Abstract
Carotenoids not only play indispensable roles in plant growth and development but also enhance nutritional value and health benefits for humans. In this study, total carotenoids progressively decreased during fruit ripening. Fifty-four genes involving in mevalonate (MVA), 2-C-methyl-D-erythritol 4-phosphate (MEP), carotenoid biosynthesis and catabolism pathway were identified. The expression levels of most of the carotenoid metabolism related genes kept changing during fruit ripening generating a metabolic flux toward carotenoid synthesis. Down regulation of VvDXS, VvDXR, VvGGPPS and VvPSY and a dramatic increase in the transcription levels of VvCCD might be responsible for the reduction of carotenoids content. The visible correlation between carotenoid content and gene expression profiles suggested that transcriptional regulation of carotenoid biosynthesis pathway genes is a key mechanism of carotenoid accumulation. In addition, the decline of carotenoids was also accompanied with the reduction of chlorophyll content. The reduction of chlorophyll content might be due to the obstruction in chlorophyll synthesis and acceleration of chlorophyll degradation. These results will be helpful for better understanding of carotenoid biosynthesis in grapevine fruit and contribute to the development of conventional and transgenic grapevine cultivars for further enrichment of carotenoid content.
Introduction
The isoprenoids, also known as terpenoids and terpenes, are the largest class of plant secondary metabolites and have numerous biochemical functions in plants. They play pivotal role as photosynthetic pigments (e.g., carotenoids and phytol), plant hormones (e.g., gibberellins, strigolactones and brassinosteroids), electron carriers (e.g., plastoquinone), and as plant defense compounds as well as attractants for pollinators (monoterpenes, sesquiterpenes, and diterpenes)1, 2. Carotenoids are a sungroup of isoprenoid molecules with more than 750 members occurring throughout the natural world and participate in various physiological and developmental processes in plants3, 4. In photosynthetic green tissues, carotenoids play an essential role in photosynthesis for photosystem assembly, light harvesting, and photoprotection5. In non-photosynthetic tissues, carotenoids provide bright colors and produce aromas and flavors to attract insects and animals for pollination and seed dispersal6. Carotenoids also serve as precursors for two important phytohormones, abscisic acids (ABA) and strigolactones, which are key regulators for plant development and stress response7–9. In addition, increasing interest is devoted to carotenoid content and composition of food crops because of their important roles in human nutrition and health10, 11.
Like all isoprenoids, carotenoids are synthesized from the five carbon units isopentenyl diphosphate (IPP) and its double-bond isomer dimethylallyl diphosphate (DMAPP)12, 13. Two pathways exist in plant cells for the production of these prenyl diphosphate precursors, but carotenoids are mainly synthesized from IPP and DMAPP produced by MEP pathway, as shown in Supplementary Fig. S1. The first committed step of carotenoid biosynthesis is the production of 40-carbon phytoene from condensation of two GGPP molecules (Fig. S1). This reaction, catalyzed by the enzyme phytoene synthase (PSY), is considered the main bottleneck in the carotenoid pathway12.Phytoene is then desaturated and isomerized to all-trans-lycopene through the action of two desaturases and two isomerases: phytoene desaturase (PDS), ζ-carotene desaturase (ZDS), prolycopene isomerase (CRTISO) and ζ-carotene isomerase (ZISO). The formation of δ-carotene and γ-carotene from lycopene are catalyzed by lycopene ε-cyclase (LCYE) and β-cyclase (LCYB), and then the orange α-carotene and β-carotene are synthetized by LCYB. Finally, these carotenes are transformed into lutein and zeaxanthin by heme and non-heme β-carotene hydroxylases (CYP97 and CHYB). Zeaxanthin is converted to violaxanthin by the action of zeaxanthin epoxidase (ZEP) and further to neoxanthin by the action of the neoxanthin synthase (NXS). These two xanthophylls are cleaved by 9-cis-epoxycarotenoid dioxygenase (NCED), a key enzyme in the biosynthesis of ABA4–6, 14–16.
Grapevine (Vitis) is one of the most commonly consumed and widely cultivated fruit crop worldwide17–19. Due to its important nutritional values and health benefits, grapevine becomes the most popular and important in the diets of people throughout the world. The carotenoid content and composition of grape berry has received considerable attention due to their potential precursors to a group of potent aroma compounds (C13-norisoprenoids) in grapevines and wines17. To maximize the health-promoting benefits of carotenoids through increased consumption, characterization of carotenoid synthesis and accumulation in important food crops such as grapevine is essential. A complete understanding of the carotenogenesis genes is fundamental for elucidating the mechanisms of carotenoid biosynthesis in grapevine, as well as for the breeding of new grapevine varieties with rich carotenoids, which are good for human health. With the release of the grapevine genome sequence and with the increasing affordability of high throughput analysis tools20, 21, there will be a better opportunity to systematically study the carotenogenesis genes in grapevine. In this study, 54 carotenoid biosynthetic genes were identified in grapevine genomic. Transcriptome were used to profile the expression of carotenoid biosynthetic/catabolic genes during grape berry development and ripening. Carotenoid concentrations were also determined at three distinct stages of berry development: green, véraison and ripe/harvest stages. The systematic analysis of carotenoid biosynthesis genes in grapevine will improve our understanding of the genetic mechanisms of carotenoid biosynthesis and carotenoid accumulation in grapevine.
Results
Identification of 54 genes involving in grapevine carotenoid biosynthetic and catabolic pathway
To determine the molecular basis and mechanism of the carotenoid biosynthetic/catabolic during grapevine fruit development, the genes involving in grapevine carotenoid biosynthetic/catabolic pathway were identified from the current genome. A total of fifty-four carotenoid biosynthesis-related genes were identified via BLAST-P search in NCBI using the Arabidopsis gene sequences as queries1, 15. Subsequently, to verify the reliability of the initial results, a survey was conducted to confirm these genes using functional annotation of the grapevine transcriptome in three distinct stages of berry development (NCBI GEO Accession: GSE77218)19. As a result, 54 non-redundant carotenoid biosynthesis-related genes were identified and each carotenoid biosynthetic gene in grapevine was named based on the enzymatic reaction, similar to those given in the A. thaliana carotenoid biosynthetic pathway. Detailed information about each carotenoid biosynthetic and catabolic gene was showed in Table 1, including protein length, isoelectric points (pI), molecular weights, aliphatic index, grand average of hydropathicity (GRAVY) and subcellular localizations (Table 1).
Table 1.
Carotenoid metabolic genes in grapevine.
| Name | Accession no. | Protein Len | Chrom | Chr srart | Chr end | Mol.wt (kDa) | pI | Aliphatic index | GRAVY | Loca |
|---|---|---|---|---|---|---|---|---|---|---|
| MVA pathway | ||||||||||
