Abstract
Indole, as a signal molecule, is involved in multiple physiological behavior including biofilm formation, antibiotic resistance and virulence. In this study, we demonstrated that indole was involved in iron deficient and H2O2 stress response in Muricauda olearia Th120. Transcriptome analysis showed that totally 206 genes were regulated by exogenous indole. Besides, momL-suf gene cluster, consisting of quorum quenching enzyme coding gene momL and iron-sulfur biosynthetic genes suf, were involved in indole-induced stress response pathway. The result indicated that indole not only up-regulated momL-suf gene cluster, but also enhanced the MomL secretion and the growth rates of MomL-bearing strains in H2O2 stress and iron deficient culture conditions. Co-incubation of M. olearia Th120 and Pectobacterium carotovorum subsp. carotovorum under H2O2 condition revealed that M. olearia Th120 bearing MomL possessed an increased competitive advantage, whereas its competitor had a reduced survival. The phenomenon that quorum quenching enzyme is triggered by stress factor has been rarely reported. The study also opens a new clue to explore the indole function towards quorum quenching factor in bacteria.
Introduction
Quorum sensing (QS) is a population-dependent behavior that enables bacteria to communicate with each other and to regulate downstream gene expression in response to environmental changes1–5. During growth, bacteria produce a type of small molecule known as an autoinducer (AI), which can bind to receptor proteins and trigger signal pathway cascades when the AI concentrations reach a threshold value. At present, several types of AIs have been discovered, such as the N-acyl homoserine lactones (AHLs) common in many Gram-negative bacteria1, 6, oligopeptides in Gram-positive bacteria and AI-2 in both Gram-negative and Gram-positive bacteria7–9. Indole is another signal molecule which has recently received increased attention as a putative QS signal molecule10, 11. Many Gram-positive and Gram-negative bacteria encode a single copy of the tnaA gene in their chromosome and produce indole. It plays diverse biological roles including biofilm formation, sporulation and virulence10, 12–14. It has also been shown that indole, via two component signal transduction systems, increases antibiotic resistance by inducing exporter and transporter genes in E. coli 10. Hence, it is possible that these two-component signal systems can be used as indole sensors. However, the indole signaling pathway and its function in stress response have not been comprehensively studied, especially in beneficial microorganisms.
Previous studies have demonstrated that QS systems are closely related to bacterial disease. Interfering with QS systems, named quorum quenching (QQ), is a promising control strategy for bacterial diseases because it could decrease the expression of virulence factors of pathogens15. Normally, virulence factor is the weapon for pathogens during adverse situation, so theoretically, QQ could weaken pathogens. The marine bacteria Muricauda olearia Th120 exhibits strong QQ activity, which makes this strain a potential biocontrol agent16. In our previous study, a novel AHL lactonase, MomL, was identified from M. olearia Th12017. MomL belongs to the metallo-β-lactamase superfamily and shows the highest identity (56.8%) with Aii20J from Tenacibaculum sp. 20J18. Besides, MomL possesses 54.4% and 24.5% identity with FiaL and well-studied AiiA from Flaviramulus ichthyoenteri Th78T and Bacillus sp. 240B1 respectively19–21. As a novel AHL lactonase in marine isolates, MomL shows promising activity against C6-homoserine lactone (C6-HSL), reaching a catalytic efficiency of 2.9 × 105 s−1M−1.
Iron-sulfur (Fe/S) proteins, characterized by the presence of an iron-sulfur cluster, play crucial roles in multiple cellular processes in both prokaryotic and eukaryotic cells22. The biogenesis of iron-sulfur clusters has been extensively studied in Escherichia coli 22, 23, in which there are two iron-sulfur cluster biosynthesis gene operons: the ISC (iron-sulfur cluster) and Suf (sulfur assimilation) systems23. ISC genes are organized as iscSUA-hscBA-fdx, whereas Suf genes are organized as sufABCDSE. The ISC gene cluster is thought to play housekeeping roles, including the formation of a variety of Fe/S proteins, whereas the Suf system is active under stress conditions, such as iron starvation22, 23.
Interestingly, momL is located within the stress-response suf operon and is closely linked to sufB and sufC. This gene organization has not been previously reported in other organisms. Also, little is known about how the QQ factor momL is regulated by QS in M. olearia Th120. In this study, we focused on the regulation mechanism of signal molecular indole towards the stress-response genes suf and QQ gene momL, and it firstly described the triggering of the expression of a QQ enzyme by stress factors.
Material and Methods
Bacterial strains, media and growth conditions
M. olearia Th120 was cultured in marine broth 2216 at 28 °C. The AHL biosensor Chromobacterium violaceum CV026 was cultured in LB medium at 28 °C17. Indole (Sigma-Aldrich, St. Louis, Missouri, USA) stock solutions were prepared in methanol, and the 2,2′-dipyridyl (Solarbio) stock solution was prepared in ethanol.
H2O2 sensitivity assay
M. olearia Th120 was grown in marine broth 2216 at 28 °C overnight. The cultures were then diluted with fresh medium to a final optical density at 600 nm (OD600) of 0.1. The cell suspensions were mixed with H2O2 or H2O2 with 0.2 mM indole at final concentrations of 0.5, 1 or 2 mM and were incubated at 28 °C for 20 min. The cells were serially diluted and spread onto marine agar plates, and the colony numbers were counted from each of the plates after 48–72 hours of incubation24.
