Abstract
Most of the biological understanding of mechanisms underlying coronary artery disease (CAD) derives from studies of mouse models. The identification of multiple CAD loci and strong candidate genes in large human genome-wide association studies (GWAS) presented an opportunity to examine the relevance of mouse models for the human disease. We comprehensively reviewed the mouse literature, including 827 literature-derived genes, and compared it to human data. First, we observed striking concordance of risk factors for atherosclerosis in mice and humans. Second, there was highly significant overlap of mouse genes with human genes identified by GWAS. In particular, of the 46 genes with strong association signals in CAD-GWAS that were studied in mouse models all but one exhibited consistent effects on atherosclerosis-related phenotypes. Third, we compared 178 CAD-associated pathways derived from human GWAS with 263 from mouse studies and observed that over 50% were consistent between both species.
Introduction
Over the past 35–40 years, the mouse has become by far the most widely used animal model for atherosclerosis research. Mice and humans are diverged by about 80 million years and have undoubtedly been subjected to many different selective pressures. Nevertheless, their genomes have remained very similar, with about 95% sharing of their protein coding genes, although non-coding genes and regulatory regions are considerably less conserved. The latter may be of particular relevance to the topic since human variants affecting atherosclerosis risk do so most often by modulating gene regulation rather than protein structure (Nikpay et al., 2015; Samani et al., 2007; Seifert et al., 2016).
The purpose of this review is to comprehensively evaluate the consistency of findings by which gene alterations lead to atherosclerosis-related phenotypes in mice and humans. We briefly discuss the development of mouse models and summarize results from histological and genetic studies of atherosclerosis using mouse models. Altogether, we review a total of over 9,000 publications. We then compare the results in mice with what is currently known about factors affecting disease risk in humans. During the past decade, in particular, dramatic advances have been made in mapping and functionally characterizing loci contributing to common forms of CAD (Ghosh et al., 2015b; Makinen et al., 2014; McPherson and Tybjaerg-Hansen, 2016; Nikpay et al., 2015; Nurnberg et al., 2016; Samani et al., 2007; Seifert et al., 2016). These comparisons are performed on three levels. First, we examine the consistency between the mouse results and the various risk factors that have been identified in human epidemiologic and genetic studies. Second, we examine the overlap between findings in mouse and recent results from human genome-wide association studies (GWAS). Third, in addition to gene-based comparisons, we construct 263 mouse atherosclerosis pathways based on our literature search of genes and compared these to 178 known human CAD pathways based on GWAS. Each of these comparisons has limitations, which we discuss. Based on our findings, we make some recommendations about the most useful research strategies to move forward.
Background: Mouse models in atherosclerosis research
Historical perspective
Since the early 1900s, the mouse has been the classic animal model for genetic analysis. Research on the mouse as a model of atherosclerosis began in the late 1970s and early 1980s with efforts to characterize plasma lipoprotein metabolism and develop diets that promote atherosclerotic lesions development in mice (LeBoeuf et al., 1983; Lusis et al., 1983; Paigen et al., 1987; Reue et al., 1984; Roberts and Thompson, 1976). Mice have very low levels of atherogenic lipoproteins and unlike rabbits, hamsters and some other animal models, do not develop significant lesions when fed a western-type high fat, high cholesterol diet. However, it was found that the addition of cholic acid to such diets did result in aortic lesions that corresponded to early, fatty streak lesions in humans (Paigen et al., 1987; Roberts and Thompson, 1976). In the early 1990s mouse models exhibiting very high cholesterol levels and relatively advanced lesions were created through genetic engineering, including apolipoprotein E knockout (Apoe−/−) and LDL receptor knockout (Ldlr−/−) mice (Ishibashi et al., 1994; Li et al., 1993; Plump et al., 1992; Warden et al., 1993).
Two approaches were utilized to identify genetic factors contributing to atherosclerosis in the mouse. The first involved genetic analysis of natural variation in the mouse using quantitative trait locus (QTL) analysis, an analytic method for mapping genes underlying complex traits. Such studies revealed the considerable genetic complexity of atherosclerosis, but the poor resolution of QTL mapping made gene identification difficult (Rodriguez et al., 2013). The second approach involved the analysis of genes hypothesized to play a role in atherosclerosis based on known biology, known as “candidate genes”. In most such studies the expression of the candidate genes was modified through genetic engineering (for example, gene targeting), usually followed by analysis on a hyperlipidemic background to assess lesion development. Such studies have now been performed for many hundreds of genes (Stylianou et al., 2012). Below, we ask how the results of these studies fit with what is known about human atherosclerosis, particularly the results of human GWAS.
Pathology
In experimental models such as Apoe−/− or Ldlr−/− mice fed “Western” diets, the development of atherosclerotic lesions is similar to that observed in pathologic studies of the human disease. The first discernible event is the accumulation of aggregated apolipoprotein B-containing lipoprotein particles in the intimal space. This is followed by the recruitment of blood monocytes to the intima at sites of lipoprotein accumulation and subsequent transformation to macrophages. These cells then proceed to take up the lipoproteins through scavenger receptors or phagocytosis to give rise to cholesterol loaded “foam cells” that are the hallmark of early atherosclerosis. As lesions grow in size, a necrotic core comprised of foam cells, cholesterol crystals, and apoptotic cells forms, and a “fibrous cap” containing smooth muscle cells (migrated from media to intima) and extracellular matrix proteins such as collagen develops (Lusis, 2000). In addition to monocytes, T-lymphocytes are recruited to the lesions, and in certain genetic backgrounds, intimal and medial calcification occurs (Rattazzi et al., 2005).
There are, however, some distinct differences as compared to human atherosclerosis. While, as in humans, lesions in mice tend to occur in regions of disturbed blood flow, the primary sites of lesion development in mice are the aorta and carotids, not the coronary arteries (Hu et al., 2005). Also, while intimal thickening usually occurs in early lesions in humans, this is not the case in mice (Schwartz et al., 1995). Moreover, the mouse models rarely show evidence of lesion rupture, a particularly significant point since about three quarters of heart attacks result from plaque rupture followed by thrombosis. It should be noted that certain mouse models, notably the combined Srb-1−/−/Apoe−/− mouse, develop significant coronary lesions, although these have not been widely used in research, presumably because of the difficulty of breeding engineered alleles onto a double knockout background (Zhang et al., 2005). Also, lesions in certain vessels, such as the innominate artery that branches from the aorta, exhibit greater complexity than aortic lesions, including the presence of fibrin within lesions, although they do not appear to be a good model for studies of mechanisms of plaque rupture in humans (Schwartz et al., 2007).
Most studies in mice have evaluated only lesion sizes, not composition. It is clear that in mice the genetic background as well as environmental factors can influence the relative content in macrophages, smooth muscle cells, lymphocytes, calcified deposits, collagen, and necrotic debris (Bennett et al., 2006; Shaposhnik et al., 2007). Given its importance in plaque vulnerability, the quantitative evaluation of lesion composition is clearly of interest (Schwartz et al., 2000). On the other hand, many human studies often lack any pathological characterization of atherosclerotic lesions. Specifically, large-scale GWAS use often clinical definitions for atherosclerotic disease manifestations ranging from symptoms (e.g. angina pectoris) and findings in imaging studies (e.g. coronary angiography) to clinical manifestations (e.g. myocardial infarction, large artery stroke).
Natural genetic variation
The studies of natural variation in mice have focused on the overall genetic architecture of atherosclerosis and molecular signatures associated with lesion development (Hsu and Smith, 2013; Kayashima et al., 2015; Rodriguez et al., 2013; Welch, 2012). For example, a large cross between strains C3H/HeJ and C57BL/6J on an Apoe−/− background identified numerous loci significantly associated with atherosclerosis, vascular calcification, and traits such as obesity, insulin levels, and plasma lipids. Pathway analysis using Ingenuity revealed a number of pathways strongly associated with atherosclerosis such as cholesterol metabolism, mitochondrial oxidative phosphorylation and inflammation (Wang et al., 2007).
Because of the poor resolution of linkage analysis in mice (and humans), very few novel genes contributing to complex forms of atherosclerosis susceptibility were identified based on genetic crosses (Broeckel et al., 2002; Erdmann et al., 2013). With the development of technologies for high density genotyping, however, relatively high resolution association mapping of complex traits has become possible in mice as well as humans. Such association analyses can be performed using outbred mouse populations or using inbred mouse population following correction for population structure (Bennett et al., 2010; Valdar et al., 2006). An advantage of the use of inbred mice for such analyses is that many different phenotypes can be examined across the same genetic backgrounds (Bennett et al., 2015; Orozco et al., 2012; Parks et al., 2013).
Studies of natural variation in mice have revealed that atherosclerosis is a highly complex trait, involving many genes contributing to a variety of risk factors. A recent study of a large set of inbred strains known as the Hybrid Mouse Diversity Panel (HMDP) revealed strong evidence of association with inflammation, nitric oxide signaling and levels of trimethylamine-N-oxide (TMAO) as well as traditional risk factors such as plasma lipid levels. Moreover, the study provided strong evidence of gene-by-gene as well as gene-by-environment interactions (Bennett et al., 2015).
Engineered models and drug studies
To investigate mechanisms contributing to atherosclerosis susceptibility, hundreds of “candidate genes” have been examined for their effects on atherosclerosis in mice. Such candidate genes were chosen based on hypotheses generated from the understanding of human atherosclerosis and are thus inherently biased. When relating the results of such studies to human atherosclerosis genetics, a number of other caveats should be considered, including differences in effect size, the problem of “passenger genes” and some statistical considerations. Such confounding factors are discussed in a later section.
We comprehensively reviewed over 9000 Pubmed publications that examined genes contributing to atherosclerosis in mice. These included a total of 827 mouse atherosclerosis genes as defined by a significant effect on atherosclerotic lesion size or composition when perturbed in a mouse model. To our knowledge this represents the most comprehensive collection of mouse genes which have significant effects on atherosclerosis in mouse models. Citations for all included genes are provided in Table S1. The mouse genes were classified into three groups:
Engineered models of reduced gene expression (knockdown or knockout)
Engineered models of gene overexpression (primarily transgenic mice)
Models of drug treatment
Comparisons of mouse and human data on a gene level have to consider genetic differences. To assess overlap of genes between the species, the most recent ENSEMBL homologs were considered (Kersey et al., 2016). After species-specific adjustments, 19,583 protein coding human genes could be matched to 20,738 mouse genes. Thus, about 98.8% (19,583 of 19,815) of human genes are represented in the mouse genome and 94.5% (20,738 of 21,936) of mouse genes are represented in the human genome (Harrow et al., 2012; Mudge and Harrow, 2015).
Following the classification of mouse models, our research revealed a total of 827 mouse genes of which 535 (64.5%) were models of reduced gene expression, 48 (5.8%) were models of overexpressed genes, and 244 (29.5%) were models of drug treatment. Genes that showed significant alterations in lesion size (808 genes) or plaque stability (19) were considered as having an effect on atherosclerosis. Altogether 805 of our 827 literature derived mouse genes have a human ortholog (97.3%). These 805 mouse genes were matched to 797 unique genes in humans. Ten mouse genes (Ang, Ang2, Ang4, Ang5, Ang6, Bglap, Bglap2, Bglap3, Ces3a, Ces3b) were represented by 3 human genes (ANG, BGLAP, CES). Mice with deletion of Apoe (68.6% of studies) or Ldlr (27.2%) served most frequently as models for atherosclerosis studies. Thus, the vast majority of all genetic mouse models were explored on the background of excessive hypercholesterolemia, a condition that is similar to familial hypercholesterolemia in man. Of all 827 genes, 58.6% (485) of genes were described to mediate pro-atherogenic effects, 41.2% (341) anti-atherogenic effects, and Pecam1 is a gene affecting atherosclerosis in a site specific manner either positively or negatively.(Goel et al., 2008)
Using the gene enrichment tool DAVID we observed that the top disease categories corresponding to the 827 mouse genes were strongly associated with atherosclerosis. Among the top ten of human associated conditions, six are directly related to either CAD or atherosclerosis in humans and two represent known CAD risk factors. The two remaining conditions are Alzheimer’s disease and longevity (Table 1).
Table 1. Association of human-mouse orthologs with human disease databases.
Listed are the top 10 enriched DAVID terms associated with 827 literature derived mouse atherosclerosis genes. This table shows that genes involved in atherosclerosis in mice also account for CAD and related conditions in humans.
| Term | Fold Change | Mouse Genes | FDR* | P-Value** |
|---|---|---|---|---|
| Coronary artery disease | 4.5 | 53 | 2.70E-23 | 1.10E-27 |
| Ischemic heart disease | 4.4 | 32 | 1.20E-12 | 5.40E-17 |
| Restenosis | 4.3 | 47 | 2.90E-19 | 1.30E-23 |
| Longevity | 4 | 47 | 8.40E-17 | 4.90E-21 |
| Coronary atherosclerosis | 3.9 | 100 | 5.80E-39 | 3.60E-43 |
| Coronary heart disease | 3.9 | 35 | 1.40E-11 | 8.40E-16 |
| Alzheimer’s disease | 3.3 | 89 | 6.10E-12 | 3.30E-15 |
| Myocardial infarction | 3 | 62 | 5.30E-15 | 5.70E-19 |
| Hypertension | 2.6 | 79 | 5.40E-15 | 7.80E-19 |
| Diabetes, type 2 | 2.1 | 104 | 2.00E-13 | 3.00E-17 |
adjusted for multiple comparison,
calculated using Fisher’s exact test.
