In Fig. 1, 2, and 7, below, an L1 expression plasmid, p16L1-GFP, designed to express GFP under control of the simian virus 40 early promoter was used. Thus, in addition to expressing L1, p16L1-GFP can also serve as a target reporter plasmid suitable for packaging. HPV16 L1 constructs pC175S and pC428S were generated by PCR mutagenesis of p16L1-GFP. Production of neutralization assay stock was performed by cotransfecting 293TT cells with p16L1-GFP (or pC175S), p16L2h, and pYSEAP (26).
FIG. 3.
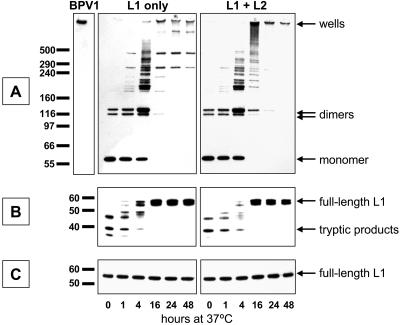
Western blots of transfected cell lysates. Cells transfected only with a plasmid expressing HPV16 L1 (left panels) or cotransfected with plasmids expressing both HPV16 L1 and L2 (right panels) were lysed in PBS supplemented with Brij 58. The lysate was then incubated for the time shown at the bottom of the figure. (A) The lysate was separated on a nonreducing sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel and then subjected to anti-L1 Western blotting. (B) The lysate was briefly digested with trypsin and then subjected to conventional reducing Western blotting to detect L1. (C) Untreated lysate was subjected to reducing Western blotting to detect L1.
FIG. 6.
Western blots of transfected cell lysates supplemented with GSSG. Cells were cotransfected with HPV16 L1 and L2 and then lysed with Brij 58. Immediately after harvest of the time zero sample, the lysate was spiked with 5 mM GSSG and then incubated for the times shown at the bottom of the figure. (A) Nonreducing Western blot of the samples; (B and C) reducing Western blots of trypsin-digested or untreated lysate, respectively.
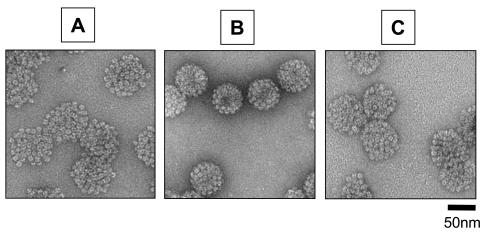
FIG. 1.
Electron micrographs of HPV16 capsids. (A and B) Lysates of cells transfected with HPV16 L1 and L2 expression vectors were incubated for 1 or 24 h, respectively, and then subjected to purification through Optiprep gradients. The samples were then examined by transmission EM. (C) The purified material used in panel A was incubated at 37°C for 24 h and then subjected to EM using the same grid preparation methods and magnification.
FIG. 2.
DNA content of Optiprep-purified HPV16 capsids. Nuclease-resistant DNA was extracted from Optiprep-fractionated capsids subjected to prior incubation at 37°C for the time shown. During the extraction, the samples were spiked with 0.5 ng of a 1-kb DNA marker fragment to monitor recovery. The purified DNA samples were digested with BglII, separated on an agarose gel, and stained with SYBR Green I. It was necessary to dilute the sample recovered from the material incubated for 24 h in order to compare it to the 1- and 4-h samples. In the chart at the bottom of the figure, the total titer of the starting crude cell lysate is compared to the titer recovered in the fractionated material. Markers (annotated in kilobase pairs) are shown in the far left lane.
FIG. 7.
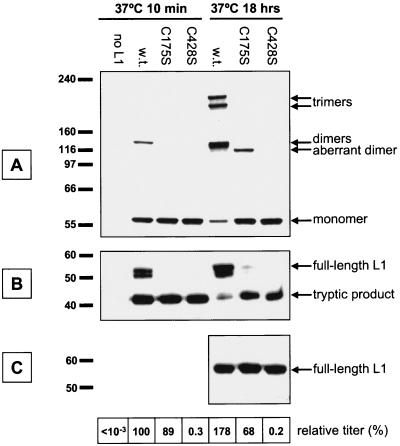
(A to C) Western blots of cells transfected with cysteine mutant L1 constructs. Cells were cotransfected with a construct expressing HPV16 L2 together with wild-type or cysteine mutant L1 constructs C175S or C428S. The cells were lysed and then incubated at 37°C for 10 min or 18 h. The chart at the bottom of the figure shows the titer of the lysates standardized to the titer found in the wild-type L1 lysate after 10 min of incubation at 37°C.
Cell culture and capsid production.