RNA extraction, reverse transcription PCR, and real-time-PCR
M. olearia Th120 cells were collected after reaching an OD600 of 0.8, and the RNA was extracted using an RNA extraction kit (Omega) according to the manufacturer’s instructions. For indole treatments, samples with different cultivation conditions were harvested as described above. For the H2O2 treatment assay, mid-log-phase cultures of M. olearia Th120 were harvested. After a 30-min treatment with 300 μM H2O2, the RNA was extracted and was reverse-transcribed to cDNA. Quantitative real-time PCR was performed in a total reaction volume of 20 μl containing 250 nM primers, 10 μl of SYBR green PCR master mix, 8.5 μl of RNase-free water, and 0.5 μl of a 10-fold-diluted cDNA template. The 16S rRNA was used as a reference gene25. Relative gene expression level was analyzed using 2−ΔΔCt method (ΔΔCt = ΔCt(test) − ΔCt(calibrator), ΔCt(test) = Ct(target, test) − Ct(reference, test) and ΔCt(calibrator) = Ct(target, calibrator) − Ct(reference, calibrator)). The primers for real-time PCR are listed in Table S1. Real-time PCR was performed with a StepOne Real-Time PCR System (AB Applied Biosystems). The program was designed as follows: pre-incubation at 95 °C for 10 min followed by 35 cycles of 15 seconds at 95 °C, 30 seconds at 55 °C and 30 seconds at 72 °C. After 2 hours, cycle threshold (CT) values and melting curves of each reaction were analyzed using StepOne Software v2.2.
Extracellular secretion detection of MomL
M. olearia Th120 cells were grown for 48 hours, and the OD600 values of the samples (treated and untreated) were measured using a spectrophotometer. The cultures were diluted with fresh medium to the same OD600 value. Filter-sterilized (0.22 μm pore size) supernatant and cells were collected, followed by a 10-min centrifugation at 4 °C at 6000 × g. The AHL biosensor Chromobacterium violaceum CV02626, which could produce violacein induced by AHL, was cultured on LB medium at 28 °C to an OD600 of 0.1, and 1 mL of culture and C6-HSL at a final concentration of 0.5 μM were added to solid LB medium. The extracellular secretions of samples from different culture conditions were added to the medium, which was placed in an incubator at 28 °C. After 24 hours of culture, transparent circles were observed if AHL was degraded and violacein synthesis was inhibited. The area of transparent circle was calculated and the mean value of three repeated experiment was used.
Bioinformatic analysis
Protein domains were analyzed by SMART software (http://smart.embl-heidelberg.de/smart/set_mode.cgi?NORMAL=1) for online analysis. The suf and momL sequences of the different strains were analyzed by BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Annotation and bioinformatic analysis were performed on the flanking genes of momL by genome sequencing and EMBOSS (The European Molecular Biology Open Software Suite) (http://emboss.open-bio.org/). DNAMAN software was used to compare the three copies of the OxyR protein. Primers for real-time fluorescent quantitative PCR were designed using Primer Premier 527.
Co-cultures of Pectobacterium carotovorum subsp. carotovorum (Pcc) and E. coli
The two types of bacteria were cultured in LB medium separately, and cells were collected by centrifugation when the OD600 reached 0.8. The two types of bacteria were mixed and cultured in 10% tryptic soy broth (TSB) medium with 2 mM H2O2 for 6 hours. The bacterial co-culture was serially diluted. At each concentration, dilutions were spread onto two LB plates, one of which contained 25 µg/mL Km. The survival of Pcc was the difference between the bacterial counts of the two LB plates (NumbersLB − NumbersLB+Km).
Co-cultures of P. carotovorum subsp. carotovorum (Pcc) and M. olearia Th120
Pcc and M. olearia Th120 were cultured in LB and MA media28, 29 separately until the OD600 reached 0.8. Cultures of the two bacteria were centrifuged, and the two types of cells were collected and mixed together. Then, the mixture was inoculated into 10% TSB medium containing 2 mM H2O2 and cultured for 6 hours. During the culturing, various concentrations of MomL protein were added into the medium. After culturing, the cultures were serially diluted and spread onto MA and LB plates, separately (MA is the regular medium for marine bacterium M. olearia Th120, and LB is for Pcc. Besides, M. olearia Th120 can not survive on LB medium, while, Pcc can not live on MA medium). Subsequently, changes in the survival of the two bacteria in the different conditions were observed.
Transcriptome sequencing and analysis
Transcriptome sequencing of M. olearia Th120 (treated with and without indole) was performed by the Biozeron company in Shanghai, China. Total RNA of M. olearia Th120 was extracted using TRIzol reagent (Invitrogen) and treated with DNase I (Takara). RNA quality was determined using a Bioanalyser 2100 (Agilent), after which the RNA was quantified using a NanoDrop 2000. RNA transcriptome libraries were constructed following a TruSeq RNA sample preparation kit from Illumina (San Diego, CA), using 5 μg of total RNA. rRNA was removed using a RiboZero rRNA removal kit (Epicenter) and fragmented with fragmentation buffer. cDNA synthesis, end repair, A-base addition and ligation of the Illumina-indexed adaptors was performed following Illumina’s protocol. Libraries were then size selected for cDNA target fragments of 200–300 bp on 2% Low Range Ultra Agarose followed by PCR amplified using Phusion DNA polymerase (NEB) for 15 PCR cycles. After generating the clusters, library sequencing was performed on an Illumina Hiseq platform, to create paired-end reads with lengths of 150 bp. The raw paired-end reads were trimmed with SeqPrep (https://github.com/jstjohn/SeqPrep) and quality controlled with Sickle (https://github.com/najoshi/sickle). Then, clean reads were aligned to the reference genome using Rockhopper (http://cs.wellesley.edu/~btjaden/Rockhopper/). Rockhopper was a user-friendly software for bacterial RNA-seq data anaylsis. This software was used to calculate gene expression level as RPKM (reads per kilobase transcriptome per million mapped reads) with default parameters. RPKM = (106 * C)/(N * L), with C = Number of reads mapped to a gene; N = Total mapped reads in the experiment; L = gene length in base-pairs for a gene (Kb) EdgeR30 was used for differential gene expression analysis (https://bioconductor.org/packages/release/bioc/html/edgeR.html). To study the functions of the differentially expressed genes, GO functional enrichment and KEGG pathway analysis were carried out by Goatools (https://github.com/tanghaibao/Goatools) and KOBAS (http://kobas.cbi.pku.edu.cn/home.do) respectively. Gene expression changes higher than 2-fold with P-values less than 0.05 were regarded as reliable and marked differences. The sequencing data were deposited in NCBI Sequence Read Archive with accession number PRJNA375720.