Risk factors: Concordance of mouse and human studies
Epidemiologic studies of human populations have identified a number of genetic or environmental “risk” factors, that are significantly associated with the prevalence of CAD (Table 2). Here we have comprehensively evaluated the consistency of findings in mice and humans in terms of the effects of these risk factors on atherosclerosis.
Table 2.
Human CAD risk factors and their effect on atherosclerosis in mouse models*
| Consistency of human CAD risk factors in atherosclerosis mouse models | ||
|---|---|---|
|
| ||
| Concordant risk factors | Effect on atherosclerosis | Reference |
| Hypercholesterolemia | ↑ | (Plump et al., 1992; van Ree et al., 1994) |
| Elevated lipoproteins levels | ||
| LDL | ↑ | (Huszar et al., 2000; Powell-Braxton et al., 1998) |
| VLDL | ↑ | (Knouff et al., 2004; VanderLaan et al., 2009) |
| HDL | ↓ | (Berard et al., 1997; Feig et al., 2014) |
| Lp(a) | ↑ | (Callow et al., 1995) (Schneider et al., 2005) (Pedersen et al., 2010) |
| Hypertriglyceridemia | ↑ | (Voyiaziakis et al., 1998) |
| Hypertension | ↑ | (Leong et al., 2015; Weiss et al., 2001; Wiesel et al., 1997) |
| Inflammatory diseases | ||
| Arthritis | ↑ | (Rose et al., 2013) |
| Lupus | ↑ | (Ma et al., 2008) |
| Psoriasis | ↑ | (Karbach et al., 2014) |
| Smoking | ↑ | (Boue et al., 2012; Gairola et al., 2001; Lietz et al., 2013) |
| Air pollution | ↑ | (Araujo, 2010; Soares et al., 2009; Sun et al., 2005) |
| T1D | ↑ | (In’t Veld, 2014; Kunjathoor et al., 1996; Shen and Bornfeldt, 2007) |
| T2D | ↑ | (Jun et al., 2011; King, 2012; Renard et al., 2004; Schreyer et al., 1998) |
| Aging | ↑ | (Merat et al., 2000; Rosenfeld et al., 2000) |
| Disstress | ↑ | (Kumari et al., 2003; Najafi et al., 2013; Roth et al., 2015) |
| TMAO | ↑ | (Gregory et al., 2015; Hartiala et al., 2014; Wang et al., 2011) |
| Thrombosis | ↑ | (Schafer et al., 2003) |
| Lack of physical activity | ↑ | (Meissner et al., 2011; Pellegrin et al., 2009) |
| Bacterial presence | ↑ | (Gibson et al., 2004; Lalla et al., 2003) |
| Renal failure | ↑ | (Bro et al., 2003; Hewitson et al., 2015; Neven and D’Haese, 2011) |
| Metabolic Syndrome | ↑ | (Kennedy et al., 2010) |
|
| ||
| Discordant risk factors | Effect on athero in Human/Mouse | Reference |
|
| ||
| Obesity | ↑/- (Lepob/ob-mice) | (Chiba et al., 2008; Nishina et al., 1994; Plummer and Hasty, 2008) |
| Gender | male ↑/female ↑ | (Venegas-Pino et al., 2016) |
↑ indicates that the factor is positively associated with atherosclerosis, ↓ indicates a negative association, - indicates no association with atherosclerosis
Dyslipidemia - high levels of plasma LDL-C, VLDL-C and triglycerides and low levels of HDL-C are strongly related with human coronary artery disease (Expert Panel on Detection and Treatment of High Blood Cholesterol in, 2001; Grundy et al., 2004). To mimic human hypercholesterolemia in mouse models, genetic and dietary adaptions were necessary, as circulating cholesterol is predominantly based on HDL in mice and LDL in humans. Also cholesteryl ester transfer protein (CETP), which transfers cholesteryl esters and triglycerides between lipoproteins, is not present in mice. Moreover, absorption of dietary cholesterol is low in mice. Summarizing results from several genetically modified mouse models that address lipid and lipoprotein metabolism, the data are most consistent for increased atherosclerosis in mice with hypercholesterolemia, hypertriglyceridemia and singular elevations of LDL or VLDL plasma levels (Huszar et al., 2000; Knouff et al., 2004; Plump et al., 1992; Powell-Braxton et al., 1998; van Ree et al., 1994; VanderLaan et al., 2009; Voyiaziakis et al., 1998). Elevated levels of HDL, on the other hand, reduced the progression of atherosclerosis (Berard et al., 1997), and in atherosclerotic lesions lacking calcification elevated levels of HDL even led to partial regression of lesion size (Feig et al., 2014).
High blood pressure is a well-known risk factor of human coronary artery disease (Stamler et al., 1993). Induction of high blood pressure in mouse models was initiated by excessive salt intake, reninangiotensin-aldosterone-system-upregulation, genetic perturbations or mechanistic constriction of renal arteries. Although the causes of hypertension in those models are different, acceleration of atherosclerosis was a consistent finding (Leong et al., 2015; Weiss et al., 2001; Wiesel et al., 1997).
Human systemic inflammatory disorders such as rheumatoid arthritis, lupus and psoriasis are strongly associated with human CAD, albeit the genetic underpinnings of the two disorders may be different (del Rincon et al., 2001; Jansen et al., 2015; Wang et al., 2012; Westerweel et al., 2007). In agreement, mouse models of human rheumatoid arthritis, lupus and psoriasis also showed increased atherosclerotic lesions in mouse models (Cao et al., 2013; Karbach et al., 2014; Ma et al., 2008; Rose et al., 2013).
Smoking accelerates the development of CAD in humans (Kannel et al., 1968). In mouse models exposure to mainstream cigarette smoke led to perturbations of lipid metabolism, arterial endothelial cell function and the development of atherosclerotic lesions (Boue et al., 2012). Cessation of smoke exposure led to decelerated plaque progression and decreased accumulation of pro-atherogenic lipids (Lietz et al., 2013).
Air pollution is a risk factor for accelerated human atherosclerosis (Araujo, 2010). Mouse models exposed to long term concentrated ambient particles smaller than 2.5μm (mimicking polluted air) showed acceleration of atherosclerosis and vascular inflammation (Sun et al., 2005).
Type 1 diabetes (T1D) promotes progression of human CAD (Schnell et al., 2013). T1D in mouse models was induced by streptozotocin or via viral infection. Both methods resulted in accelerated lesion formation (Shen and Bornfeldt, 2007).
Type 2 diabetes (T2D) is a risk factor of human CAD (Reaven, 2003). Most mouse T2D models are based on a leptin deficient model, fat or carbohydrate feeding or genetically induced dysfunction of beta cells, and most showed that T2D accelerates atherosclerosis (Jun et al., 2011; King, 2012; Renard et al., 2004; Schreyer et al., 1998). There were some contradictory results possibly due to the involvement of the immune system (Taleb et al., 2007) or atheroprotective effects of leptin deficiency itself (Lepob/ob) (Chiba et al., 2008).
Distress, acute and chronic, is a risk factor for development and progression of human CAD (Holmes et al., 2006). In a mouse model of chronic stress mice were repeatedly exposed to iced water. The animals developed more advanced and complex atherosclerotic lesions (Heidt et al., 2014; Najafi et al., 2013).
Trimethylamine-N-oxide (TMAO), derived from dietary choline or carnitine through the action of gut microbiota, has recently been shown to be a risk factor for CAD as well as heart failure (Koeth et al., 2013). Inbred strains of mice exhibit variations in TMAO levels when maintained on a Western diet and these are significantly associated with atherosclerosis (Bennett et al., 2015). Experimental mouse models revealed that dietary supplementation of choline (which can be processed to TMAO) or TMAO itself promoted atherosclerosis (Wang et al., 2011).
Thrombosis is significantly related to human coronary artery disease (Davies, 1996). An investigation of plasminogen activation inhibitor 1 (Pai1) in a mouse model, showed a pro-thrombotic phenotype with accelerated lesion formation (Schafer et al., 2003).
Lack of physical activity is a well-known risk factor of human CAD (Warburton et al., 2006). Studies in mouse models revealed decelerated development of atherosclerosis in mice subjected to exercise (Meissner et al., 2011).
Undue bacteremia is associated with CAD. Periodontal disease provides the most common entrance for such pathogens (Beck et al., 1996). Models of oral infection with periodontal pathogens showed accelerated atherosclerosis in mice (Lalla et al., 2003).
Renal failure accelerates the progression of human CAD, especially when the burden of calcification is high (Yerkey et al., 2004). Genetically engineered mouse models of renal failure (Fgf23−/− or Klotho−/) or mice in which induction was initiated with uremic toxins, such as indoxyl sulfate, exhibited accelerated atherosclerosis and highly calcified lesions (Neven and D’Haese, 2011).
Metabolic syndrome, characterized by insulin resistance, obesity, elevated blood pressure and dyslipidemia, is a risk factor for development of human coronary artery disease (Isomaa et al., 2001). A number of mouse models of metabolic syndrome exhibit accelerated lesion formation (Kennedy et al., 2010).
Obesity is an established risk factor for CAD in human. Obesity is also positively associated with atherosclerosis in mice on high fat, high cholesterol diet (Kennedy et al., 2010). Mice deficient for leptin (Lepob/ob) have a distinct obese phenotype but are protected from atherosclerosis (Chiba et al., 2008; Nishina et al., 1994; Plummer and Hasty, 2008). This effect may be related to increased size of lipoproteins in Lepob/ob obese mice, leading to reduced penetration of artery walls (Nishina et al., 1994) and the regulation of blood pressure (Huby et al., 2016). Deficiency of the leptin receptor (Lepdb/db), in contrast to leptin deficiency, led to increased plasma levels of leptin and pronounced atherosclerosis, suggesting that leptin itself promotes atherosclerosis (Wu et al., 2005).
Sex differences
In humans, men display earlier onset of CAD as compared to women. In mice, on the other hand, atherosclerosis is generally more prominent in females, although this depends on the mouse model and diet used. The increased susceptibility to atherosclerosis in female mice has been attributed in part to genes encoded by the sex chromosomes (Arnold et al., 2016; Link et al., 2015) and also to differences in the metabolism of TMAO, a potent risk factor for atherosclerosis (Bennett et al., 2013). TMAO is produced by oxidation of circulating trimethylamine, catalyzed by the enzyme flavin monooxygenase 3 (FMO3). In mice, FMO expression is dramatically suppressed by testosterone and, hence, TMAO levels are much lower in males as compared to females (Bennett et al., 2013; Kloss et al., 1982). In contrast, the levels of hepatic FMO3 expression are similar or only slightly lower in men as compared to women, and circulating TMAO levels are similar in males and females (Tang et al., 2013).
Human genome-wide association results: Overlap with mouse studies
Over the past decade GWAS have robustly mapped thousands of loci for various common diseases. Generally, the identified loci individually exhibit a very modest effect size and together they explain a small fraction of the genetic component (heritability) of disease, although some loci display allelic series ranging from common, genome-wide significantly associated alleles to rare mutations giving rise to mendelian traits, e.g. LDLR or GUCY1A3 deficiency (Erdmann et al., 2013; Goldstein and Brown, 2015). In the case of atherosclerosis, large meta-analyses have now identified a total of 57 loci that meet a Bonferroni-adjusted threshold level of significance and about 100 additional loci exhibit suggestive association (Consortium et al., 2013a; Consortium, 2011; McPherson and Tybjaerg-Hansen, 2016; Nikpay et al., 2015; Schunkert et al., 2011). In contrast to mendelian traits (in which about 85% of the causal variants affect coding sequences), common complex disease traits appear to be due primarily to variations in noncoding regions that affect gene expression (Nikpay et al., 2015; Seifert et al., 2016). In general, within the region of linkage disequilibrium (LD) the loci contain multiple genes. The causal gene may even lie outside the region of LD if it is regulated by an enhancer within the region associated with disease. Thus, a challenge at present is to identify the causal genetic variant(s) and the causal gene(s) at each locus. For our comparison with mice we have used two different human GWAS gene sets, described below (Table S2).
The first set of genes (244 GWAS Genes) is based on annotations of 169 known significant and suggestive GWAS loci, incorporating knowledge and data driven approaches (Braenne et al., 2015; Consortium et al., 2013b; Nikpay et al., 2015; Schunkert et al., 2011). This list of genes is a conservatively curated gene set, which is likely to provide the most reliable human CAD genes based on the GWAS approach.