Production and purification of papillomavirus-based gene transfer vectors and VLPs using 293TT cells were accomplished using previously reported methods (2), with minor modifications. Briefly, 293TT cells were cotransfected (Lipofectamine 2000; Invitrogen) with codon-modified L1 and L2 expression plasmids and, in some instances, target plasmid pfwB. Two days after transfection, the cells were suspended at 108 cells per ml in Dulbecco's phosphate-buffered saline (PBS) with calcium and magnesium (DPBS; Invitrogen) supplemented with 9.5 mM MgCl2, 0.2% Brij 58 (Sigma), 0.2% Benzonase (Sigma), and 0.1% Plasmid Safe exonuclease (Epicentre). The resulting cell lysate was incubated at 37°C and then chilled, adjusted to 0.8 M NaCl, and clarified by centrifugation at 2,000 × g for 15 min at 4°C.
To purify capsids, the clarified cell lysates were ultracentrifuged through Optiprep gradients, as previously described (2). Core gradient fractions with appreciable amounts of L1 were pooled for analysis. Electron microscopic (EM) analysis of purified capsids was performed on nitrocellulose-carbon grids stained with 1% uranyl acetate using a Philips CM120 microscope at 33,000× magnification.
Titration of lysates and purified stocks was performed using 293TT cells as targets. A total of 2 × 105 cells were preplated several hours in advance in 0.5 ml per well in 24-well plates. The cells were treated with 1 μl of papillomaviral vector stock or stock diluted in DPBS-0.8 M NaCl and then incubated for 48 h. Expression of GFP was detected using a FACSCalibur device (Becton Dickinson), and the titer was calculated using the following formula: (2 × 105 cells) × (1,000 μl/ml) × (dilution factor) × (fraction of cells expressing GFP) = transducing units per milliliter.
Western blotting.
Because of the high apparent molecular weight of disulfide-bonded L1 species, nonreducing Western assays were performed using precast 3-to-8% Tris-acetate (sodium dodecyl sulfate-polyacrylamide gel electrophoresis) gels (Invitrogen), which allow for effective separation of large proteins. Cell lysates were prepared for nonreducing separation by 1:40 dilution into 1× Nupage load dye (Invitrogen) supplemented with 25 mM α-iodoacetamide (Calbiochem). The lysate was then denatured at 65°C for 10 min.
Reducing Western assays used 10% Nupage bis-Tris-morpholinepropanesulfonic acid gels (Invitrogen). Trypsin digestion was performed by mixing cell lysate samples with an equal volume of 0.25% trypsin stock (Invitrogen) followed by incubation at 37°C for 10 min. The digested lysate was then chilled and diluted 1:40 with 1× Nupage load dye supplemented with 5% 2-mercaptoethanol and 1× Complete Mini protease inhibitor cocktail (Roche). Trypsin-digested samples and untreated lysates diluted 1:40 in 1× Nupage load dye with 2-mercaptoethanol were denatured at 65°C for 10 min.
Both types of gel were transferred onto nitrocellulose membranes (Bio-Rad) using Nupage transfer buffer (Invitrogen) according to the manufacturers' instructions. Unstained protein standards (HiMark or BenchMark; Invitrogen) were detected by staining the blot with Ponceau S solution (Sigma). The blot was then blocked in Tris-buffered saline (Bio-Rad) with 0.1% Tween 20 (TBST) supplemented with 5% nonfat dry milk (Carnation). BPV1 blots were probed with MAB837 (Chemicon) diluted 1:2,000 in TBST. HPV16 and HPV18 blots were probed with Camvir-1 (Pharmingen) diluted 1:10,000 in TBST. Goat anti-mouse-horseradish peroxidase secondary antibody (Bio-Rad) was detected by chemiluminescence (Perkin-Elmer).
DNA analysis.
Nuclease-resistant DNA was extracted from Optiprep fractions using a Qiaquick PCR purification kit (QIAGEN). Prior to extraction, the sample was spiked with a purified 1-kb BspHI (NEB) fragment of pBluescript II KS(+) (Stratagene) in order to monitor the efficiency of DNA recovery. Nuclease-resistant DNA was extracted from cell lysates by phenol chloroform extraction and ethanol precipitation as previously described (2). Nicking of packaged plasmid DNA (see Fig. 5, below) was likely the result of incomplete nuclease inactivation, since plasmids extracted from Optiprep-fractionated capsids were predominantly supercoiled (data not shown) (2). A control sample of purified target plasmid DNA was nicked using the enzyme N.BstNBI (NEB). In some instances, plasmids were linearized by digestion with the restriction enzyme BglII (NEB). DNA samples were separated on 0.9% Tris-acetate-EDTA agarose gels and then stained with SYBR Green I (Sigma). A 1-kb Plus DNA ladder (Invitrogen) was used as a standard.
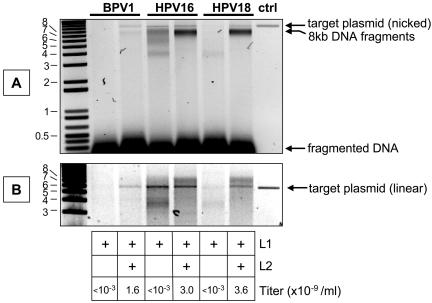
FIG. 5.