Results
Indole improved survival state of M. olearia Th120 in H2O2 stress and iron deficient conditions
Previous studies indicated that indole is responsible for multiple physiological behaviors, especially in enteric bacteria. Besides, it was reported that indole could leads bacterial persistence formation by activating stress responses31. However, the indole function towards stress responses in beneficial bacteria M. olearia Th120 is largely unknown. Our experiments indicate that indole can improve the survival of M. olearia Th120 in the presence of H2O2. Specifically, the survival of cells in 0.5, 1, and 2 mM H2O2 were 80%, 70%, and 45%, respectively, in the presence of 200 μM indole. However, the survival of the cells were 60%, 45%, and 15% in the presence of H2O2 but the absence of indole (Fig. 1A). Besides, the survival state under iron deficient was also detected. The growth rate assay indicated that 200 μM exogenous indole caused M. olearia Th120 to have an improved growth rate in lag and exponential phases under ferric-deficient conditions. No obvious difference was observed after stationary phase. What is more, indole did not affect the cell yield of M. olearia Th120 (Fig. 1B and Fig. S1).
Figure 1.
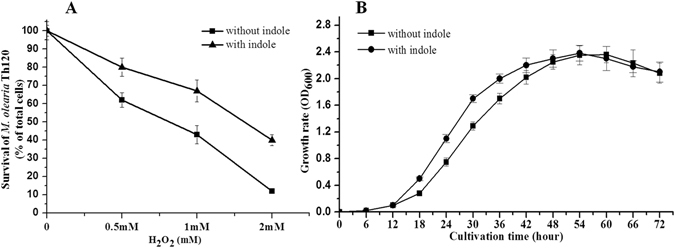
Function of indole for M. olearia Th120 in iron deficient and H2O2 conditions. (A) The survival of M. olearia Th120 upon H2O2 treatment with or without indole. (B) M. olearia Th120 growth rate assay under conditions of ferric infertility (100 μM 2,2′-dipyridyl). Error bars represent the standard deviations of three replicates.
suf-momL was involved in indole-induced stress responses in a co-transcriptional manner
In E. coli, the suf operon is responsible for resistance to iron starvation23. We focused on the study of whether suf of M. olearia Th120 was involved in indole-induced iron deficient and H2O2 stress response. Interestingly, genome sequencing and bioinformatic studies indicated that the suf genes of M. olearia Th120 is co-localized within the momL gene, an arrangement that has not been reported in similar well-studied genes, such as aiiA. The classic suf locus contains sufA, sufB, sufC, sufD, sufS, and sufE (Fig. S2). SufS and SufE are desulfurase homologs, whereas SufC is an ATPase. SufS and SufE are responsible for passing the sulfur that dissociates from cysteine to the Fe-S assembly complex, which is composed of two SufC units and single copies of both SufB and SufD. With the participation of the cofactor FADH2, the Suf proteins assemble iron and sulfate into Fe-S clusters. SufA is responsible for transporting Fe-S clusters to the aconitase, AcnB. In previous studies, suf was always observed as an intact gene cluster with no other unrelated genes among the suf genes. In M. olearia Th120, momL is inserted between sufB and sufC, and these three genes are linked closely to each other.
To analyze the transcriptional features of momL and suf, orfE/orf1/sufA/momL/sufD/orf10 and orfF were chosen for real-time PCR analysis. In normal condition, the sufA, momL, sufD, orf10 and orfF transcription levels were highly similar, whereas the transcript level of orfE, whose expression was approximately 2-fold higher compared with that of sufA, differed greatly from the expression level of sufA-orfF. Moreover, the expression level of orf1 was very low: only 20% of that of sufA (Fig. 2A). These differences suggest that suf and momL are co-transcribed, whereas orf1 and orfE are expressed independently of suf-momL. In addition, orf10, which encodes a novel Fe-S cluster synthesis protein, and orfF, which encodes an unknown protein, are also co-transcribed with suf-momL.
Figure 2.
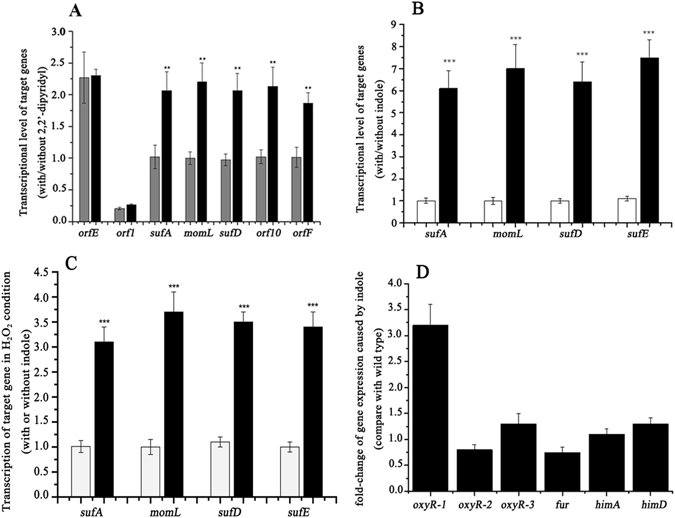
The transcriptional analysis (Ct values) of related genes. (A) Transcription levels of genes in the suf-momL operon with (black) or without (gray) the iron-chelating agent 2,2′-dipyridyl. (B) Transcription levels of suf-momL genes in iron-deficient condition with (black) or without indole (transparent). (C) indole-induced transcriptional change in H2O2 condition (black, with indole; transparent, without indole). (D) The effects of indole on a series of suf regulators. Error bars represent the standard deviations of three replicates. For statistical analysis, ***/**/* means P < 0.001, P < 0.01, and P < 0.05, respectively.
