Abstract
Chicken anemia virus (CAV) is a small circular single-stranded DNA virus with a single promoter-enhancer region containing four consensus cyclic AMP response element sequences (AGCTCA), which are similar to the estrogen response element (ERE) consensus half-sites (A)GGTCA. These sequences are arranged as direct repeats, an arrangement that can be recognized by members of the nuclear receptor superfamily. Transient-transfection assays which use a short CAV promoter construct that ended at the transcription start site and drive expression of enhanced green fluorescent protein (EGFP) showed high basal activity in DF-1, LMH, LMH/2A, and primary theca and granulosa cells. The estrogen receptor-enhanced cell line, LMH/2A, had significantly greater expression than LMH cells, and this expression was significantly increased with estrogen treatment. A long promoter construct which included GGTCA-like sequences downstream of the first CAV protein translation start site was found to have significantly less EGFP expression in DF-1 cells than the short promoter, which was largely due to decreased RNA transcription. DNA-protein binding assays indicated that proteins recognizing a consensus ERE palindrome also bind GGTCA-like sequences in the CAV promoter. Estrogen receptor and other members of the nuclear receptor superfamily may provide a mechanism to regulate CAV activity in situations of low virus copy number.
Chicken infectious anemia virus (CAV) is a small DNA virus of the family Circoviridae and has recently been classified as the sole member of the genus Gyrovirus (44). CAV is present in virtually all commercial chicken operations and is highly resistant to disinfectants and heat (67). Infection is acquired by oral or vertical transmission. Neutralizing antibodies develop 3 to 6 weeks postinfection and eliminate overt virus replication. Development of CAV antibodies often occurs after the onset of sexual maturity in specific-pathogen-free (SPF) flocks with low levels of viral exposure and limited stress (29). CAV infects T-cell precursors in the thymic cortex, CD8+ splenic lymphocytes, and hemocytoblasts in the bone marrow (1, 16, 39) and causes apoptosis of infected cells due to viral protein 3 (VP3), also called apoptin (35). Clinical disease is limited to chickens less than 3 weeks of age and is characterized by anemia, thymic and splenic atrophy, and secondary infections, including dermatitis (12, 55, 66). Older chickens do not develop clinical disease but have decreased immune responses, as evidenced by poor vaccine responses, increased susceptibility to other infections (1), and decreased cell-mediated immune responses (25, 26). Although the virus is mostly lymphotropic, there is also evidence that CAV can be present in the reproductive system. Cardona et al. (5, 6) have shown that the presence of antibodies for up to 12 months did not eliminate CAV DNA from the reproductive tissues or prevent vertical transmission. Viral DNA could also be detected in organs of all embryos from antibody-positive hens, with the highest incidence in embryonal spleens and reproductive tissues (28). Experimentally infected males can transmit CAV through the semen until the development of antibodies (13).
The CAV genome consists of a 2.3-kb negative-sense, single-stranded DNA with one promoter-enhancer region (33). A single unspliced, polycistronic mRNA is transcribed, encoding three viral proteins (VP1, VP2, and VP3) through the use of alternate start codons (34). VP1 is the only protein found in the capsid (59). VP2 has been shown to have protein phosphatase activity (41) and may have a role in capsid formation such as a scaffold or chaperone for proper folding of VP1, since both VP1 and VP2 proteins are required to induce protective immunity (36). VP3, called apoptin, causes apoptosis (68) and has recently been shown to bind to DNA (22, 23).
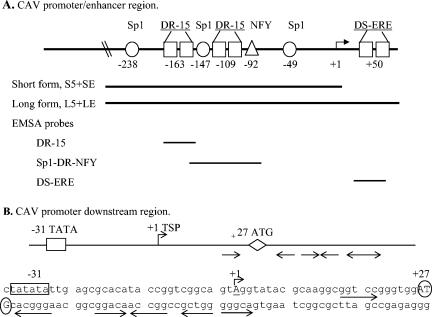
Sequences in the 5′ nontranscribed region of the CAV genome are believed to be the sole promoter enhancer for CAV (Fig. 1A) (38). Dependent on the strain of CAV, this region has four or five 21-bp direct repeat (DR) regions with a 12-bp insert between the first two (or three) and the last two DR. Both the 12-bp insert region and at least two of the DR are necessary for efficient viral transcription and replication (37, 38). These DR regions have a consensus binding site for cyclic AMP response element binding protein, but this site binds an unidentified nuclear protein and not cyclic AMP response element binding protein (38), while the 12-bp insert region was found to bind Sp1.
FIG. 1.
(A) Schematic diagram of the CAV promoter region with the areas used for the short and long promoter expression vectors indicated by the bars under the diagram (see text for primer sequences). The regions used for EMSA are indicated by bars, and the sequences are listed in Table 2. The CAV promoter 21-bp direct repeat sequences are TGTACAGGGGGGTACGTCACCCGTACAGGGGGGTACGTCACA, with the ERE-like elements in boldface underlined type. The ERE-like sequences arranged as direct repeats with a 15-bp separation are indicated in the diagram as boxes labeled DR-15. Known or putative transcription factor binding sites are indicated as circles for Sp1, squares for ERE-like, and triangles for NFY. (B) Schematic diagram and sequence of the downstream region of the CAV promoter. Numbering is based on the transcription start point (TSP) as +1, indicated by the right arrow, and corresponds to nt 333 for the 2,298-bp CIA-1 genome (GenBank accession no. L14767) (48) or nt 354 for the longer 2,319-bp Cux-1 sequence (GenBank accession no. M55918) (33). The TATA box is indicated by the boxed sequences, and the ATG is circled. Arrows underline GGTCA-like sequences and indicate orientation.
Due to the common occurrence of seroconversion at the time of sexual maturation in SPF flocks, the presence of viral DNA in male and female gonads, and in freshly laid fertilized eggs, we hypothesized that CAV gene regulation is effected by the regulation of the reproductive system. The reproductive system has many genes that require spatial and time-specific expression. A large part of this regulation is at the level of transcription with gene expression regulated by promoter-specific sequences in a cell-specific manner, and many of these genes are regulated by nuclear receptors of the steroid/thyroid receptor superfamily. These ligand-activated transcription factors regulate target genes by binding to specific DNA sequences or response elements and recruiting coactivators or corepressors. Estrogen receptor (ER), a type I nuclear receptor, binds as homodimers to the estrogen response element (ERE) that consists of a palindrome with a pentamer or hexamer half-site sequence, (A)GGTCA, and a 3-bp space between the half-sites. There is, however, flexibility in ER binding to half-site elements. Most estrogen-responsive genes contain imperfect EREs that vary from the consensus by up to 3 nucleotides (nt), and widely spaced direct repeats of the GGTCA sequence have been shown to be active in controlling numerous estrogen-regulated genes (2, 19-21).
The type II nuclear receptors, such as thyroid receptor, retinoic acid receptor, retinoid X receptor, and vitamin D receptor, bind pentamer or hexamer elements resembling the ERE half-site (A)GGTCA arranged as DR regions with varying spacing but may also recognize palindromes and inverted or everted repeats (7). These nuclear receptors often bind as heterodimers with retinoid X receptor, but homodimers and monomers are also possible (7, 45, 56) and have been shown to bind to the consensus ERE palindrome (20). Unlike type I nuclear receptors, type II receptors can constitutively bind to their response element and influence gene expression in the presence or absence of ligand, usually by activation or repression, respectively. There is also a diverse group of nuclear receptors with no known ligand that recognize similar elements and are referred to as orphan nuclear receptors. The best characterized is chicken ovalbumin upstream promoter transcription factor (40). Together, ER and the type II nuclear receptors can bind a wide range of AGGTCA-like motifs and often compete for binding to the same DNA sequence. The presence of ligand, relative abundance of heterodimerization partners, and competition for binding sites are an integral part of the precise regulation of many genes.
The cyclic AMP response element sites (ACGTCA) in the CAV promoter direct repeat regions are an imperfect half-site sequence that varies from the consensus (AGGTCA) by 1 nt and are arranged as direct repeats with a 15-bp separation between half-sites (DR-15) (Fig. 1A). Nearby known or potential Sp1 and NFY binding sites may cooperatively interact with ER, as has been shown for many hormone-regulated genes (52, 54, 61). This led us to hypothesize that ER or other nuclear receptors could be involved in the regulation of CAV at the transcription level.
MATERIALS AND METHODS
Animals.
Forty-to-sixty-week-old hens were used as the source for ovarian follicle cell cultures and were obtained from the SPF flocks maintained by the Department of Microbiology and Immunology at Cornell University. All animal procedures were approved by the Institutional Animal Care and Use Committee of Cornell University.