Nuclease-resistant DNA in transfected cell lysates. Cells transfected with L1 alone or cotransfected with L1 and L2 were lysed in the presence of a nuclease cocktail. DNA was then extracted from the lysate, separated on an agarose gel, and stained with SYBR Green I. (A) Untreated nuclease-resistant DNA samples; (B) the same samples digested with BglII. Purified target plasmid control is shown in the far right lanes.
Neutralization assays.
Neutralization assays (26) were performed using crude vector stocks diluted 1:10,000. Antibody H16.V5 was provided by Neil Christensen (5). Neutralizing sera from several vaccinees in a phase I clinical trial of HPV16 VLPs (14) or from HPV16-infected women (21) were selected based on moderate or high neutralizing activity (26). Serum from rabbits immunized with HPV16 L2 was provided by Richard Roden (Johns Hopkins Medical Institutions).
RESULTS
Stabilization of HPV16 capsids.
Production of papillomaviral vectors can be accomplished by cotransfection of codon-modified versions of the papillomavirus L1 and L2 genes, together with a reporter plasmid, into a 293 cell line (293TT) engineered to overexpress simian virus 40 T antigen. The assembly of L1 and L2 around cotransfected reporter plasmids is promiscuous and does not require DNA packaging sequences in cis or expression of any papillomavirus early genes in trans (2).
Although the production methods were originally developed for BPV1-based vectors, the methods can also be used to make crude HPV16 vector stocks with titers comparable to BPV1 stocks (2). Our initial report detailed methods for efficient single-step purification of BPV1 vectors by ultracentrifugation through an Optiprep velocity-density gradient. However, subsequent attempts to apply the purification method to HPV16- and HPV18-based vectors resulted in >95% losses of input titer (26). The titer of BPV1 vector stocks was also more resistant to other physical insults, such as sonication or repeated freeze-thaw cycles (data not shown).
Using resistance to freeze-thaw to assay stability, we examined a variety of harvest procedures to find a method that might stabilize the titer of HPV16 and HPV18 vector stocks. One method that successfully stabilized the titer of HPV16 and HPV18 vector stocks was lysis of vector-producing cells in PBS supplemented with the nonionic detergent Brij 58 and a nuclease cocktail, followed by overnight incubation of the lysate at 37°C.
EM analysis showed that HPV16 capsids stabilized by overnight incubation reproducibly exhibited decreased size and increased regularity compared to capsids purified without prior maturation (Fig. 1A and B). EM analysis of unpurified mature and immature capsids in clarified cell lysate revealed comparable morphological differences (data not shown), suggesting that the extremely open configuration of immature HPV16 capsids was not simply an artifact of the purification process. Interestingly, incubating purified immature HPV16 capsids at 37°C overnight did not effectively convert the capsids to a fully mature appearance (Fig. 1C). This failure to mature might be due to irreversible distortion of the immature capsid during purification. Alternatively, the poor maturation of purified HPV16 capsids could imply a need for components present in cell lysates, as has recently been described for in vitro assembly of polyomavirus capsids (6).
In addition to producing morphological changes in HPV16 capsids, overnight incubation of crude lysates dramatically improved titer recovery following Optiprep purification of HPV16 and HPV18 vector stocks (data not shown) (see the chart at the bottom of Fig. 2). Interestingly, incubation of the lysate at 37°C for 4 h was insufficient for stabilizing the titer of the HPV16 vector stock. The titer loss seen following purification of immature vector stocks correlated with a substantial loss of encapsidated reporter plasmid DNA (Fig. 2). This suggests that the titer loss following purification of immature stocks was due to evisceration of the immature particles during the Optiprep purification, perhaps due to the high shear forces and hydrostatic pressure associated with ultracentrifugation.
Capsid maturation is associated with disulfide bonding and trypsin resistance.
Intermolecular disulfide bonds are known to contribute to the stability of papillomavirus capsids (23, 30). To study the kinetics of inter-L1 disulfide bond formation during maturation, we performed nonreducing Western blotting on lysates of cells transfected with HPV16 L1 or cotransfected with L1 and L2. As seen in Fig. 3A, monomeric L1 predominated immediately after cell lysis. Incubation of the lysates at 37°C led to a progressive increase in multimeric L1 forms and a concomitant loss of L1 monomers. Accumulation of multimeric species required many hours of incubation and was not influenced by the coexpression of L2.
We interpret the bands with apparent molecular masses of roughly 195 and 215 kDa as distinct species of disulfide-linked L1 trimers. Close inspection of lightly exposed films revealed that the dimer band (apparent molecular mass of 125 kDa) is actually composed of a poorly resolved doublet seen at the 16-, 24-, and 48-h time points. The slower-migrating dimer and the faster-migrating trimer species both appear earlier in the maturation process, suggesting that disulfide bonding in the capsid occurs in an ordered sequence of events.