The second set of genes (880 Extended GWAS Genes) is also based on GWAS loci (Consortium et al., 2013b; Nikpay et al., 2015) but additionally incorporates functional aspects using eQTLs and ENCODE annotations, such that additional genes not meeting a genome-wide level of significance (p < 5E-08) are incorporated. The extension of genes increases the probability of including false positive genes, but offers a better opportunity to infer biological mechanisms and pathways contributing to CAD (Zhao et al., 2016).
Experimental perturbation in mice
The 797 human orthologs (Table S2) of the mouse genes identified in engineered mouse models significantly overlapped with the human CAD gene sets described above. Thus, 45 (18.4%) of the CAD GWAS Genes (p=1.13E-17) and 72 (8.2%) of the Extended GWAS Genes (p=1.27E-08) overlapped with the list of 797 mouse orthologs. The number of genes that overlap is necessarily small because only a small fraction of human genes has been identified (as judged to the fraction of heritability explained) and some of these are likely to be false positives.
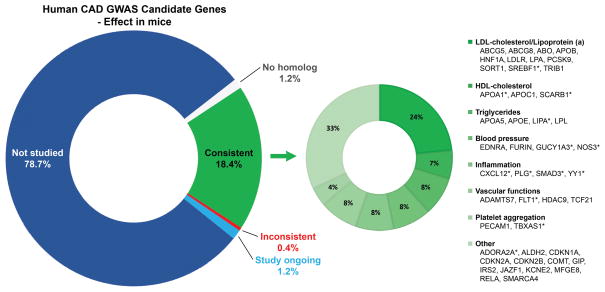
Perhaps a more meaningful analysis is to ask what fraction of human GWAS genes that have been studied in mouse models yield a consistent result. Since the human GWAS results were very recently published, only a fraction of the human candidates has been studied in mice. Moreover, we should note that a negative result in the mouse may very well not be published. Nevertheless, we found that of the 244 significantly or suggestive human GWAS genes, 46 have already been studied for their effect on lesion size in mice and 45 of these significantly affected atherosclerosis in mouse models (Fig. 1, Fig. 2), leaving around 190 human candidate genes that have not yet been studied (Fig. S1). Detailed information is provided in Table S2. Given that many of the human GWAS genes in our list are unlikely to be causal, such a high degree of overlap seems surprising. However, most of the overlapping genes were previously (before GWAS) considered to be strong candidates based on their roles in lipid metabolism, blood pressure, or inflammation, and were, therefore, studied in mice (Figure 1).
Figure 1. Human CAD GWAS candidate genes – focus on genes already validated in mice.
The pie chart (left) shows human CAD GWAS candidate genes grouped into those not yet studied in mice (blue), studied in experimental models with consistent effects (green) or inconsistent effects (red) and genes with no human homolog (white). The second pie chart shows the percentage of genes contributing to CAD associated traits or functions (right). Genes with pleiotropic effects are arranged according to their most dominant contribution.
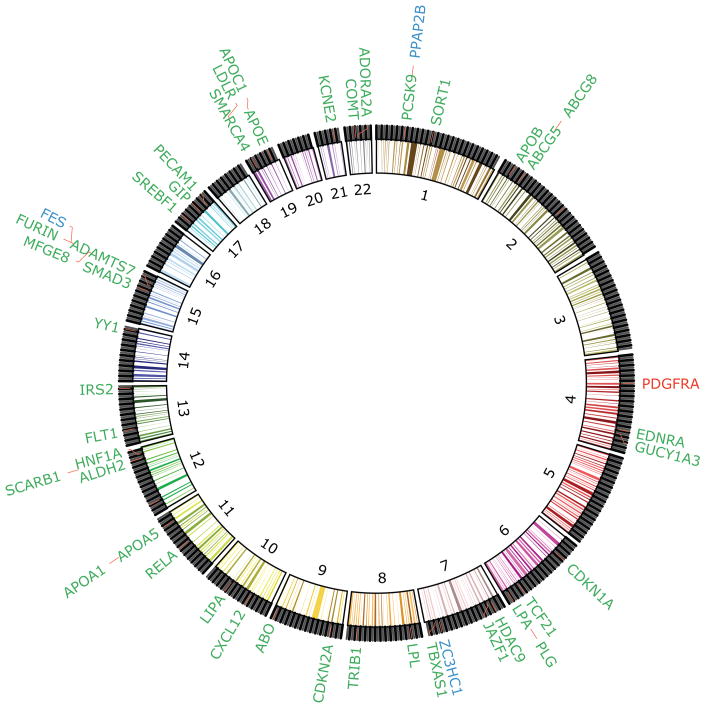
Figure 2. Human CAD GWAS candidate genes that have been tested in mouse animal models.
This circular plot shows a fraction of 244 human CAD GWAS candidate genes that have been tested in animal models. The numbers within the circle represent the 22 human autosome pairs. Candidate genes are arranged according to GWAS peak SNPS. Genes labeled green have already been studied in mouse models and show significant effects on atherosclerosis. Lipoprotein(a) (LPA), which has no ortholog in mice, was studied in a transgene mouse model. Red indicates that experimental perturbation of the gene in mice showed no significant effect on atherosclerosis. Genes labeled blue are the subject of ongoing research in mouse models.
Natural variation across inbred mouse strains
There are some potential problems with the use of gene knockouts in mice to model human atherosclerosis. Importantly, such studies are biased by the choice of the gene. Moreover, the effect may depend on the genetic background and most knockouts studied to date may be compromised by a “passenger gene” effect (see below) (Lusis et al., 2007). Finally, knockouts have a much larger effect size than the subtle quantitative or structural genetic variations of most GWAS genes. These issues with knockouts can be avoided by comparing human GWAS results with results from studies of natural variations in mice. The most comprehensive study of atherosclerosis among inbred strains of mice was performed in a set of about 100 highly diverse, commercially available inbred strains of mice known as the Hybrid Mouse Diversity Panel (HMDP)(Bennett et al., 2015). Altogether about 1000 mice were studied on the background of combined ApoE-Leiden and human CETP transgenes. Atherosclerosis susceptibility varied several hundred-fold among the strains (Bennett et al., 2015).
We first asked if the mouse orthologs of the human GWAS genes, exhibited significant correlation (p<0.05) with atherosclerosis across the HMDP. Of the 244 GWAS Genes, 66 (27.0%) exhibited significant correlation (in the form of expression), with lesion size in aorta. Additionally, expression levels of 27 (11.1%) genes in the liver were correlated significantly with increased lesion size in mouse aorta (Table 3). In addition to the CAD GWAS, there have been large GWAS studies of blood lipids, with altogether 274 candidate genes identified (Goodarzi, 2016; Welter et al., 2014). We tested whether the mouse orthologs of these genes exhibited significantly correlation (p<0.05) in expression in livers or adipose tissue with plasma lipids in mice. Overall 55.1% (151 of 274) of these genes exhibited significant correlation with plasma lipids in the HMDP (Table 3). Results of both comparisons are presented in Table S3. We conclude that the GWAS genes for both atherosclerosis and plasma lipids exhibit highly consistent variations when studied across different inbred strains of mice.
Table 3.
Correlation of gene sets with lesion size (top) and blood lipids (bottom) in HMDP mice**
| Aorta | P-value* | Liver | P-value* | |
|---|---|---|---|---|
| Mouse Atherosclerosis Genes (827) | 202 (24.4%) | 2.08E-109 | 156 (18.9%) | 3.67E-66 |
| CAD GWAS Genes (244) | 66 (27.0%) | 5.38E-36 | 27 (11.1%) | 2.54E-6 |
| CAD Extended GWAS Genes (880) | 152 (17.3%) | 6.17E-65 | 98 (11.1%) | 1.75E-25 |
| Adipose | P-value* | Liver | P-value* | |
|
| ||||
| LIPID GWAS Genes (274) | 116 (42.3%) | 6.21E-97 | 126 (46.0%) | 3.77E-111 |
calculated using Fisher’s exact test.
Given is the name and size of each gene set, the tissue in which the gene expression was measured and the number of genes significantly correlated to lesion size in aorta and liver and to lipids in adipose tissue and liver. Calculations were based on expression levels of mouse genes.
Pathway modeling: Concordance of human and mouse studies
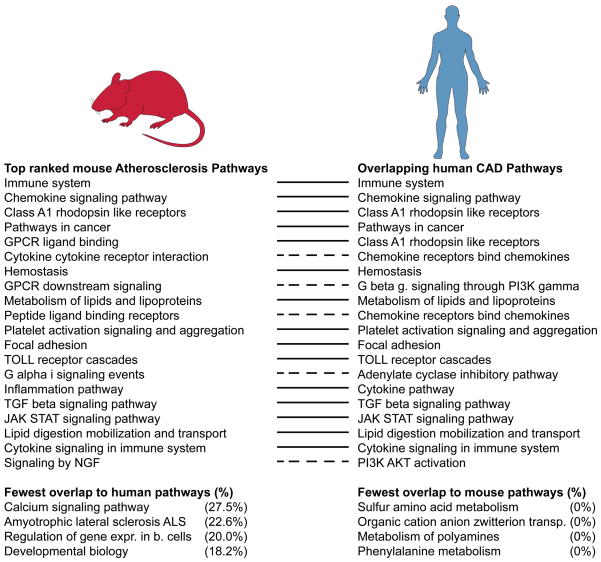
Three separate studies have modeled CAD pathways based on data of the Coronary Artery Disease Genome-Wide Replication and Meta-Analysis Consortium (CARDIoGRAM) (Ghosh et al., 2015b; Makinen et al., 2014; Zhao et al., 2016). Mäkinen et al. utilized a SNP set enrichment analysis (SSEA) that first mapped CARDIoGRAM GWAS to genes based on expression quantitative trait loci (eQTLs), and then assessed the enrichment of CAD signals in tissue-specific co-expression networks and canonical pathways from Biocarta, KEGG and Reactome databases. The study revealed 79 significant pathways for human CAD (Makinen et al., 2014). Gosh et al. used an alternative gene mapping strategy which was based on chromosomal distance between SNPs and genes and an alternative gene set enrichment analysis to identify 32 significant CAD pathways derived from the Reactome database (Ghosh et al., 2015a). Zhao et al. curated different candidate gene lists based on peak GWAS SNPs and implemented gene regulatory networks and expression patterns to identify 93 CAD relevant pathways based on canonical pathways from Biocarta, KEGG, and Reactome (Zhao et al., 2016). When combined, these three studies identified a total of 178 unique human CAD GWAS associated pathways (Table S4).
To examine the concordance of human and mouse pathways (Fig. 3), we modeled atherosclerosis pathways in mice using similar methods based on the set of 827 genes shown to contribute to atherosclerosis using experimental perturbation (see above). We applied knowledge-based canonical pathways from KEGG, Reactome, and Biocarta databases to define over-represented gene sets. We used Fisher Exact test and defined significance of over-representation as p<0.01 after Bonferroni correction. We focused on 263 pathways showing significant overrepresentation of atherosclerotic mouse genes. Pathways for lipid metabolism, immune system, toll-like receptor cascade, focal adhesion, hemostasis, platelet activation signaling and aggregation were highly overrepresented.
Figure 3. Top ranked mouse atherosclerosis pathways – overlap with human CAD pathways.
Listed are most significant (p-value) mouse atherosclerosis pathways. Identical and highly overlapping pathways between species are represented by continuous lines. Dashed lines indicate human pathways contributing with >90% of their genes to a mouse pathway. The least overlapping pathways of both species are shown below.
Of the 178 human CAD pathways 72 (40.2%) pathways were identical with our mouse pathways. A limiting factor is the different nomenclature of Biocarta, KEGG and Reactome makes a direct comparison impossible. To address this limitation, we analyzed the overlap of genes included in these pathways and considered those pathways highly significant that mutually shared over 50% of their genes or that included at least 90% of their genes in a pathway of the other species. Using these criteria, we observed that 55.1% (98 unique pathways) of human CAD associated pathways overlapped with atherosclerosis pathways of the mouse and 53.2% (140) of mouse pathways overlapped with human CAD GWAS associated pathways.
For pathways based on mouse data, only 5 overlapped less than 30% with human pathways. These mouse-specific pathways included diabetes, developmental biology and regulation of gene expression in beta cells. The last pathway, for example, consists of 20 genes and shares Akt1, Akt2, Akt3, Foxo1, Hnf4a, Hnf4g and insulin with known human CAD associated pathways, whereas the genes Foxa2, Foxa3, Gck, Iapp, Mafa, Neurod1, Nkx2-2, Nkx6-1, Pax6, Pdx1, Pklr and Slc2a2 were not present in the human pathways. Altogether, 37 human pathways showed less than 30% overlap. These included metabolism of polyamines, organic cation anion zwitterion transport, pantothenate and CoA biosynthesis, phenylalanine metabolism, sulfur amino acid metabolism and tRNA aminoacylation. Two unexpected pathways, the neuroactive ligand receptor interaction pathway and neurotrophin signaling pathway, exhibited around 50% overlap between mice and humans. Since most mouse models do not develop myocardial infarctions, it was of interest to examine whether the species differed in relevant pathways such as coagulation or collagen degradation. However, no such significant differences were observed. Overall, these findings highlight a high biological consistency between the species, as only 42 (11.4%) of 369 (human and mouse) unique atherosclerosis related pathways shared less than 30% of genes between species.