Cell cultures and hormones.
The chicken fibrocytic cell line DF-1 was obtained from the American Type Culture Collection (Rockville, Md.) and grown in Dulbecco's modified Eagle medium with 10% fetal bovine serum (FBS), 1.5 g of NaHCO3/liter, and 1% antibiotic-antimycotic mix (Invitrogen Corp., Carlsbad, Calif.). The chicken hepatocyte cell lines LMH (4) and LMH/2A were also obtained from the American Type Culture Collection. The LMH/2A cell line overexpresses ER 150 times compared with the parent LMH cell line (58). These cells were maintained in Waymouth's MB752/1 medium (Invitrogen) supplemented with 10% FBS, penicillin (100 U/ml), and streptomycin (100 μg/ml) and grown on 0.1% gelatin-coated flasks.
For ovarian cell cultures, the 5 largest yolk-filled ovarian follicles (F1 to F5 for largest to smallest follicles) were removed from 2 hens and placed in ice-cold phosphate-buffered saline (PBS), pH 7. The granulosa and theca layers were separated by identifying the thecal layer from a stab incision of the follicle and carefully peeling the theca cells from the granulosa layer that remained with the yolk. Granulosa cells were collected and pooled from the three largest follicles (F1 to F3) and dispersed as described by Davis et al. (9). Briefly, cells were placed in dispersion medium consisting of M199 medium with 0.1% collagenase type IV (Gibco), 0.2% bovine serum albumin, and 0.01% trypsin inhibitor (Gibco). Cells were digested at 37°C for 15 min with occasional pipetting and plated onto 25- or 75-cm2 flasks. The theca layers from the two largest (F1 and F2) and the two smallest (F4 and F5) yellow follicles were collected and maintained separately for these assays. The theca layers were minced with scalpel blades and then dispersed by using dispersion media with a 5× collagenase concentration (0.5%) for 60 min at 37°C, with pipetting at 20-min intervals. Theca and granulosa cells were maintained in M199 with Earle's salts, l-glutamine, 2,200 mg of NaHCO2/liter, and 25 mM HEPES (Invitrogen) and supplemented with 5% FBS, 100 U of penicillin/ml, 100 μg of streptomycin/ml, and 0.5 μg of insulin (Sigma-Aldrich Corp., St. Louis, Mo.)/ml. All cells were grown at 37°C with 5% CO2.
Plasmid and promoter constructs.
The CAV short and long promoter sequences were generated by standard PCR with primers designed based on CIA-1 strain (GenBank accession no. L14767) as follows. Both the short and long promoters were amplified with the same forward primer, 5′-GTTACTATTCCATCACCATTCTA-3′ (nt 13 to 35), with the reverse primer 5′-GCACATACCGGTCGGCAGT-3′ (nt 314 to 332) for the short promoter sequence and the reverse primer 5′-CAGTGAACGGCGCTTAGCC-3′ (nt 394 to 413) for the long promoter sequence. The long promoter construct included sequences downstream of the first viral protein translation start site and was designed to be in reading frame with the enhanced green fluorescent protein (EGFP) start codon. PCR products were cloned into the pCR2.1 TOPO vector (Invitrogen) and subcloned into the pEGFP-1 expression vector (BD Biosciences Clontech Laboratories, Inc., Palo Alto, Calif.) encoding the gene for EGFP but lacking a promoter. Two forms were made for both the short and long promoters. The pEGFP-S5 and pEGFP-L5 vectors were subcloned by using the SacI/ApaI sites (see Fig. 3), and the EcoRI site was used for the pEGFP-SE and pEGFP-LE vectors. Constructs were verified by sequencing (Biotechnology Resource Center, Cornell University). The expression plasmid pEGFP-N1 (BD Biosciences Clontech Laboratories, Inc.) with the immediate-early cytomegalovirus (CMV) promoter and the promoterless pEGFP-1 were used as the positive and negative controls, respectively. The pEF1/V5-His/lacZ vector (Invitrogen Corp.) containing the gene for β-galactosidase under the control of the EF-1α promoter was used as a cotransfection vector to normalize for transfection efficiency. However, in assays with high transfection efficiency, the β-galactosidase activity was inversely related to EGFP expression, indicating competition for transcription machinery. Due to this problem, samples were normalized for transfection efficiency by dividing by the positive control vector, and results were reported as percentages of CMV-driven expression. All assays had >10% transfection efficiency, as estimated by counting fluorescent cells.
FIG. 3.
The short and long CAV promoter regions were generated by PCR and cloned into pCR2.1 and then subcloned into pEGFP by using the 5′ SacI and 3′ ApaI sites to give the pEGFP-S5 and pEGFP-L5 expression vectors for the short and long promoter forms, respectively. The CAV promoter sequence is in uppercase letters, regular type, and the complete long promoter sequence is shown with a asterisk, indicating the length used for the short form. The pCR2.1 ends are shown in boldface underlined type, and the pEGFP sequences up to the ATG are in lowercase letters. Sequences after the viral ATG are shown as triplets. The 3′ pCR2.1 end contains a potential ATG start codon that is not in reading frame and would result in a nonsense translated message and an in-frame premature stop codon that would eliminate transcription originating from the viral start site. ATG codons are shown in boxes, and stop codons are circled.
Transient transfection.
Plasmid DNAs were grown in Escherichia coli permissive cells and purified with QIAGEN midi-prep columns (QIAGEN, Inc., Valencia, Calif.). Concentrations were determined by spectrophotometer (SmartSpec 3000; Bio-Rad, Hercules, Calif.) readings at a λ of 260. Aliquots of each plasmid used in transfection assays were electrophoresed through a 1% agarose gel to verify relative concentrations and equal percentages of supercoiled and relaxed forms between plasmids.
For transactivation assays, cells were seeded onto 6- or 12-well plates 1 day before transfection and grown in phenol-red-free medium supplemented with 5% charcoal-stripped FBS. Cells were transfected at 60 to 80% confluence with Lipofectamine Plus or Lipofectamine 2000 (Invitrogen). Primary or secondary theca and granulosa cells were transfected 24 to 48 h after plating. For 12-well plates, 1 μg of DNA was used per well, consisting of 0.5 μg of EGFP reporter plasmid and 0.5 μg of pEF1a/His/lacZ. The transfection mix was removed after 3 to 4 h, and medium was replaced with or without specific hormones. The following hormones were used (all from Sigma-Aldrich Corp.): 17β-estradiol (E2, 10 nM to 1 μM), dexamethasone (1 μM), 9-cis-retinoic acid (10 nM), all-trans-retinoic acid (10 nM), 3,3′,5-triiodo-l-thyronine sodium salt (T3, 100 nM), progesterone (10 nM), and testosterone (10 nM). For LMH and LMH/2A cells, serum supplementation included 2% chicken serum as has been reported previously (4, 31, 58). Twelve-well plates were used for EGFP protein measurement by fluorometer, with each treatment run in triplicate wells and each assay repeated three times or as indicated in the figure legends. For real-time quantitative (qPCR) and quantitative reverse transcription (qRT)-PCR, cells were grown in six-well plates, with cells from each well split for DNA and RNA extraction. Six-well plates were also used for fluorescence-activated cell sorter (FACS) analysis. Transfected cells were harvested 24 to 28 h posttransfection.
Fluorometric assays.
Twenty-four to 28 h after transfection, cells in 12-well plates were washed once with cold PBS and then harvested in 200 μl of 1× reporter lysis buffer (Promega, Madison, Wis.). Cells were frozen at −80°C, allowed to return to room temperature for one freeze-thaw cycle, then vortexed for 15 s, and centrifuged at maximum speed for 5 min to pellet cellular debris, and supernatants were used for EGFP measurement with a TD 360 fluorometer (Turner BioSystems, Inc., Sunnyvale, Calif.) that was calibrated with recombinant green fluorescent protein (BD Biosciences Clontech).
RNA and DNA extraction.
Total cellular RNA was extracted 24 h after transfection with RNAzol (Tel-Test, Inc., Friendswood, Tex.) and DNase treated with the DNA-free kit (Ambion, Inc., Austin, Tex.). Samples were frozen at −80°C until RT-PCR assay. Cells used for DNA were suspended in digestion buffer (100 mM NaCl, 10 mM Tris-HCl [pH 8.0], 0.5% sodium dodecyl sulfate, 25 mM EDTA, and 200 μg of proteinase K/ml) and mixed at room temperature for 3 to 4 h. DNA was extracted by the phenol-chloroform method (53), precipitated with 100% ethanol, washed once with 70% ethanol, and dissolved in sterile glass-distilled water. DNA was stored at 4°C until use.