Previous reports have shown that treatment of assembled capsids with reducing agents results in the exposure of trypsin-sensitive sites in L1 (23). Conversely, it would be predicted that formation of inter-L1 disulfide bonds within the capsid might lead to increased resistance to trypsin digestion. Figure 3B shows that the increased inter-L1 disulfide bonding seen after prolonged incubation of the lysate at 37°C correlates with increased trypsin resistance. Incorporation of L2 in the capsids had little impact on the kinetics of acquisition of trypsin resistance. Analysis of cells transfected with HPV18 L1 or HPV18 L1 and L2 showed a nearly identical accumulation of L1 multimers and trypsin resistance (data not shown).
In contrast to HPV16 and HPV18, the maturation of BPV1 capsids progressed through a wide variety of L1 multimer species, ending in species with apparent molecular weights large enough to prevent exit from the wells of nonreducing gels (Fig. 4A). BPV1 virions extracted from infected bovine tissue (7) also failed to exit the wells. Despite the more extensive disulfide cross-linking of BPV1 L1, the acquisition of trypsin resistance followed kinetics similar to that with HPV16 (compare Fig. 3B and 4B). Coexpression of BPV1 L2 did not have a substantial impact on L1 disulfide bonding or trypsin resistance.
FIG. 4.
Western blots of transfected cell lysates. Panels A to C are arranged in the same way as in Fig. 3, except that the far left lane of panel A shows a sample of BPV1 virions purified from infected bovine tissue.
DNA content of BPV1, HPV16, and HPV18 capsids.
In a previous report involving purified HPV33 capsids, the presence of encapsidated DNA was found to correlate with increased L1 disulfide cross-linking and trypsin resistance (12). We have previously found that L2 expression is strictly required for encapsidation of DNA within BPV1 capsids (2). Since coexpression of L2 is not required for disulfide cross-linking or trypsin resistance of BPV1 L1 capsids (Fig. 4), it can be inferred that BPV1 capsid maturation is not significantly influenced by the presence of encapsidated DNA. As noted above, incorporation of HPV16 and HPV18 L2 did not appreciably influence capsid maturation. However, it is not known whether HPV16 or HPV18 L1 can encapsidate DNA in the absence of L2.
To determine whether L2 expression is required for DNA encapsidation in HPV16 and HPV18 capsids, we examined the nuclease-resistant DNA content of cells transfected with L1 alone or cotransfected with L1 and L2 of BPV1, HPV16, or HPV18. The results confirmed that, for BPV1, L2 coexpression is strictly required for DNA encapsidation (Fig. 5). In contrast, cells transfected with HPV16 L1 alone contained substantial amounts of nuclease-resistant DNA.
Although significant amounts of encapsidated target plasmid DNA were present in HPV16 L1-only and L1/L2 particles, the majority of the encapsidated DNA in both types of HPV16 capsid appeared to be comprised of linear DNA species (compare Fig. 5A and B). These linear DNA species presumably represent partially encapsidated loops of cellular DNA that were trimmed by the nucleases added to the lysis buffer. Cells cotransfected with HPV16 L1 and L2 yielded nuclease-resistant DNA fragments similar in size to the ∼8-kb genome of HPV16. In contrast, cells transfected with HPV16 L1 alone had a variety of smaller nuclease-resistant DNA species.
HPV18 showed a phenotype intermediate between those of BPV1 and HPV16 in that expression of HPV18 L2 was required for encapsidation of DNA, but the encapsidated DNA in the L1/L2-transfected cells was predominantly comprised of linear ∼8-kb cellular DNA fragments. In all cases, L2 was strictly required for transduction of target cells with GFP (chart at the bottom of Fig. 5). The observations show that encapsidated DNA is not required for capsid maturation.
Oxidation accelerates maturation.
The above experiments were performed using cells lysed at high density (100 million cells per ml) in detergent-supplemented PBS. To address the possibility that the slow kinetics of disulfide bond formation might be an artifact of the lysis conditions, we performed kinetic analyses of disulfide bond formation in lower-density lysates (10 million cells per ml), in lysates using various buffer systems (Dulbecco's modified Eagle's medium, HEPES-KCl, PBS [pH 5], PBS supplemented with a protease inhibitor cocktail, or 10 mM ATP), or in lysates produced using nondetergent cell disruption methods (sonication, freeze-thaw, or osmotic shock). In all cases we observed protracted intermolecular L1 disulfide bonding kinetics (data not shown). On the other hand, addition of the oxidizing agents flavin adenine dinucleotide (data not shown) or oxidized glutathione (GSSG) to the cell lysate dramatically accelerated disulfide bonding (Fig. 6A). GSSG-treated lysates also showed faster acquisition of L1 resistance to trypsin digestion (Fig. 6B). As seen in panel C of Fig. 6, addition of GSSG ultimately resulted in partial proteolytic degradation of L1 in the lysate. This could be due either to activation of cellular proteases in the lysate or to partial misfolding of the capsid during the accelerated maturation process.
