Abstract
FOXO1 protein inactivation in the nucleus is one of targets for the treatment of diabetes mellitus. Annona muricata leaves contain flavonoid and phenolic compound alkaloids that were known to be able to increase pancreatic β cell proliferation in animal experiment. This research aimed to predict the active compound ability of the Annona muricata leaves to bind and inhibit FOXO1 protein through in silico study. Analysis of molecular docking was performed by using Autodock Vina PyRx. this research proved that anonaine, rutin, muricatocin a, isolaureline, xylopine, and kaempferol 3-O-rutinoside had an equal or smaller free binding energy compared to the control compound. Rutin and Muricatocin A had the same binding ability toward 66% amino acid residues, compared to control compound with hydrogen bond type, while xylopine, anonaine, isolaureline, kaempferol 3-O-rutinoside had a similar binding ability towards 33% amino acid residues compared to control compound with hydrogen bond type.
Keywords: Anonaine, Active compound, Annona muricata leaves, FOXO1, In silico, Isolaureline, Kaempferol 3-O-rutinoside, Muricatocin A, Rutin, Xylopine
Introduction
Currently, the development of DM research is focused on the improvement of the regeneration and suppression of the pancreatic β cells apoptosis. A mechanism that is thought to play an important role in the regulation of pancreatic β cell regeneration is the FOXO1 (forkhead box protein O1) transcription factors. One of the mechanisms to prevent the occurrence of apoptosis and to increase pancreatic β cells proliferation is the prevention FOXO1 transcription factors migration to the nucleus or the FOXO1 inactivation in the nucleus (Martinez et al. 2006; Gross et al. 2008).
FOXO1 is a transcription factor that is normally found in the cytoplasm. FOXO1 is activated through MAPK (mitogen-activated protein kinase), AKT, and Pdx1 (promoting gene) pathway. FOXO1 phosphorylation in the cytoplasm activates cyclin D1 and Cdk4 (cyclin dependent kinase) protein that activates the cell cycle from G0 to G1 phase in preparation for the DNA synthesis (deoxyribonucleic acid) (Gross et al. 2008).
Previous studies reported that ethanol extract from Annona muricata leaves has the ability to improve β cells of animal studies that have been damaged by STZ (streptozotocin) induction as well as control the glucose levels in the blood to be within the normal limits (Adeyemi et al. 2010). However, the work mechanism of active ingredients, which is contained in the extract, is still unexplained.
Leaves, fruits, roots, tree barks, and seeds of Annona muricata have become a lot of research subjects as an anti-diabetic material (Adeyemi et al. 2010; Malviya et al. 2010). The main active compounds of Annona muricata are acetogenin, flavonoids, phenolic compound alkaloids, and tannins (Gajalakshmi et al. 2012). Those active compounds are believed to have a biological effect for the cell.
In the last few years, in silico studies have been developed as a method to predict the ability of an active compound to cause a biological effect that will be used in the disbursement of new drugs. The purpose of these studies is to reduce the cost and the length that are required for an active ingredient to turn into a drug that can be marketed to the public (Wadood et al. 2013). Research results of the pharmaceutical industry in the United States indicate that, in 2001, the cost of drug manufacture from an active ingredient to a marketable drug would cost around $880 million with approximately 12 years of manufacturing. In addition to the cost and duration, several factors can cause the failure to a drug manufacture process, such as the low effectiveness of the drugs, the incidence of toxic effects, and also the obstacle in the marketing process (Hileman 2006).
One of the in silico methods that is widely used is molecular docking (interaction networks). The principle of this technique is to predict the ability of an active compound (ligand) to bind with a target protein (e.g. receptor) to form a stable complex. Several tools can be used for molecular docking methods such as ArgusDock, DOCK, FRED, eHITS, Auto Dock and FTDock. The ability of a ligand to binds with the active site of a receptor is then tested to assess the activation or inhibition strength (Pedro and Lei 2010).
Based on the facts above, this study aims to predict the ability of the active compound contained in Annona muricata leaves to regenerate a damaged β cell by assessing the inhibition ability of the active compound against FOXO1 protein through an in silico study.