suf-momL was involved in indole-induced stress responses pathway. The transcription of suf-momL was analyzed under indole-induced iron-deficient and H2O2 condition. Without indole, the sufA, sufD, orf10 and orfF expression levels were approximately 2-fold higher in an environment containing the intracellular iron (II) chelator 2,2′-dipyridyl (100 μM), whereas the orfE and orf1 expression levels were not significantly altered in this iron-deficient condition (Fig. 2A). While, with 200 μM indole, the transcription levels of sufA, sufD, and sufE increased 6-, 6-, and 7-fold, respectively, and the transcription level of momL increased nearly 7-fold in this iron-deficient condition (Fig. 2B). Similarly, in H2O2 stress condition, indole up-regulated sufA, sufD, sufE and momL to 3–4 fold changes respectively (Fig. 2C). Previous studies have indicated that three proteins are involved in regulating the suf genes: OxyR, a peroxide-sensing positive regulator; Fur, a ferric uptake regulator; and HimA/D, a bacterial nucleoid DNA-binding protein that induces increased suf expression during oxidative stress23, 32, 33. A bioinformatic analysis of M. olearia Th120 revealed that the genome includes not only the fur and him genes but also three copies of the oxyR gene, designated oxyR1/2/3. These oxyR genes contain well-conserved DNA-binding sites (Fig. S3). Transcriptional analysis showed that there were no marked changes in the levels of oxyR2, oxyR3, fur, or himA in the presence of indole, whereas oxyR1 expression increased significantly at 3.2-fold over normal conditions (Fig. 2D).
MomL secretion ability in different iron conditions was also studied using the C. violaceum mutant CV026 as an indicator strain. MomL secretion decreased in an environment of adequate iron nutrition, whereas it was enhanced in iron deficient conditions (Fig. 3A). In iron deficient condition, MomL secretion increased with the addition of indole, which increased 1.2-fold in the presence of 10 and 50 μM indole. At a concentration of 200 μM indole, MomL secretion increased approximately 2-fold in iron deficient condition (Fig. 3B). Besides, under H2O2 condition, indole could also increase MomL secretion significantly (Fig. 3C). Because of MomL known as AHL lactonase, to analyze whether the AHL lactonase activity plays role in H2O2 condition, co-cultivation assays of different bacterial species were performed. P. carotovorum subsp. carotovorum (Pcc), once classified as Erwinia carotovorum and regarded as a ubiquitous plant pathogen producing virulence factors through the induction of the signal molecule AHL, was used in this experiment34. In 10% TSB medium with 2 mM H2O2, the survival of Pcc were 43% and 40% when Pcc cells were cultured alone or co-cultured with E. coli-pET24a, respectively. Because the survival of Pcc was calculated by formula NumbersLB − NumbersLB+Km (method part for detailed information), in order to exclude the possibility of spontaneous Km antibiotic resistance of Pcc, control experiment was performed. The result indicated that Km is completely lethal for Pcc (Table S2). However, the Pcc survival was significantly reduced to 21% when the cells were co-cultured with E. coli-momL, the E. coli strain containing heterologous gene momL (Fig. 4A). Moreover, Pcc and M. olearia Th120 co-culture assays were also performed. Pcc had a similar survival with M. olearia Th120 under H2O2 conditions. However, adding exogenous purified MomL protein led to a decreased survival of Pcc, and correspondingly, an increased survival of M. olearia Th120. As a control assay, boiled inactived MomL had no function (Fig. 4B and Table S3). Transcriptional analysis indicated that the exogenous MomL protein can significantly reduce the expression and the production of the pectate lyase gene, the major virulence factor of Pcc (Fig. 4C and D). To study the effect of MomL on pectate lyase expression, four related genes (pectate lyase gene pel, pectate lyase precursor 1 gene pelp1, pectate lyase precursor 2 gene pelp2 and pectin lyase regulator gene pelr) were detected by real-time PCR. The expression levels of pel, pelp1 and pelp2 decreased significantly in the presence of MomL. However, the regulator gene pelr was up-regulated, leading to speculation that pelr plays a negative regulatory role in pectin lyase expression (Fig. 4C).
Figure 3.
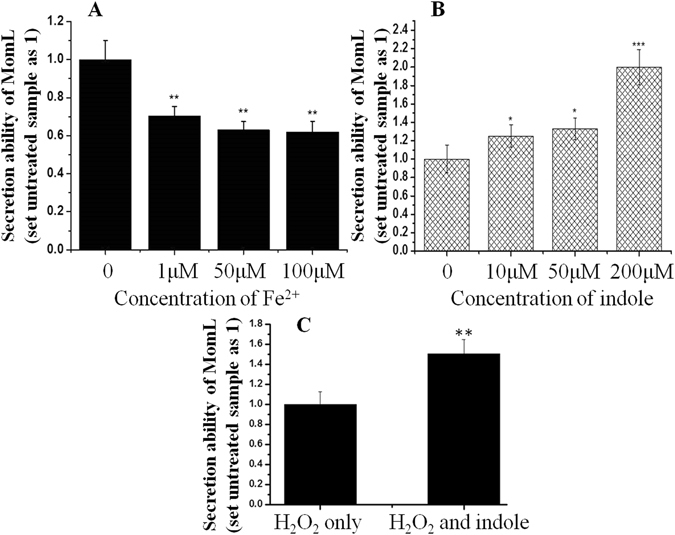
MomL secretion capacity analysis. (A) The secretion capacity of MomL in different iron environments. (B) Indole’s function for the MomL secretion capacity in iron deficient condition. (C) Indole’s function for the MomL secretion capacity in H2O2 condition.