One way to visualize concordance of atherosclerosis associated pathways between human and mouse is to summarize them as clusters. We used a principal component analysis to reduce the high-dimensionality of the data to group the pathways according to the overlap of their constituent genes (Fig. S2). In this representation, the closer the pathways, the higher the percentage of shared genes. We divided clusters according to the functions of their genes, with the following categories: antigen, apoptosis, cancer, cell-cycle, focal adhesion, immune system, inflammation, lipid and lipoprotein metabolism, platelet function and proliferation and transcription.
Confounding Issues
In assessing the relevance of studies in mice for human CAD, there are a number of potential confounders that need to be considered. Below we briefly discuss the most significant.
Large versus small effect size genetic perturbations
Whereas common forms of CAD result from interactions of many different genetic and environmental factors, the majority of mouse studies are carried out using large genetic or environmental perturbations, such as knockout mice or extremely high cholesterol diets. Such extreme perturbations may affect many pathways and hundreds or thousands of genes, quite possible influencing atherosclerosis in kind of a bystander manner rather than directly. The genes identified in GWAS studies on the other hand, generally exhibit very modest functional variation.
The problem of passenger genes
Until the past several years, most gene targeted mice were generated on a strain 129/J genetic background and then transferred to the strain C57BL/6J by a series of backcrosses. While 5–10 generations of backcrossing will successfully eliminate the majority of genetic 129/5 contribution, regions immediately flanking the targeted gene will remain, since rare recombination events will be required to substitute C57BL/6J alleles. Thus, hundreds of 129/J genes including many with functional differences, will typically contaminate such studies. Since many different biologic pathways converge on atherosclerosis, the possibility of a passenger gene effect is significant. Similarly, transgenic mice are often generated on backgrounds other than C57BL/J (Lusis et al., 2007). Recent studies have examined the passenger gene issue in studies of immune functions and bone metabolism (Ackert-Bicknell and Rosen, 2015; Vanden Berghe et al., 2015).
The candidate gene approach is inherently biased
Candidate genes are generally selected for study based on what is known from human studies of atherosclerosis or from other biologic data. For example, some studies are directed at investigating mechanisms contributing to risk factors such as high blood pressure or hypercholesterolemia. Clearly, this could result in an artificial overlap of results in mouse-human comparisons. Studies of natural variations in mice, on the other hand, do not suffer from this bias and are more likely to reflect meaningful overlap.
Statistical considerations
Phenomena such as “winners curse”, the concept that the first report is likely to a have a larger effect size than subsequent studies, and “publication bias” the concept that the literature will be biased because positive results are more likely to be published than negative results, have undoubtedly skewed the results of studies of candidate genes. For example, beginning in the 1980’s with the development of methods for examining genetic variation at the DNA level, many studies of relatively small numbers of patients and controls (typically, hundreds) reported “significant” associations of DNA variants with common diseases. Few of the many hundreds of reported findings could be replicated in the much larger and unbiased GWAS studies (Altshuler et al., 2008). A similar situation may apply to investigations of candidate genes in mice using knockout or transgene technologies, particularly given the very large non-genetic variation of atherosclerosis lesion assays in mice (Daugherty and Rateri, 2012). Indeed there are numerous examples of the failure to replicate findings for such studies, such as those involving Abcg1, SRA and CD36 (Moore et al., 2005; Witztum, 2005).
Lesion analyses in mice and humans
Comparisons of different studies in mice are difficult given the lack of a standardized protocol. Variables between studies include the model, the method (en face, frozen sections, cholesterol content), the staining procedure (lipid, immune), the diet, the length of the study, the age of the mice, the vivarium conditions, and the number of mice studied. Because of the small size of mouse lesions, quantitative analyses of cell composition and gene expression are difficult (Erbilgin et al., 2013; Zhao et al., 2007). On the other hand, most human studies used clinical definitions for atherosclerosis.
Discussion
Most of our current understanding of the mechanisms underlying atherosclerosis derives from studies with mouse models. An important issue relates to the relevance of mouse models for human pathobiology, particularly since mouse data are being used to develop drug targets, and mouse models are also used to test therapeutic compounds in preclinical studies. The recent identification of human loci associated with CAD or plasma lipids in large GWAS studies (Consortium, 2011; Consortium et al., 2013b; Goodarzi, 2016; Nikpay et al., 2015; Samani et al., 2007; Schunkert et al., 2011; Welter et al., 2014) provided an opportunity to perform systematic gene comparisons between the two species. We first carried out a comprehensive survey of genes that have been significantly associated with atherosclerosis in mice using experimental perturbation, identifying a total of 827 genes that have been reported to influence lesion development. We also surveyed results from studies of natural genetic variation affecting atherosclerosis in mice. We then performed three different comparison analyses. The first addressed the consistency of atherosclerosis risk factors between the species. The second addressed the overlap of human CAD GWAS genes with results from mouse studies. And the third examined the overlap of 178 human CAD pathways and 263 mouse atherosclerosis pathways. We observed strong overlap in all three analyses. In particular, nearly all the human GWAS genes that have been studied in mice (45 of 46) show consistent effects on atherosclerosis. This includes lipoprotein(a), which was studied as a transgenic but does not have a homolog in mouse.
Our conclusions differ from a recent study by Pasterkamp and colleagues in which the investigators concluded that, with the exception of lipoprotein metabolism, mouse studies are likely to be of very limited value in understanding the human disease (Pasterkamp et al., 2016). These investigators performed a comprehensive survey of mouse studies of candidate genes (in total 659) and focused on the overlap with human GWAS genes. We confirmed about 75% of their mouse candidate genes in our literature search, but did not find clear evidence for the remaining 25% in the literature. This may have been caused by the selection of the human ortholog names, different search criteria and more stringent inclusion of genes in our study (for example, several of their genes exhibited only a suggestive effect on atherosclerosis in mice). However, in addition to the number and interpretation of mouse genes identified in the literature, there were further significant differences between the studies. First, our analysis included additional atherosclerosis risk factors while theirs focused only on lipids. Second, we used a more refined set of human genes based not only on distance to the peak SNP, but also functional information, particularly expression quantitative trait loci data. Using GWAS data we performed additional comparisons, such as examining the GWAS genes that have been tested in mouse models for consistency. Third, we did not restrict our comparison to genes from engineered mouse models, but included data from studies of naturally occurring variations in mice. This is perhaps the strongest line of evidence supporting the relevance of mouse models because it avoids some of the biases inherent in the other comparisons. Finally, the pathway databases used in the two studies are different: the previous study used Ingenuity, a compilation of broad literature data from multiple species and tissues, while we used the Biocarta, KEGG and Reactome databases. In contrast to the previous report, we conclude that the genes and pathways uncovered from the mouse atherosclerosis studies reflect highly overlapping biology with that revealed by human GWAS of coronary artery disease. This does not mean that there were no significant differences. For example, the most impactful human GWAS locus, on chromosome 9p21, is probably mediated by changes in the expression of a long noncoding RNA, ANRIL, that is not conserved in the mouse (Holdt et al., 2013). Also, while the conservation of disease pathways between mice and humans is highly significant, it is far from complete.
The relevance of animal models for common human disorders has been questioned, largely based on the failure of drug targets developed in animal models to show efficacy in human trials (Greek and Hansen, 2013; Mak et al., 2014; Perrin, 2014; Richmond and Su, 2008). One explanation of course is that there are significant differences between the species. Another plausible explanation is that there are several false positives in the mouse data, as discussed above. A third relates to experimental design. In particular, it is not clear that knockout mice are appropriate for modeling the subtle genetic variations that underlie common forms of atherosclerosis. Finally, most mouse studies were performed on a single genetic background (generally, strain C57BL/6J), which may severely limit the breadth of the conclusions that can be drawn (Gasch et al., 2016). Despite these limitations in mouse studies, our comparison between species revealed many converging pathways and supports the value of mouse models in revealing critical mechanistic processes of atherogenesis.
Focusing on the pathogenic pathways rather than individual genes is likely to be a more productive way to translate information from animal models to human studies. Of immediate interest is to further determine the factors regulating these pathways and to identify the tissues in which they contribute to atherosclerosis. Additionally, the convergent and divergent pathways between species revealed from our comparative review may help to guide the selection of the appropriate models in future preclinical studies. In addition to lesion progression, some studies have suggested an overlap of genes controlling lesion regression in mice and humans, suggesting a possible avenue for therapeutic investigation (Ramsey et al., 2014). Given the many advantages of mice for studying atherosclerosis, including access to tissues and the ability to control the environment, studies on mice will probably continue to lead the way to mechanistic understanding.
Supplementary Material
Acknowledgments
This work was supported by grants from the Fondation Leducq (CADgenomics: Understanding CAD Genes, 12CVD02), the German Federal Ministry of Education and Research (BMBF) within the framework or the e:Med research and funding concept (e:AtheroSysMed, grant 01ZX1313A-2014), the European Union Seventh Framework Program FP7/2007–2013 under grant agreement n° HEALTH-F2-2013-601456 (CVgenes-at-target), and from the DFG as part of the Sonderforschungsbereich CRC 1123 (B2).