Materials and methods
Analytical-Descriptive Research Design was used to determine the potential of the Annona muricata leaves active compounds and to determine the binding affinity of the active compounds toward FOXO1 protein computationally. The compound was obtained from the PubChem (https://pubchem.ncbi.nlm.nih.gov) server with a recorded CID. While FOXO1 receptor, that was the inhibitor, was obtained from the data bank (http://www.rcsb.org/pdb/home/home.do) server by ID 3CO6. Inhibitor reference that can inhibit the activity of FOXO1 proteins was taken from calbiochem by CID AS1842856 (Nagashima et al. 2010). Furthermore, the docking process was performed by using PyRx program (Autodock vina) (Dallakyan and Olson 2015). The docking process was done specifically with ‘the compound’ as ligand and inhibitor reference (AS1842856). Compound that has the smallest or most negative binding energy was chosen because it showed the best complex conformation. Docking results were stored and visualized by using Ligandscout and PyMOL ligand (DeLano 2002). The visualization of docking results by using LigPlot showed the interaction between these two molecules (FOXO1-ligand compound or inhibitor reference) (Wallace et al. 1995). LigPlot results showed hydrophobic bonding and hydrogen bonding that occurred within the complex. Potential inhibition can be seen from the amino acid on FOXO1 active side that was bound by the active compound.
Results
Potential analysis of the Annona muricata leaves active compound as FOXO1 protein inhibitors with in silico method
Various in vitro and in vivo studies demonstrate that the transcription factor of FOXO1 protein contribute to the proliferation process of pancreatic β cell. It is said that the inhibition of FOXO1 protein translocation from the cytoplasm to the nucleus or the inactivation of FOXO1 protein in the nucleus is the key to inhibit DM apoptosis in animal experiment and to induce pancreatic β cell proliferation (Martinez et al. 2006; Gross et al. 2008). Based on the data above, this research attempted to examine the potential of Annona muricata leaves active compound as the inhibitor of FOXO1 protein in silico, compared to the drug control that have been known to inhibit FOXO1 protein. An active compound that has the potential to inhibit FOXO1 protein is a compound that has equal or smaller free binding energy than the control. Analysis results of the potential of Annona muricata leaves active compound on the inhibition of FOXO1 protein can be seen in Table 1.
Table 1.
Analysis of inhibiting potency of Annona muricata leaves active compounds against Foxo1 receptor with in silico method
| Compound | CID | Receptor | Binding affinity (kcal/mol) |
|---|---|---|---|
| Anonaine | 160597 | Foxo1 | −6.9 |
| Xylopine | 160503 | Foxo1 | −6.8 |
| Kaempferol 3-O-rutinoside | 20055286 | Foxo1 | −6.6 |
| Isolaureline | 12311076 | Foxo1 | −6.4 |
| Inhibitor Foxo1 | AS1842856—calbiochem | Foxo1 | −6.3 |
| Rutin | 5280805 | Foxo1 | −6.3 |
| Muricatocin A | 44584143 | Foxo1 | −6.3 |
| Chlorogenic acid | 1794427 | Foxo1 | −6.1 |
| Muricatocin B | 44584144 | Foxo1 | −6.1 |
| Muricatocin c | 44584147 | Foxo1 | −6.1 |
| Kaempferol | 5280863 | Foxo1 | −6.1 |
| Quercetin | 5280343 | Foxo1 | −6.0 |
| Epicatechine | 182232 | Foxo1 | −5.7 |
| Catechine | 1203 | Foxo1 | −5.7 |
| Roseoside | 9930064 | Foxo1 | −5.6 |
| Loliolide | 100332 | Foxo1 | −5.2 |
| Annopentocin A | 5319155 | Foxo1 | −5.2 |
| Annopentocin C | 5319173 | Foxo1 | −5.2 |
| Annomuricin C | 11758463 | Foxo1 | −5.1 |
| Annomuricin E | 10371584 | Foxo1 | −5.1 |
| Annomuricin B | 44575650 | Foxo1 | −5.0 |
| Murihexocin | 44559048 | Foxo1 | −5.0 |
| Annocatalin | 44566987 | Foxo1 | −5.0 |
| Annomuricin A | 157682 | Foxo1 | −4.9 |
| Muricapentocin | 44559053 | Foxo1 | −4.9 |
| Gigantetronenin | 5470005 | Foxo1 | −4.9 |
| Cis-corossolone | 11093061 | Foxo1 | −4.9 |
| (+)-Epiloliolide | 44511808 | Foxo1 | −4.9 |
| Muricoreacin | 44559047 | Foxo1 | −4.8 |
| Annonacin A | 44575507 | Foxo1 | −4.7 |
| Annopentocin B | 5319163 | Foxo1 | −4.6 |
| Annomutacin | 44337973 | Foxo1 | −4.4 |
| Vomifoliol | 440244 | Foxo1 | −4.4 |
| Gallic acid | 370 | Foxo1 | −4.3 |
Table 1 showed that Annonain, rutin, muricatocin A, isolaureline, xylopine, and kaempferol 3-O-rutinoside active compounds in Annona muricata leaves have an equal or smaller free binding energy than the control, so with an in silico method, those active compounds potentially can inhibit FOXO1 protein. Energy binding control to FoxO1 protein is −6.3. The amount of free binding energy (∆G) is an indicator of the ability to bind the active compound to the target protein. In any spontaneous process, protein–ligand binding occurs only when the change in Gibbs free energy (∆G) of the system is negative when the system reaches an equilibrium state at constant pressure and temperature. Because the protein–ligand association extent is determined by the magnitude of the negative ∆G, it can be considered that ∆G determines the stability of any given protein–ligand complex, or, alternatively, the binding affinity of a ligand to a given acceptor (Gilson and Zhou 2007). Based on that statement that annonain, rutin, muricatocin a, isolaureline, xylopine, and kaempferol 3-O-rutinoside active compounds in Annona muricata predicted spontaneous bond to active side of FOXO1 protein receptor form stable molecule protein–ligand complex.