| VvAACT1 | VIT_00s0531g00050.t01 | 440 | chrUn | 31408786 | 31413407 | 45958.9 | 8.54 | 99.59 | 0.187 | C |
| VvAACT2 | VIT_12s0057g01200.t01 | 415 | chr12 | 9967097 | 9973038 | 42339.6 | 5.82 | 100.19 | 0.261 | — |
| VvHMGS | VIT_02s0025g04580.t01 | 464 | chr2 | 4131692 | 4138165 | 51004.0 | 5.92 | 76.70 | −0.192 | — |
| VvHMGR1 | VIT_18s0122g00610.t01 | 593 | chr18 | 494573 | 498105 | 63636.3 | 6.83 | 92.45 | 0.072 | M |
| VvHMGR2 | VIT_04s0044g01740.t01 | 561 | chr4 | 23597424 | 23599575 | 59777.0 | 7.86 | 94.39 | 0.175 | — |
| VvHMGR3 | VIT_03s0038g04100.t01 | 575 | chr3 | 2970538 | 2972585 | 61190.6 | 6.07 | 97.36 | 0.160 | — |
| VvMVK | VIT_14s0128g00330.t01 | 388 | chr14 | 2969463 | 2974536 | 41209.6 | 5.96 | 102.81 | 0.259 | S |
| VvpMVK | VIT_02s0012g02530.t01 | 508 | chr2 | 10033966 | 10045076 | 55167.9 | 5.67 | 91.04 | −0.100 | — |
| VvMDC | VIT_13s0106g00790.t01 | 422 | chr13 | 10644213 | 10652294 | 46640.1 | 6.48 | 85.07 | −0.300 | — |
| VvFPS | VIT_19s0015g01010.t01 | 341 | chr19 | 9094526 | 9098457 | 39064.9 | 5.53 | 95.43 | −0.201 | — |
| MEP pathway | ||||||||||
| VvDXS1 | VIT_05s0020g02130.t01 | 716 | chr5 | 3851283 | 3856072 | 77091.0 | 6.36 | 88.04 | −0.078 | C |
| VvDXS2 | VIT_11s0052g01730.t01 | 735 | chr11 | 19555292 | 19558837 | 78975.0 | 7.13 | 86.83 | −0.133 | C |
| VvDXS3 | VIT_04s0008g04970.t01 | 719 | chr4 | 4457279 | 4468718 | 78929.2 | 6.53 | 88.71 | −0.018 | C |
| VvDXS4 | VIT_11s0052g01780.t01 | 690 | chr11 | 19599532 | 19602913 | 74142.5 | 6.32 | 88.10 | −0.119 | C |
| VvDXS5 | VIT_00s0218g00110.t01 | 718 | chrUn | 14030385 | 14035390 | 77179.6 | 7.92 | 88.87 | −0.106 | C |
| VvDXS6 | VIT_11s0052g01240.t01 | 731 | chr11 | 18968097 | 18972989 | 78991.5 | 7.22 | 90.26 | −0.121 | C |
| VvDXR | VIT_17s0000g08390.t01 | 471 | chr17 | 9583558 | 9590206 | 51176.1 | 6.03 | 99.64 | −0.016 | C |
| VvMCT | VIT_12s0035g01950.t01 | 308 | chr12 | 22263637 | 22269775 | 33830.0 | 6.54 | 100.88 | −0.095 | C |
| VvCMK | VIT_06s0009g02320.t01 | 394 | chr6 | 14652972 | 14664719 | 43473.3 | 8.05 | 81.22 | −0.270 | C |
| VvMDS | VIT_02s0025g00380.t01 | 168 | chr2 | 486537 | 487271 | 17970.5 | 6.74 | 91.85 | −0.008 | C |
| VvHDS | VIT_06s0004g02900.t01 | 740 | chr6 | 3629087 | 3636025 | 82345.3 | 5.91 | 90.49 | −0.255 | C |
| VvHDR | VIT_03s0063g02030.t01 | 465 | chr3 | 5305933 | 5311191 | 52522.5 | 5.32 | 80.88 | −0.399 | C |
| VvIDI | VIT_04s0023g00600.t01 | 293 | chr4 | 16802127 | 16809421 | 33557.4 | 6.16 | 87.54 | −0.318 | C |
| Carotenoid biosynthetic pathway | ||||||||||
| VvGGPPS1 | VIT_04s0023g01210.t01 | 368 | chr4 | 17612949 | 17614055 | 39801.9 | 5.91 | 97.80 | −0.049 | C |
| VvGGPPS2 | VIT_18s0001g12 000.t01 | 371 | chr18 | 10227177 | 10228292 | 39683.4 | 5.78 | 92.59 | −0.058 | M |
| VvGGPPS-LS | VIT_05s0020g01240.t01 | 276 | chr5 | 2987258 | 2988130 | 29744.2 | 4.93 | 101.45 | 0.039 | — |
| VvGGPPS-SS | VIT_19s0090g00530.t01 | 298 | chr19 | 6656884 | 6657780 | 32785.3 | 6.19 | 84.87 | −0.307 | C |
| VvPSY1 | VIT_04s0079g00680.t01 | 437 | chr4 | 11495290 | 11499126 | 49643.7 | 8.42 | 82.61 | −0.330 | — |
| VvPSY2 | VIT_12s0028g00960.t01 | 396 | chr12 | 1467727 | 1470354 | 45152.8 | 9.15 | 84.72 | −0.328 | C |
| VvPSY3 | VIT_06s0004g00820.t01 | 357 | chr6 | 935500 | 940685 | 41210.1 | 6.25 | 82.02 | −0.331 | — |
| VvPDS | VIT_09s0002g00100.t01 | 582 | chr9 | 70786 | 92779 | 65495.5 | 6.58 | 92.97 | −0.183 | M |
| VvZ-ISO | VIT_05s0062g01110.t01 | 365 | chr5 | 19770533 | 19774697 | 40610.1 | 8.83 | 101.78 | 0.323 | C |
| VvZDS | VIT_14s0030g01740.t01 | 552 | chr14 | 6600703 | 6613497 | 61181.9 | 7.07 | 87.99 | −0.178 | — |
| VvCRTISO1 | VIT_08s0032g00800.t01 | 641 | chr8 | 4370241 | 4386309 | 70659.6 | 8.50 | 91.83 | −0.054 | — |
| VvCRTISO2 | VIT_12s0035g01080.t01 | 570 | chr12 | 20842885 | 20849118 | 62034.1 | 7.20 | 89.65 | −0.094 | C |
| VvLCYE | VIT_11s0016g01880.t01 | 529 | chr11 | 1519660 | 1526200 | 59261.3 | 6.58 | 89.58 | −0.064 | C |
| VvLCYB | VIT_08s0007g05690.t01 | 508 | chr8 | 19614960 | 19620336 | 57101.1 | 7.20 | 91.32 | −0.094 | — |
| VvCHYB1 | VIT_02s0025g00240.t01 | 299 | chr2 | 360270 | 361917 | 33057.2 | 8.40 | 90.03 | 0.032 | C |
| VvCHYB2 | VIT_16s0050g01090.t01 | 306 | chr16 | 18017218 | 18018829 | 34217.5 | 9.58 | 80.95 | −0.080 | C |
| VvCYP97C | VIT_08s0007g04530.t01 | 546 | chr8 | 18452646 | 18457877 | 61121.1 | 5.78 | 93.90 | −0.107 | C |
| VvCYP97A | VIT_04s0023g00080.t01 | 638 | chr4 | 15934070 | 15957007 | 70813.5 | 6.56 | 92.92 | −0.163 | C |
| VvZEP1 | VIT_07s0031g00620.t01 | 658 | chr7 | 16796162 | 16803660 | 72087.5 | 7.93 | 83.43 | −0.174 | — |
| VvZEP2 | VIT_13s0156g00350.t01 | 475 | chr13 | 24068514 | 24072138 | 53119.2 | 8.74 | 91.52 | −0.150 | — |
| VvZEP3 | VIT_00s0533g00020.t01 | 479 | chrUn | 31461160 | 31463786 | 52399.3 | 8.65 | 94.82 | −0.103 | C |
| VvVDE | VIT_04s0043g01010.t01 | 479 | chr4 | 15682355 | 15687333 | 54596.4 | 6.03 | 77.29 | −0.397 | C |
| VvNXS | VIT_14s0006g02880.t01 | 246 | chr14 | 21063839 | 21068184 | 27562.4 | 9.76 | 92.44 | 0.242 | C |
| VvCCS | VIT_06s0080g00810.t01 | 497 | chr6 | 20814283 | 20815776 | 56254.1 | 8.47 | 86.90 | −0.167 | C |
| Carotenoid catabolism | ||||||||||
| VvCCD1 | VIT_13s0064g00840.t01 | 546 | chr13 | 22673076 | 22681735 | 61634.8 | 6.13 | 81.90 | −0.271 | — |
| VvCCD4a | VIT_02s0087g00910.t01 | 599 | chr2 | 18560792 | 18562591 | 65900.3 | 6.88 | 82.35 | −0.236 | C |
| VvCCD4b | VIT_02s0087g00930.t01 | 589 | chr2 | 18588968 | 18590737 | 65607.2 | 6.63 | 82.41 | −0.187 | C |
| VvCCD8 | VIT_04s0008g03380.t01 | 546 | chr4 | 2785955 | 2788636 | 60526.9 | 6.99 | 79.80 | −0.324 | C |
| VvNCED1 | VIT_19s0093g00550.t01 | 609 | chr19 | 17645484 | 17647313 | 67132.0 | 6.38 | 76.04 | −0.317 | C |
| VvNCED2 | VIT_10s0003g03750.t01 | 605 | chr10 | 6374536 | 6376353 | 67339.1 | 6.36 | 78.00 | −0.365 | C |
| VvNCED6 | VIT_05s0051g00670.t01 | 575 | chr5 | 11589343 | 11591070 | 63137.5 | 8.24 | 87.13 | −0.202 | C |
aThe subcellular location result of grapevine carotenoid metabolic genes by TargetP. ‘C’: Chloroplast; ‘M’: Mitochondrion; ‘S’: Secretory pathway; ‘—’ was any other location without chloroplast, mitochondrion, secretory pathway in cell.
Among the 54 carotenoid biosynthetic genes in grapevine, 10, 13, 24 and 7 genes were involved in MVA, MEP, carotenoid biosynthetic and catabolic pathways, respectively (Tables 1 and S1). These genes were members of 32 different gene families, and 22 genes were identified as single gene copy (Table 1). All these genes, only two (VvPSY3 and VvCCD8) were not expressed with a very low number of reads sequenced and an extremely low RPKM (reads per kilo base of exon model per million reads) value during three ripening stages. One transcript of VvMVK showed no change in expression level (|log2 fold-change (log2FC)| < 0.25). 20 transcripts were observed as up-regulated or down-regulated slightly (0.25 < |log2FC| < 1). Thirty-one transcripts were considered as significantly differentially-expressed genes (|log2FC|) ≥ 1 (Table S1).
Chromosomal distribution of grapevine biosynthetic genes
As shown in Fig. 1, 51 of 54 grapevine carotenoid biosynthetic genes were distributed unevenly throughout the 17 out of the 19 chromosomes, with eight genes located in Chromosome 4, while none of the carotenoid biosynthetic/catabolic genes mapped in Chromosomes 1 and 15 (Fig. 1). The remaining gene (VvDXS5, VvAACT1 and VvZEP3) had not yet been assembled to any chromosome according to the current genome.
Figrue 1.
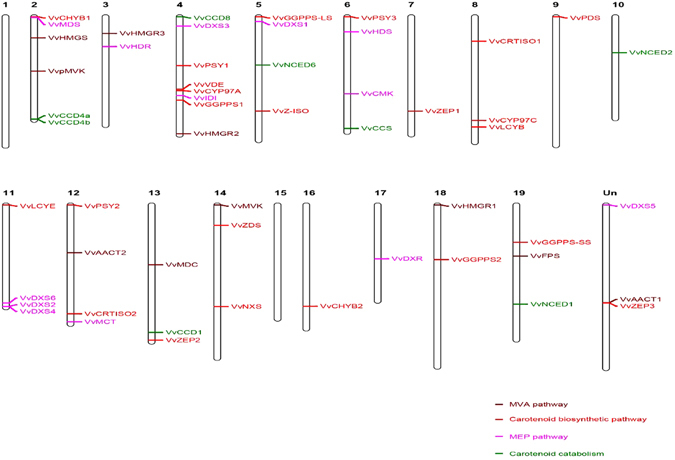
Chromosomal location of grapevine carotenoid metabolic genes.