Mutation of cysteine 175 or 428 prevents maturation.
Cysteines homologous to HPV16 L1 cysteines 175 and 428 have previously been shown to participate in intermolecular disulfide bonds within papillomavirus capsids (23, 29). We therefore mutated these cysteines to serine (mutants C175S and C428S) to observe the effect on capsid maturation and infectivity. After overnight maturation, the C175S mutant displayed low levels of a 115-kDa aberrant dimer species, whereas the C428S mutant remained almost exclusively monomeric (Fig. 7A). As seen in panel B of Fig. 7, mutation of cysteine 175 or 428 prevented the acquisition of trypsin resistance after overnight incubation, confirming that formation of disulfide bonds is essential for capsid maturation.
Surprisingly, crude vector stock made using the C175S mutant displayed nearly wild-type GFP transduction levels (chart at the bottom of Fig. 7). However, vector stocks generated using the C175S mutant were physically unstable and attempts to purify C175S mutant capsids yielded severely distorted noninfectious particles (data not shown).
Vector stocks made using the C428S mutant were also competent for transduction, but at levels of only 0.3% of the wild-type stock (Fig. 7). Despite its low transduction competence, the crude C428S vector preparation contained encapsidated DNA at levels nearly identical to that of stocks made using wild-type L1 (data not shown). Attempts to purify capsids from overnight-matured C428S stock were unsuccessful, as the L1 in the lysate failed to penetrate the gradient, presumably due to dissociation of the capsids into capsomers (23, 29). The results show that mutant HPV16 capsomers with deficient disulfide bonding form fragile, transduction-competent particles.
In BPV1 L1, cysteine 426 is homologous to HPV16 cysteine 428. Since BPV1 L1 showed evidence of more extensive disulfide bonding compared to HPV16 (Fig. 3 and 4), we constructed a BPV1 C426S mutant. Cells transfected with the BPV1 L1 C426S mutant exhibited a regular ladder of L1 multimers in nonreducing Western assays (data not shown). This result reinforces the notion that the BPV1 capsid can form additional inter-L1 disulfide bonds that do not involve cysteine 426.
Antibody neutralization of mature and immature vector stocks.
Papillomaviral vectors have significant utility as tools for detecting HPV-neutralizing antibodies in vaccinated and naturally infected individuals (26, 27). We therefore sought to determine whether mature and immature vector stocks could be similarly neutralized by various types of antibodies. In a panel of experiments involving the neutralizing monoclonal antibody H16.V5 (5), sera from VLP-vaccinated women (14), sera from naturally HPV16-infected women (21), or polyclonal anti-HPV16 L2 rabbit serum, we found essentially no difference in the 50% neutralization cutoffs for crude stocks of mature, immature, and C175S mutant HPV16 vectors (data not shown).
DISCUSSION
During their assembly, the capsids of many types of viruses have been shown to undergo maturation through an ordered process of conformational changes (reviewed in reference 1). In many instances, the maturation process is triggered by proteolytic processing of virion components (reviewed in references 3, 16, 17, and 34). In other cases, capsid maturation steps coincide with entry of the viral genome (9) or ejection of viral scaffolding proteins (reviewed in reference 8). In all cases where normal capsid maturation kinetics have been studied, the process has been thought to occur on a time scale of seconds or minutes. Our results show that, unlike many other types of viral capsids, papillomavirus capsids undergo an extremely slow maturation process that is driven by the formation of disulfide bonds between L1 molecules. Maturation of the capsid results in increased physical stability, increased resistance to tryptic proteolysis, and a decrease in particle size in electron micrographs.
It is possible that the relative slowness of papillomavirus capsid maturation is a reflection of the long overall time scale of the viral life cycle in vivo. Unlike conventionally lytic nonenveloped viruses, papillomaviruses are thought to be released by the gradual process of desquamation typically seen near the surface of the epithelium. Thus, the papillomavirus capsid is probably exposed to partially oxidizing conditions for several days prior to release into the environment. Despite their physical fragility, our data show that immature virions can be nearly as infectious as fully mature virions, leaving open the possibility that immature virions released from deeper layers of the epithelia could play a role in natural infection.
It has been shown that treatment of assembled papillomavirus capsids with reducing agents can cause either expansion of the capsid (23) or complete disassembly of the capsid into capsomers (30). However, the forward process of capsid maturation has not previously been examined in detail (36). It is possible that the maturation of papillomavirus capsids has been largely overlooked, because L1 is usually expressed using lytic systems, such as baculovirus, vaccinia virus, or Semliki Forest virus vectors (13, 15, 20, 39). In such systems, viral cytopathic effects probably induce an altered redox potential (31) in a fraction of the cells, resulting in a mixture of immature and mature capsids. Such mixed populations may also be typical of capsids isolated from papillomavirus-induced lesions, perhaps reflecting harvest of immature capsids derived from deeper layers of the lesion (11, 35). Our nonlytic mammalian expression system has allowed the synchronous study of capsid maturation beginning with a uniformly immature population of capsids.