Footnotes
Author contributions
MVS performed the literature search of mouse genes. All authors participated in the analyses of the data and critically reviewed the manuscript, written by MVS, HS, and AJL.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ackert-Bicknell CL, Rosen CJ. Passenger Gene Mutations: Unwanted Guests in Genetically Modified Mice. J Bone Miner Res. 2015 doi: 10.1002/jbmr.2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altshuler D, Daly MJ, Lander ES. Genetic mapping in human disease. Science. 2008;322:881–888. doi: 10.1126/science.1156409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araujo JA. Particulate air pollution, systemic oxidative stress, inflammation, and atherosclerosis. Air Qual Atmos Health. 2010;4:79–93. doi: 10.1007/s11869-010-0101-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold AP, Reue K, Eghbali M, Vilain E, Chen X, Ghahramani N, Itoh Y, Li J, Link JC, Ngun T, Williams-Burris SM. The importance of having two X chromosomes. Philos Trans R Soc Lond B Biol Sci. 2016:371. doi: 10.1098/rstb.2015.0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck J, Garcia R, Heiss G, Vokonas PS, Offenbacher S. Periodontal disease and cardiovascular disease. J Periodontol. 1996;67:1123–1137. doi: 10.1902/jop.1996.67.10s.1123. [DOI] [PubMed] [Google Scholar]
- Bennett BJ, Davis RC, Civelek M, Orozco L, Wu J, Qi H, Pan C, Packard RR, Eskin E, Yan M, Kirchgessner T, Wang Z, Li X, Gregory JC, Hazen SL, Gargalovic PS, Lusis AJ. Genetic Architecture of Atherosclerosis in Mice: A Systems Genetics Analysis of Common Inbred Strains. PLoS Genet. 2015;11:e1005711. doi: 10.1371/journal.pgen.1005711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett BJ, de Aguiar Vallim TQ, Wang Z, Shih DM, Meng Y, Gregory J, Allayee H, Lee R, Graham M, Crooke R, Edwards PA, Hazen SL, Lusis AJ. Trimethylamine-N-oxide, a metabolite associated with atherosclerosis, exhibits complex genetic and dietary regulation. Cell metabolism. 2013;17:49–60. doi: 10.1016/j.cmet.2012.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett BJ, Farber CR, Orozco L, Kang HM, Ghazalpour A, Siemers N, Neubauer M, Neuhaus I, Yordanova R, Guan B, Truong A, Yang WP, He A, Kayne P, Gargalovic P, Kirchgessner T, Pan C, Castellani LW, Kostem E, Furlotte N, Drake TA, Eskin E, Lusis AJ. A high-resolution association mapping panel for the dissection of complex traits in mice. Genome Res. 2010;20:281–290. doi: 10.1101/gr.099234.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett BJ, Scatena M, Kirk EA, Rattazzi M, Varon RM, Averill M, Schwartz SM, Giachelli CM, Rosenfeld ME. Osteoprotegerin inactivation accelerates advanced atherosclerotic lesion progression and calcification in older ApoE−/− mice. Arterioscler Thromb Vasc Biol. 2006;26:2117–2124. doi: 10.1161/01.ATV.0000236428.91125.e6. [DOI] [PubMed] [Google Scholar]
- Berard AM, Foger B, Remaley A, Shamburek R, Vaisman BL, Talley G, Paigen B, Hoyt RF, Jr, Marcovina S, Brewer HB, Jr, Santamarina-Fojo S. High plasma HDL concentrations associated with enhanced atherosclerosis in transgenic mice overexpressing lecithin-cholesteryl acyltransferase. Nat Med. 1997;3:744–749. doi: 10.1038/nm0797-744. [DOI] [PubMed] [Google Scholar]
- Boue S, Tarasov K, Janis M, Lebrun S, Hurme R, Schlage W, Lietz M, Vuillaume G, Ekroos K, Steffen Y, Peitsch MC, Laaksonen R, Hoeng J. Modulation of atherogenic lipidome by cigarette smoke in apolipoprotein E-deficient mice. Atherosclerosis. 2012;225:328–334. doi: 10.1016/j.atherosclerosis.2012.09.032. [DOI] [PubMed] [Google Scholar]
- Braenne I, Civelek M, Vilne B, Di Narzo A, Johnson AD, Zhao Y, Reiz B, Codoni V, Webb TR, Foroughi Asl H, Hamby SE, Zeng L, Tregouet DA, Hao K, Topol EJ, Schadt EE, Yang X, Samani NJ, Bjorkegren JL, Erdmann J, Schunkert H, Lusis AJ Leducq Consortium C.A.D.G.d. Prediction of Causal Candidate Genes in Coronary Artery Disease Loci. Arterioscler Thromb Vasc Biol. 2015;35:2207–2217. doi: 10.1161/ATVBAHA.115.306108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broeckel U, Hengstenberg C, Mayer B, Holmer S, Martin LJ, Comuzzie AG, Blangero J, Nurnberg P, Reis A, Riegger GA, Jacob HJ, Schunkert H. A comprehensive linkage analysis for myocardial infarction and its related risk factors. Nat Genet. 2002;30:210–214. doi: 10.1038/ng827. [DOI] [PubMed] [Google Scholar]
- Cao LY, Soler DC, Debanne SM, Grozdev I, Rodriguez ME, Feig RL, Carman TL, Gilkeson RC, Orringer CE, Kern EF, McCormick TS, Cooper KD, Korman NJ. Psoriasis and cardiovascular risk factors: increased serum myeloperoxidase and corresponding immunocellular overexpression by Cd11b(+) CD68(+) macrophages in skin lesions. Am J Transl Res. 2013;6:16–27. [PMC free article] [PubMed] [Google Scholar]
- Chiba T, Shinozaki S, Nakazawa T, Kawakami A, Ai M, Kaneko E, Kitagawa M, Kondo K, Chait A, Shimokado K. Leptin deficiency suppresses progression of atherosclerosis in apoE-deficient mice. Atherosclerosis. 2008;196:68–75. doi: 10.1016/j.atherosclerosis.2007.01.040. [DOI] [PubMed] [Google Scholar]
- Consortium CAD; Deloukas P, Kanoni S, Willenborg C, Farrall M, Assimes TL, Thompson JR, Ingelsson E, Saleheen D, Erdmann J, Goldstein BA, Stirrups K, Konig IR, Cazier JB, Johansson A, Hall AS, Lee JY, Willer CJ, Chambers JC, Esko T, Folkersen L, Goel A, Grundberg E, Havulinna AS, Ho WK, Hopewell JC, Eriksson N, Kleber ME, Kristiansson K, Lundmark P, Lyytikainen LP, Rafelt S, Shungin D, Strawbridge RJ, Thorleifsson G, Tikkanen E, Van Zuydam N, Voight BF, Waite LL, Zhang W, Ziegler A, Absher D, Altshuler D, Balmforth AJ, Barroso I, Braund PS, Burgdorf C, Claudi-Boehm S, Cox D, Dimitriou M, Do R, Doney AS, El Mokhtari N, Eriksson P, Fischer K, Fontanillas P, Franco-Cereceda A, Gigante B, Groop L, Gustafsson S, Hager J, Hallmans G, Han BG, Hunt SE, Kang HM, Illig T, Kessler T, Knowles JW, Kolovou G, Kuusisto J, Langenberg C, Langford C, Leander K, Lokki ML, Lundmark A, McCarthy MI, Meisinger C, Melander O, Mihailov E, Maouche S, Morris AD, Muller-Nurasyid M, Mu TC, Nikus K, Peden JF, Rayner NW, Rasheed A, Rosinger S, Rubin D, Rumpf MP, Schafer A, Sivananthan M, Song C, Stewart AF, Tan ST, Thorgeirsson G, van der Schoot CE, Wagner PJ, Wells GA, Wild PS, Yang TP, Amouyel P, Arveiler D, Basart H, Boehnke M, Boerwinkle E, Brambilla P, Cambien F, Cupples AL, de Faire U, Dehghan A, Diemert P, Epstein SE, Evans A, Ferrario MM, Ferrieres J, Gauguier D, Go AS, Goodall AH, Gudnason V, Hazen SL, Holm H, Iribarren C, Jang Y, Kahonen M, Kee F, Kim HS, Klopp N, Koenig W, Kratzer W, Kuulasmaa K, Laakso M, Laaksonen R, Lee JY, Lind L, Ouwehand WH, Parish S, Park JE, Pedersen NL, Peters A, Quertermous T, Rader DJ, Salomaa V, Schadt E, Shah SH, Sinisalo J, Stark K, Stefansson K, Tregouet DA, Virtamo J, Wallentin L, Wareham N, Zimmermann ME, Nieminen MS, Hengstenberg C, Sandhu MS, Pastinen T, Syvanen AC, Hovingh GK, Dedoussis G, Franks PW, Lehtimaki T, Metspalu A, Zalloua PA, Siegbahn A, Schreiber S, Ripatti S, Blankenberg SS, Perola M, Clarke R, Boehm BO, O’Donnell C, Reilly MP, Marz W, Collins R, Kathiresan S, Hamsten A, Kooner JS, Thorsteinsdottir U, Danesh J, Palmer CN, Roberts R, Watkins H, Schunkert H, Samani NJ Consortium D, Consortium C, Wellcome Trust Case Control, C. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet. 2013a;45:25–33. doi: 10.1038/ng.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium CADG. A genome-wide association study in Europeans and South Asians identifies five new loci for coronary artery disease. Nat Genet. 2011;43:339–344. doi: 10.1038/ng.782. [DOI] [PubMed] [Google Scholar]
- Consortium CD; Deloukas P, Kanoni S, Willenborg C, Farrall M, Assimes TL, Thompson JR, Ingelsson E, Saleheen D, Erdmann J, Goldstein BA, Stirrups K, Konig IR, Cazier JB, Johansson A, Hall AS, Lee JY, Willer CJ, Chambers JC, Esko T, Folkersen L, Goel A, Grundberg E, Havulinna AS, Ho WK, Hopewell JC, Eriksson N, Kleber ME, Kristiansson K, Lundmark P, Lyytikainen LP, Rafelt S, Shungin D, Strawbridge RJ, Thorleifsson G, Tikkanen E, Van Zuydam N, Voight BF, Waite LL, Zhang W, Ziegler A, Absher D, Altshuler D, Balmforth AJ, Barroso I, Braund PS, Burgdorf C, Claudi-Boehm S, Cox D, Dimitriou M, Do R, Doney AS, El Mokhtari N, Eriksson P, Fischer K, Fontanillas P, Franco-Cereceda A, Gigante B, Groop L, Gustafsson S, Hager J, Hallmans G, Han BG, Hunt SE, Kang HM, Illig T, Kessler T, Knowles JW, Kolovou G, Kuusisto J, Langenberg C, Langford C, Leander K, Lokki ML, Lundmark A, McCarthy MI, Meisinger C, Melander O, Mihailov E, Maouche S, Morris AD, Muller-Nurasyid M, Mu TC, Nikus K, Peden JF, Rayner NW, Rasheed A, Rosinger S, Rubin D, Rumpf MP, Schafer A, Sivananthan M, Song C, Stewart AF, Tan ST, Thorgeirsson G, van der Schoot CE, Wagner PJ, Wells GA, Wild PS, Yang TP, Amouyel P, Arveiler D, Basart H, Boehnke M, Boerwinkle E, Brambilla P, Cambien F, Cupples AL, de Faire U, Dehghan A, Diemert P, Epstein SE, Evans A, Ferrario MM, Ferrieres J, Gauguier D, Go AS, Goodall AH, Gudnason V, Hazen SL, Holm H, Iribarren C, Jang Y, Kahonen M, Kee F, Kim HS, Klopp N, Koenig W, Kratzer W, Kuulasmaa K, Laakso M, Laaksonen R, Lee JY, Lind L, Ouwehand WH, Parish S, Park JE, Pedersen NL, Peters A, Quertermous T, Rader DJ, Salomaa V, Schadt E, Shah SH, Sinisalo J, Stark K, Stefansson K, Tregouet DA, Virtamo J, Wallentin L, Wareham N, Zimmermann ME, Nieminen MS, Hengstenberg C, Sandhu MS, Pastinen T, Syvanen AC, Hovingh GK, Dedoussis G, Franks PW, Lehtimaki T, Metspalu A, Zalloua PA, Siegbahn A, Schreiber S, Ripatti S, Blankenberg SS, Perola M, Clarke R, Boehm BO, O’Donnell C, Reilly MP, Marz W, Collins R, Kathiresan S, Hamsten A, Kooner JS, Thorsteinsdottir U, Danesh J, Palmer CN, Roberts R, Watkins H, Schunkert H, Samani NJ Consortium D, Consortium C, Wellcome Trust Case Control, C. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet. 2013b;45:25–33. doi: 10.1038/ng.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugherty A, Rateri DL. Do vivarium conditions influence atherosclerotic lesion size? Arterioscler Thromb Vasc Biol. 2012;32:2339–2340. doi: 10.1161/ATVBAHA.112.300117. [DOI] [PubMed] [Google Scholar]
- Davies MJ. The contribution of thrombosis to the clinical expression of coronary atherosclerosis. Thromb Res. 1996;82:1–32. doi: 10.1016/0049-3848(96)00035-7. [DOI] [PubMed] [Google Scholar]
- del Rincon ID, Williams K, Stern MP, Freeman GL, Escalante A. High incidence of cardiovascular events in a rheumatoid arthritis cohort not explained by traditional cardiac risk factors. Arthritis Rheum. 2001;44:2737–2745. doi: 10.1002/1529-0131(200112)44:12<2737::AID-ART460>3.0.CO;2-%23. [DOI] [PubMed] [Google Scholar]
- Erbilgin A, Siemers N, Kayne P, Yang WP, Berliner J, Lusis AJ. Gene expression analyses of mouse aortic endothelium in response to atherogenic stimuli. Arterioscler Thromb Vasc Biol. 2013;33:2509–2517. doi: 10.1161/ATVBAHA.113.301989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann J, Stark K, Esslinger UB, Rumpf PM, Koesling D, de Wit C, Kaiser FJ, Braunholz D, Medack A, Fischer M, Zimmermann ME, Tennstedt S, Graf E, Eck S, Aherrahrou Z, Nahrstaedt J, Willenborg C, Bruse P, Braenne I, Nothen MM, Hofmann P, Braund PS, Mergia E, Reinhard W, Burgdorf C, Schreiber S, Balmforth AJ, Hall AS, Bertram L, Steinhagen-Thiessen E, Li SC, Marz W, Reilly M, Kathiresan S, McPherson R, Walter U, Ott J, Samani NJ, Strom TM, Meitinger T, Hengstenberg C, Schunkert H CardioGram. Dysfunctional nitric oxide signalling increases risk of myocardial infarction. Nature. 2013;504:432–436. doi: 10.1038/nature12722. [DOI] [PubMed] [Google Scholar]