Figure 4.
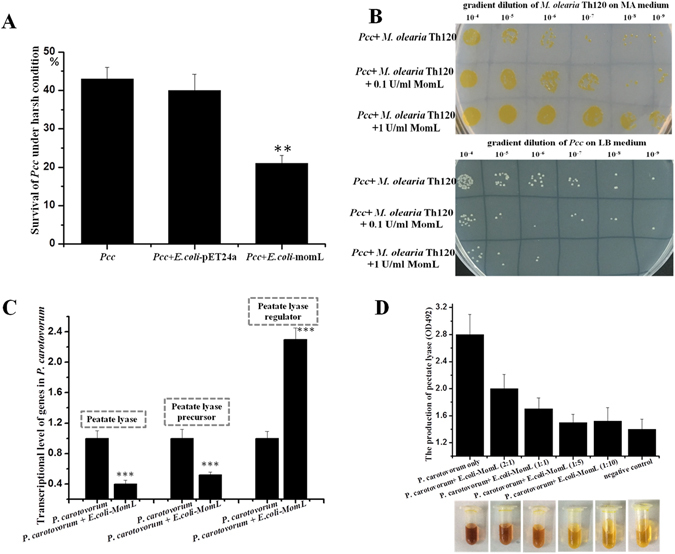
Co-cultivation assays of different bacterial species. (A) P. carotovorum subsp. carotovorum (Pcc) and E. coli-momL co-cultivation. E. coli and E. coli- pET24a are used for the negative control. (B) Pcc and Muricauda olearia Th120 co-culture on 10% TSB medium containing 2 mM H2O2. After culturing, the cultures were serially diluted and spread onto MA and LB plates, separately (MA and LB were medium for M. olearia Th120 and Pcc respectively. What is more, marine bacterium M. olearia Th120 can not survive on LB medium, while, Pcc can not live on MA medium). (C) Transcriptional analysis of pectate lyase genes of M. olearia Th120. (D) Detection of pectate lyase production.
Transcriptome analysis of M. olearia Th120 under indole conditions
To further analyze the function of indole, we performed transcriptome sequencing of M. olearia Th120 with and without 200 μM exogenous indole. Illumina deep sequencing produced approximately 38 M total clean reads from the 5.5 Gbp clean sequence data for each sample. Around 99% of the clean reads had quality scores over the Q20 value. Over 97.9% of the clean reads were mapped to the reference genome (PRJNA244114). In total, 2,885 genes were detected and quantified among all of the samples.
To test the reliability of the transcriptome assay, we randomly chose 20 genes to perform real-time PCR and found that compared with the transcriptome data, the R 2 value (Pearson correlation coefficient) was 0.9185 (Fig. S4), indicating a good correlation. Moreover, consistent with real-time PCR assay, suf-momL gene cluster and oxyR1 were identified as indole-induced genes in the transcriptome assay (Table S4). Volcano plots, which integrated both the P value and fold change of each transcript, were constructed to present the general scattering of the transcripts and filter the differentially expressed genes for indole treatment. Genes regulated by indole were pooled together (Fig. 5, pink and blue color). A total of 206 genes were regulated more than 2-fold by exogenous indole (P > 0.05), among which 104 genes were up-regulated and 102 were down-regulated.
Figure 5.
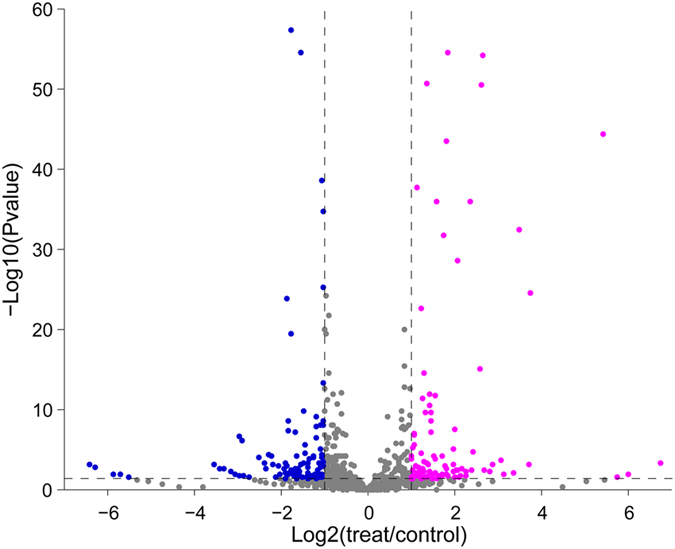
Volcano plots of the overall scatter of gene transcription of M. olearia Th120 (with or without indole treatment). P value < 0.05 and |log2 (foldchange)| > 1 were selected as the criteria to filter genes that were significantly regulated (down-regulated genes blue dots, up-regulated genes pink dots).
The differentially expressed genes were functionally annotated after being input into the GO analysis and were divided into three major categories: biological process, molecular function and cellular component (Fig. 6). For the sample treated by indole, the most affected biological process were associated with cellular process and metabilic process. Interestingly, multiple genes for response to stimulus and signaling were also affected by indole in M. olearia Th120. Furthermore, for cellular component, the significantly regulated part were membrane and membrane-related part. For molecular function, transporter and binding activity were mainly affected by indole. The KOBAS analysis revealed the enriched KEGG pathways under indole treatement (Table S5). Four gene categories, that is, lysine biosynthesis, vancomycin resistance, C5-branched dibasic acid metabolism and two-component system were significantly enriched.