Analysis of interaction between ligand of Annona muricata leaves active compound and FOXO1 protein with in silico method
Analysis of potential as FOXO1 protein inhibitors showed that anonaine, rutin, muricatocin A, isolaureline, xylopine, and kaempferol 3-O-rutinoside active compounds have an equal or smaller free bonding energy than the control, which is predicted to have the potential as inhibitors of FOXO1 protein. Thus, the interaction between ligand and protein in silico was analyzed to determine the strength of the bond between those active compounds. An active compound is predicted to have strong ties to the target protein if it is capable to bind strongly through hydrogen bonds with the same amino acid residue compared to the control or inhibitor reference. Analysis results of the ligand interaction on Annona muricata leave active compound and FOXO1 protein can be seen in Table 2 (Fig. 1).
Table 2.
Analysis of ligands interaction between active compound of Annona muricata leaves with Foxo1 receptors with in silico method
| Ligand ID | Interaction |
|---|---|
| Xylopine | Hydrogen bond: Ala159, Tyr196, Ser212
Hydrophobic bond: Asn158, Trp160, Tyr165, Trp209 |
| Anonaine | Hydrogen bond: Tyr196, Ser212
Hydrophobic bond: Asn158, Trp160, Tyr165, Gly208, Trp209 |
| Rutin | Hydrogen bond: Asn158, Ala159, Tyr165, Tyr196, Ser205 Hydrophobic bond: Phe197, Lys200, Trp209, Gly208, Ser212 |
| Isolaureline | Hydrogen bond: Tyr165, Tyr196 Hydrophobic bond: Arg157, Asn158, Ala159, Trp160, Ser212 |
| Kaempferol 3-O-rutinoside | Hydrogen bond: Ala159, Tyr196, Ser205, Trp209, Ser212, Asn216 Hydrophobic bond: Asn158, Trp160, Tyr165, Lys200, Gly208, His215. |
| Muricatocin A | Hydrogen bond: Asn158, Asn216,Gly208, Ser212, Hydrophobic bond: Tyr165, Trp160, Tyr196, Lys200, Trp209, Asn211 |
| Inhibitor reference/AS1842856 | Hydrogen bond: Asn158, Tyr165, Ser212
Hydrophobic bond: Ala159, Ser164, Trp160, Tyr196, Phe197, Ser205, Trp209 |
Bold and underline letter indicate that the complex (FOXO1-ligands) has the same interaction with the FOXO1-inhibitor reference
Fig. 1.
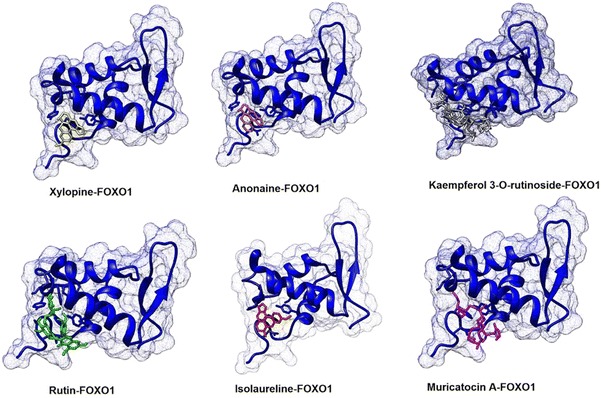
Interaction visualization between ligand and protein target
Table 2 showed that the inhibitor reference was bound to the active side of the target protein through hydrogen bonding with the following types of amino acids Asn158, Tyr165, Ser212 and bound through a hydrophobic bond with the following amino acids Ala159, Ser164, Trp160, Tyr196, Phe197, Ser205, and Trp209.