Standard RT-PCR and PCR analysis.
RT was performed as previously described (14). Briefly, cDNA was copied by using 1 μg of RNA, the GeneAmp Gold RNA PCR core kit (Applied Biosystems, Foster City, Calif.), and random hexamer primers in a 20-μl total volume. The reaction mixture was incubated for 10 min at 25°C, followed by 99°C for 5 min. PCR was then performed by using 3 μl of the RT-PCR product with AmpliTaq Gold DNA polymerase and the touchdown PCR procedure previously described (63). For detection of ER mRNA, the primers were as follows: forward, 5′-ATGATCGGCTTAGTCTGGCTCTC-3′; reverse, 5′-GGTAGTACGCTGCTGGTTTCTC-3′. The primers for the cellular housekeeping gene, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) have been described previously (14). PCR products were resolved on a 1% agarose gel and visualized with an Eagle Eye II still video system (Stratagene, La Jolla, Calif.).
qPCR and RT-PCR.
Real-time qPCR and qRT-PCR assays were performed by using the TaqMan PCR core reagent kit and TaqMan PCR one-step reagent kit (Perkin-Elmer Applied Biosystems, Wellesley, Mass.) for DNA and RNA samples, respectively. Samples were run on the GeneAmp 9700 PCR system, recorded, and analyzed with the ABI 7700 prism sequence detection system (Perkin-Elmer Applied Biosystems). Samples were run with 500 ng of RNA or 50 ng of DNA, and results were normalized for equal loading by using the cellular genes GAPDH and inducible nitric oxide synthase (iNOS) for RNA and DNA, respectively, as previously described (15). Tenfold serial dilutions of known standards were run with each plate for generation of a standard curve and calculation of the copy number of the unknown samples by using the threshold value as previously described (15, 24).
qPCR and RT-PCR primers, probes, and standards.
Primers and probes for qPCR and RT-PCR assays were designed by using Primer Express software (Perkin Elmer Applied Biosystems) and were supplied by Integrated DNA Technologies, Inc. (Coralville, Iowa). The probes were 5′ end labeled with 6-carboxy fluorescein (FAM) for EGFP or with tetrachloro-6-carboxyfluoroscein for the cellular housekeeping genes, GAPDH and iNOS for RNA and DNA, respectively. The 3′-end-label quencher was 6-carboxy-tetramethyl rhodamine (TAMRA). Sequences for GAPDH and iNOS primers and probes were used as reported previously (15). The EGFP primers and probe are as follows: forward, 5′-AGCAAAGACCCCAACGAGAA-3′; reverse, 5′-GTCCATGCCGAGAGTGATCC-3′; probe, 5′-FAM-CGCGATCACATGGTCCTGCTGG-TAMRA-3′. Samples for the EGFP product and its cellular housekeeping gene were run as same-well duplex reactions. The plasmid pEGFP-1 was used to generate the standard curve for EGFP DNA. The EGFP RNA standard was generated by cloning the PstI-NotI fragment from pEGFP-1 into PstI-NotI-digested pCR4-TOPO (Invitrogen) to make pCR4-EGFP. This plasmid was then linearized with NotI, and the EGFP RNA was transcribed by using the MEGAscript T3 kit (Ambion) according to the directions. EGFP results were normalized for equal loading by dividing by the cellular housekeeping genes, and EGFP RNA results were normalized for equal transfection of expression vector DNA copies.
Immunofluorescent cell staining.
LMH and LMH/2A cells were grown on coverslips in media with or without E2 for 24 h. Coverslips were then rinsed with PBS, fixed in acetone for 15 min, and stored at −20°C until use. The anti-ER antibody Ab-14 (NeoMarkers, Fremont, Calif.) was diluted 1:100 and applied to the cells for 30 min at room temperature. Coverslips were then rinsed three times for 15 min in PBS. A fluorescein isothiocyanate-conjugated goat anti-mouse secondary antibody (SouthernBiotech, Birmingham, Ala.) diluted 1:500 was then applied, incubated for 30 min at 37°C, rinsed three times for 15 min in PBS, and visualized with a fluorescence microscope with epi-illumination.
FACS analysis.
Cells transfected in six-well plates were gently washed with PBS and then dispersed with 500 μl of 1× saline-trypsin-versene (0.85% NaCl, 0.04% KCl, 0.1% glucose, 0.035% NaHCO3, 0.025% EDTA, 0.005% trypsin, 100 U of penicillin G/ml, 100 μg of streptomycin/ml). Cells were pelleted at 3,000 × g for 5 min, washed twice with 1 ml of PBS, then resuspended in 500 μl of freshly prepared 3% paraformaldehyde, fixed for 30 min at room temperature, washed twice with PBS, and resuspended in 0.5 to 1.0 ml of PBS for FACS analysis. A total of 104 cells were counted for each treatment, including cells transfected with the negative and positive control vectors, pEGFP-1 and pEGFP-N1, respectively. The mean fluorescence intensity was calculated for cells gated greater than the negative control, and the results were calculated as percentages of the positive control.
Preparation of nuclear proteins.
Cell cultures grown in 60-mm-diameter dishes were washed once with cold wash buffer (PBS, 1% FBS, 10 μM leupeptin) and then incubated on ice with 100 μl of buffer A (10 mM HEPES [pH 7.8], 1.5 mM MgCl, 10 mM KCl, 0.5 mM phenylmethylsulfonyl fluoride, 10 μM leupeptin, 1 mM dithiothreitol [DTT]) for 30 min. Cells were then scraped from the plates and placed in 1.5-ml Eppendorf tubes and centrifuged at 16,000 × g for 30 s. The supernatant was discarded, and the nuclear pellet was resuspended in 70 μl of buffer C (25% glycerol, 20 mM HEPES [pH 7.8], 0.42 M NaCl, 1.5 mM MgCl, 0.2 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride, 10 μM leupeptin, 1 mM DTT) and incubated on ice with a stir bar for 60 min. Debris was pelleted by centrifugation at 12,000 × g for 5 min, and the supernatant was assayed for protein concentration with a Coomassie protein assay kit (Pierce Biotechnology, Rockford, Ill.). Nuclear proteins were used immediately or frozen at −80°C until use.
EMSA analysis.
Electrophoretic mobility shift assays (EMSA) were performed with nuclear protein extracts from LMH and LMH/2A cells and double-stranded DNA oligonucleotides designed from the CAV promoter as labeled probes (Fig. 1A). In addition, oligonucleotides containing the consensus sequences for the ERE, the ERE half-site, Sp1, and an irrelevant GC-rich sequence from the chicken β-actin gene were used as controls or in competition assays. Forward-strand sequences for the probes and competitors are listed in Table 1. Oligonucleotides were purchased from QIAGEN, and the single-stranded oligonucleotides were gel purified on a 5% nondenaturing polyacrylamide gel in 0.5× Tris-borate-EDTA buffer (45 mM Tris base, 45 mM boric acid, 1 mM EDTA), annealed, and labeled with [γ-32P]ATP (Amersham Biosciences Corp., Piscataway, N.J.) by using a 5′-end-labeling kit (Amersham). Ten micrograms of nuclear protein extracts was incubated with 1 μg of poly(dI-dC) acid (Sigma) in binding buffer (50 mM Tris-HCl [pH 7.6], 50 mM KCl, 10% glycerol, 20 nM ZnSO4, 0.5 mM DTT) for 15 min at room temperature before addition of the labeled probe (100,000 to 200,000 cpm). Unlabeled competitor sequences at 50-, 100-, and 200-fold excess or 1 to 2 μg of anti-ERα antibody ER Ab-10 (NeoMarkers) were added before the labeled probe in competition and supershift assays, respectively. Samples were incubated for 15 min at 37°C and 15 min at room temperature. Protein-DNA complexes were resolved on 5% polyacrylamide gels with 0.5× Tris-borate-EDTA running buffer at 170 V for 2 h and visualized by autoradiography.
TABLE 1.
Oligonucleotides used in EMSA
| Name | nta | Sequence (sense strand, 5′-3′) |
|---|---|---|
| DR-15b | 144-185 | TGTACAGGGGGGTACGTCATCCGTACAGGGGGGTACGTCACA |
| DS-EREb | 368-395 | AACGGCGGACAACCGGCCGCTGGGGGCA |
| Sp1-DR-NFYb | 186-247 | AAGAGGCGTTCCCGTACAGGGGGGTACGTCACGCGTACAGGGGGGTACGTCACAGCCAATCA |
| EREconc | TAATGGAAGGTCACAGTGACCTGAGCAC | |
| ERE half-sitec | ATAGTCAGGTCACAGAGTTGACATAA | |
| Nonspecificd | TGCAAGGCCGGTTTCGCCGGGGACGATGCC | |
| Sp1 consensuse | ATACGATCGGGGCGGGGCTACGTTACG |
Sequences from the CAV promoter region are illustrated in Fig. 1.