Tandem and segmental duplications have been suggested to be two of the main causes for gene family expansion in plants20. The tandem duplication events were defined according to the methods of Holub21, where a chromosomal region within 200 kb containing two or more genes is defined as a tandem duplication event. There were five genes (VvDXS2/VvDXS4/VvDXS6 and VvCCD4a/VvCCD4b) clustered into two tandem duplication event regions on grape chromosome 2 and 11 (Table 1). Besides the tandem duplication events, 3 segregation duplication events (VvHMGR1/ VvHMGR3, VvGGPPS1/VvGGPPS2 and VvCHYB1/VvCHYB2) were also identified (Fig. 2), indicating that some grapevine carotenoid biosynthetic genes were possibly generated by gene duplication. Moreover, the segregation duplication events can also provide a reference for the carotenoid biosynthetic gene evolutionary relationship and functional prediction.
Figrue 2.
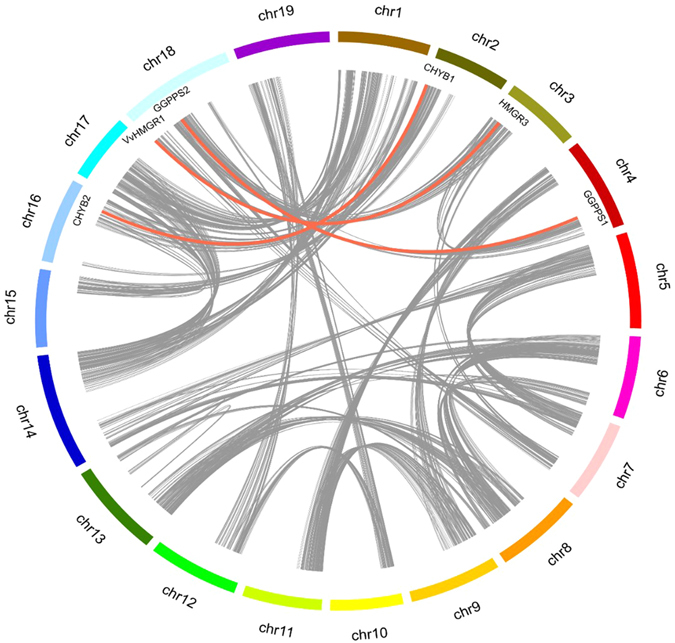
Synteny analysis of grapevine carotenoid metabolic genes. Gray lines indicate all synteny blocks in the grapevine genome, whereas the red lines suggest duplicated gene pairs.
Exon-intron organization of grapevine carotenoid biosynthetic gene
The exon-intron structure analysis was performed to gain more insight into the grapevine carotenoid biosynthetic genes. High variation was observed in numbers of exons and introns among grapevine carotenoid biosynthetic genes (Fig. 3). Nine genes (VvGGPPS1, VvGGPPS2, VvGGPPS-SS, VvCCS, VvCCD4a, VvCCD4b, VvNCED1, VvNCED2 and VvNCED6) had no intron in the grapevine genome. Most carotenoid metabolic genes contained 6 to 12 exons in their coding DNA sequences. However, VvHDS, the most exons contained in this study, had 19 exons in its coding DNA sequences. VvCYP97A was the longest carotenoid metabolic gene with 22 kb genomic sequence. The large variation in the structures of carotenoid metabolic genes indicated that the grapevine genomic had changed significantly during its extensive evolutionary processes.
Figrue 3.
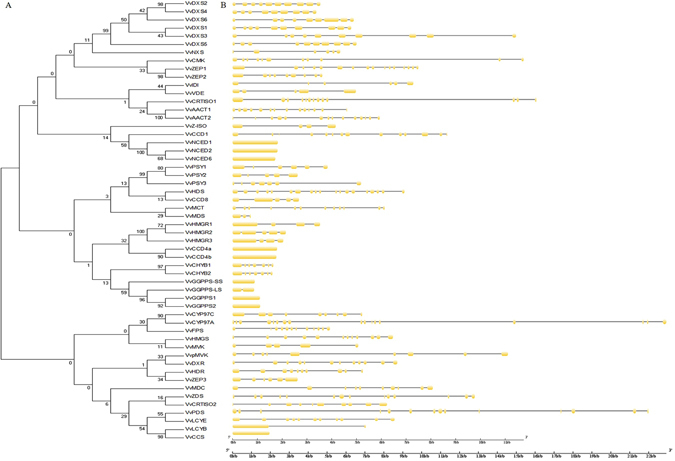
Structure analysis of carotenoid metabolic genes. (A) NJ phylogenetic tree of carotenoid metabolic proteins. (B) Exon-intron structure of carotenoid metabolic genes. Two different legends were used, 4 genes with long genomic sequence (VvPDS, VvZDS, VvCRTISO1 and VvCYP97A) used the long legends, and the others genes used the short legends. Yellow indicates exons; black indicates introns.
Variation of carotenoid and chlorophyll content during berry developing and ripening
In order to understand carotenoid and chlorophyll accumulation profiles in grapevine, the concentrations of carotenoid and chlorophyll in grape flesh were monitored at the three sampling time-points during fruit development and ripening (Fig. 4). It was observed that the amounts of carotenoid and chlorophyll decreased significantly throughout fruit ripening stages. As expected, the total amount of carotenoids (expressed as μg/g fw) progressively decreased from 7.51 μg/g fw at the green stage to 2.19 μg/g fw at the ripe stage (Fig. 4). Similarly, the total contents of chlorophyll a and chlorophyll b decreased 3.7 and 3.2-fold during ripening, respectively (Fig. 4). The ratios of carotenoids/chlorophylls (~0.18) as well as the ratio of chlorophyll a/b (~2.0) remained constant throughout the sampling stages. Our results were consistent with the previous finding that carotenoid and chlorophyll contents of grape berries decreased with ripening especially from veraison to harvest22–24.
Figrue 4.
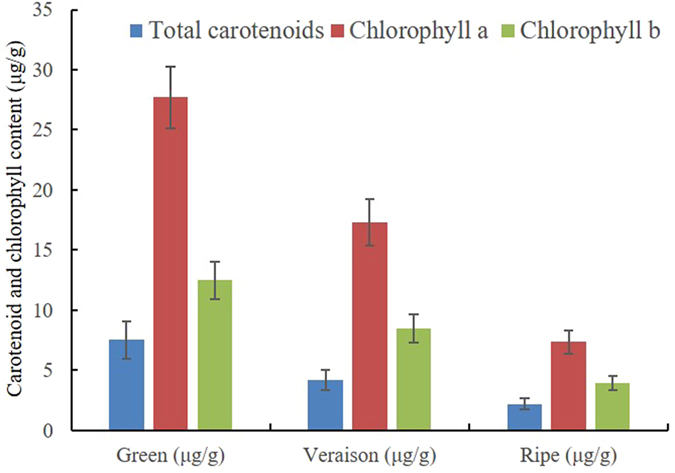
Total carotenoid and chlorophyll content during grapevine fruit ripening.
Characterization and expression analysis of genes involving in the MVA pathway
Nine genes involved in six enzymatic steps were found in MVA pathway, which led to the formation of IPP and DMAPP. In addition, there was one FPS gene which catalyzed the synthesis of farnesyl diphosphate (FPP), the major substrate used by cytosolic and mitochondrial branches of the isoprenoid pathway25, 26.
Acetyl-CoA acetyltransferase (AACT) catalyses the condensation of two acetyl-CoA subunits to form acetoacetyl-CoA thus directing this central metabolite to the MVA pathway. In this study, VvAACT1 and VvAACT2 showed the highest expression levels at veraison and ripe stages, respectively. Interestingly, VvACAT2 mRNA expression (RPKM = 321.8 ± 71, the average value of the three ripening stages) had an approximate 5.8-fold higher than VvAACT1 (RPKM = 55.0 ± 13, also the average value of the three ripening stages) during berry developmental stages, suggesting that this enzyme may divert the metabolic flux of acetyl-CoA from the biosynthesis of fatty acids and amino acids toward the synthesis of isoprenoids (Fig. 5 and Table S1). 3-Hydroxy-3-methylglutaryl-CoA reductase (HMGR), the first committed step of the MVA pathway for isoprenoid biosynthesis, catalyses the formation of MVA from 3-hydroxy-3-methyl-glutaryl-CoA (HMG-CoA)27. Three HMGR genes (VvHMGR1, VvHMGR2 and VvHMGR3) were identified in grapevine and two members (VvHMGR1 and VvHMGR3) were generated by gene duplication (Fig. 2). This was in agreement with previous reports that plant HMGR genes had arisen by gene duplication and subsequent sequence divergence28. Three members of VvHMGR exhibited significant differential expression patterns during fruit development. VvHMGR1 and VvHMGR2 showed the expression peak at green berry stage, whereas the highest expression of VvHMGR3 was detected at ripe stage (Fig. 5 and Table S1), suggesting that VvHMGR might play a complicated role in the MVA pathway. Furthermore, the last three enzymes were encoded by single gene in grapevine. The expression of three genes exhibited no or slight changes throughout the berry developmental stages. These results indicated that the last three enzymatic steps of the MVA pathway might not be important control points in terpene biosynthesis.
Figrue 5.