- Expert Panel on Detection, E., and Treatment of High Blood Cholesterol in, A. Executive Summary of The Third Report of The National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, And Treatment of High Blood Cholesterol In Adults (Adult Treatment Panel III) JAMA. 2001;285:2486–2497. doi: 10.1001/jama.285.19.2486. [DOI] [PubMed] [Google Scholar]
- Feig JE, Hewing B, Smith JD, Hazen SL, Fisher EA. High-density lipoprotein and atherosclerosis regression: evidence from preclinical and clinical studies. Circ Res. 2014;114:205–213. doi: 10.1161/CIRCRESAHA.114.300760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP, Payseur BA, Pool JE. The Power of Natural Variation for Model Organism Biology. Trends Genet. 2016;32:147–154. doi: 10.1016/j.tig.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Vivar J, Nelson CP, Willenborg C, Segre AV, Makinen VP, Nikpay M, Erdmann J, Blankenberg S, O’Donnell C, Marz W, Laaksonen R, Stewart AF, Epstein SE, Shah SH, Granger CB, Hazen SL, Kathiresan S, Reilly MP, Yang X, Quertermous T, Samani NJ, Schunkert H, Assimes TL, McPherson R. Systems Genetics Analysis of Genome-Wide Association Study Reveals Novel Associations Between Key Biological Processes and Coronary Artery Disease. Arterioscler Thromb Vasc Biol. 2015a;35:1712–1722. doi: 10.1161/ATVBAHA.115.305513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Vivar J, Nelson CP, Willenborg C, Segrè AV, Mäkinen VP, Nikpay M, Erdmann J, Blankenberg S, O’Donnell C, März W, Laaksonen R, Stewart AF, Epstein SE, Shah SH, Granger CB, Hazen SL, Kathiresan S, Reilly MP, Yang X, Quertermous T, Samani NJ, Schunkert H, Assimes TL, McPherson R. Systems Genetics Analysis of Genome-Wide Association Study Reveals Novel Associations Between Key Biological Processes and Coronary Artery Disease. Arterioscler Thromb Vasc Biol. 2015b;35:1712–1722. doi: 10.1161/ATVBAHA.115.305513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel R, Schrank BR, Arora S, Boylan B, Fleming B, Miura H, Newman PJ, Molthen RC, Newman DK. Site-specific effects of PECAM-1 on atherosclerosis in LDL receptor-deficient mice. Arterioscler Thromb Vasc Biol. 2008;28:1996–2002. doi: 10.1161/ATVBAHA.108.172270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein JL, Brown MS. A century of cholesterol and coronaries: from plaques to genes to statins. Cell. 2015;161:161–172. doi: 10.1016/j.cell.2015.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodarzi MO. Genetics of Common Endocrine Disease: The Present and the Future. J Clin Endocrinol Metab. 2016;101:787–794. doi: 10.1210/jc.2015-3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greek R, Hansen LA. Questions regarding the predictive value of one evolved complex adaptive system for a second: exemplified by the SOD1 mouse. Prog Biophys Mol Biol. 2013;113:231–253. doi: 10.1016/j.pbiomolbio.2013.06.002. [DOI] [PubMed] [Google Scholar]
- Grundy SM, Cleeman JI, Merz CN, Brewer HB, Jr, Clark LT, Hunninghake DB, Pasternak RC, Smith SC, Jr, Stone NJ National Heart, L., Blood, I., American College of Cardiology, F., and American Heart, A. Implications of recent clinical trials for the National Cholesterol Education Program Adult Treatment Panel III guidelines. Circulation. 2004;110:227–239. doi: 10.1161/01.CIR.0000133317.49796.0E. [DOI] [PubMed] [Google Scholar]
- Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, Barnes I, Bignell A, Boychenko V, Hunt T, Kay M, Mukherjee G, Rajan J, Despacio-Reyes G, Saunders G, Steward C, Harte R, Lin M, Howald C, Tanzer A, Derrien T, Chrast J, Walters N, Balasubramanian S, Pei B, Tress M, Rodriguez JM, Ezkurdia I, van Baren J, Brent M, Haussler D, Kellis M, Valencia A, Reymond A, Gerstein M, Guigo R, Hubbard TJ. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidt T, Sager HB, Courties G, Dutta P, Iwamoto Y, Zaltsman A, von Zur Muhlen C, Bode C, Fricchione GL, Denninger J, Lin CP, Vinegoni C, Libby P, Swirski FK, Weissleder R, Nahrendorf M. Chronic variable stress activates hematopoietic stem cells. Nat Med. 2014;20:754–758. doi: 10.1038/nm.3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdt LM, Hoffmann S, Sass K, Langenberger D, Scholz M, Krohn K, Finstermeier K, Stahringer A, Wilfert W, Beutner F, Gielen S, Schuler G, Gabel G, Bergert H, Bechmann I, Stadler PF, Thiery J, Teupser D. Alu elements in ANRIL non-coding RNA at chromosome 9p21 modulate atherogenic cell functions through trans-regulation of gene networks. PLoS Genet. 2013;9:e1003588. doi: 10.1371/journal.pgen.1003588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes SD, Krantz DS, Rogers H, Gottdiener J, Contrada RJ. Mental stress and coronary artery disease: a multidisciplinary guide. Prog Cardiovasc Dis. 2006;49:106–122. doi: 10.1016/j.pcad.2006.08.013. [DOI] [PubMed] [Google Scholar]
- Hsu J, Smith JD. Genetic-genomic replication to identify candidate mouse atherosclerosis modifier genes. J Am Heart Assoc. 2013;2:e005421. doi: 10.1161/JAHA.112.005421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Polinsky P, Sadoun E, Rosenfeld ME, Schwartz SM. Atherosclerotic lesions in the common coronary arteries of ApoE knockout mice. Cardiovasc Pathol. 2005;14:120–125. doi: 10.1016/j.carpath.2005.02.004. [DOI] [PubMed] [Google Scholar]
- Huby AC, Otvos L, Jr, Belin de Chantemele EJ. Leptin Induces Hypertension and Endothelial Dysfunction via Aldosterone-Dependent Mechanisms in Obese Female Mice. Hypertension. 2016;67:1020–1028. doi: 10.1161/HYPERTENSIONAHA.115.06642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huszar D, Varban ML, Rinninger F, Feeley R, Arai T, Fairchild-Huntress V, Donovan MJ, Tall AR. Increased LDL cholesterol and atherosclerosis in LDL receptor-deficient mice with attenuated expression of scavenger receptor B1. Arterioscler Thromb Vasc Biol. 2000;20:1068–1073. doi: 10.1161/01.atv.20.4.1068. [DOI] [PubMed] [Google Scholar]
- Ishibashi S, Goldstein JL, Brown MS, Herz J, Burns DK. Massive xanthomatosis and atherosclerosis in cholesterol-fed low density lipoprotein receptor-negative mice. J Clin Invest. 1994;93:1885–1893. doi: 10.1172/JCI117179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isomaa B, Almgren P, Tuomi T, Forsen B, Lahti K, Nissen M, Taskinen MR, Groop L. Cardiovascular morbidity and mortality associated with the metabolic syndrome. Diabetes Care. 2001;24:683–689. doi: 10.2337/diacare.24.4.683. [DOI] [PubMed] [Google Scholar]
- Jansen H, Willenborg C, Schlesinger S, Ferrario PG, Konig IR, Erdmann J, Samani NJ, Lieb W, Schunkert H. Genetic variants associated with celiac disease and the risk for coronary artery disease. Mol Genet Genomics. 2015;290:1911–1917. doi: 10.1007/s00438-015-1045-3. [DOI] [PubMed] [Google Scholar]
- Jun JY, Ma Z, Segar L. Spontaneously diabetic Ins2(+/Akita):apoE-deficient mice exhibit exaggerated hypercholesterolemia and atherosclerosis. Am J Physiol Endocrinol Metab. 2011;301:E145–154. doi: 10.1152/ajpendo.00034.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannel WB, Castelli WP, McNamara PM. Cigarette smoking and risk of coronary heart disease. Epidemiologic clues to pathogensis. The Framingham Study. Natl Cancer Inst Monogr. 1968;28:9–20. [PubMed] [Google Scholar]
- Karbach S, Croxford AL, Oelze M, Schuler R, Minwegen D, Wegner J, Koukes L, Yogev N, Nikolaev A, Reissig S, Ullmann A, Knorr M, Waldner M, Neurath MF, Li H, Wu Z, Brochhausen C, Scheller J, Rose-John S, Piotrowski C, Bechmann I, Radsak M, Wild P, Daiber A, von Stebut E, Wenzel P, Waisman A, Munzel T. Interleukin 17 drives vascular inflammation, endothelial dysfunction, and arterial hypertension in psoriasis-like skin disease. Arterioscler Thromb Vasc Biol. 2014;34:2658–2668. doi: 10.1161/ATVBAHA.114.304108. [DOI] [PubMed] [Google Scholar]
- Kayashima Y, Makhanova NA, Matsuki K, Tomita H, Bennett BJ, Maeda N. Identification of aortic arch-specific quantitative trait loci for atherosclerosis by an intercross of DBA/2J and 129S6 apolipoprotein E-deficient mice. PLoS One. 2015;10:e0117478. doi: 10.1371/journal.pone.0117478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy AJ, Ellacott KL, King VL, Hasty AH. Mouse models of the metabolic syndrome. Dis Model Mech. 2010;3:156–166. doi: 10.1242/dmm.003467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersey PJ, Allen JE, Armean I, Boddu S, Bolt BJ, Carvalho-Silva D, Christensen M, Davis P, Falin LJ, Grabmueller C, Humphrey J, Kerhornou A, Khobova J, Aranganathan NK, Langridge N, Lowy E, McDowall MD, Maheswari U, Nuhn M, Ong CK, Overduin B, Paulini M, Pedro H, Perry E, Spudich G, Tapanari E, Walts B, Williams G, Tello-Ruiz M, Stein J, Wei S, Ware D, Bolser DM, Howe KL, Kulesha E, Lawson D, Maslen G, Staines DM. Ensembl Genomes 2016: more genomes, more complexity. Nucleic Acids Res. 2016;44:D574–580. doi: 10.1093/nar/gkv1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King AJ. The use of animal models in diabetes research. Br J Pharmacol. 2012;166:877–894. doi: 10.1111/j.1476-5381.2012.01911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloss MW, Rosen GM, Rauckman EJ, Padilla GM. Androgenic suppression of mouse hepatic FAD-containing monooxygenase activity. Life Sci. 1982;31:1037–1042. doi: 10.1016/0024-3205(82)90176-x. [DOI] [PubMed] [Google Scholar]
- Knouff C, Briand O, Lestavel S, Clavey V, Altenburg M, Maeda N. Defective VLDL metabolism and severe atherosclerosis in mice expressing human apolipoprotein E isoforms but lacking the LDL receptor. Biochim Biophys Acta. 2004;1684:8–17. doi: 10.1016/j.bbalip.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, Britt EB, Fu X, Wu Y, Li L, Smith JD, DiDonato JA, Chen J, Li H, Wu GD, Lewis JD, Warrier M, Brown JM, Krauss RM, Tang WH, Bushman FD, Lusis AJ, Hazen SL. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med. 2013;19:576–585. doi: 10.1038/nm.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalla E, Lamster IB, Hofmann MA, Bucciarelli L, Jerud AP, Tucker S, Lu Y, Papapanou PN, Schmidt AM. Oral infection with a periodontal pathogen accelerates early atherosclerosis in apolipoprotein E-null mice. Arterioscler Thromb Vasc Biol. 2003;23:1405–1411. doi: 10.1161/01.ATV.0000082462.26258.FE. [DOI] [PubMed] [Google Scholar]
- LeBoeuf RC, Puppione DL, Schumaker VN, Lusis AJ. Genetic control of lipid transport in mice. I. Structural properties and polymorphisms of plasma lipoproteins. J Biol Chem. 1983;258:5063–5070. [PubMed] [Google Scholar]
- Leong XF, Ng CY, Jaarin K. Animal Models in Cardiovascular Research: Hypertension and Atherosclerosis. Biomed Res Int. 2015;2015:528757. doi: 10.1155/2015/528757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Reddick RL, Maeda N. Lack of apoA-I is not associated with increased susceptibility to atherosclerosis in mice. Arterioscler Thromb. 1993;13:1814–1821. doi: 10.1161/01.atv.13.12.1814. [DOI] [PubMed] [Google Scholar]