An oligonucleotide containing an ERE consensus 15-bp palindrome or a single half-site. Bold and underlined sequences correspond to ERE-like half-sites.
A nonspecific irrelevant GC-rich sequence from the chicken β-actin gene.
An oligonucleotide containing a consensus Sp1 recognition sequence.
Statistical methods.
Results are presented as averages ± standard deviations or standard errors of the means, as indicated. Transient-transfection results were normalized relative to CMV-driven EGFP (pEGFP-N1) expression. Statistical comparisons are made by using two-tailed Student's t test, and differences were considered significant if the P value was <0.05.
RESULTS
Analysis of CAV promoter-enhancer region.
Examination of the CAV promoter region, including sequences downstream of the transcription start site, found potential half-site recognition sequences for ER, type II nuclear receptors, or related orphan receptors. The ACGTCA sequences in the direct repeats of the previously identified promoter region (38) resembled imperfect half-sites arranged as direct repeats with a 15-nt separation. The use of the Signal Scan program (42, 43) identified an additional imperfect ERE half-site (GGTCA), starting at nt +42 relative to the transcription start point by using the CIA-1 sequence (Fig. 1B) (GenBank accession no. L14767) (48). Other published CAV sequences and the sequencing of promoter constructs found GGACA at this site, which is the preferred half-site for the thyroid receptor (32). Further examination of this area revealed a clustering of nearby GGTCA-like sequences that varied from the consensus by 1 nt and were arranged as inverted and direct repeats (Fig. 1B).
Analysis of cell lines for ER expression.
RT-PCR and immunofluorescence were used to determine relative ER content in LMH, LMH/2A, and DF-1 cells. All cell types had detectable ER mRNA, although there was very little in DF-1 cells. The LMH/2A cell line was derived from LMH cells stably transfected to overexpress the estrogen receptor (58), and this overexpression was confirmed by both RT-PCR and immunofluorescence (data not shown).
Transient-transfection assays.
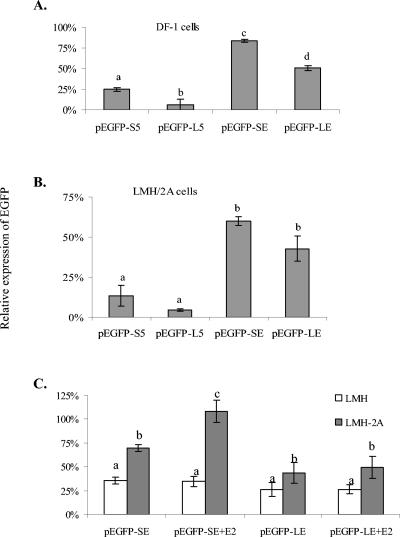
The LMH, LMH/2A, and DF-1 cell lines were used to examine the response of the CAV promoter constructs. In DF-1 and LMH/2A cells, expression of EGFP after transfection with the long CAV promoter constructs pEGFP-L5 and pEGFP-LE was lower than with the short promoter constructs pEGFP-S5 and pEGFP-SE (Fig. 2A and B). The long promoter constructs included the first viral protein translation start codon and were in frame with the EGFP start codon to avoid decreased expression due to nonsense translation. Reexamination of the vector sequence for pEGFP-S5 and pEGFP-L5 identified potential start and stop codons that were derived from the pCR2.1 cloning vector digested with ApaI (Fig. 3). These codons were 3′ of the transcription start site and could affect protein translation efficiency. To avoid this problem, new constructs were made by using the proximal EcoRI sites to generate pEGFP-SE and pEGFP-LE for the short and long promoters, respectively. These constructs included only 6 nt from the transfer vector without intervening start or stop codons between the CAV start codon and the EGFP start site. The removal of the extra sequences from pCR2.1 resulted in significantly enhanced EGFP expression for pEGFP-SE and pEGFP-LE compared to pEGFP-S5 and pEGFP-L5 (Fig. 2A and B). The expression of EGFP with the pEGFP-S5 and pEGFP-SE promoter constructs was higher than the long constructs pEGFP-L5 and pEGFP-LE, and the differences were significant in DF-1 cells (Fig. 2A and B).
FIG. 2.
EGFP expression in DF-1 (A) and LMH/2A (B) cells transfected with pEGFP-S5, pEGFP-SE, pEGFP-L5, and pEGFP-LE. The pEGFP-S5 and pEGFP-SE expression vectors contain the CAV short promoter construct cloned in the SacI-ApaI and EcoRI sites, respectively, while pEGFP-L5 and pEGFP-LE contain the long promoter construct cloned into the SacI-ApaI and EcoRI sites, respectively. pEGFP-S5 and pEGFP-L5 contain additional start and stop codons 3′ to the transcription start point. (C) EGFP expression compared in LMH and LMH/2A cells transfected with pEGFP-SE and pEGFP-LE, with or without 10−7 M 17β-estradiol (E2). Results are the average of 3 or more independent experiments, each with triplicate wells. EGFP expression was measured by fluorometer and normalized as a percentage of the positive-control CMV promoter-driven vector. Error bars indicate standard deviations. Columns within a panel with different letters are significantly different, with a P value of <0.05 by two-tailed t test analysis.
The ER-enhanced cell line LMH/2A (58) was used to examine the response of the CAV promoter constructs to estrogen. Expression of EGFP was significantly higher in LMH/2A than LMH cells after transfection with pEGFP-SE and pEGFP-LE (Fig. 2C). There was a modest but significant increase of expression in LMH/2A cells transfected with pEGFP-SE in the presence of 10−7 M E2 (1.6 times) but not in LMH cells (Fig. 2C).
FACS analysis.
Results from FACS analysis to measure EGFP expression gave results comparable to those for EGFP measured by fluorometer, and this technique was used to screen responses to different hormone treatments with primary or secondary cell cultures of theca and granulosa cells from the hen ovary. These cells have been previously found to be positive for CAV by in situ PCR assay (6). Granulosa cells had significantly more expression then theca cells, and expression was not influenced by hormone treatment in these cell types (data not shown).
qPCR and RT-PCR.
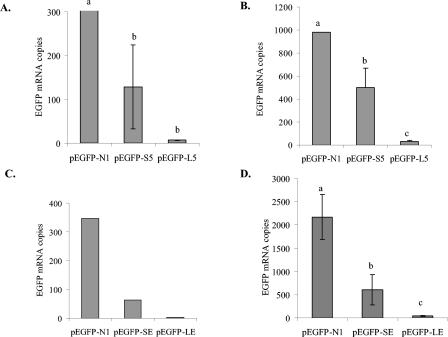
qPCR and qRT-PCR assays were used to measure EGFP mRNA in LMH/2A and DF-1 cells 24 h after transient transfection with pEGFP-S5, pEGFP-L5, pEGFP-SE, or pEGFP-LE. In addition, cells were transfected with pEGFP-N1 or pEGFP-1 as positive and negative controls, respectively. Cells transfected with pEGFP-N1 had three- to fivefold more EGFP transcripts than cells transfected with pEGFP-S5 or pEGFP-SE. Cells transfected with pEGFP-S5 or pEGFP-SE had 5 to 16 times more EGFP transcripts than cells transfected with the long promoter constructs (Fig. 4). The use of the promoterless pEGFP-1 resulted in negligible amounts of EGFP transcripts in all assays (data not shown).
FIG. 4.
Measurement of EGFP mRNA in LMH/2A and DF-1 cells transfected with the positive-control CMV promoter vector pEGFP-N1 or one of the CAV promoter constructs, pEGFP-S5, pEGFP-L5, pEGFP-SE, or pEGFP-LE. The number of DNA copies transfected into the cells and the mRNA transcripts were measured by qPCR or qRT-PCR. (A and B) LMH/2A (A) and DF-1 (B) transfected with pEGFP-N1, pEGFP-S5, or pEGFP-L5. Results are from one experiment with five separate transfected wells. (C) LMH/2A cells transfected with pEGFP-N1, pEGFP-SE, or pEGFP-LE from one assay, with no analysis of significance due to only one sample per treatment. (D) DF-1 cells transfected with pEGFP-N1, pEGFP-SE, or pEGFP-LE from two separate experiments with one to four separate wells per experiment. Results are reported as mRNA copy number and are normalized for transfection efficiency by dividing by DNA copies of the expression plasmid. RNA and DNA EGFP copies were normalized to the cellular genes GAPDH and iNOS, respectively. Columns within a panel with different letters are significantly different, with a P value of <0.05.