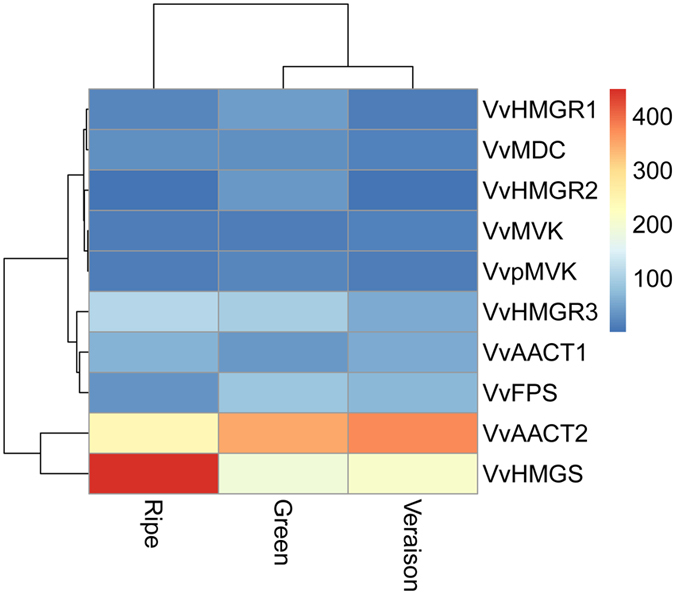
Expression level of MVA pathway genes during grapevine ripening. Data were obtained by Illumina RNA Sequencing using two biological replicas and are expressed as Reads Per Kilobase of exon model per Million mapped reads (RPKM).
Characterization and expression analysis of genes involving in the MEP pathway
In plants, IPP and DMAPP entering in the biosynthesis of carotenoids are mainly synthesized by the MEP pathway in the plastids12, 29, 30. All these 13 grapevine genes were most possibly located on chloroplasts (Table 1) through protein subcellular localization prediction, which were consistent with the previous reports about the MEP pathway enzymes of Arabidopsis and Salvia31, 32.
The expressions of 11 genes involving in MEP pathway were declined during grapevine fruit development and ripening, only VvHDR and VvIDI showed an increasing expression during growth (Fig. 6 and Table S1). 1-Deoxy-D-xylulose-5-phosphate synthase (DXS) catalyzes the first committed step of the MEP pathway and carotenoid biosynthesis. Six VvDXS members were identified in grapevine and displayed identical expression profiles, with the lowest and highest expression levels at veraison and green stage, respectively. Interestingly, the expression level of VvDXS1 was significantly higher than all other members (Table S1), suggesting that VvDXS1 might play predominant role in driving grapevine fruit carotenoid accumulation. 1-deoxy-D-xylulose-5-phosphate (DXP) is converted to MEP by the enzyme DXP reductoisomerase (DXR) encoded in grapevine by single gene. The expression profile of VvDXR was similar to VvDXS with its lowest and highest expression levels at veraison and green berry stages, respectively. MEP is subsequently converted into IPP and DMAPP by the consecutive action of five independent enzymes: 4-diphosphocytidyl-2-C-methyl-D-erythritol synthase (MCT), 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase (CMK), 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase (MDS), 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate synthase (HDS), and 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate reductase (HDR)33. VvMCT, VvCMK, VvMDS and VvHDS showed a slight decrease in expression levels during ripening, whereas VvHDR demonstrated increased mRNA expression during grapevine fruit ripening (Fig. 6 and Table S1). Isomerization of IPP and DMAPP is catalyzed by isopentenyl diphosphate isomerase (IDI) encoded in grapevine by single gene whose mRNA expression profile remained strong and increased gradually throughout grapevine fruit ripening.
Figrue 6.
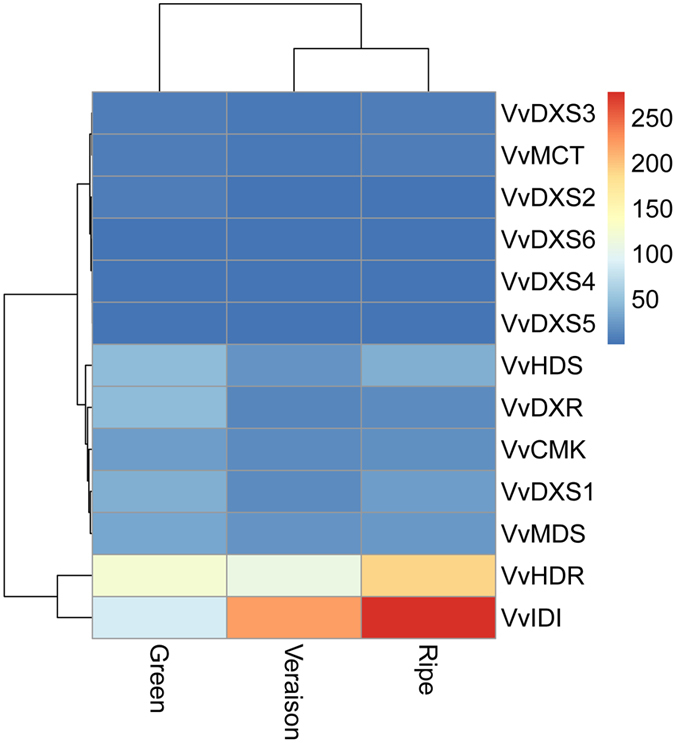
Expression level of MEP pathway genes during grapevine ripening. Data were obtained by Illumina RNA Sequencing using two biological replicas and are expressed as Reads Per Kilobase of exon model per Million mapped reads (RPKM).
Characterization and expression analysis of carotenoid biosynthetic genes during grapevine berry development and ripening
A total of 24 genes were involved in carotenoid biosynthesis pathway, which led to the formation of carotenoids from the precursor geranylgeranyl diphosphate (GGPP). GGPP, the universal precursor for all plastid isoprenoids, is generated by geranylgeranyl diphosphate synthase (GGPPS) that catalyses the condensation of three IPP and one DMAPP units34. In our study, four VvGGPPS genes were identified and showed a steady decrease during grapevine fruit development (Fig. 7 and Table S1). VvGGPPS2 was predominantly expressed in grapevine fruit (Fig. 7 and Table S1), in agreement with the phenomenon that GGPPS2 was specific for chromoplasts in tomato flowers and/or fruits35. Phytoene synthase (PSY) the main bottleneck in the carotenoid pathway, catalyzes the condensation of two GGPP molecules to produce 15-cisphytoene36, 37. Three VvPSY genes were identified and with different exprseeion patterns in grapevine. VvPSY1 was low at green fruit stage but started increasing at veraison, then decreased sharply at the ripening stage. In contrast, VvPSY2 maintained low expression throughout fruit development, but increased sharply at ripening stage. VvPSY3 was no expressed in grapevine fruit (Fig. 7 and Table S1). These results indicated that VvPSY genes showed a tissue-specific expression pattern.
Figrue 7.
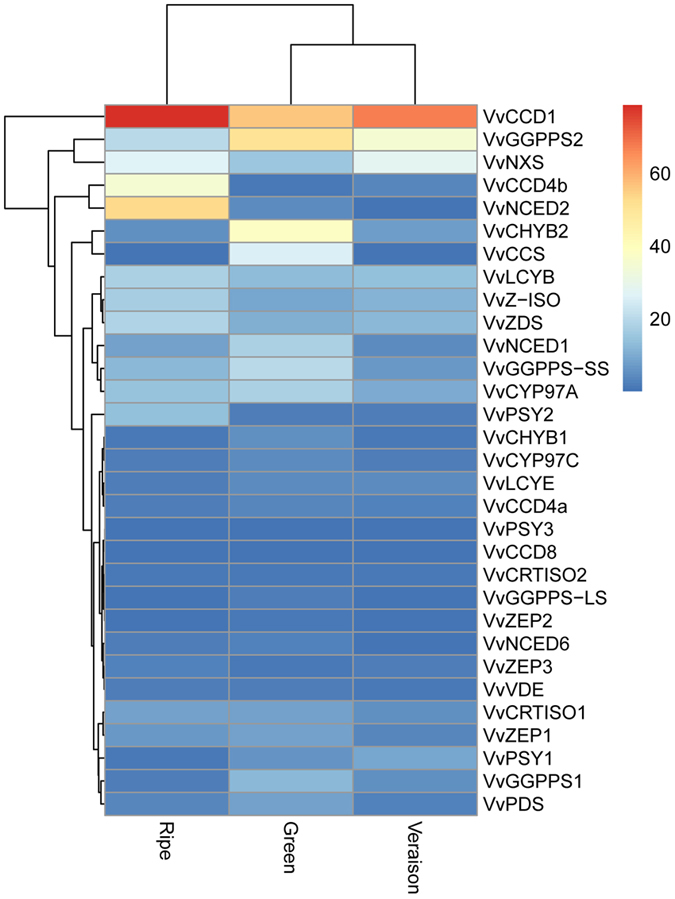
Expression level of carotenoid biosynthetic and catabolic pathway genes during grapevine ripening. Data were obtained by Illumina RNA Sequencing using two biological replicas and are expressed as Reads Per Kilobase of exon model per Million mapped reads (RPKM).
The expression level of VvPDS depicted highest at green fruit stage, and then showed decreasing trend from veraison till to ripe stage. The expressions of VvZ-ISO, VvZDS and VvCRTISO kept increasing during fruit development and reached their picks at ripening stage. Additionally, VvCRTISO1 was much more highly expressed than VvCRTISO2 did with the progresses in growth season (Table S1), indicating VvCRTISO1 may have a more direct role in formation of lycopene. Furthermore, two different lycopene cyclases, lycopene β-cyclas (VvLCYB) and lycopene ε-cyclase (VvLCYE) were identified and showed an opposite expression pattern. Transcripts of VvLCYE decreased from veraison to harvest, whereas VvLCYB slightly increased through three periods of development (Table S1). The high expression levels of VvLCYB at harvest stage may help to maintain the amounts of γ- and β-carotene as intermediate metabolites for other compounds. Two VvCHYB were identified with a similar but quantitatively different expression pattern (Table S1). Neoxanthin synthase (VvNXS) and capsanthin-capsorubin synthase (VvCCS), two downstream genes of VvCHYB, also exhibited an opposite expression pattern. VvNXS were high expression at veraison and harvest stages, and VvCCS showed a very low expression. The expression pattern of two genes could be correlated to changes in ABA levels in grapevine berries that ABA levels typically peak at or around veraison and is thought to be responsible for the control of grape berry ripening38, 39.