Our results using nonreducing Western blotting confirm observations by other groups that cysteine 428 of HPV16 L1 (or its homolog in other HPV types) is critically involved in inter-L1 disulfide bond formation in the capsid (19, 23, 25, 29). However, our finding that the HPV16 capsid apparently contains two species of L1 trimer and two species of L1 dimer is inconsistent with models suggesting that only L1 cysteines 175 and 428 participate in intermolecular disulfide bonds (25, 29). The presence of multiple dimeric and trimeric species is potentially consistent with more recent data from Ishii and colleagues suggesting that intermolecular disulfide bonds involving cysteine 185 may also be present in the capsid (19). Since cysteines 175 and 185 are thought to share an external loop (4), it is possible that the quasi-atomic invading arm model of the papillomavirus capsid recently proposed by Modis and colleagues (25) could accommodate 185-428 disulfide bonds with only minor rearrangements. Thus, one simple explanation of our observations would be that 175-428 bonds are a prerequisite for formation of dimeric L1 species, while trimeric species involve a mixture of 175-428 and 185-428 bonds. To further examine the issue, we constructed a panel of L1 mutants targeting each cysteine in L1. The C185S mutant could not be evaluated because its steady-state expression levels were too low to be reliably detected by Western blotting. All of the other cysteine-to-serine mutants exhibited a normal pattern of dimers and trimers after maturation (unpublished observation). The observation confirms the theory that cysteines other than 175, 185, and 428 are dispensable for dimer and trimer formation but leaves open the question of precisely how cysteines 175, 185, and 428 might participate in disulfide bonding within the mature capsid.
The BPV1 capsid exhibits a higher degree of disulfide cross-linking than the HPV16 or HPV18 capsids. This difference in potential for cross-linking might explain why immature BPV1-based vectors are more robust than immature HPV16 and HPV18 vectors. We speculated that the higher degree of cross-linking in the BPV1 capsid might be an evolutionary adaptation to its environmentally exposed mode of transmission. To address the question, we examined the L1 disulfide cross-linking status of several cutaneous papillomavirus types for which we have recently constructed codon-modified expression plasmids (unpublished data). The mature capsids of cottontail rabbit papillomavirus and HPVs 1, 2, and 6 showed a pattern of L1 dimers and trimers nearly indistinguishable from the pattern seen for HPVs 16 and 18 (unpublished result). Thus, extensive disulfide cross-linking is not a typical feature of cutaneous papillomavirus types.
We also noted heterogeneity in the degree to which different papillomavirus capsids exclude cellular DNA. Nuclease-treated HPV16 and HPV18 L1/L2 vector stocks contained substantial amounts of encapsidated cellular DNA (Fig. 5), reminiscent of early reports of adventitious uptake of cellular DNA fragments by polyomaviruses (24, 33) and more recent reports of encapsidation of cellular DNA by HPV33 capsids (12). The presence of encapsidated cellular DNA fragments implies that, prior to cell lysis, papillomavirus procapsids can exist in a partially assembled state around loops of cellular DNA. BPV1-based vector stocks contained relatively little encapsidated cellular DNA, suggesting that comparable BPV1 procapsids either fail to exclude nucleases or are less abundant at steady state.
The role of the minor capsid protein, L2, in papillomavirus morphogenesis is poorly understood. Some previous reports have suggested that the L2s of various papillomavirus types are required for encapsidation of DNA (32, 38), but other reports have found substantial amounts of DNA within L1-only VLPs (12). In this report, we have shown that the discrepant results may stem in part from biological differences between different papillomavirus capsids. In our system, HPV16 L1 is competent for uptake of small DNA species when expressed in the absence of L2, whereas BPV1 and HPV18 L1-only particles contain very little encapsidated DNA. Interestingly, the presence of HPV16 L2 influences the average size of encapsidated cellular DNA loops, implying some role for L2 in DNA packaging, even for HPV16 capsids. Since coexpression of L2 had little effect on capsid maturation for BPV1 or HPV18, it is possible to conclude that encapsidated DNA is dispensable for capsid maturation.
We did not observe a major difference in the antibody neutralization profiles for mature and immature HPV16 vector stocks. The results suggest that immunodominant neutralizing epitopes on the virion surface are formed prior to maturation, consistent with the ability of disassembled capsomers to induce robust neutralizing antibody responses (28). The results imply that papillomavirus vaccines currently in development will have similar efficacy against mature and immature virions.