DNA-protein interaction assays.
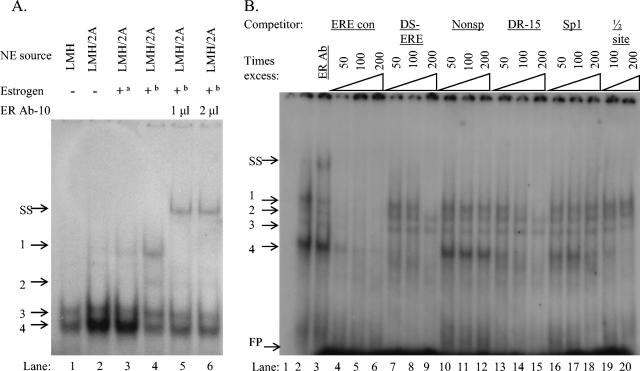
EMSA was used to detect DNA-protein interactions. Synthetic double-stranded oligonucleotides (Table 1) were labeled with [γ-32P]ATP, as a probe, and incubated with nuclear proteins from LMH/2A and LMH cells. DNA-protein complexes were first analyzed by using a consensus ERE 15-bp palindrome (EREcon) and nuclear proteins from LMH and LMH/2A cells. Two complexes were visible with both cell types (Fig. 5A, bands 3 and 4), while two additional complexes were visible with nuclear extracts from the ER-enhanced LMH/2A cell line (Fig. 5A, bands 1 and 2). The slowest-migrating band (Fig. 5A, band 1) was only present with the LMH/2A cell line and was ER specific, as shown by supershifting with the addition of ERα antibody ER Ab-10 (Fig. 5A, lanes 5 to 6, and B lane 3). The intensity of this band was enhanced when E2 was added to the EMSA binding reaction and was further enhanced when LMH/2A cells were grown in medium supplemented with E2 for 15 to 30 min before harvesting of the cells (Fig. 5A, compare lanes 2 to 4).
FIG. 5.
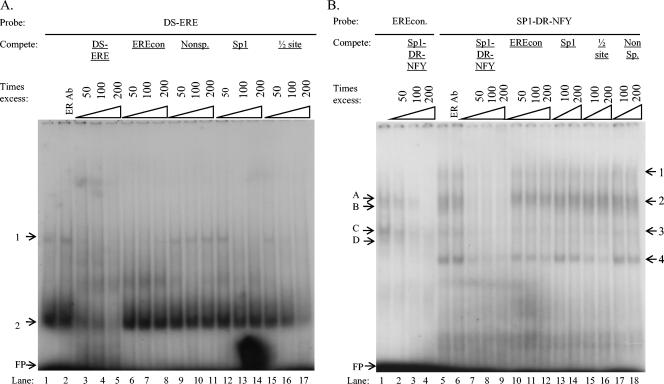
(A) EMSA reaction with the [γ-32P]ATP-labeled ERE consensus oligonucleotides (Table 1) (EREcon) as the probe and nuclear extracts from LMH cells (lane 1) or LMH/2A cells (lanes 2 to 6). Nuclear extracts used in lanes 2 were harvested from LMH/2A cells grown in complete medium without E2 supplementation, and E2 was added to the EMSA binding reaction for lane 3. Nuclear extracts used in lanes 4 to 6 were from LMH/2A cells grown in complete medium supplemented with 10−7 M E2 for 30 min before harvest. The ERα-specific antibody ER-Ab10 was added to the nuclear extracts in lane 6 and 7. a, E2 added to the EMSA binding reaction; b, nuclear extracts from cells grown in medium supplemented with 10−7 M E2 30 min prior to harvesting for nuclear extracts; +, present; −, absent. (B) Nuclear extracts from LMH/2A cells with E2 added to the medium 30 min before harvesting were incubated with the EREcon probe. Lane 1 has no nuclear extracts added as a negative control, ER-AB10 was added to lane 3, and unlabeled EREcon oligonucleotides were added at 50-, 100-, and 200-fold excesses in lanes 4 to 6. Unlabeled oligonucleotides from the CAV promoter, DS-ERE, and DR-15 (Table 1) were added at 50-, 100-, and 200-fold excesses in lanes 7 to 9 and 13 to 15, respectively. Unlabeled nonspecific and Sp1 consensus oligonucleotides (Table 1) (Nonsp and Sp1) were added at 50-, 100-, and 200-fold excesses in lanes 10 to 12 and 16 to 18, respectively, and an unlabeled single half-site response element (Table 1) ([1/2] site) was added at 100- and 200-fold excesses in lanes 19 to 20. Protein-DNA complexes were resolved on 5% polyacrylamide gels and visualized by autoradiography. Protein-DNA complexes 1 to 4 are indicated to the left of the gels. FP, unbound free probe; SS, supershifted band.
Competition assays with the EREcon probe and nuclear extracts from E2-supplemented LMH/2A cells indicated that all complexes were specific, as shown by elimination of bands when unlabeled EREcon was added (Fig. 5B, lanes 4 to 6). Unlabeled oligonucleotides from the CAV promoter region, DS-ERE and DR-15 (Table 1), were also able to compete in a dose-dependent manner for proteins, forming the ER-specific band and the fastest-migrating band (Fig. 5B, bands 1 and 4 in lanes 7 to 9 and 13 to 15), while an irrelevant GC-rich sequence (Table 1) had no effect (Fig. 5B, lanes 10 to 12). The fastest-migrating band (band 4) was also competed by an unlabeled oligonucleotide containing a half-site sequence (Table 1), suggesting that this protein is binding as a monomer (Fig. 5B, lanes 19 to 20). The use of a longer probe from the CAV promoter that includes Sp1, ERE-like repeats, and a putative NFY site (Table 1) competes for all four complexes formed with the EREcon probe in a dose-dependent manner, with three complexes eliminated and the fourth complex diminished (Fig. 6B, lane 2 to 4).
FIG. 6.
(A) Nuclear extracts from LMH/2A cells, with E2 added to the medium 30 min before harvesting, were incubated with [γ-32P]ATP-labeled DS-ERE probe (Table 1) from the CAV promoter. The ERα-specific antibody ER-Ab10 was added to the nuclear extracts in lane 2, and unlabeled DS-ERE oligonucleotides were added at 50-, 100-, and 200-fold excesses in lanes 3 to 5. Unlabeled competitor oligonucleotides EREcon, nonspecific (Nonsp.), Sp1, and half-site ( site) (Table 1) were added at 50-, 100-, and 200-fold excesses in lanes 6 to 8, 9 to 11, 12 to 14, and 15 to 17, respectively. (B) Nuclear extracts from LMH/2A cells, with E2 added to the medium 30 min before harvesting, were incubated with the [γ-32P]ATP-labeled EREcon probe in lanes 1 to 4 or with Sp1-DR-NFY-labeled probe from the CAV promoter (Table 1) in lanes 5 to 18. Unlabeled Sp1-DR-NFY oligonucleotides were added at 50-, 100-, and 200-fold excesses as competition with the EREcon probe in lanes 2 to 5. The Sp1-DR-NFY probe was competed with unlabeled Sp1-DR-NFY and EREcon oligonucleotides at 50-, 100-, and 200-fold excesses in lanes 7 to 9 and 10 to 12, respectively. Unlabeled oligonucleotides Sp1, half-site ( site), and nonspecific (Nonsp.) were added at 100- and 200-fold excesses in lanes 13 to 14, 15 to 16, and 17 to 18, respectively. Protein-DNA complexes were resolved on 5% polyacrylamide gels and visualized by autoradiography and are indicated by the numbers 1 to 4 or the letters A to D to the left and right of the gels. FP, unbound free probe.
In reciprocal experiments, DS-ERE and SP1-ERE-NFY (Table 1) were labeled as probes and EREcon, the ERE half-site, and Sp1 (Table 1) were used as competitors. The DS-ERE probe formed two specific bands which were not supershifted with ER antibody (Fig. 6A, lanes 1 to 2). Both bands were absent in the presence of the unlabeled half-site sequence (Fig. 6A, lane 15 to 17), while EREcon and Sp1 competed with the slowest-migrating band (Fig. 6A, lanes 6 to 8 and 12 to 14). There was no effect with the nonspecific competitor (Fig. 6A, lanes 9 to 11). The Sp1-DR-NFY probe formed four complexes and no visible supershift with ER antibody (Fig. 6B, lanes 5 to 6). All bands were specific, as shown by competition with itself (lanes 7 to 9). The fastest-migrating band was diminished by EREcon and ERE half-site competition (lanes 10 to 12 and 15 to 16) but not by Sp1 or nonspecific competition (lanes 13, 14, 17, and 18). The results of the competition assays are summarized in Table 2.