Characterization and expression analysis of carotenoid catabolic pathways during grapevine development and ripening
The steady-state level of carotenoids is dependent on the metabolic equilibrium between biosynthesis and degradation of carotenoids along with storage40. In our study, seven carotenoid cleavage dioxygenase (VvCCD) gene family members were identified and divided into two groups: four VvCCDs and three 9-cis-epoxycarotenoiddioxygenase (VvNCEDs). One member of VvCCD family (VvCCD1), predominantly expressed during fruit development, was found to increase and reached the highest value in the fruit ripen stage. Similar to VvCCD1, but to a lesser extent, VvCCD4b increased dramatically during grapevine development and ripening (Fig. 7 and Table S1). The strong expression of two genes may help to control carotenoid turnover and contribute to the colors or aromas of grapevine fruit. Furthermore, three VvNCEDs showed different expression profiles in grapevine fruit. For example, VvNCED1 and VvNCED6 had the highest expression at green fruit stage, while VvNCED2 displayed the highest expression at ripe stage. The results agree with earlier studies that the expression pattern of VvNCEDs could not be correlated to changes in ABA levels in grapevine berries40. These results support the previous studies stating that different CCDs and NCEDs recognized different carotenoid substrates and cleaved at different sites, producing various apocarotenoids41.
Characterization and expression analysis of chlorophyll metabolism genes during grapevine fruit development and ripening
Color change often accompanies the onset of fruit ripening and it is one of the most noticeable characteristics of grape fruit ripening. Color change in fruit typically involves chlorophyll loss and an increase in production of yellow, orange, red or purple pigments. The chlorophyll metabolic pathway can be divided into three distinct phases: (1) synthesis of chlorophyll a from glutamate; (2) interconversion between chlorophyll a and b (chlorophyll cycle); and (3) degradation of chlorophyll a into anon-fluorescent chlorophyll catabolite (Fig. S2)42. In this study, 29 genes involving in the three chlorophyll metabolic phases were identified (Table S2).
Twenty-one genes in chlorophyll biosynthesis, which catalyzed the formation of chlorophyll a from glutamate, exhibited gradual decrease during grapevine fruit ripening, except coproporphyrinogen III oxidase (VvCPOX) and protochlorophyllide reductase C (VvPORC) (Fig. 8 and Table S2). These results were consistent with a previous study indicating that chlorophyll degradation represented a dramatic change in metabolism at the onset of fruit ripening43. Further, four genes were identified in chlorophyll cycle pathway. With the exception of chlorophyll b reductase (VvNYC1), which was highly expressed at ripe stage, the other three genes also showed decreased expression patterns from the green fruit stage to the veraison/ripe stage. In chlorophyll degradation pathway, four genes were identified and the expression of three genes (chlorophyllase 2, VvCLH2; pheophytinase, VvPPH and pheophorbide a oxygenase, VvPaO) increased steadily during grapevine fruit ripening. VvPaO, a gene converting pheophorbide a into red chlorophyll catabolite, was highly expressed and showed a significant increase throughout the grapevine berry developmental stages, indicating that VvPaO may have an important role in the chlorophyll degradation. Transcriptomic datas demonstrated that the chlorophyll decline during grapevine fruit ripening was due to a acceleration in chlorophyll degradation and/or the blocking of chlorophyll synthesis.
Figrue 8.
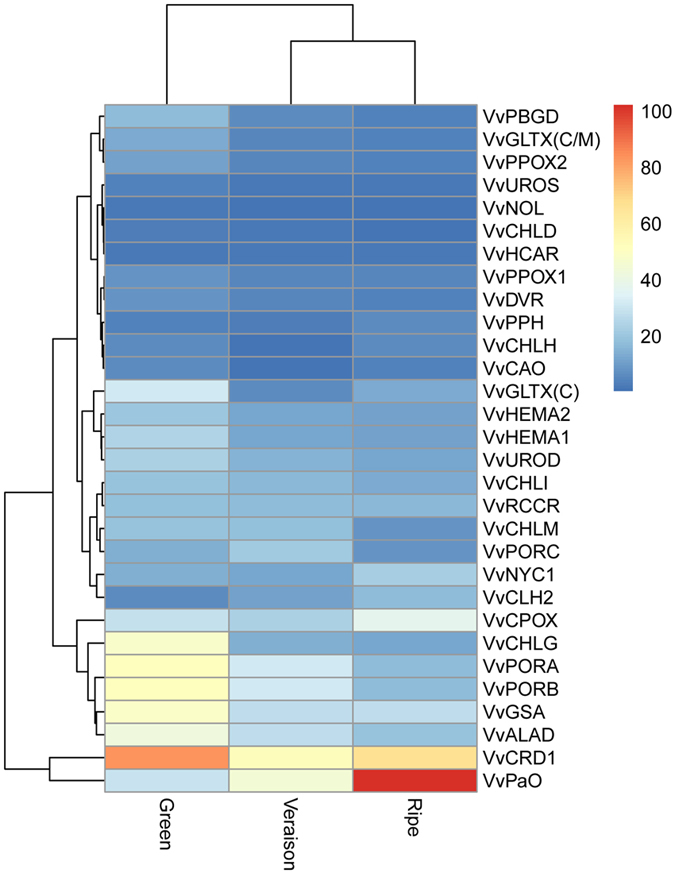
Expression level of chlorophyll metabolism genes during grapevine ripening. Data were obtained by Illumina RNA Sequencing using two biological replicas and are expressed as Reads Per Kilobase of exon model per Million mapped reads (RPKM).
Quantitative real-time PCR validation
Eight genes involved in MVA, MEP, carotenoid biosynthesis and catabolism pathway were chosen for quantitative real-time PCR (qRT-PCR). Transcript abundance patterns were calculated over the entire course of berry development. The qRT-PCR results generally agreed (87.5%) with the changes in transcript abundance determined by RNA-seq (Fig. S3). Only VvDXS1, which showed lowest expression level in ripe stage, were different from that observed in RNA-seq. The qRT-PCR results suggested the reliability of the RNA-seq data.
Discussion
Carotenoid metabolism has received much attention due to the production of the phytohormones ABA and strigolactone; as well as the formation of norisoprenoids, impact flavour and aroma compounds in a number of commercially important fruits and flowers. In grapevine, carotenoids are not only responsible for the visual appeal of grape fruit, but also enhance nutritional value and health benefits for humans44, 45. Thus, carotenoid content has become an important grapevine breeding objective and the study on grapevine carrotenoid metabolism has been focused. In this study, an integrative study combining carotenoid content and gene expression profiles was performed to gain insight into the synthesis and accumulation of carotenoids in grapevine during fruit development and ripening.
A toal of 54 carotenoid metabolism-related genes were identified and distributed on 17 of 19 chromosomes in grapevine, in agreement with the previous study in Brassica rapa 37 that the distribution of carotenoid metabolism genes was fractionation status at the whole-genome level. Furthermore, gene duplication and divergence events have been suggested to be the main contributors to evolutionary momentum46. In the current study, 11 of 54 carotenoid metabolism-related genes have arisen through either tandem or segmental duplication. Unlike Brassica rapa, two pair of tandem duplication events, VvDXS2/VvDXS4/VvDXS6 and VvCCD4a/VvCCD4b, were observed in the grapevine carotenoid genes (Fig. 2). Three pairs of segmental duplication (VvHMGR1/VvHMGR3, VvGGPPS1/VvGGPPS2 and VvCHYB1/VvCHYB2) were also identified (Fig. 2). Taken together, both tandem and segmental duplications existed in grapevine carotenoid metabolism-related genes, implying that gene duplications play a major role in genomic rearrangements and expansions of grapevine carotenoid metabolism. In addition, expression differentiation was also found between duplicated genes (Table S1). For example, in three pairs of segmental duplication, one copy was much more highly expressed than the other. In other genes, VvCCD4a and VvCCD4b showed the opposite expression (Table S1). The expression differentiation between duplicate genes has been reported and it increases the probability of the retention of duplicated genes in a genome47, 48. These duplicated gene expression variations may be signs of subfunctionalization among different tissues and contribute to an increased complexity in the regulatory networks of the carotenoid pathway in grapevine.
Carotenoids are derived from the condensation of the 5-carbon precursors IPP and DMAPP, which are produced via the MEP pathway in plastids49. So far, both DXS and DXR are thought to be important rate-controlling steps of the MEP pathway and play important role in carotenoid flux regulation6, 50. In grapevine, six VvDXS genes were divided into three distinct classes (Fig. S4), suggested that each VvDXS gene may play different role in terpenoid biosynthesis. VvDXS1, the member of the DXS1 clade, was highly and predominantly expressed compared with those of other VvDXS genes, indicating that VvDXS1 might play a predominant role in driving grapevine fruit carotenoid accumulation. The results are also in agreement with the previous study that Genes in the DXS1 clade played housekeeping roles and were probably involved in primary metabolism, such as the biosynthesis of carotenoids and the phytol chain of chlorophyll51–53. Futthermore, a positive correlation was found between the accumulation of DXR transcript and carotenoid production in Arabidopsis seedlings50 or apocarotenoids in mycorrhizal roots from monocots54. Similar to Arabidopsis, the expression of VvDXR was also consistent with the situation of VvDXS1 with wide and high level expression during grapevine fruit development and ripening. In addition, the wide and high level expression of VvDXS1 and VvDXR may help to maintain the amounts of IPP and DMAPP as intermediate metabolites for carotenoid compounds and contribute to the colors or aromas of grapevine fruit.