Figure 6.
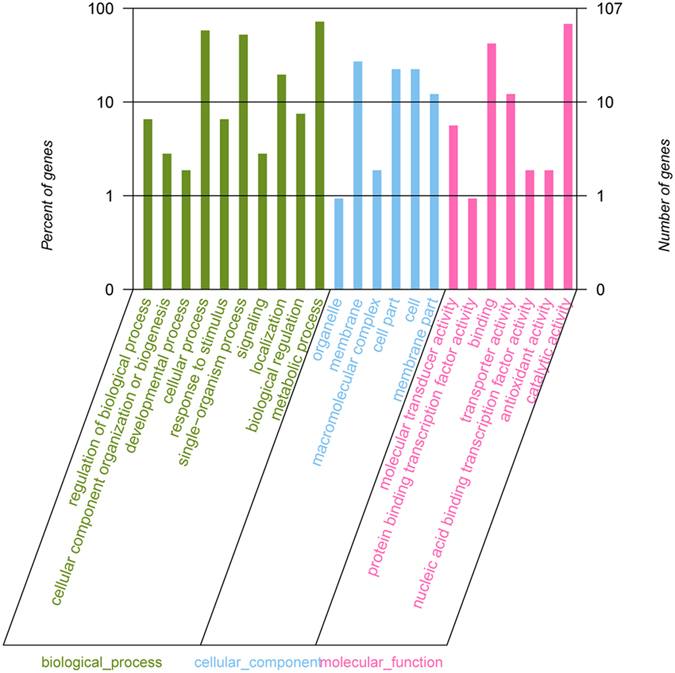
Functional categories of the regulated genes, broadly separated into ‘biological process’, ‘cellular component’, and ‘molecular function’, based on Gene Ontology.
Besides KEGG and GO analysis, our transcriptome results also revealed that five TonB system genes responded to indole, and all of them were up-regulated. Among them, four were noted as TonB-dependent receptors, and their changes in expression ranged from 2 to 8 times the expression levels of indole-free samples. One TonB protein-coding gene was up-regulated 6.5-fold (Table S6). TonB systems can function in iron acquisition. Additionally, 24 membrane proteins, containing transport systems and two component regulatory system proteins, were regulated by indole, among which 21 were up-regulated and only 3 were down-regulated.
Discussion
M. olearia Th120, which was previously isolated from the body surface mucus of Paralichthys olivaceus, produces enzymes capable of degrading signal molecules. Located in the genome of M. olearia Th120, momL encodes an AHL lactonase. The location of the momL gene is unique, as it is located within the iron-sulfur protein-coding suf genes, with momL inserted between sufB and sufC. The combination of QQ gene(s) and stress-response gene(s) has not previously been observed. The location of the momL gene does not disrupt any of the genes in the operon, suggesting that the momL location is an adaptation mechanism and not a random insertion. In addition, the momL gene is co-transcribed with suf under iron deficient and H2O2 conditions. It is promising to further study whether the AHL lactonase role or other novel function of MomL is needed for the phenotypes. Based on this study, one possible reason is that MomL is secreted into extracellular spaces to degrade the AHL signal molecules of competitors and we speculate that this mechanism might possibly damage the competitors’ QS systems, ultimately decrease the virulence factor “weapon” production and the survival of the competitor. We are constructing MomL mutant with no enzymatic activity using directed evolution strategy to explore the involved mechanism. Moreover, the results above also suggest that suf and momL are not combined together randomly, but rather, that this special arrangement allows two otherwise unrelated survival mechanisms to be activated by the same environmental stimuli, enhancing cell survival. However, because of the lack of genetic operation system of M. olearia Th120, we can not deleted momL, and there is currently no direct evidence to prove how momL contributes to increased survival of cells under H2O2 or iron limitation stress. Besides, lacking of Pcc genetic operation system at this stage make us unable to delete related genes and unclear about how quorum quenching enzyme MomL decreased the survival of Pcc. We are undergoing these experiments in our group now.
The QQ activity of M. olearia Th120 can be used in biocontrol for the control and prevention of aquatic animal diseases17. As MomL expression is low in cells, it is important to improve its expression for future applications. Signal molecules regulate a series of genes, affecting the physiological activities of a variety of cell types31, 35, 36. Therefore, a variety of signal molecules have been tested for improving MomL expression levels in this study. Interestingly, an well-studied signal molecule, indole, had a notable effect on suf-momL transcription. Although it is well-studied that indole, as a QS signal molecular, plays important roles in bacteria including biofilm formation, antibiotic resistance and virulence, the mutual regulation between QS and QQ in bacterial is largely unknown. This study demonstrated that indole could not only trigger stress-repsonse genes suf, but also regulate QQ enzyme factor momL. This is also the first time description that QQ enzyme is triggered by stress factor. Experiments suggested that oxyR1, which is regulated by indole, correlated with suf-momL expression and MomL secretion. Correspondingly, the addition of indole increases the survival of cells during oxidative stress. We speculate that indole may act as a signaling molecule to regulate the oxyR1 gene in an unknown way when it senses changes in environmental conditions (Fig. 7). In addition, it is hypothesized that the three oxyR genes of M. olearia have different functions to allow for more versatile responses to the environment. This discovery of a fundamental response system opens new ways to make this strain more useful as a beneficial bacterium for disease control in the future.
Figure 7.