- Lietz M, Berges A, Lebrun S, Meurrens K, Steffen Y, Stolle K, Schueller J, Boue S, Vuillaume G, Vanscheeuwijck P, Moehring M, Schlage W, De Leon H, Hoeng J, Peitsch M. Cigarette-smoke-induced atherogenic lipid profiles in plasma and vascular tissue of apolipoprotein E-deficient mice are attenuated by smoking cessation. Atherosclerosis. 2013;229:86–93. doi: 10.1016/j.atherosclerosis.2013.03.036. [DOI] [PubMed] [Google Scholar]
- Link JC, Chen X, Prien C, Borja MS, Hammerson B, Oda MN, Arnold AP, Reue K. Increased high-density lipoprotein cholesterol levels in mice with XX versus XY sex chromosomes. Arterioscler Thromb Vasc Biol. 2015;35:1778–1786. doi: 10.1161/ATVBAHA.115.305460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusis AJ. Atherosclerosis. Nature. 2000;407:233–241. doi: 10.1038/35025203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusis AJ, Taylor BA, Wangenstein RW, LeBoeuf RC. Genetic control of lipid transport in mice. II. Genes controlling structure of high density lipoproteins. J Biol Chem. 1983;258:5071–5078. [PubMed] [Google Scholar]
- Lusis AJ, Yu J, Wang SS. The problem of passenger genes in transgenic mice. Arterioscler Thromb Vasc Biol. 2007;27:2100–2103. doi: 10.1161/ATVBAHA.107.147918. [DOI] [PubMed] [Google Scholar]
- Ma Z, Choudhury A, Kang SA, Monestier M, Cohen PL, Eisenberg RA. Accelerated atherosclerosis in ApoE deficient lupus mouse models. Clin Immunol. 2008;127:168–175. doi: 10.1016/j.clim.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak IW, Evaniew N, Ghert M. Lost in translation: animal models and clinical trials in cancer treatment. Am J Transl Res. 2014;6:114–118. [PMC free article] [PubMed] [Google Scholar]
- Makinen VP, Civelek M, Meng Q, Zhang B, Zhu J, Levian C, Huan T, Segre AV, Ghosh S, Vivar J, Nikpay M, Stewart AF, Nelson CP, Willenborg C, Erdmann J, Blakenberg S, O’Donnell CJ, Marz W, Laaksonen R, Epstein SE, Kathiresan S, Shah SH, Hazen SL, Reilly MP, Coronary ADGWR, Meta-Analysis C, Lusis AJ, Samani NJ, Schunkert H, Quertermous T, McPherson R, Yang X, Assimes TL. Integrative genomics reveals novel molecular pathways and gene networks for coronary artery disease. PLoS Genet. 2014;10:e1004502. doi: 10.1371/journal.pgen.1004502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson R, Tybjaerg-Hansen A. Genetics of Coronary Artery Disease. Circ Res. 2016;118:564–578. doi: 10.1161/CIRCRESAHA.115.306566. [DOI] [PubMed] [Google Scholar]
- Meissner M, Lombardo E, Havinga R, Tietge UJ, Kuipers F, Groen AK. Voluntary wheel running increases bile acid as well as cholesterol excretion and decreases atherosclerosis in hypercholesterolemic mice. Atherosclerosis. 2011;218:323–329. doi: 10.1016/j.atherosclerosis.2011.06.040. [DOI] [PubMed] [Google Scholar]
- Moore KJ, Kunjathoor VV, Koehn SL, Manning JJ, Tseng AA, Silver JM, McKee M, Freeman MW. Loss of receptor-mediated lipid uptake via scavenger receptor A or CD36 pathways does not ameliorate atherosclerosis in hyperlipidemic mice. J Clin Invest. 2005;115:2192–2201. doi: 10.1172/JCI24061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudge JM, Harrow J. Creating reference gene annotation for the mouse C57BL6/J genome assembly. Mamm Genome. 2015;26:366–378. doi: 10.1007/s00335-015-9583-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najafi AH, Aghili N, Tilan JU, Andrews JA, Peng X, Lassance-Soares RM, Sood S, Alderman LO, Abe K, Li L, Kolodgie FD, Virmani R, Zukowska Z, Epstein SE, Burnett MS. A new murine model of stress-induced complex atherosclerotic lesions. Dis Model Mech. 2013;6:323–331. doi: 10.1242/dmm.009977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neven E, D’Haese PC. Vascular calcification in chronic renal failure: what have we learned from animal studies? Circ Res. 2011;108:249–264. doi: 10.1161/CIRCRESAHA.110.225904. [DOI] [PubMed] [Google Scholar]
- Nikpay M, Goel A, Won HH, Hall LM, Willenborg C, Kanoni S, Saleheen D, Kyriakou T, Nelson CP, Hopewell JC, Webb TR, Zeng L, Dehghan A, Alver M, Armasu SM, Auro K, Bjonnes A, Chasman DI, Chen S, Ford I, Franceschini N, Gieger C, Grace C, Gustafsson S, Huang J, Hwang SJ, Kim YK, Kleber ME, Lau KW, Lu X, Lu Y, Lyytikainen LP, Mihailov E, Morrison AC, Pervjakova N, Qu L, Rose LM, Salfati E, Saxena R, Scholz M, Smith AV, Tikkanen E, Uitterlinden A, Yang X, Zhang W, Zhao W, de Andrade M, de Vries PS, van Zuydam NR, Anand SS, Bertram L, Beutner F, Dedoussis G, Frossard P, Gauguier D, Goodall AH, Gottesman O, Haber M, Han BG, Huang J, Jalilzadeh S, Kessler T, Konig IR, Lannfelt L, Lieb W, Lind L, Lindgren CM, Lokki ML, Magnusson PK, Mallick NH, Mehra N, Meitinger T, Memon FU, Morris AP, Nieminen MS, Pedersen NL, Peters A, Rallidis LS, Rasheed A, Samuel M, Shah SH, Sinisalo J, Stirrups KE, Trompet S, Wang L, Zaman KS, Ardissino D, Boerwinkle E, Borecki IB, Bottinger EP, Buring JE, Chambers JC, Collins R, Cupples LA, Danesh J, Demuth I, Elosua R, Epstein SE, Esko T, Feitosa MF, Franco OH, Franzosi MG, Granger CB, Gu D, Gudnason V, Hall AS, Hamsten A, Harris TB, Hazen SL, Hengstenberg C, Hofman A, Ingelsson E, Iribarren C, Jukema JW, Karhunen PJ, Kim BJ, Kooner JS, Kullo IJ, Lehtimaki T, Loos RJ, Melander O, Metspalu A, Marz W, Palmer CN, Perola M, Quertermous T, Rader DJ, Ridker PM, Ripatti S, Roberts R, Salomaa V, Sanghera DK, Schwartz SM, Seedorf U, Stewart AF, Stott DJ, Thiery J, Zalloua PA, O’Donnell CJ, Reilly MP, Assimes TL, Thompson JR, Erdmann J, Clarke R, Watkins H, Kathiresan S, McPherson R, Deloukas P, Schunkert H, Samani NJ, Farrall M Consortium CAD. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet. 2015;47:1121–1130. doi: 10.1038/ng.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishina PM, Naggert JK, Verstuyft J, Paigen B. Atherosclerosis in genetically obese mice: the mutants obese, diabetes, fat, tubby, and lethal yellow. Metabolism. 1994;43:554–558. doi: 10.1016/0026-0495(94)90195-3. [DOI] [PubMed] [Google Scholar]
- Nurnberg ST, Zhang H, Hand NJ, Bauer RC, Saleheen D, Reilly MP, Rader DJ. From Loci to Biology: Functional Genomics of Genome-Wide Association for Coronary Disease. Circ Res. 2016;118:586–606. doi: 10.1161/CIRCRESAHA.115.306464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco LD, Bennett BJ, Farber CR, Ghazalpour A, Pan C, Che N, Wen P, Qi HX, Mutukulu A, Siemers N, Neuhaus I, Yordanova R, Gargalovic P, Pellegrini M, Kirchgessner T, Lusis AJ. Unraveling inflammatory responses using systems genetics and gene-environment interactions in macrophages. Cell. 2012;151:658–670. doi: 10.1016/j.cell.2012.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paigen B, Mitchell D, Reue K, Morrow A, Lusis AJ, LeBoeuf RC. Ath-1, a gene determining atherosclerosis susceptibility and high density lipoprotein levels in mice. Proc Natl Acad Sci U S A. 1987;84:3763–3767. doi: 10.1073/pnas.84.11.3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks BW, Nam E, Org E, Kostem E, Norheim F, Hui ST, Pan C, Civelek M, Rau CD, Bennett BJ, Mehrabian M, Ursell LK, He A, Castellani LW, Zinker B, Kirby M, Drake TA, Drevon CA, Knight R, Gargalovic P, Kirchgessner T, Eskin E, Lusis AJ. Genetic control of obesity and gut microbiota composition in response to high-fat, high-sucrose diet in mice. Cell Metab. 2013;17:141–152. doi: 10.1016/j.cmet.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasterkamp G, van der Laan SW, Haitjema S, Asl HF, Siemelink MA, Bezemer T, van Setten J, Dichgans M, Malik R, Worrall BB, Schunkert H, Samani NJ, de Kleijn DP, Markus HS, Hoefer IE, Michoel T, de Jager SC, Bjorkegren JL, den Ruijter HM, Asselbergs FW. Human Validation of Genes Associated With a Murine Atherosclerotic Phenotype. Arterioscler Thromb Vasc Biol. 2016 doi: 10.1161/ATVBAHA.115.306958. [DOI] [PubMed] [Google Scholar]
- Perrin S. Preclinical research: Make mouse studies work. Nature. 2014;507:423–425. doi: 10.1038/507423a. [DOI] [PubMed] [Google Scholar]
- Plummer MR, Hasty AH. Atherosclerotic lesion formation and triglyceride storage in obese apolipoprotein AI-deficient mice. J Nutr Biochem. 2008;19:664–673. doi: 10.1016/j.jnutbio.2007.08.009. [DOI] [PubMed] [Google Scholar]
- Plump AS, Smith JD, Hayek T, Aalto-Setala K, Walsh A, Verstuyft JG, Rubin EM, Breslow JL. Severe hypercholesterolemia and atherosclerosis in apolipoprotein E-deficient mice created by homologous recombination in ES cells. Cell. 1992;71:343–353. doi: 10.1016/0092-8674(92)90362-g. [DOI] [PubMed] [Google Scholar]
- Powell-Braxton L, Veniant M, Latvala RD, Hirano KI, Won WB, Ross J, Dybdal N, Zlot CH, Young SG, Davidson NO. A mouse model of human familial hypercholesterolemia: markedly elevated low density lipoprotein cholesterol levels and severe atherosclerosis on a low-fat chow diet. Nat Med. 1998;4:934–938. doi: 10.1038/nm0898-934. [DOI] [PubMed] [Google Scholar]
- Ramsey SA, Vengrenyuk Y, Menon P, Podolsky I, Feig JE, Aderem A, Fisher EA, Gold ES. Epigenome-guided analysis of the transcriptome of plaque macrophages during atherosclerosis regression reveals activation of the Wnt signaling pathway. PLoS Genet. 2014;10:e1004828. doi: 10.1371/journal.pgen.1004828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattazzi M, Bennett BJ, Bea F, Kirk EA, Ricks JL, Speer M, Schwartz SM, Giachelli CM, Rosenfeld ME. Calcification of advanced atherosclerotic lesions in the innominate arteries of ApoE-deficient mice: potential role of chondrocyte-like cells. Arterioscler Thromb Vasc Biol. 2005;25:1420–1425. doi: 10.1161/01.ATV.0000166600.58468.1b. [DOI] [PubMed] [Google Scholar]
- Reaven G. Type 2 diabetes and coronary heart disease: we keep learning how little we know. Arterioscler Thromb Vasc Biol. 2003;23:917–918. doi: 10.1161/01.ATV.0000077249.35122.19. [DOI] [PubMed] [Google Scholar]
- Renard CB, Kramer F, Johansson F, Lamharzi N, Tannock LR, von Herrath MG, Chait A, Bornfeldt KE. Diabetes and diabetes-associated lipid abnormalities have distinct effects on initiation and progression of atherosclerotic lesions. J Clin Invest. 2004;114:659–668. doi: 10.1172/JCI17867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reue KL, Quon DH, O’Donnell KA, Dizikes GJ, Fareed GC, Lusis AJ. Cloning and regulation of messenger RNA for mouse apolipoprotein E. J Biol Chem. 1984;259:2100–2107. [PubMed] [Google Scholar]
- Richmond A, Su Y. Mouse xenograft models vs GEM models for human cancer therapeutics. Dis Model Mech. 2008;1:78–82. doi: 10.1242/dmm.000976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A, Thompson JS. Inbred mice and their hypbrids as an animal model for atherosclerosis research. Adv Exp Med Biol. 1976;67:313–327. doi: 10.1007/978-1-4614-4618-7_18. [DOI] [PubMed] [Google Scholar]
- Rodriguez JM, Wolfrum S, Robblee M, Chen KY, Gilbert ZN, Choi JH, Teupser D, Breslow JL. Altered expression of Raet1e, a major histocompatibility complex class 1-like molecule, underlies the atherosclerosis modifier locus Ath11 10b. Circ Res. 2013;113:1054–1064. doi: 10.1161/CIRCRESAHA.113.302052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose S, Eren M, Murphy S, Zhang H, Thaxton CS, Chowaniec J, Waters EA, Meade TJ, Vaughan DE, Perlman H. A novel mouse model that develops spontaneous arthritis and is predisposed towards atherosclerosis. Ann Rheum Dis. 2013;72:89–95. doi: 10.1136/annrheumdis-2012-201431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, Mayer B, Dixon RJ, Meitinger T, Braund P, Wichmann HE, Barrett JH, Konig IR, Stevens SE, Szymczak S, Tregouet DA, Iles MM, Pahlke F, Pollard H, Lieb W, Cambien F, Fischer M, Ouwehand W, Blankenberg S, Balmforth AJ, Baessler A, Ball SG, Strom TM, Braenne I, Gieger C, Deloukas P, Tobin MD, Ziegler A, Thompson JR, Schunkert H Wtccc, and the Cardiogenics, C. Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357:443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer K, Muller K, Hecke A, Mounier E, Goebel J, Loskutoff DJ, Konstantinides S. Enhanced thrombosis in atherosclerosis-prone mice is associated with increased arterial expression of plasminogen activator inhibitor-1. Arterioscler Thromb Vasc Biol. 2003;23:2097–2103. doi: 10.1161/01.ATV.0000097766.36623.DF. [DOI] [PubMed] [Google Scholar]