The cytoplasmic MVA pathway also contribute to the synthesis of IPP and DMAPP (Fig. S1), which provide precursors for the biosynthesis of sesquiterpenes, polyterpenes, sterols, dolichol, and for ubiquinone formation in mitochondria. In our sthdy, the expression of VvAACT2 was significantly higher than that of VvAACT1 (5.8-fold), suggesting that VvAACT2 may play a more important role in cytosolic isoprenoid biosynthesis during grapevine berry development and ripening. Similar to grapevine, Arabidopsis AACT2 (AtAACT2) was six times more efficient than AtAACT1 did in complement the erg10 yeast mutant lacking AACT 55. Three VvHMGRs, the rate-limiting step in MVA pathway, showed a completely differential expression pattern throughout the berry developmental stages, in agreement with previous studies that differential expressions of HMGR isozymes had a critical regulatory role for channeling and counter balancing carbon flux to the different downstream pathways of development or stress response56.
GGPPS is an important branch point enzyme in terpenoid biosynthesis since their enzymatic product GGPP is the direct precursor not only carotenoids, but also diterpenes, ABA and other compounds57. In grapevine, four divergent VvGGPPS isoforms showed different subcellular localization consistent with the subcellular compartmentalization of the diverse GGPP-dependent terpenoid pathways. VvGGPPS2 showed significantly higher expression level than any other member, indicating that VvGGPS2 had essential functions in carotenoid biosynthetic pathway during grapevine fruit development. A decreasing expression of VvGGPS2 from green to the red ripe stage had a significant positive correlation with a decrease of the total carotenoid content. PSY is a rate-limiting enzyme of carotenoid biosynthesis and is frequently regarded as the major bottleneck in carbon flux to carotenoids15. For example, over-expressing tomato PSY1 (SlPSY1) significantly increased the carotenoid content in tomato fruit58. In this study, three VvPSY exhibited different expression pattern. Unlike SlPSY1 with most highly expressed in ripe tomato fruits59, VvPSY1 was highly expressed in green and veraison stages, whereas VvPSY2 showed the highest expression in ripe grapevine fruits, confirming the complex regulation of the pathway. VvPSY3, a homologly with tomato SlPSY3 (Fig. S5), was not expressed in fruit, suggesting that VvPSY3 should be associated with root carotenogenesis, because SlPSY3 mainly expressed in root60. Furthermore, the cyclization of lycopene is a central branch point in the carotenoid biosynthetic pathway (Fig. S1). The expression of VvLCYB gradually increased and was significantly higher than that of VvLCYE during grapevine fruit ripening, especially in ripe stage of VvLCYB (Table S1). The induction of VvLCYB during grapevine fruit development and ripening might be responsible for maintain the amounts of β-carotene as intermediate metabolites for other compounds, such as ABA and C13-norisoprenoids. In grapevine, ABA concentrations increase and peak at the onset of ripening61. The gene expression profiles of VvNXS and VvCCS seem to support the phenomenon. The low expression of VvCCS at veraison and harvest stages created a blockade downstream, and the high expression of VvNXS might help to accumulate the neoxanthin, which is an intermediate in the biosynthesis of the plant hormone ABA.
The steady-state levels of carotenoid is dependent on the balance between biosynthesis and degradation along with storage15. Consistent with Arabidopsis CCD 62, seven VvCCD family genes were divided into two groups, including four VvCCDs and three VvNCEDs (Table S1). VvCCD1 and VvCCD4b were found to increase stably and reached the highest values in the ripe stage. The significantly negative correlation between carotenoid content and gene expression profiles indicated that VvCCD1 and VvCCD4b preferentially involved in volatile compound production from different carotenoid substrates during grapevine fruit development and ripening. This negative correlation between the expression of CCD1 and CCD4 and carotenoid levels also exists in chrysanthemum flowers, potato, and strawberry63–65. Taken together, these results suggested that expression profile of genes coding for carotenoid biosynthetic pathway associated with the accumulation of carotenoid in grapevine fruit.
Conclusions
Fifty-four carotenoid biosynthetic genes were identified in grapevine genome. The composition of carotenoid biosynthetic genes could explain the metabolic profiles of carotenoid accumulation and help to elaborate the genetic mechanism of carotenoid biosynthesis in grapevine. The expression analysis of carotenoid biosynthetic genes showed that down regulation of VvDXS, VvDXR, VvGGPPS and VvPSY and a dramatic increase in the transcription levels of VvCCD were responsible for the reduction of carotenoids. The visible correlation between carotenoid content and gene expression profiles also suggested that transcriptional regulation of carotenoid biosynthesis pathway genes is a key mechanism of carotenoid accumulation. These results represent an important reference study for further characterisation of carotenoid biogenesis and accumulation in grapevine.
Materials and Methods
Plant material
Three-years-old ‘Fujiminori’ grapevine trees, grown in the standard field conditions at the Nanjing Agricultural University fruit farm, Nanjing, China, were chosen as the experimental material. Berries samples were collected at three time points: green stage (40 DAF), veraison (65 DAF) and ripe/harvest stages (90 DAF) throughout the growing season. Furthermore, in order to capture a representative biological selection of transcripts at each time-point, RNA for Illumina sequencing was purified from tissues of 40 berries sampled from 20 bunches.
Identification and analysis of the carotenoid biosynthesis-related genes in grapevine
Carotenoid biosynthesis-related genes were identified via BLAST-P search in NCBI using the Arabidopsis gene sequences as queries. Gene sequences of Arabidopsis involved in the carotenoid biosynthetic pathway were acquired from the TAIR database (www.arabidopsis.org) and KEGG pathway database (http://www.genome.jp/kegg/pathway.html). The grapevine genome and a set of annotated gene sequences from Grape genome database (http://genomes.cribi.unipd.it/grape/) were used to identify the carotenoid biosynthetic genes in grapevine. Subsequently, to verify the reliability of the initial results, a survey was conducted to confirm these genes using functional annotation of the grapevine transcriptome in three distinct stages of berry development (NCBI GEO Accession: GSE77218)19.
Determination of carotenoid and chlorophyll content
Accurately weighted 0.5 g of fresh grapevine sample was taken, and homogenized in tissue homogenizer with 10 ml of 95% ethanol. Homoginized sample mixture was centrifuge for 10,000 rpm for 15 min at 4 °C. The supernatant were separated and 0.5 ml of it is mixed with 4.5 ml of the 95% ethanol. The solution mixture was analyzed for Chlorophyll a, Chlorophyll b and carotenoids content in spectrophotometer (Parkin). The chlorophyll a, Chlorophyll b and carotenoid are respectively determined by spectrophotometric measurement at 664, 649and 470 nm as previous research66.
Sequence feature analysis of grapevine carotenoid biosynthesis-related genes
The genomic sequence for each carotenoid metabolism gene was extracted from the whole genomic sequence according to gene location in the chromosome in the annotation file using a programmed Perlscript. Intron/exon structures were predicted using the Gene Structure Display Server (http://gsds.cbi.pku.edu.cn/chinese.php). The molecular weight and theoretical isoelectric point were predicted using the Prot-Param analyses (http://cn.expasy.org/tools/protparam.html)67 on the basis of their sequence. Conserved domains were searched using the Conserved Domain Database (http://www. ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). The localizations of deduced proteins were predicted on the TargetP1.1 server (http://www.cbs.dtu.dk/services/TargetP/)68.
Chromosomal locations, gene duplication and phylogenetic analysis of grapevine carotenoid biosynthesis-related genes
The information of chromosomal position of each carotenoid biosynthesis-related genes was obtained from the grapevine genome (http://www.genoscope.cns.fr/externe/GenomeBrowser/Vitis/), and then the map was drafted using MapInspect software (http://www.plantbreeding.wur.nl/uk/software-mapinspect.html). MCScanX software (http://chibba.pgml.uga.edu/mcscan2/) was used to detect the gene duplication events, with the E-value set below 1 × 10−5 following the description in a previous study69. The diagrams were generated by the program Circos version 0.63 (http://circos.ca/). Phylogenetic trees were constructed in MEGA7.0 software using the neighbor-joining (NJ) method and maximum likelihood (ML) method with 1000 bootstrap replications.
Expression analysis of grapevine carotenoid biosynthesis-related genes
The expression patterns of carotenoid biosynthetic genes in grapevine were acquired from gene expression omnibus (GEO) database of NCBI (GSE77218), which measured using RNA-Seq data19. Differential expression was analyzed and calculated based on the count values of each transcript between libraries using edgeR (the Empirical analysis of Digital Gene Expression in R) software70. The thresholds for judging significant differences in transcript expression were “FDR < 0.001” and “|log2 fold-change (log2FC)| ≥ 1”. Transcripts with |log2FC| < 0.25 were assumed to no change in expression levels. Other transcripts (0.25 < |log2FC| < 1) were considered as “up-regulated slightly” or “down-regulated slightly”.
qRT-PCR validation
qRT-PCR was performed to verify the expression patterns revealed by the RNA-seq study. Total RNA samples of three stages of grapevine fruit development were extracted using Trizol reagent (Invitrogen, Carlsbad, CA, USA). Purified RNA samples were reverse-transcribed using the PrimeScript RT Reagent Kit with gDNA Eraser (Takara, Dalian, China) as per the manufacturer’s protocol. Eight transcripts were selected for the qRT-PCR assay. Gene specific qRT-PCR primers were designed using Primer3 software (http://primer3.ut.ee/) (Table S3). qRT-PCR was carried out using an ABI PRISM 7500 real-time PCR system (Applied Biosystems, USA). Each reaction mix was composed of 10 μl 2 × SYBR Green Master Mix Reagent (Applied Biosystems, USA), 2.0 μl cDNA sample, and 400 nM of gene-specific primers in a final volume of 20 μl. PCR conditions were: 2 min at 95 °C, followed by 40 cycles of denaturation at 95 °C for 10 s and annealing at 60 °C for 40 s. The relative mRNA level for each gene was calculated using the 2−ΔΔCT formula71. A primer pair was also designed for TC81781 (The Institute for Genomic Research, Release 6.0), encoding an actin protein (housekeeping gene). At least three replicates of each cDNA sample were performed for qRT-PCR analysis.