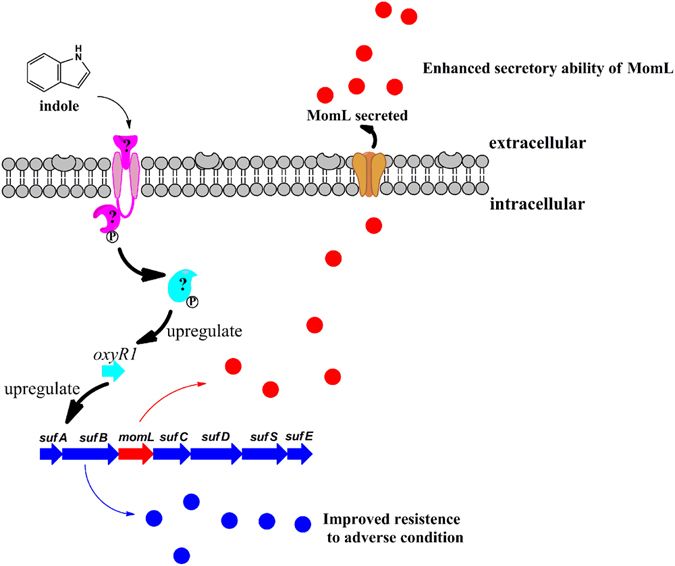
Schematic diagram of the indole and suf-momL signaling cascade pathway.
Through transcriptome analyses, it was found that indole regulated more than 200 genes in M. olearia; these genes displayed a variety of functions. GO analysis indicated that multiple genes for response to stimulus were affected by indole. It is speculated that these genes were either involved in indole-suf signal cascade pathway or among other stress response pathway. Besides, for cellular component, the significantly regulated part were membrane-related genes. Interestingly, most of the membrane proteins were up-regulated under the regulation of indole, including transporters, permeases and TonB systems. Based on this result, we hypothesize that cell membrane proteins play important roles when signaling molecules enter cells. In addition, previous studies have shown that indole could affect the resistance of bacteria to antibiotics31. Our transcriptome study showed consistent result. The KOBAS analysis suggested that four KEGG pathways were regulated by indole treatement. Among the four enriched pathways, vancomycin resistance and two-component system were significantly enriched. We suggest that indole can either affect the transport capacity of the cell or change cell permeability to alter the resistance to antibiotics.
Electronic supplementary material
Acknowledgements
We are very grateful to Dr. Liangcheng Du and Mr. Stephen Wright in University of Nebraska-Lincoln for making suggestions and language modification. Thanks to Mr. Liang Zeng in the Biozeron company in Shanghai for discussion about Transcriptome sequencing. Thanks to Dr. Kaihao Tang in Ocean University of China for valuable discussion and information. This work was supported by projects from the National Natural Science Foundation of China (No. 31571970, 41276141 and 41506160) and Natural Science Foundation of Shandong Province (No. 2014ZRE29014).
Author Contributions
Conceived and designed the experiments: Y.W., X.Z., H.L. Performed the experiments: Y.W., H.L., X.C. Analyzed the data: X.Z., Y.W., H.L., X.C. Wrote the paper: Y.W., X.Z., H.L.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-04606-8
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ng WL, Bassler BL. Bacterial quorum-sensing network architectures. Annual review of genetics. 2009;43:197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Waters CM, Bassler BL. Quorum sensing: cell-to-cell communication in bacteria. Annual review of cell and developmental biology. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 3.Davies DG, et al. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science. 1998;280:295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- 4.Gambello MJ, Iglewski BH. Cloning and characterization of the Pseudomonas aeruginosa lasR gene, a transcriptional activator of elastase expression. J Bacteriol. 1991;173:3000–3009. doi: 10.1128/jb.173.9.3000-3009.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nealson KH, Platt T, Hastings JW. Cellular control of the synthesis and activity of the bacterial luminescent system. J Bacteriol. 1970;104:313–322. doi: 10.1128/jb.104.1.313-322.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biswa P, Doble M. Production of acylated homoserine lactone by gram-positive bacteria isolated from marine water. FEMS microbiology letters. 2013;343:34–41. doi: 10.1111/1574-6968.12123. [DOI] [PubMed] [Google Scholar]
- 7.Saenz HL, et al. Inducible expression and cellular location of AgrB, a protein involved in the maturation of the staphylococcal quorum-sensing pheromone. Archives of microbiology. 2000;174:452–455. doi: 10.1007/s002030000223. [DOI] [PubMed] [Google Scholar]
- 8.Johnson MR, et al. Population density-dependent regulation of exopolysaccharide formation in the hyperthermophilic bacterium Thermotoga maritima. Molecular microbiology. 2005;55:664–674. doi: 10.1111/j.1365-2958.2004.04419.x. [DOI] [PubMed] [Google Scholar]
- 9.Pereira CS, Thompson JA, Xavier KB. AI-2-mediated signalling in bacteria. FEMS microbiology reviews. 2013;37:156–181. doi: 10.1111/j.1574-6976.2012.00345.x. [DOI] [PubMed] [Google Scholar]
- 10.Lee JH, Lee J. Indole as an intercellular signal in microbial communities. FEMS microbiology reviews. 2010;34:426–444. doi: 10.1111/j.1574-6976.2009.00204.x. [DOI] [PubMed] [Google Scholar]
- 11.Li X, Yang Q, Dierckens K, Milton DL, Defoirdt T. RpoS and indole signaling control the virulence of Vibrio anguillarum towards gnotobiotic sea bass (Dicentrarchus labrax) larvae. PloS one. 2014;9:e111801. doi: 10.1371/journal.pone.0111801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martino PD, Fursy R, Bret L, Sundararaju B, Phillips RS. Indole can act as an extracellular signal to regulate biofilm formation of Escherichia coli and other indole-producing bacteria. Canadian journal of microbiology. 2003;49:443–449. doi: 10.1139/w03-056. [DOI] [PubMed] [Google Scholar]
- 13.Lee J, Jayaraman A, Wood TK. Indole is an inter-species biofilm signal mediated by SdiA. BMC microbiology. 2007;7:42. doi: 10.1186/1471-2180-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirakawa H, Inazumi Y, Masaki T, Hirata T, Yamaguchi A. Indole induces the expression of multidrug exporter genes in Escherichia coli. Molecular microbiology. 2005;55:1113–1126. doi: 10.1111/j.1365-2958.2004.04449.x. [DOI] [PubMed] [Google Scholar]