Rutin and muricatocin A had the same binding ability of 66% amino acid residues compared to the control with hydrogen bonds, whereas xylopine, anonaine, isolaureline, kaempferol 3-O-rutinoside had the same binding ability of 33% amino acid residues compared to the control with hydrogen bonds. An active compound is predicted to have the ability to inhibit the receptor to approach control if they have binding similarity between the amino acid residue and control. Based on the result showed that rutin and muricatocin A predicted more smaller ability to inhibit FOXO1 receptor than control.
Discussion
Potential analysis of the Annona muricata leaves active compound as FOXO1 protein inhibitors with in silico method
Results from the previous study showed that the active compound of Annona muricata leaves can improve pancreatic β cell proliferation in rat that was induced by STZ (Adeyemi et al. 2010). However, what active compound and how the mechanism works were unknown. Thus, an in silico analysis was performed to estimate the role of the active compounds and the mechanism of β cells regeneration.
Verification regarding the predictive binding and inhibiting ability of the Annona muricata leaves active compound and FOXO1 protein was done through molecular docking method, which was the binding simulation between Annona muricata leaves active compound and the FOXO1 protein, and then it was compared to the control that has been known to inhibit FOXO1 protein.
Several transcription factors were associated with the proliferation of pancreatic β cells, such as Pdx1, Pax4, FOXO1, and FoxM1. FOXO1 is a subfamily of a transcription factor from Forkhead Transcription factors, which includes: FOXO1, Foxo3, and Foxo4 protein in mammals. The activation of these proteins is regulated by phosphorylation through the phosphoinositide 3 kinase-Akt. Normally, FOXO1 can be found in the cytoplasm. Phosphorylation of FOXO1 protein in the cytoplasm activates cyclin D1 and Cdk4 protein, and thus activates the cell cycle from G0 to G1 phase for the preparation of DNA synthesis. However, several factors have been reported to cause FOXO1 protein migration to the nucleus, such as high glucose level in blood, hyperlipidemia, and the aging process (Gross et al. 2008).
The role of FOXO1 protein in pancreatic β cell proliferation process has been proved by some previous researchers. Research conducted by Martinez et al. proved that the administration of palmitic acid in a culture of mouse insulinoma cells (MIN6) can increase the activity of FOXO1 protein in the nucleus (Martinez et al. 2008). It activates the target genes transcription (CHOP) and thus induces the apoptosis. The inhibition of the FOXO1 protein active side was proved to be capable of inhibiting MIN6 cell apoptosis that was induced by palmitic acid (Martinez et al. 2008). Nagashima et al. (2010) proved that the inhibition of the FOXO1 protein activity in the nucleus causes suppression to the gluconeogenesis process in the liver that increases insulin sensitivity in the peripheral tissues and liver of DM mice. Another study conducted by Talchai et al. (2012) proved that FOXO1 protein can affect the de-differentiation process of pancreatic β cell. Research results proved that the experimental animal that does not have FOXO1 protein in the β cell of cytoplasm experiences hyperglycemia and decreased β cell mass after a stress induction. In vitro method demonstrated that the reduction in β cell mass is not caused by the death cell processes, but due to the de-differentiation of β cell into an embryonic progenitor cell that expresses Neurogenin 3, Oct 4, Nanog, and L-Myc. In addition, it demonstrated that the loss of FOXO1 protein in the cytoplasm leads to an increase of α, δ, and PP cells, while the decrease in β cell is characterized by a decrease of gene activity in the insulin synthesize, GLUT transporter and glucokinase enzyme (Talchai et al. 2012). Results of these previous studies showed that FOXO1 protein can be one of the research targets in the development of DM treatment. Inactivation of FOXO1 protein in the nucleus or prevention of FOXO1 protein transcription factor translocation from the cytoplasm to the nucleus is a way to prevent the occurrence of apoptosis or a decrease in pancreatic β cells.
In this study, active compounds of the Annona muricata leaves and the ligand were obtained from literature studies. Results of the literature studied found 33 kinds of active compounds in Annona muricata leaves. The active compounds were included in the group of alkaloids, annonaceous acetogenin, megastigmane, flavonols triglicoside, phenolic and cyclopeptide (Moghamtousi et al. 2015). Then, a simulation to bind all of the active compounds with the active side of FOXO1 protein was performed computationally and was compared to the drug control that inhibits FOXO1protein.