TABLE 2.
Summarized results for competition EMSA
| Probe | Result with competitora:
|
|||||
|---|---|---|---|---|---|---|
| ERE | DR-15 | Sp1-DR-NFY | DS-ERE | Sp1 | Half-site | |
| ERE | ++ | + | + | + | − | + |
| DR-15 | − | ++ | NT | NT | − | − |
| Sp1-DR-NFY | + | NT | ++ | NT | − | + |
| DS-ERE | + | NT | NT | ++ | + | + |
++, all bands were competed; +, one or more, but not all, bands were competed; −, no bands were competed; NT, not tested.
DISCUSSION
In this study, we tested the hypothesis that the imperfect ERE-like sites in the 5′ promoter-enhancer region of CAV serve as recognition sequences for one or more of the nuclear hormone receptors that bind to AGGTCA half-sites and whether promoter activity can be modulated by their hormones. The ER-enhanced cell line LMH/2A had significantly higher expression of EGFP under control of the CAV promoter than the LMH parent cell line. This expression was modestly but significantly increased with estrogen treatment, suggesting either low-affinity ER-DNA binding or a secondary response. In contrast, treatment with retinoic acid or thyroid hormone did not increase expression of EGFP in transfected cell lines. However, transfection assays overexpress the promoter-reporter gene and often overwhelm limited amounts of endogenous nuclear receptors, requiring cotransfection of the receptor to detect a response (60). There was a lower expression of EGFP in all cell types tested when pEGFP-L5 and pEGFP-LE were used than when pEGFP-S5 and pEGFP-SE were used, which was significant in DF-1 cells (Fig. 2A and B). The longer constructs of the viral promoter include several potential half-sites and the first protein translation start site. Direct measurement of mRNA transcripts showed that the decreased expression was at least in part due to decreased transcription. However, we cannot exclude decreased stability of the mRNA transcript, as is reported for the apolipoprotein II, vitellogenin, and iron regulatory protein 1 genes (3, 8, 18).
DNA-protein binding assays indicated that nonpurified nuclear proteins that bind to a consensus ERE sequence also bind to areas of the CAV promoter that contain GGTCA-like half-sites. The ERE consensus probe formed four protein-DNA complexes, the largest of which was identified as ER with anti-ER antibody (Fig. 5). This complex was only detected in the ER-enhanced LMH/2A cell line, although the other cell types used were also ER positive by more sensitive techniques of RT-PCR and immunofluorescence. ER binding was not detected with CAV promoter probes and nuclear extracts from LMH/2A cells; however, these promoter sequences did completely eliminate the ER-specific band when present as unlabeled competitors at 100- and 200-fold excesses. The fastest-migrating complex formed with the ERE consensus probe was also eliminated by CAV promoter competition and may be a protein monomer, since it is also eliminated by half-site competition. The remaining complexes may be members of the nuclear receptor superfamily, including type II nuclear receptors and orphan nuclear receptors, that can bind to an ERE consensus palindrome (20, 30). Further assays are needed with purified proteins and monoclonal antibodies to identify these proteins.
The lower expression from the long promoter constructs is an interesting new finding, suggesting that sequences at or near the transcription start site act as repressors of transcription. Repression of viral gene expression is a strategy used by many latent viruses to limit protein production, avoid immune detection (27), and regulate gene expression in a stage-specific manner. Some of these viruses have been found to have hormone response elements that are a part of this regulation. Examples include the simian virus 40 major late promoter (10, 11, 17, 62, 69, 70), hepatitis B virus (46, 47, 64, 65), and human immunodeficiency virus (49-51, 57).
The CAV promoter has strong basal expression, and this is likely to be the main controlling factor when chickens are exposed to high levels of virus, which is the prevalent situation in commercial poultry operations. The data reported in this paper suggest that this expression can be modulated by members of the nuclear receptor superfamily. Unlike the abundant and ubiquitous transcription factors, such as Sp1, these modulating factors are likely to be present in limited amounts with cell- and stage-specific distribution and could be controlling factors when the viral DNA copy numbers are low. The presence of CAV in the reproductive organs and the interactions between specific nuclear receptors and the virus promoter-enhancer region could be responsible for successful latency and localized enhanced expression, resulting in viral passage to the embryo and, hence, the next generation.
Spatial and temporal restriction of gene expression is important for a successful latent viral presence, both to avoid immune detection and to allow for successful passage to the next host or generation. Since CAV has a very limited genome with only three viral proteins, such regulation would be dependent on the regulation of cellular genes. In SPF flocks with low stress and low virus exposure, transcription would proceed at a slow rate and be controlled by cellular transcription factors with specific activation or repression dependent on cell type and development stage. We have previously shown the long-term presence of viral DNA in the reproductive organs (6, 28), a system that is tightly regulated by a complex interplay between hormone receptors and/or orphan nuclear receptors and their ligands. The presence of CAV in the reproductive tissues would allow the virus to replicate and expand in the developing embryo along with the development of the reproductive organs and then, like these quiescent organs, become dormant until reactivated at the time of reproductive maturity. This would give the virus an ideal location for stage-specific activity, ensure the likelihood of passage to the next generation, and explain the frequent occurrence of seroconversion to antibody-positive status at the time of sexual maturity in SPF flocks (55). This situation differs from the situation of commercial flocks, which are exposed to large numbers of virus particles and numerous other infectious and environmental stressors. High-dose exposure would result in viral presence above the threshold for control by limited repressive factors that would help maintain latency and result in the rapid onset of disease in young chicks and seroconversion in older birds. The field situation of frequent immune challenge (i.e., T cells activated by frequent pathogen and vaccine challenges) may also result in an abundance of cellular factors that can enhance transcription, protein synthesis, and replication. In both cases, the virus has adapted very well to coexistence with its host as is apparent with its ubiquitous presence.
Acknowledgments
This work was supported in part by gifts from SPAFAS Charles River Laboratories, Wilmington, Mass. M.M.M. was supported by Institutional NRSA award no. 5 T32 AI07618-01 and -02 and Ruth L. Kirschstein NRSA award no. T32 RR07059-08.