Electronic supplementary material
Acknowledgements
This work was supported by grants the Natural Science Foundation of China (NSFC) (No. 31672131), the Science and Technology Support-Plan of Jiangsu province (BE2013431), the China Postdoctoral Science Foundation (2015M581811) and the Project of International cooperation and exchange (X2017037).
Author Contributions
J.G. Fang and X.P. Leng designed the study and guided the research. X.P. Leng and P.P. Wang performed research and wrote the main manuscript text. X.D. Zhu and X.P. Li analysised the terpenoid metabolic expression profiling. C. Wang and H.Y. Li analysised the carotenoid metabolism expression profiling and performed the qRT-PCR. Q. Mu performed the chlorophyll metabolism expression profiling analysis. A. Li and Z.J. Liu determinated the concentrations of carotenoid and chlorophyll. All authors reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Xiangpeng Leng and Peipei Wang contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-04004-0
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Laule O, et al. Crosstalk between cytosolic and plastidial pathways of isoprenoid biosynthesis in Arabidopsis thaliana. Proc Natl Acad Sci. 2003;100:6866–6871. doi: 10.1073/pnas.1031755100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seemann M, Tse Sum Bui B, Wolff M, Miginiac-Maslow M, Rohmer M. Isoprenoid biosynthesis in plant chloroplasts via the MEP pathway: direct thylakoid/ferredoxin-dependent photoreduction of GcpE/IspG. FEBS Lett. 2006;580:1547–1552. doi: 10.1016/j.febslet.2006.01.082. [DOI] [PubMed] [Google Scholar]
- 3.Cazzonelli CI. Carotenoids in nature: insights from plants and beyond. Funct Plant Biol. 2011;38:833–847. doi: 10.1071/FP11192. [DOI] [PubMed] [Google Scholar]
- 4.Lu S, Li L. Carotenoid metabolism: biosynthesis, regulation, and beyond. J Integr Plant Biol. 2008;50:778–785. doi: 10.1111/j.1744-7909.2008.00708.x. [DOI] [PubMed] [Google Scholar]
- 5.Yuan H, Zhang JX, Nageswaran D, Li L. Carotenoid metabolism and regulation in horticultural crops. Hortic Rev. 2015;2:15036. doi: 10.1038/hortres.2015.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nisar N, Li L, Lu S, Khin NC, Pogson BJ. Carotenoid metabolism in plants. Mol Plant. 2015;8:68–82. doi: 10.1016/j.molp.2014.12.007. [DOI] [PubMed] [Google Scholar]
- 7.Milborrow BV, Lee HS. Endogenous biosynthetic precursors of (+)-abscisic acid. VI. Carotenoids and ABA are formedby the ‘non-mevalonate’ triose-pyruvate pathway in chloroplasts. Aust J Plant Physiol. 1998;25:507–512. doi: 10.1071/PP98006. [DOI] [Google Scholar]
- 8.Auldridge ME, McCarty DR, Klee HJ. Plant carotenoid cleavage oxygenases and their apocarotenoid products. Curr Opin Plant Biol. 2006;9:315–321. doi: 10.1016/j.pbi.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 9.Havaux M. Carotenoid oxidation products as stress signals in plants. Plant J. 2014;79:597–606. doi: 10.1111/tpj.12386. [DOI] [PubMed] [Google Scholar]
- 10.Cooper DA. Carotenoids in health and disease: resent scientific evaluations, research recommendations and the consumer. J Nutr. 2004;134:221S–224S. doi: 10.1093/jn/134.1.221S. [DOI] [PubMed] [Google Scholar]
- 11.Fiedor J, Burda K. Potential role of carotenoids as antioxidants in human health and disease. Nutrients. 2014;6:466–488. doi: 10.3390/nu6020466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vranová E, Coman D, Gruissem W. Structure and dynamics of the isoprenoid pathway network. Mol Plant. 2012;5:318–333. doi: 10.1093/mp/sss015. [DOI] [PubMed] [Google Scholar]
- 13.Vranová E, Coman D, Gruissem W. Network analysis of the MVA and MEP pathways for isoprenoid synthesis. Annu Rev Plant Biol. 2013;64:665–700. doi: 10.1146/annurev-arplant-050312-120116. [DOI] [PubMed] [Google Scholar]
- 14.Young PR, et al. The genes and enzymes of the carotenoid metabolic pathway in Vitis vinifera L. BMC Genomics. 2012;13:243. doi: 10.1186/1471-2164-13-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ruiz-Solaa MA, Rodríguez-Concepción M. Carotenoid Biosynthesis in Arabidopsis: A Colorful Pathway. Arabidopsis Book. 2012;10:e0158. doi: 10.1199/tab.0158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu LH, Shao ZY, Zhang M, Wang QM. Regulation of carotenoid metabolism in tomato. Mol Plant. 2015;8:28–39. doi: 10.1016/j.molp.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 17.Leng XP, et al. Comparative transcriptome analysis of grapevine in response to copper stress. Sci Rep. 2015;5:17749. doi: 10.1038/srep17749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leng XP, et al. Transporters, chaperones and P-type ATPases controlling grapevine copper homeostasis. Funct Integr Genomic. 2015;15:673–684. doi: 10.1007/s10142-015-0444-1. [DOI] [PubMed] [Google Scholar]
- 19.Shangguan LF, et al. RNA-sequencing reveals biological networks during table grapevine (‘Fujiminori’) fruit development. PLoS ONE. 2017;12:e0170571. doi: 10.1371/journal.pone.0170571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo CL, et al. Evolution and expression analysis of the grape (Vitis vinifera L.) WRKY gene family. J Exp Bot. 2014;65:1513–1528. doi: 10.1093/jxb/eru007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holub EB. The arms race is ancient history in Arabidopsis, the wildflower. Nature Reviews Genetics. 2001;2:516–527. doi: 10.1038/35080508. [DOI] [PubMed] [Google Scholar]
- 22.Bureau SM, Razungles AJ, Baumes RL, Bayonove CL. Effect of qualitative modification of light on the carotenoid contents in Vitis vinifera L. cv. Syrah berries. Sci Aliment. 1998;18:485–495. [Google Scholar]
- 23.Bureau S, Baumes R, Razungles A. Effects of vine or bunch shading on the glycosylated flavor precursors of Vitis vinifera L. cv. Syrah. J Agric Food Chem. 2000;48:1290–1297. doi: 10.1021/jf990507x. [DOI] [PubMed] [Google Scholar]
- 24.Oliveira C, Ferreira AC, Costa P, Guerra J, Guedes de Pinho P. Effect of some viticultural parameters on the grape carotenoid profile. J Agric Food Chem. 2004;52:4178–4184. doi: 10.1021/jf0498766. [DOI] [PubMed] [Google Scholar]
- 25.Newman JD, Chappell J. Isoprenoid biosynthesis in plants: carbon partitioning within the cytoplasmic pathway. Crit Rev Biochem Mol Biol. 1999;34:95–106. doi: 10.1080/10409239991209228. [DOI] [PubMed] [Google Scholar]
- 26.Closa M, et al. The Arabidopsis thaliana FPP synthase isozymes have overlapping and specific functions in isoprenoid biosynthesis, and complete loss of FPP synthase activity causes early developmental arrest. Plant J. 2010;63:512–525. doi: 10.1111/j.1365-313X.2010.04253.x. [DOI] [PubMed] [Google Scholar]
- 27.Antolín-Llovera M, et al. Modulation of plant HMG-CoA reductase by protein phosphatase 2A: positive and negative control at a key node of metabolism. Plant Signal Behav. 2011;6:1127–31. doi: 10.4161/psb.6.8.16363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Friesen JA, Rodwell VW. The 3-hydroxy-3-methylglutaryl coenzyme-A (HMG-CoA) reductases. Genome Biology. 2004;5:248. doi: 10.1186/gb-2004-5-11-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Banerjee A, Sharkey TD. Methylerythritol 4-phosphate (MEP) pathway metabolic regulation. Nat Prod Rep. 2014;31:1043–1055. doi: 10.1039/C3NP70124G. [DOI] [PubMed] [Google Scholar]
- 30.Cordoba E, Salmi M, León P. Unravelling the regulatory mechanisms that modulate the MEP pathway in higher plants. J Exp Bot. 2009;60:2933–2943. doi: 10.1093/jxb/erp190. [DOI] [PubMed] [Google Scholar]
- 31.Hsieh MH, Chang CY, Hsu SJ, Chen JJ. Chloroplast localization of methylerythritol 4-phosphate pathway enzymes and regulation of mitochondrial genes in ispD and ispE albino mutants in Arabidopsis. Plant Mol Biol. 2008;66:663–673. doi: 10.1007/s11103-008-9297-5. [DOI] [PubMed] [Google Scholar]
- 32.Ma YM, et al. Genome-wide identification and characterization of novel genes involved in terpenoid biosynthesis in Salvia miltiorrhiza. J Exp Bot. 2012;63:2809–2823. doi: 10.1093/jxb/err466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grassi S, et al. Comparative genomics reveals candidate carotenoid pathway regulators of ripening watermelon fruit. BMC Genomics. 2013;14:781. doi: 10.1186/1471-2164-14-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coman D, Altenhoff A, Zoller S, Gruissem W, Vranová E. Distinct evolutionary strategies in the GGPPS family from plants. Front Plant Sci. 2014;5:230. doi: 10.3389/fpls.2014.00230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ament K, Van Schie CC, Bouwmeester HJ, Haring MA, Schuurink RC. Induction of a leaf specific geranylgeranyl pyrophosphate synthase and emission of (E,E)-4,8,12-trimethyltrideca-1,3,7,11-tetraene in tomato are dependent on both jasmonic acid and salicylic acid signaling pathways. Planta. 2006;224:1197–1208. doi: 10.1007/s00425-006-0301-5. [DOI] [PubMed] [Google Scholar]