The drug that was used as a control is the active compound of a small molecule that were identified to be 5-amino-7-(cyclohexyl-amino)-1-ethyl-6-fluoro-4-oxo-1,4-dihydroquinoline-3-carboxylic acid (AS1842856). The drug was able to bind with the active side of human FOXO1 protein and inhibit FOXO1 trans-activation activity. In vitro administration of AS1842856 on the hepatic cell line was proved to cause a decrease in glucose production. The result of the in vivo study from the AS1842856 administration in DM mice was proven to reduce glucose level of the fasting blood (Nagashima et al. 2010).
Results of docking analysis indicated that the active compound of Annona muricata leaves was anonaine, rutin, muricatocin A, isolaureline, xylopine, and kaempferol 3-O-rutinoside, which have lower or equal free bonding energy to the drug control.
When a protein binds to a ligand, there will be interaction between the ligand and protein, apart from its bond with water. The bond between the protein and ligand forms a stable bond. To form this stable bond, ligand will occupy the protein binding pocket that is composed of amino acids.
The amount of free binding energy is an indicator of the ability to bind the active compound to the target protein. Free binding energy is the enthalpy change that is required to break a particular bond in one mole of gas molecules. If the binding energy is high then the molecular bonds tend to be stronger, more stable and less reactive. A more reactive compound has a lower binding energy (Chang 2006). The active compound is predicted to have the ability to bind to the target protein and interact spontaneously if it has a lower or equal free bonding energy than the control (Zukhurullah et al. 2012).
Based on the data above, it can be concluded that the active compounds of Annona muricata leaves were anonaine, rutin, muricatocin A, isolaureline, xylopine, and kaempferol 3-O-rutinoside and they were able to interact with the active side of FOXO1 protein spontaneously. The interaction restrained the work of FOXO1 protein.
Analysis of interaction between ligand of Annona muricata leaves active compound and FOXO1 protein with in silico method
An active compound is predicted to have a strong bond to the target receptor if it can bind strongly via hydrogen bonds and can bind with one of the same amino acid residues of the active side compared to a control or inhibitor reference (Zukhurullah et al. 2012). An active compound is predicted to have stronger ability to inhibit the receptor if it has more amino acid residues binding similarity with the control.
Hydrogen bonding is the electrostatic interaction between hydrogen atoms that is attached to an electronegative atom with another electronegative atom. The hydrogen bond strength is under the covalent bond, but its presence is very important. Its presence contributes to molecule structures and characteristics. In medicine, hydrogen bond plays a role in studying the design and interaction between drug molecules and metabolic system in the body (Jeffrey 1997).
In silico analysis results indicated that the inhibitor reference was bound to the active side of the receptor through hydrogen bond with the Asn158, Tyr165, Ser212 amino acids and was bound through hydrophobic bond with Ala159, Ser164, Trp160, Tyr196, Phe197, Ser205, Trp209 amino acids. Rutin and Muricatocin A had the same binding ability of 66% amino acid residues compared to the control with hydrogen bonds, whereas xylopine, anonaine, isolaureline, kaempferol 3-O-rutinoside had the same binding ability of 33% amino acid residues compared to the control with hydrogen bonds. Thus, it can be predicted that the xylopine, anonaine, isolaureline, kaempferol 3-O-rutinoside. Rutin and muricatocin A compound of Annona muricata leaves can increase pancreatic β cell proliferation through the inhibition of FOXO1protein.
Conclusion
Results from in silico analysis on the binding and inhibiting ability of active compounds in Annona muricata leaves to the FOXO1 protein showed that xylopine, anonaine, isolaureline, kaempferol 3-O-rutinoside, rutin and muricatocin A compound have the ability to strongly and spontaneously bound with the active side of FOXO1 protein, and it also have the ability inhibit the activity FOXO1 protein.
Acknowledgements
We thank to Prof. Dr. M. Aris Widodo MS, Sp.FK, PhD, DR. Dra. Sri Winarsih, Apt, Mkes and Prof. DR. Dr. Achmad Rudijanto, Sp.PD, KEMD for their advice to conduct this study.
Author contributions
First author (DSD) conducted research and wrote paper, second author (DHU) designed study and reviewed paper, third author (CK) designed study and reviewed paper.
Compliance with ethical standards
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Conflict of interest
The authors have no competing interest for this research.