REFERENCES
- 1.Baker, T. S., N. H. Olson, and S. D. Fuller. 1999. Adding the third dimension to virus life cycles: three-dimensional reconstruction of icosahedral viruses from cryo-electron micrographs. Microbiol. Mol. Biol. Rev. 63:862-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buck, C. B., D. V. Pastrana, D. R. Lowy, and J. T. Schiller. 2004. Efficient intracellular assembly of papillomaviral vectors. J. Virol. 78:751-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bukrinskaya, A. G. 2004. HIV-1 assembly and maturation. Arch. Virol. 149:1067-1082. [DOI] [PubMed] [Google Scholar]
- 4.Chen, X. S., R. L. Garcea, I. Goldberg, G. Casini, and S. C. Harrison. 2000. Structure of small virus-like particles assembled from the L1 protein of human papillomavirus 16. Mol. Cell 5:557-567. [DOI] [PubMed] [Google Scholar]
- 5.Christensen, N. D., J. Dillner, C. Eklund, J. J. Carter, G. C. Wipf, C. A. Reed, N. M. Cladel, and D. A. Galloway. 1996. Surface conformational and linear epitopes on HPV-16 and HPV-18 L1 virus-like particles as defined by monoclonal antibodies. Virology 223:174-184. [DOI] [PubMed] [Google Scholar]
- 6.Chromy, L. R., J. M. Pipas, and R. L. Garcea. 2003. Chaperone-mediated in vitro assembly of polyomavirus capsids. Proc. Natl. Acad. Sci. USA 100:10477-10482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Day, P. M., D. R. Lowy, and J. T. Schiller. 2003. Papillomaviruses infect cells via a clathrin-dependent pathway. Virology 307:1-11. [DOI] [PubMed] [Google Scholar]
- 8.Dokland, T. 1999. Scaffolding proteins and their role in viral assembly. Cell Mol. Life Sci. 56:580-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dokland, T., and H. Murialdo. 1993. Structural transitions during maturation of bacteriophage lambda capsids. J. Mol. Biol. 233:682-694. [DOI] [PubMed] [Google Scholar]
- 10.Donello, J. E., J. E. Loeb, and T. J. Hope. 1998. Woodchuck hepatitis virus contains a tripartite posttranscriptional regulatory element. J. Virol. 72:5085-5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Favre, M., F. Breitburd, O. Croissant, and G. Orth. 1975. Structural polypeptides of rabbit, bovine, and human papillomaviruses. J. Virol. 15:1239-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fligge, C., F. Schafer, H. C. Selinka, C. Sapp, and M. Sapp. 2001. DNA-induced structural changes in the papillomavirus capsid. J. Virol. 75:7727-7731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hagensee, M. E., N. Yaegashi, and D. A. Galloway. 1993. Self-assembly of human papillomavirus type 1 capsids by expression of the L1 protein alone or by coexpression of the L1 and L2 capsid proteins. J. Virol. 67:315-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harro, C. D., Y. Y. Pang, R. B. Roden, A. Hildesheim, Z. Wang, M. J. Reynolds, T. C. Mast, R. Robinson, B. R. Murphy, R. A. Karron, J. Dillner, J. T. Schiller, and D. R. Lowy. 2001. Safety and immunogenicity trial in adult volunteers of a human papillomavirus 16 L1 virus-like particle vaccine. J. Natl. Cancer Inst. 93:284-292. [DOI] [PubMed] [Google Scholar]
- 15.Heino, P., J. Dillner, and S. Schwartz. 1995. Human papillomavirus type 16 capsid proteins produced from recombinant Semliki Forest virus assemble into virus-like particles. Virology 214:349-359. [DOI] [PubMed] [Google Scholar]
- 16.Hendrix, R. W., and R. L. Duda. 1998. Bacteriophage HK97 head assembly: a protein ballet. Adv. Virus Res. 50:235-288. [DOI] [PubMed] [Google Scholar]
- 17.Heymann, J. B., N. Cheng, W. W. Newcomb, B. L. Trus, J. C. Brown, and A. C. Steven. 2003. Dynamics of herpes simplex virus capsid maturation visualized by time-lapse cryo-electron microscopy. Nat. Struct. Biol. 10:334-341. [DOI] [PubMed] [Google Scholar]
- 18.Howley, P. M., and D. R. Lowy. 2001. Papillomaviruses and their replication, p. 2197-2229. In P. M. Howley and D. M. Knipe (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 19.Ishii, Y., K. Tanaka, and T. Kanda. 2003. Mutational analysis of human papillomavirus type 16 major capsid protein L1: the cysteines affecting the intermolecular bonding and structure of L1-capsids. Virology 308:128-136. [DOI] [PubMed] [Google Scholar]