- 36.Fraser PD, et al. Evaluation of transgenic tomato plants expressing an additional phytoene synthase in a fruit-specific manner. Proc Natl Acad Sci. 2002;99:1092–1097. doi: 10.1073/pnas.241374598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li PR, et al. Carotenoid biosynthetic genes in Brassica rapa: comparative genomic analysis, phylogenetic analysis, and expression profiling. BMC Genomics. 2015;16:492. doi: 10.1186/s12864-015-1655-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun LA, et al. Reciprocity between abscisic acid and ethylene at the onset of berry ripening and after harvest. BMC Plant Biol. 2010;10:257–268. doi: 10.1186/1471-2229-10-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuhn N, et al. Berry ripening: recently heard through the grapevine. J Exp Bot. 2014;65:4543–4559. doi: 10.1093/jxb/ert395. [DOI] [PubMed] [Google Scholar]
- 40.Wheeler S, Loveys B, Ford C, Davies C. The relationship between the expression of abscisic acid biosynthesis genes, accumulation of abscisic acid and the promotion of Vitis vinifera L. berry ripening by abscisic acid. Aust J Grape and Wine Res. 2009;15:195–204. doi: 10.1111/j.1755-0238.2008.00045.x. [DOI] [Google Scholar]
- 41.Walter MH, Strack D. Carotenoids and their cleavage products: biosynthesis and functions. Nat Prod Rep. 2011;28:663–692. doi: 10.1039/c0np00036a. [DOI] [PubMed] [Google Scholar]
- 42.Tanaka A, Tanaka R. Chlorophyll metabolism. Curr Opin Plant Biol. 2006;9:248–255. doi: 10.1016/j.pbi.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 43.Barry CS, McQuinn RP, Chung MY, Besuden A, Giovannoni JJ. Amino acid substitutions in homologs of the STAY-GREEN protein are responsible for the green-flesh and chlorophyll retainer mutations of tomato and pepper. Plant Physiol. 2008;147:179–187. doi: 10.1104/pp.108.118430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oliveira C, et al. Carotenoid compounds in grapes and their relationship to plant water status. J Agric Food Chem. 2003;51:5967–5971. doi: 10.1021/jf034275k. [DOI] [PubMed] [Google Scholar]
- 45.Mendes-Pinto MM, Silva Ferreira AC, Caris-Veyrat C, Guedes de Pinho P. Carotenoid, chlorophyll, and chlorophyll-derived compounds in grapes and port wines. J Agric Food Chem. 2005;53:10034–10041. doi: 10.1021/jf0503513. [DOI] [PubMed] [Google Scholar]
- 46.Chothia C, Gough J, Vogel C, Teichmann SA. Evolution of the protein repertoire. Science. 2003;300:1701–1703. doi: 10.1126/science.1085371. [DOI] [PubMed] [Google Scholar]
- 47.Duarte JM, et al. Expression pattern shifts following duplication indicative of subfunctionalization and neofunctionalization in regulatory genes of Arabidopsis. Mol Biol Evol. 2006;23:469–478. doi: 10.1093/molbev/msj051. [DOI] [PubMed] [Google Scholar]
- 48.Blanc G, Wolfe KH. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell. 2004;16:1679–1691. doi: 10.1105/tpc.021410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rodriguez-Concepcion M. Supply of precursors for carotenoid biosynthesis in plants. Arch Biochem Biophys. 2010;504:118–122. doi: 10.1016/j.abb.2010.06.016. [DOI] [PubMed] [Google Scholar]
- 50.Carretero-Paulet L, et al. Enhanced flux through the methylerythritol 4-phosphate pathway in Arabidopsis plants overexpressing deoxyxylulose 5-phosphate reductoisomerase. Plant Mol Biol. 2006;62:683–695. doi: 10.1007/s11103-006-9051-9. [DOI] [PubMed] [Google Scholar]
- 51.Walter MH, Hans J, Strack D. Two distantly related genes encoding 1-deoxy-D-xylulose 5-phosphate synthases: differential regulation in shoots and apocarotenoid-accumulating mycorrhizal roots. Plant J. 2002;31:243–254. doi: 10.1046/j.1365-313X.2002.01352.x. [DOI] [PubMed] [Google Scholar]
- 52.Phillips MA, et al. Functional identification and differential expression of 1-deoxy-D-xylulose5-phosphate synthase in induced terpenoid resin formation of Norway spruce (Picea abies) Plant Mol Biol. 2007;65:243–257. doi: 10.1007/s11103-007-9212-5. [DOI] [PubMed] [Google Scholar]
- 53.Cordoba E, et al. Functional characterization of the three genes encoding 1-deoxy-D-xylulose5-phosphate synthase in maize. J Exp Bot. 2011;62:2023–2038. doi: 10.1093/jxb/erq393. [DOI] [PubMed] [Google Scholar]
- 54.Walter MH, Fester T, Strack D. Arbuscular mycorrhizal fungi induce the non-mevalonate methylerythritol phosphate pathway of isoprenoid biosynthesis correlated with accumulation of the “yellow pigment” and other apocarotenoids. Plant J. 2000;21:571–578. doi: 10.1046/j.1365-313x.2000.00708.x. [DOI] [PubMed] [Google Scholar]
- 55.Jin H, Song Z, Nikolau BJ. Reverse genetic characterization of two paralogous acetoacetyl CoA thiolase genes in Arabidopsis reveals their importance in plant growth and development. Plant J. 2012;70:1015–1032. doi: 10.1111/j.1365-313X.2012.04942.x. [DOI] [PubMed] [Google Scholar]
- 56.Kim YJ, Lee OR, Oh JY, Jang MG, Yang DC. Functional analysis of 3-hydroxy-3-methylglutaryl coenzyme a reductase encoding genes in triterpene saponin-producing ginseng. Plant Physiol. 2014;165:373–387. doi: 10.1104/pp.113.222596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tholl D, Lee S. Terpene specialized metabolism in Arabidopsis thaliana. Arabidopsis Book. 2011;9:e0143. doi: 10.1199/tab.0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fraser PD, et al. Manipulation of phytoene levels in tomato fruit: effects on isoprenoids, plastids, and intermediary metabolism. Plant Cell. 2007;19:3194–3211. doi: 10.1105/tpc.106.049817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sato S, et al. The tomato genome sequence provides insights into fleshy fruit evolution. Nature. 2012;485:635–641. doi: 10.1038/nature11119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li F, Vallabhaneni R, Wurtzel ET. PSY3, a new member of the phytoene synthase gene family conserved in the Poaceae and regulator of abiotic stress-induced root carotenogenesis. Plant Physiol. 2008;146:1333–1345. doi: 10.1104/pp.107.111120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Castellarin SD, et al. Characterization of major ripening events during softening in grape: turgor, sugar accumulation, abscisic acid metabolism, colour development, and their relationship with growth. J Exp Bot. 2016;67:709–722. doi: 10.1093/jxb/erv483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tan BC, et al. Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J. 2003;35:44–56. doi: 10.1046/j.1365-313X.2003.01786.x. [DOI] [PubMed] [Google Scholar]
- 63.Ohmiya A, Kishimoto S, Aida R, Yoshioka S, Sumitomo K. Carotenoid cleavage dioxygenase (CmCCD4a) contributes to white color formation in chrysanthemum petals. Plant Physiol. 2006;142:1193–1201. doi: 10.1104/pp.106.087130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Garcia-Limones C, et al. Functional characterization of FaCCD1: a carotenoid cleavage dioxygenase from strawberry involved in lutein degradation during fruit ripening. J Agric Food Chem. 2008;56:9277–9285. doi: 10.1021/jf801096t. [DOI] [PubMed] [Google Scholar]
- 65.Campbell R, et al. The metabolic and developmental roles of carotenoid cleavage dioxygenase 4 from potato. Plant Physiol. 2010;154:656–664. doi: 10.1104/pp.110.158733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sumanta N, Haque CI, Nishika J, Suprakash R. Spectrophotometric analysis of chlorophylls and carotenoids from commonly grown fern species by using various extracting solvents. Res J Chem Sci. 2014;4:63–69. [Google Scholar]
- 67.Guruprasad K, Reddy BV, Pandit MW. Correlation between stability of a protein and its dipeptide composition: a novel approach for predicting in vivo stability of a protein from its primary sequence. Protein Eng. 1990;4:155–161. doi: 10.1093/protein/4.2.155. [DOI] [PubMed] [Google Scholar]
- 68.Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- 69.Wang Y, et al. MCScanX: a tool kit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40:e49. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