References
- Adeyemi DO, Komolafe OA, Adewole OS, Obuotor EM, Abiodun AA, Adenowo TK. Histomorphological and morpholometric studies of the pancreatic islet cell of diabetic rats treated with extracts of Annona muricata. Folia Morphol. 2010;69:92–100. [PubMed] [Google Scholar]
- Chang R. Kimia dasar: konsep-konsep inti. 1. Jakarta: Erlangga; 2006. [Google Scholar]
- Dallakyan S, Olson AJ. Small-molecule library screening by docking with PyRx. Methods Mol Biol. 2015;1263:243–250. doi: 10.1007/978-1-4939-2269-7_19. [DOI] [PubMed] [Google Scholar]
- DeLano WL (2002) The PyMOL molecular graphics system. DeLano Scientific, Palo Alto. http://www.pymol.org. Accessed 28 July 2016
- Gajalakshmi S, Vijayalakshmi D, Rajeswari V. Phytochemical and pharchological of properties of Annona muricata: a review. Int J Pharm Pharm Sci. 2012;4:3–6. [Google Scholar]
- Gilson MK, Zhou HX. Calculation of protein-ligand binding affinities. Annu Rev Biophys Biomol Struct. 2007;36:21–42. doi: 10.1146/annurev.biophys.36.040306.132550. [DOI] [PubMed] [Google Scholar]
- Gross DN, van den Heuvel AP, Birnbaum MJ. The role of FoxO in the regulation of metabolism. Oncogene. 2008;27:2320–2336. doi: 10.1038/onc.2008.25. [DOI] [PubMed] [Google Scholar]
- Hileman B. Accounting for R&D, many doubt the $800 million pharmaceutical price tag. Chem Eng News. 2006;84:50. [Google Scholar]
- Jeffrey GA. An introduction to hydrogen bonding. Oxford: Oxford University Press; 1997. [Google Scholar]
- Malviya N, Jain S, Malviya S. Antidiabetic potential of medicinal plants. Acta Pol Pharm. 2010;67:113–118. [PubMed] [Google Scholar]
- Martinez SC, Cras-Méneur C, Bernal-Mizrachi E, Permutt MA. Glucose regulates Foxo1 through insulin receptor signaling in the pancreatic islet beta-cell. Diabetes. 2006;55:1581–1591. doi: 10.2337/db05-0678. [DOI] [PubMed] [Google Scholar]
- Martinez SC, Tanabe K, Cras-Méneur C, Abumrad NA, Bernal-Mizrachi E, Permutt MA. Inhibition of Foxo1 protects pancreatic islet beta-cells against fatty acid and endoplasmic reticulum stress-induced apoptosis. Diabetes. 2008;57:846–859. doi: 10.2337/db07-0595. [DOI] [PubMed] [Google Scholar]
- Moghamtousi SZ, Fadaeinasab M, Nikyad S, Mohan G, Ali HM, Kadir HA. Annona muricata (Annonaceae): a review of traditional uses, isolated acetogenins and biological activities. Int J Mol Sci. 2015;16:15625–15658. doi: 10.3390/ijms160715625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagashima T, Shigematsu N, Maruki R, Urano Y, Tanaka H, Shimaya A, Shimokawa T, Shibasaki M. Discovery of novel forkhead box O1 inhibitors for treating type 2 diabetes: improvement of fasting glycemia in diabetic db/db mice. Mol Pharmacol. 2010;78:961–970. doi: 10.1124/mol.110.065714. [DOI] [PubMed] [Google Scholar]
- Pedro F, Lei H. A systematic review on computer-aided drug design: docking and scoring. J Macao Politech Inst. 2010;4:47–51. [Google Scholar]
- Talchai C, Xuan S, Lin HV, Sussel L, Domenico A. Pancreatic β-cell dedifferentiation as mechanism of diabetic β-cell failure. Cell. 2012;150:1223–1234. doi: 10.1016/j.cell.2012.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadood A, Ahmed N, Shah L, Ahmad A, Hassan H, Shams S. In-silico drug design: an approach which revolutionarised the drug discovery process. OA Drug Des Deliv. 2013;1:1–3. [Google Scholar]
- Wallace AC, Laskowski RA, Thornton JM. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- Zukhurullah M, Aswad M, Subehan S. Kajian beberapa senyawa antiinflamsi: docking terhadap siklooksigenase-2 secara in silico. Majalah Farmasi dan Famakologi. 2012;16:37–44. [Google Scholar]