- 20.Kirnbauer, R., F. Booy, N. Cheng, D. R. Lowy, and J. T. Schiller. 1992. Papillomavirus L1 major capsid protein self-assembles into virus-like particles that are highly immunogenic. Proc. Natl. Acad. Sci. USA 89:12180-12184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kjaer, S. K., A. J. van den Brule, J. E. Bock, P. A. Poll, G. Engholm, M. E. Sherman, J. M. Walboomers, and C. J. Meijer. 1996. Human papillomavirus—the most significant risk determinant of cervical intraepithelial neoplasia. Int. J. Cancer 65:601-606. [DOI] [PubMed] [Google Scholar]
- 22.Leder, C., J. A. Kleinschmidt, C. Wiethe, and M. Muller. 2001. Enhancement of capsid gene expression: preparing the human papillomavirus type 16 major structural gene L1 for DNA vaccination purposes. J. Virol. 75:9201-9209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, M., P. Beard, P. A. Estes, M. K. Lyon, and R. L. Garcea. 1998. Intercapsomeric disulfide bonds in papillomavirus assembly and disassembly. J. Virol. 72:2160-2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Michel, M. R., B. Hirt, and R. Weil. 1967. Mouse cellular DNA enclosed in polyoma viral capsids (pseudovirions). Proc. Natl. Acad. Sci. USA 58:1381-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Modis, Y., B. L. Trus, and S. C. Harrison. 2002. Atomic model of the papillomavirus capsid. EMBO J. 21:4754-4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pastrana, D. V., C. B. Buck, Y. Y. Pang, C. D. Thompson, P. E. Castle, P. C. FitzGerald, S. Kruger Kjaer, D. R. Lowy, and J. T. Schiller. 2004. Reactivity of human sera in a sensitive, high-throughput pseudovirus-based papillomavirus neutralization assay for HPV16 and HPV18. Virology 321:205-216. [DOI] [PubMed] [Google Scholar]
- 27.Rollman, E., L. Arnheim, B. Collier, D. Oberg, H. Hall, J. Klingstrom, J. Dillner, D. V. Pastrana, C. B. Buck, J. Hinkula, B. Wahren, and S. Schwartz. 2004. HPV-16 L1 genes with inactivated negative RNA elements induce potent immune responses. Virology 322:182-189. [DOI] [PubMed] [Google Scholar]
- 28.Rose, R. C., W. I. White, M. Li, J. A. Suzich, C. Lane, and R. L. Garcea. 1998. Human papillomavirus type 11 recombinant L1 capsomeres induce virus-neutralizing antibodies. J. Virol. 72:6151-6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sapp, M., C. Fligge, I. Petzak, J. R. Harris, and R. E. Streeck. 1998. Papillomavirus assembly requires trimerization of the major capsid protein by disulfides between two highly conserved cysteines. J. Virol. 72:6186-6189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sapp, M., C. Volpers, M. Muller, and R. E. Streeck. 1995. Organization of the major and minor capsid proteins in human papillomavirus type 33 virus-like particles. J. Gen. Virol. 76:2407-2412. [DOI] [PubMed] [Google Scholar]
- 31.Senkevich, T. G., C. L. White, E. V. Koonin, and B. Moss. 2002. Complete pathway for protein disulfide bond formation encoded by poxviruses. Proc. Natl. Acad. Sci. USA 99:6667-6672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stauffer, Y., K. Raj, K. Masternak, and P. Beard. 1998. Infectious human papillomavirus type 18 pseudovirions. J. Mol. Biol. 283:529-536. [DOI] [PubMed] [Google Scholar]
- 33.Trilling, D. M., and D. Axelrod. 1970. Encapsidation of free host DNA by simian virus 40: a simian virus 40 pseudovirus. Science 168:268-271. [DOI] [PubMed] [Google Scholar]
- 34.Weber, J. 1976. Genetic analysis of adenovirus type 2 III. Temperature sensitivity of processing viral proteins. J. Virol. 17:462-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yabe, Y., H. Sadakane, and H. Isono. 1979. Connection between capsomeres in human papilloma virus. Virology 96:547-552. [DOI] [PubMed] [Google Scholar]
- 36.Zhao, Q., S. Wu, W. Manger, and S. Gadam. August. 2002. Process for making human papillomavirus virus-like particles with improved properties. U.S. patent 6,436,402.
- 37.Zhou, J., W. J. Liu, S. W. Peng, X. Y. Sun, and I. Frazer. 1999. Papillomavirus capsid protein expression level depends on the match between codon usage and tRNA availability. J. Virol. 73:4972-4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou, J., D. J. Stenzel, X. Y. Sun, and I. H. Frazer. 1993. Synthesis and assembly of infectious bovine papillomavirus particles in vitro. J. Gen. Virol. 74:763-768. [DOI] [PubMed] [Google Scholar]
- 39.Zhou, J., X. Y. Sun, D. J. Stenzel, and I. H. Frazer. 1991. Expression of vaccinia recombinant HPV 16 L1 and L2 ORF proteins in epithelial cells is sufficient for assembly of HPV virion-like particles. Virology 185:251-257. [DOI] [PubMed] [Google Scholar]