REFERENCES
- 1.Adair, B. M. 2000. Immunopathogenesis of chicken anemia virus infection. Dev. Comp. Immunol. 24:247-255. [DOI] [PubMed] [Google Scholar]
- 2.Aumais, J. P., H. S. Lee, C. DeGannes, J. Horsford, and J. H. White. 1996. Function of directly repeated half-sites as response elements for steroid hormone receptors. J. Biol. Chem. 271:12568-12577. [DOI] [PubMed] [Google Scholar]
- 3.Binder, R., D. A. Gordon, S. P. Hwang, and D. L. Williams. 1990. Estrogen-induced destabilization and associated degradation intermediates of apolipoprotein II mRNA. Prog. Clin. Biol. Res. 322:227-240. [PubMed] [Google Scholar]
- 4.Binder, R., C. C. MacDonald, J. B. Burch, C. B. Lazier, and D. L. Williams. 1990. Expression of endogenous and transfected apolipoprotein II and vitellogenin II genes in an estrogen responsive chicken liver cell line. Mol. Endocrinol. 4:201-208. [DOI] [PubMed] [Google Scholar]
- 5.Cardona, C., B. Lucio, P. O'Connell, J. Jagne, and K. A. Schat. 2000. Humoral immune responses to chicken infectious anemia virus in three strains of chickens in a closed flock. Avian Dis. 44:661-667. [PubMed] [Google Scholar]
- 6.Cardona, C. J., W. B. Oswald, and K. A. Schat. 2000. Distribution of chicken anaemia virus in the reproductive tissues of specific-pathogen-free chickens. J. Gen. Virol. 81:2067-2075. [DOI] [PubMed] [Google Scholar]
- 7.Carlberg, C. 1993. RXR-independent action of the receptors for thyroid hormone, retinoid acid and vitamin D on inverted palindromes. Biochem. Biophys. Res. Commun. 195:1345-1353. [DOI] [PubMed] [Google Scholar]
- 8.Cunningham, K. S., R. E. Dodson, M. A. Nagel, D. J. Shapiro, and D. R. Schoenberg. 2000. Vigilin binding selectively inhibits cleavage of the vitellogenin mRNA 3′-untranslated region by the mRNA endonuclease polysomal ribonuclease 1. Proc. Natl. Acad. Sci. USA 97:12498-12502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis, A. J., C. F. Brooks, and P. A. Johnson. 1999. Gonadotropin regulation of inhibin alpha-subunit mRNA and immunoreactive protein in cultured chicken granulosa cells. Gen. Comp. Endocrinol. 116:90-103. [DOI] [PubMed] [Google Scholar]
- 10.Farrell, M. L., and J. E. Mertz. 2002. Cell type-specific replication of simian virus 40 conferred by hormone response elements in the late promoter. J. Virol. 76:6762-6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farrell, M. L., and J. E. Mertz. 2002. Hormone response element in SV40 late promoter directly affects synthesis of early as well as late viral RNAs. Virology 297:307-318. [DOI] [PubMed] [Google Scholar]
- 12.Goryo, M., T. Suwa, T. Umemura, C. Itakura, and S. Yamashiro. 1989. Histopathology of chicks inoculated with chicken aneamia agent (MSB1-TK5803 strain). Avian Pathol. 18:73-89. [DOI] [PubMed] [Google Scholar]
- 13.Hoop, R. K. 1993. Transmission of chicken anaemia virus with semen. Vet. Rec. 133:551-552. [DOI] [PubMed] [Google Scholar]
- 14.Jarosinski, K. W., P. H. O'Connell, and K. A. Schat. 2003. Impact of deletions within the BamHI-L fragment of attenuated Marek's disease virus on vIL-8 expression and the newly identified transcript of open reading frame LORF4. Virus Genes 26:255-269. [DOI] [PubMed] [Google Scholar]
- 15.Jarosinski, K. W., R. Yunis, P. H. O'Connell, C. J. Markowski-Grimsrud, and K. A. Schat. 2002. Influence of genetic resistance of the chicken and virulence of Marek's disease virus (MDV) on nitric oxide responses after MDV infection. Avian Dis. 46:636-649. [DOI] [PubMed] [Google Scholar]
- 16.Jeurissen, S. H., F. Wagenaar, J. M. Pol, A. J. van der Eb, and M. H. Noteborn. 1992. Chicken anemia virus causes apoptosis of thymocytes after in vivo infection and of cell lines after in vitro infection. J. Virol. 66:7383-7388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnston, S. D., X. Liu, F. Zuo, T. L. Eisenbraun, S. R. Wiley, R. J. Kraus, and J. E. Mertz. 1997. Estrogen-related receptor alpha 1 functionally binds as a monomer to extended half-site sequences including ones contained within estrogen-response elements. Mol. Endocrinol. 11:342-352. [DOI] [PubMed] [Google Scholar]
- 18.Kaldy, P., E. Menotti, R. Moret, and L. C. Kuhn. 1999. Identification of RNA-binding surfaces in iron regulatory protein-1. EMBO J. 18:6073-6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kato, S., H. Sasaki, M. Suzawa, S. Masushige, L. Tora, P. Chambon, and H. Gronemeyer. 1995. Widely spaced, directly repeated PuGGTCA elements act as promiscuous enhancers for different classes of nuclear receptors. Mol. Cell. Biol. 15:5858-5867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klinge, C. M., D. L. Bodenner, D. Desai, R. M. Niles, and A. M. Traish. 1997. Binding of type II nuclear receptors and estrogen receptor to full and half-site estrogen response elements in vitro. Nucleic Acids Res. 25:1903-1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krieg, S. A., A. J. Krieg, and D. J. Shapiro. 2001. A unique downstream estrogen responsive unit mediates estrogen induction of proteinase inhibitor-9, a cellular inhibitor of IL-1beta-converting enzyme (caspase 1). Mol. Endocrinol. 15:1971-1982. [DOI] [PubMed] [Google Scholar]
- 22.Leliveld, S. R., R. T. Dame, M. A. Mommaas, H. K. Koerten, C. Wyman, A. A. Danen-van Oorschot, J. L. Rohn, M. H. Noteborn, and J. P. Abrahams. 2003. Apoptin protein multimers form distinct higher-order nucleoprotein complexes with DNA. Nucleic Acids Res. 31:4805-4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leliveld, S. R., R. T. Dame, J. L. Rohn, M. H. Noteborn, and J. P. Abrahams. 2004. Apoptin's functional N- and C-termini independently bind DNA. FEBS Lett. 557:155-158. [DOI] [PubMed] [Google Scholar]
- 24.Markowski-Grimsrud, C. J., M. M. Miller, and K. A. Schat. 2002. Development of strain-specific real-time PCR and RT-PCR assays for quantitation of chicken anemia virus. J. Virol. Methods 101:135-147. [DOI] [PubMed] [Google Scholar]
- 25.Markowski-Grimsrud, C. J., and K. A. Schat. 2003. Infection with chicken anaemia virus impairs the generation of pathogen-specific cytotoxic T lymphocytes. Immunology 109:283-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McConnell, C. D., B. M. Adair, and M. S. McNulty. 1993. Effects of chicken anemia virus on cell-mediated immune function in chickens exposed to the virus by a natural route. Avian Dis. 37:366-374. [PubMed] [Google Scholar]
- 27.Merezak, C., C. Pierreux, E. Adam, F. Lemaigre, G. G. Rousseau, C. Calomme, C. Van Lint, D. Christophe, P. Kerkhofs, A. Burny, R. Kettmann, and L. Willems. 2001. Suboptimal enhancer sequences are required for efficient bovine leukemia virus propagation in vivo: implications for viral latency. J. Virol. 75:6977-6988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller, M. M., K. A. Ealey, W. B. Oswald, and K. A. Schat. 2003. Detection of chicken anemia virus DNA in embryonal tissues and eggshell membranes. Avian Dis. 47:662-671. [DOI] [PubMed] [Google Scholar]
- 29.Miller, M. M., W. B. Oswald, J. Scarlet, and K. A. Schat. 2001. Patterns of chicken infectious anemia virus (CIAV) seroconversion in three Cornell SPF flocks, p. 410-417. In Proceedings of the International Symposium on Infectious Bursal Disease and Chicken Infectious Anaemia, vol. 2. Institut für Geflügelkrankheiten, Justus Liebig University, Giessen, Germany.