- 15.Clatworthy AE, Pierson E, Hung DT. Targeting virulence: a new paradigm for antimicrobial therapy. Nature chemical biology. 2007;3:541–548. doi: 10.1038/nchembio.2007.24. [DOI] [PubMed] [Google Scholar]
- 16.Tang K, et al. Evaluation of a new high-throughput method for identifying quorum quenching bacteria. Scientific reports. 2013;3:2935. doi: 10.1038/srep02935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang K, et al. MomL, a novel marine-derived N-acyl homoserine lactonase from Muricauda olearia. Applied and environmental microbiology. 2015;81:774–782. doi: 10.1128/AEM.02805-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mayer C, Romero M, Muras A, Otero A. Aii20J, a wide-spectrum thermostable N-acylhomoserine lactonase from the marine bacterium Tenacibaculum sp. 20J, can quench AHL-mediated acid resistance in Escherichia coli. Applied microbiology and biotechnology. 2015;99:9523–9539. doi: 10.1007/s00253-015-6741-8. [DOI] [PubMed] [Google Scholar]
- 19.Momb J, et al. Mechanism of the quorum-quenching lactonase (AiiA) from Bacillus thuringiensis. 2. Substrate modeling and active site mutations. Biochemistry. 2008;47:7715–7725. doi: 10.1021/bi8003704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu D, et al. Mechanism of the quorum-quenching lactonase (AiiA) from Bacillus thuringiensis. 1. Product-bound structures. Biochemistry. 2008;47:7706–7714. doi: 10.1021/bi800368y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y, et al. Genome analysis of Flaviramulus ichthyoenteri Th78(T) in the family Flavobacteriaceae: insights into its quorum quenching property and potential roles in fish intestine. BMC genomics. 2015;16:38. doi: 10.1186/s12864-015-1275-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roche B, et al. Iron/sulfur proteins biogenesis in prokaryotes: formation, regulation and diversity. Biochimica et biophysica acta. 2013;1827:455–469. doi: 10.1016/j.bbabio.2012.12.010. [DOI] [PubMed] [Google Scholar]
- 23.Outten FW, Djaman O, Storz G. A suf operon requirement for Fe-S cluster assembly during iron starvation in Escherichia coli. Molecular microbiology. 2004;52:861–872. doi: 10.1111/j.1365-2958.2004.04025.x. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y, et al. Biosynthetic mechanism for sunscreens of the biocontrol agent Lysobacter enzymogenes. PLoS One. 2013;8:e66633. doi: 10.1371/journal.pone.0066633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, et al. Mechanisms involved in the functional divergence of duplicated GroEL chaperonins in Myxococcus xanthus DK1622. PLoS genetics. 2013;9:e1003306. doi: 10.1371/journal.pgen.1003306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McClean KH, et al. Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology. 1997;143(Pt 12):3703–3711. doi: 10.1099/00221287-143-12-3703. [DOI] [PubMed] [Google Scholar]
- 27.Singh VK, Mangalam AK, Dwivedi S, Naik S. Primer premier: program for design of degenerate primers from a protein sequence. BioTechniques. 1998;24:318–319. doi: 10.2144/98242pf02. [DOI] [PubMed] [Google Scholar]
- 28.Stanier RY. Studies on Marine Agar-Digesting Bacteria. Journal of bacteriology. 1941;42:527–559. doi: 10.1128/jb.42.4.527-559.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oppenheimer CH, ZoBell CE. The growth and viability of sixty three species of marine bacteria as influenced by hydrostatic pressure. Journal of Marine Research. 1952;11:10–18. [Google Scholar]
- 30.Trapnell C, et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vega NM, Allison KR, Khalil AS, Collins JJ. Signaling-mediated bacterial persister formation. Nature chemical biology. 2012;8:431–433. doi: 10.1038/nchembio.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Granston AE, Nash HA. Characterization of a set of integration host factor mutants deficient for DNA binding. Journal of molecular biology. 1993;234:45–59. doi: 10.1006/jmbi.1993.1562. [DOI] [PubMed] [Google Scholar]
- 33.Flamm EL, Weisberg RA. Primary structure of the hip gene of Escherichia coli and of its product, the beta subunit of integration host factor. Journal of molecular biology. 1985;183:117–128. doi: 10.1016/0022-2836(85)90206-2. [DOI] [PubMed] [Google Scholar]
- 34.Gardan L, Gouy C, Christen R, Samson R. Elevation of three subspecies of Pectobacterium carotovorum to species level: Pectobacterium atrosepticum sp. nov., Pectobacterium betavasculorum sp. nov. and Pectobacterium wasabiae sp. nov. International journal of systematic and evolutionary microbiology. 2003;53:381–391. doi: 10.1099/ijs.0.02423-0. [DOI] [PubMed] [Google Scholar]
- 35.Han Y, et al. Identification of a small molecule signaling factor that regulates the biosynthesis of the antifungal polycyclic tetramate macrolactam HSAF in Lysobacter enzymogenes. Applied microbiology and biotechnology. 2015;99:801–811. doi: 10.1007/s00253-014-6120-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao M, Coggin A, Yagnik K, Teplitski M. Role of specific quorum-sensing signals in the regulation of exopolysaccharide II production within Sinorhizobium meliloti spreading colonies. PloS one. 2012;7:e42611. doi: 10.1371/journal.pone.0042611. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