- Schnell O, Cappuccio F, Genovese S, Standl E, Valensi P, Ceriello A. Type 1 diabetes and cardiovascular disease. Cardiovasc Diabetol. 2013;12:156. doi: 10.1186/1475-2840-12-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreyer SA, Wilson DL, LeBoeuf RC. C57BL/6 mice fed high fat diets as models for diabetes-accelerated atherosclerosis. Atherosclerosis. 1998;136:17–24. doi: 10.1016/s0021-9150(97)00165-2. [DOI] [PubMed] [Google Scholar]
- Schunkert H, Konig IR, Kathiresan S, Reilly MP, Assimes TL, Holm H, Preuss M, Stewart AF, Barbalic M, Gieger C, Absher D, Aherrahrou Z, Allayee H, Altshuler D, Anand SS, Andersen K, Anderson JL, Ardissino D, Ball SG, Balmforth AJ, Barnes TA, Becker DM, Becker LC, Berger K, Bis JC, Boekholdt SM, Boerwinkle E, Braund PS, Brown MJ, Burnett MS, Buysschaert I, Carlquist JF, Chen L, Cichon S, Codd V, Davies RW, Dedoussis G, Dehghan A, Demissie S, Devaney JM, Diemert P, Do R, Doering A, Eifert S, Mokhtari NE, Ellis SG, Elosua R, Engert JC, Epstein SE, de Faire U, Fischer M, Folsom AR, Freyer J, Gigante B, Girelli D, Gretarsdottir S, Gudnason V, Gulcher JR, Halperin E, Hammond N, Hazen SL, Hofman A, Horne BD, Illig T, Iribarren C, Jones GT, Jukema JW, Kaiser MA, Kaplan LM, Kastelein JJ, Khaw KT, Knowles JW, Kolovou G, Kong A, Laaksonen R, Lambrechts D, Leander K, Lettre G, Li M, Lieb W, Loley C, Lotery AJ, Mannucci PM, Maouche S, Martinelli N, McKeown PP, Meisinger C, Meitinger T, Melander O, Merlini PA, Mooser V, Morgan T, Muhleisen TW, Muhlestein JB, Munzel T, Musunuru K, Nahrstaedt J, Nelson CP, Nothen MM, Olivieri O, Patel RS, Patterson CC, Peters A, Peyvandi F, Qu L, Quyyumi AA, Rader DJ, Rallidis LS, Rice C, Rosendaal FR, Rubin D, Salomaa V, Sampietro ML, Sandhu MS, Schadt E, Schafer A, Schillert A, Schreiber S, Schrezenmeir J, Schwartz SM, Siscovick DS, Sivananthan M, Sivapalaratnam S, Smith A, Smith TB, Snoep JD, Soranzo N, Spertus JA, Stark K, Stirrups K, Stoll M, Tang WH, Tennstedt S, Thorgeirsson G, Thorleifsson G, Tomaszewski M, Uitterlinden AG, van Rij AM, Voight BF, Wareham NJ, Wells GA, Wichmann HE, Wild PS, Willenborg C, Witteman JC, Wright BJ, Ye S, Zeller T, Ziegler A, Cambien F, Goodall AH, Cupples LA, Quertermous T, Marz W, Hengstenberg C, Blankenberg S, Ouwehand WH, Hall AS, Deloukas P, Thompson JR, Stefansson K, Roberts R, Thorsteinsdottir U, O’Donnell CJ, McPherson R, Erdmann J, Samani NJ Cardiogenics, Consortium C.A. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet. 2011;43:333–338. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz SM, deBlois D, O’Brien ER. The intima. Soil for atherosclerosis and restenosis. Circ Res. 1995;77:445–465. doi: 10.1161/01.res.77.3.445. [DOI] [PubMed] [Google Scholar]
- Schwartz SM, Galis ZS, Rosenfeld ME, Falk E. Plaque rupture in humans and mice. Arterioscler Thromb Vasc Biol. 2007;27:705–713. doi: 10.1161/01.ATV.0000261709.34878.20. [DOI] [PubMed] [Google Scholar]
- Schwartz SM, Virmani R, Rosenfeld ME. The good smooth muscle cells in atherosclerosis. Curr Atheroscler Rep. 2000;2:422–429. doi: 10.1007/s11883-000-0081-5. [DOI] [PubMed] [Google Scholar]
- Seifert FU, Lammens K, Stoehr G, Kessler B, Hopfner KP. Structural mechanism of ATP-dependent DNA binding and DNA end bridging by eukaryotic Rad50. EMBO J. 2016;35:759–772. doi: 10.15252/embj.201592934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaposhnik Z, Wang X, Weinstein M, Bennett BJ, Lusis AJ. Granulocyte macrophage colony-stimulating factor regulates dendritic cell content of atherosclerotic lesions. Arterioscler Thromb Vasc Biol. 2007;27:621–627. doi: 10.1161/01.ATV.0000254673.55431.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Bornfeldt KE. Mouse models for studies of cardiovascular complications of type 1 diabetes. Ann N Y Acad Sci. 2007;1103:202–217. doi: 10.1196/annals.1394.004. [DOI] [PubMed] [Google Scholar]
- Stamler J, Stamler R, Neaton JD. Blood pressure, systolic and diastolic, and cardiovascular risks. US population data. Arch Intern Med. 1993;153:598–615. doi: 10.1001/archinte.153.5.598. [DOI] [PubMed] [Google Scholar]
- Stylianou IM, Bauer RC, Reilly MP, Rader DJ. Genetic basis of atherosclerosis: insights from mice and humans. Circ Res. 2012;110:337–355. doi: 10.1161/CIRCRESAHA.110.230854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Wang A, Jin X, Natanzon A, Duquaine D, Brook RD, Aguinaldo JG, Fayad ZA, Fuster V, Lippmann M, Chen LC, Rajagopalan S. Long-term air pollution exposure and acceleration of atherosclerosis and vascular inflammation in an animal model. JAMA. 2005;294:3003–3010. doi: 10.1001/jama.294.23.3003. [DOI] [PubMed] [Google Scholar]
- Taleb S, Herbin O, Ait-Oufella H, Verreth W, Gourdy P, Barateau V, Merval R, Esposito B, Clement K, Holvoet P, Tedgui A, Mallat Z. Defective leptin/leptin receptor signaling improves regulatory T cell immune response and protects mice from atherosclerosis. Arterioscler Thromb Vasc Biol. 2007;27:2691–2698. doi: 10.1161/ATVBAHA.107.149567. [DOI] [PubMed] [Google Scholar]
- Tang WH, Wang Z, Levison BS, Koeth RA, Britt EB, Fu X, Wu Y, Hazen SL. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N Engl J Med. 2013;368:1575–1584. doi: 10.1056/NEJMoa1109400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdar W, Flint J, Mott R. Simulating the collaborative cross: power of quantitative trait loci detection and mapping resolution in large sets of recombinant inbred strains of mice. Genetics. 2006;172:1783–1797. doi: 10.1534/genetics.104.039313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ree JH, van den Broek WJ, Dahlmans VE, Groot PH, Vidgeon-Hart M, Frants RR, Wieringa B, Havekes LM, Hofker MH. Diet-induced hypercholesterolemia and atherosclerosis in heterozygous apolipoprotein E-deficient mice. Atherosclerosis. 1994;111:25–37. doi: 10.1016/0021-9150(94)90188-0. [DOI] [PubMed] [Google Scholar]
- Vanden Berghe T, Hulpiau P, Martens L, Vandenbroucke RE, Van Wonterghem E, Perry SW, Bruggeman I, Divert T, Choi SM, Vuylsteke M, Shestopalov VI, Libert C, Vandenabeele P. Passenger Mutations Confound Interpretation of All Genetically Modified Congenic Mice. Immunity. 2015;43:200–209. doi: 10.1016/j.immuni.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanderLaan PA, Reardon CA, Thisted RA, Getz GS. VLDL best predicts aortic root atherosclerosis in LDL receptor deficient mice. J Lipid Res. 2009;50:376–385. doi: 10.1194/jlr.M800284-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voyiaziakis E, Goldberg IJ, Plump AS, Rubin EM, Breslow JL, Huang LS. ApoA-I deficiency causes both hypertriglyceridemia and increased atherosclerosis in human apoB transgenic mice. J Lipid Res. 1998;39:313–321. [PubMed] [Google Scholar]
- Wang SS, Schadt EE, Wang H, Wang X, Ingram-Drake L, Shi W, Drake TA, Lusis AJ. Identification of pathways for atherosclerosis in mice: integration of quantitative trait locus analysis and global gene expression data. Circ Res. 2007;101:e11–30. doi: 10.1161/CIRCRESAHA.107.152975. [DOI] [PubMed] [Google Scholar]
- Wang Y, Gao H, Loyd CM, Fu W, Diaconu D, Liu S, Cooper KD, McCormick TS, Simon DI, Ward NL. Chronic skin-specific inflammation promotes vascular inflammation and thrombosis. J Invest Dermatol. 2012;132:2067–2075. doi: 10.1038/jid.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, Feldstein AE, Britt EB, Fu X, Chung YM, Wu Y, Schauer P, Smith JD, Allayee H, Tang WH, DiDonato JA, Lusis AJ, Hazen SL. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton DE, Nicol CW, Bredin SS. Health benefits of physical activity: the evidence. CMAJ. 2006;174:801–809. doi: 10.1503/cmaj.051351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warden CH, Hedrick CC, Qiao JH, Castellani LW, Lusis AJ. Atherosclerosis in transgenic mice overexpressing apolipoprotein A-II. Science. 1993;261:469–472. doi: 10.1126/science.8332912. [DOI] [PubMed] [Google Scholar]
- Weiss D, Kools JJ, Taylor WR. Angiotensin II-induced hypertension accelerates the development of atherosclerosis in apoE-deficient mice. Circulation. 2001;103:448–454. doi: 10.1161/01.cir.103.3.448. [DOI] [PubMed] [Google Scholar]
- Welch CL. Beyond genome-wide association studies: the usefulness of mouse genetics in understanding the complex etiology of atherosclerosis. Arterioscler Thromb Vasc Biol. 2012;32:207–215. doi: 10.1161/ATVBAHA.111.232694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H, Klemm A, Flicek P, Manolio T, Hindorff L, Parkinson H. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerweel PE, Luyten RK, Koomans HA, Derksen RH, Verhaar MC. Premature atherosclerotic cardiovascular disease in systemic lupus erythematosus. Arthritis Rheum. 2007;56:1384–1396. doi: 10.1002/art.22568. [DOI] [PubMed] [Google Scholar]
- Wiesel P, Mazzolai L, Nussberger J, Pedrazzini T. Two-kidney, one clip and one-kidney, one clip hypertension in mice. Hypertension. 1997;29:1025–1030. doi: 10.1161/01.hyp.29.4.1025. [DOI] [PubMed] [Google Scholar]
- Witztum JL. You are right too! J Clin Invest. 2005;115:2072–2075. doi: 10.1172/JCI26130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu KK, Wu TJ, Chin J, Mitnaul LJ, Hernandez M, Cai TQ, Ren N, Waters MG, Wright SD, Cheng K. Increased hypercholesterolemia and atherosclerosis in mice lacking both ApoE and leptin receptor. Atherosclerosis. 2005;181:251–259. doi: 10.1016/j.atherosclerosis.2005.01.029. [DOI] [PubMed] [Google Scholar]
- Yerkey MW, Kernis SJ, Franklin BA, Sandberg KR, McCullough PA. Renal dysfunction and acceleration of coronary disease. Heart. 2004;90:961–966. doi: 10.1136/hrt.2003.015503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Picard MH, Vasile E, Zhu Y, Raffai RL, Weisgraber KH, Krieger M. Dietinduced occlusive coronary atherosclerosis, myocardial infarction, cardiac dysfunction, and premature death in scavenger receptor class B type I-deficient, hypomorphic apolipoprotein ER61 mice. Circulation. 2005;111:3457–3464. doi: 10.1161/CIRCULATIONAHA.104.523563. [DOI] [PubMed] [Google Scholar]
- Zhao L, Hall JA, Levenkova N, Lee E, Middleton MK, Zukas AM, Rader DJ, Rux JJ, Pure E. CD44 regulates vascular gene expression in a proatherogenic environment. Arterioscler Thromb Vasc Biol. 2007;27:886–892. doi: 10.1161/01.ATV.0000259362.10882.c5. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Chen J, Freudenberg JM, Meng Q, Rajpal DK, Yang X. Network-Based Identification and Prioritization of Key Regulators of Coronary Artery Disease Loci. Arterioscler Thromb Vasc Biol. 2016;36:928–941. doi: 10.1161/ATVBAHA.115.306725. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.