- 30.Mohan, R., and R. A. Heyman. 2003. Orphan nuclear receptor modulators. Curr. Top. Med. Chem. 3:1637-1647. [DOI] [PubMed] [Google Scholar]
- 31.Monroe, D. G., and M. M. Sanders. 2000. The COUP-adjacent repressor (CAR) element participates in the tissue-specific expression of the ovalbumin gene. Biochim. Biophys. Acta 1517:27-32. [DOI] [PubMed] [Google Scholar]
- 32.Munoz, A., and J. Bernal. 1997. Biological activities of thyroid hormone receptors. Eur. J. Endocrinol. 137:433-445. [DOI] [PubMed] [Google Scholar]
- 33.Noteborn, M. H., G. F. de Boer, D. J. van Roozelaar, C. Karreman, O. Kranenburg, J. G. Vos, S. H. Jeurissen, R. C. Hoeben, A. Zantema, G. Koch, H. van Ormondt, and A. J. van der Eb. 1991. Characterization of cloned chicken anemia virus DNA that contains all elements for the infectious replication cycle. J. Virol. 65:3131-3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Noteborn, M. H., O. Kranenburg, A. Zantema, G. Koch, G. F. de Boer, and A. J. van der Eb. 1992. Transcription of the chicken anemia virus (CAV) genome and synthesis of its 52-kDa protein. Gene 118:267-271. [DOI] [PubMed] [Google Scholar]
- 35.Noteborn, M. H., D. Todd, C. A. Verschueren, H. W. de Gauw, W. L. Curran, S. Veldkamp, A. J. Douglas, M. S. McNulty, A. J. van der Eb, and G. Koch. 1994. A single chicken anemia virus protein induces apoptosis. J. Virol. 68:346-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Noteborn, M. H., C. A. Verschueren, G. Koch, and A. J. Van der Eb. 1998. Simultaneous expression of recombinant baculovirus-encoded chicken anaemia virus (CAV) proteins VP1 and VP2 is required for formation of the CAV-specific neutralizing epitope. J. Gen. Virol. 79:3073-3077. [DOI] [PubMed] [Google Scholar]
- 37.Noteborn, M. H., C. A. Verschueren, H. van Ormondt, and A. J. van der Eb. 1998. Chicken anemia virus strains with a mutated enhancer/promoter region share reduced virus spread and cytopathogenicity. Gene 223:165-172. [DOI] [PubMed] [Google Scholar]
- 38.Noteborn, M. H., C. A. Verschueren, A. Zantema, G. Koch, and A. J. van der Eb. 1994. Identification of the promoter region of chicken anemia virus (CAV) containing a novel enhancer-like element. Gene 150:313-318. [DOI] [PubMed] [Google Scholar]
- 39.Noteborn, M. H. M., and G. Koch. 1995. Chicken anaemia virus infection: molecular basis of pathogenicity. Avian Pathol. 24:11-31. [DOI] [PubMed] [Google Scholar]
- 40.Park, J. I., S. Y. Tsai, and M. J. Tsai. 2003. Molecular mechanism of chicken ovalbumin upstream promoter-transcription factor (COUP-TF) actions. Keio J. Med. 52:174-181. [DOI] [PubMed] [Google Scholar]
- 41.Peters, M. A., D. C. Jackson, B. S. Crabb, and G. F. Browning. 2002. Chicken anemia virus VP2 is a novel dual specificity protein phosphatase. J. Biol. Chem. 277:39566-39573. [DOI] [PubMed] [Google Scholar]
- 42.Prestridge, D. S. 1996. SIGNAL SCAN 4.0: additional databases and sequence formats. Comput. Appl. Biosci. 12:157-160. [DOI] [PubMed] [Google Scholar]
- 43.Prestridge, D. S. 1991. SIGNAL SCAN: a computer program that scans DNA sequences for eukaryotic transcriptional elements. Comput. Appl. Biosci. 7:203-206. [DOI] [PubMed] [Google Scholar]
- 44.Pringle, C. R. 1999. Virus taxonomy at the XIth International Congress of Virology, Sydney, Australia, 1999. Arch. Virol. 144:2065-2069. [DOI] [PubMed] [Google Scholar]
- 45.Quack, M., and C. Carlberg. 2001. Single thyroid hormone receptor monomers are competent for co-activator-mediated transactivation. Biochem. J. 360:387-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raney, A. K., J. L. Johnson, C. N. Palmer, and A. McLachlan. 1997. Members of the nuclear receptor superfamily regulate transcription from the hepatitis B virus nucleocapsid promoter. J. Virol. 71:1058-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Raney, A. K., E. F. Kline, H. Tang, and A. McLachlan. 2001. Transcription and replication of a natural hepatitis B virus nucleocapsid promoter variant is regulated in vivo by peroxisome proliferators. Virology 289:239-251. [DOI] [PubMed] [Google Scholar]
- 48.Renshaw, R. W., C. Soine, T. Weinkle, P. H. O'Connell, K. Ohashi, S. Watson, B. Lucio, S. Harrington, and K. A. Schat. 1996. A hypervariable region in VP1 of chicken infectious anemia virus mediates rate of spread and cell tropism in tissue culture. J. Virol. 70:8872-8878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rohr, O., D. Aunis, and E. Schaeffer. 1997. COUP-TF and Sp1 interact and cooperate in the transcriptional activation of the human immunodeficiency virus type 1 long terminal repeat in human microglial cells. J. Biol. Chem. 272:31149-31155. [DOI] [PubMed] [Google Scholar]
- 50.Rohr, O., C. Schwartz, D. Aunis, and E. Schaeffer. 1999. CREB and COUP-TF mediate transcriptional activation of the human immunodeficiency virus type 1 genome in Jurkat T cells in response to cyclic AMP and dopamine. J. Cell. Biochem. 75:404-413. [PubMed] [Google Scholar]
- 51.Rohr, O., C. Schwartz, C. Hery, D. Aunis, M. Tardieu, and E. Schaeffer. 2000. The nuclear receptor chicken ovalbumin upstream promoter transcription factor interacts with HIV-1 Tat and stimulates viral replication in human microglial cells. J. Biol. Chem. 275:2654-2660. [DOI] [PubMed] [Google Scholar]
- 52.Safe, S. 2001. Transcriptional activation of genes by 17 beta-estradiol through estrogen receptor-Sp1 interactions. Vitam. Horm. 62:231-252. [DOI] [PubMed] [Google Scholar]
- 53.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 54.Saville, B., M. Wormke, F. Wang, T. Nguyen, E. Enmark, G. Kuiper, J. A. Gustafsson, and S. Safe. 2000. Ligand-, cell-, and estrogen receptor subtype (alpha/beta)-dependent activation at GC-rich (Sp1) promoter elements. J. Biol. Chem. 275:5379-5387. [DOI] [PubMed] [Google Scholar]
- 55.Schat, K. A. 2003. Chicken infectious anemia, p. 182-196. In Y. M. Saif, H. J. Barnes, A. M. Fadly, J. R. Glisson, L. R. McDougald, and D. E. Swayne (ed.), Diseases of poultry, 11th ed. Blackwell Publishing Co., Ames, Iowa.
- 56.Schrader, M., A. Wyss, L. J. Sturzenbecker, J. F. Grippo, P. LeMotte, and C. Carlberg. 1993. RXR-dependent and RXR-independent transactivation by retinoic acid receptors. Nucleic Acids Res. 21:1231-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schwartz, C., P. Catez, O. Rohr, D. Lecestre, D. Aunis, and E. Schaeffer. 2000. Functional interactions between C/EBP, Sp1, and COUP-TF regulate human immunodeficiency virus type 1 gene transcription in human brain cells. J. Virol. 74:65-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sensel, M. G., R. Binder, C. B. Lazier, and D. L. Williams. 1994. Reactivation of apolipoprotein II gene transcription by cycloheximide reveals two steps in the deactivation of estrogen receptor-mediated transcription. Mol. Cell. Biol. 14:1733-1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Todd, D., J. L. Creelan, D. P. Mackie, F. Rixon, and M. S. McNulty. 1990. Purification and biochemical characterization of chicken anaemia agent. J. Gen. Virol. 71:819-823. [DOI] [PubMed] [Google Scholar]
- 60.Wang, F., W. Porter, W. Xing, T. K. Archer, and S. Safe. 1997. Identification of a functional imperfect estrogen-responsive element in the 5′-promoter region of the human cathepsin D gene. Biochemistry 36:7793-7801. [DOI] [PubMed] [Google Scholar]
- 61.Wang, W., L. Dong, B. Saville, and S. Safe. 1999. Transcriptional activation of E2F1 gene expression by 17beta-estradiol in MCF-7 cells is regulated by NF-Y-Sp1/estrogen receptor interactions. Mol. Endocrinol. 13:1373-1387. [DOI] [PubMed] [Google Scholar]
- 62.Wiley, S. R., R. J. Kraus, F. Zuo, E. E. Murray, K. Loritz, and J. E. Mertz. 1993. SV40 early-to-late switch involves titration of cellular transcriptional repressors. Genes Dev. 7:2206-2219. [DOI] [PubMed] [Google Scholar]
- 63.Xing, Z., and K. A. Schat. 2000. Expression of cytokine genes in Marek's disease virus-infected chickens and chicken embryo fibroblast cultures. Immunology 100:70-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu, X., and J. E. Mertz. 2001. Critical roles of nuclear receptor response elements in replication of hepatitis B virus. J. Virol. 75:11354-11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yu, X., and J. E. Mertz. 1997. Differential regulation of the pre-C and pregenomic promoters of human hepatitis B virus by members of the nuclear receptor superfamily. J. Virol. 71:9366-9374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yuasa, N., T. Taniguchi, T. Imada, and H. Hihara. 1983. Distribution of chicken anemia agent (CAA) and detection of neutralizing antibody in chicks experimentally inoculated with CAA. Natl. Inst. Anim. Health Q. (Tokyo) 23:78-81. [PubMed] [Google Scholar]
- 67.Yuasa, N., T. Taniguchi, and I. Yoshida. 1979. Isolation and some characteristics of an agent inducing anemia in chicks. Avian Dis. 23:366-385. [Google Scholar]
- 68.Zhuang, S. M., J. E. Landegent, C. A. Verschueren, J. H. Falkenburg, H. van Ormondt, A. J. van der Eb, and M. H. Noteborn. 1995. Apoptin, a protein encoded by chicken anemia virus, induces cell death in various human hematologic malignant cells in vitro. Leukemia 9:S118-120. [PubMed] [Google Scholar]
- 69.Zuo, F., R. J. Kraus, T. Gulick, D. D. Moore, and J. E. Mertz. 1997. Direct modulation of simian virus 40 late gene expression by thyroid hormone and its receptor. J. Virol. 71:427-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zuo, F., and J. E. Mertz. 1995. Simian virus 40 late gene expression is regulated by members of the steroid/thyroid hormone receptor superfamily. Proc. Natl. Acad. Sci. USA 92:8586-8590. [DOI] [PMC free article] [PubMed] [Google Scholar]