Abstract
Human herpesviruses enter cells by fusion with target membranes, a process that requires three conserved glycoproteins: gB, gH, and gL. How these glycoproteins execute fusion is unknown. Neural network bioinformatics predicted a membrane α-helix contained within the ectodomain of herpes simplex virus (HSV) gH, positionally conserved in the gH of all examined herpesviruses. Evidence that it has attributes of an internal fusion peptide rests on the following lines of evidence. (i) The predicted membrane α-helix has the attribute of a membrane segment, since it transformed a soluble form of gD into a membrane-bound gD. (ii) It represents a critical domain of gH. Its partial or entire deletion, or substitution of critical residues inhibited HSV infectivity and fusion in the cell-cell fusion assay. (iii) Its replacement with the fusion peptide from human immunodeficiency virus gp41 or from vesicular stomatitis virus G partially rescued HSV infectivity and cell-cell fusion. The corresponding antisense sequences did not. (iv) The predicted α-helix located in the varicella-zoster virus gH ectodomain can functionally substitute the native HSV gH membrane α-helix, suggesting a conserved function in the human herpesviruses. We conclude that HSV gH exhibits features typical of viral fusion glycoproteins and that this property is likely conserved in the Herpesviridae family.
Herpesviruses enter cells by fusion with target membranes, a process that requires three or more glycoproteins. The glycoproteins that carry out fusion have been known for years and for most human herpesviruses include the trio of gB, gH, and gL (11, 18, 28, 37) Their specialized activities are largely unknown (8, 37). In particular, the entry of herpes simplex virus (HSV) into cells requires gD as the receptor-binding glycoprotein that interacts with alternate protein receptors, nectin1, nectin2, or herpesvirus entry mediator (10, 15, 22, 27, 39). Most likely, the role played by the interaction of gD with one of its receptors is to trigger fusion through the involvement of its membrane-proximal proline-rich profusion domain; this is a functionally separate and physically distinct domain relative to the receptor-binding domain (9), which ultimately leads to the recruitment-activation of the trio of gB, gH, and gL. These are the real executors of fusion and the puzzling performers of this complex process; their conservation in the Herpesviridae family testifies to their critical role. Thus, while the glycoprotein deputed to receptor recognition differs from one herpesvirus to the other—accounting for the specific cells and tissues that each herpesvirus infects—the mechanism of fusion appears to be conserved.
Key questions that need to addressed are “how do gB, gH, and gL execute fusion?” and “what are the respective roles in the execution of fusion?” Structurally, gB and gH are type 1 membrane glycoproteins, whereas gL is the soluble chaperone of gH, required for its folding and trafficking to the plasma membrane (5, 17, 19, 31). Deletion of any one of the quartet of gD, gB, gH, and gL renders HSV noninfectious (6, 8, 13, 21, 34). When transiently expressed in transfected cells, the quartet is sufficient to induce fusion of cells, a process that is a relatively faithful surrogate of the fusion that leads to virus entry into the cell. This property has been exploited to develop a readily quantifiable cell-cell fusion assay (25, 33, 38).
In an attempt to decipher the roles of gH, gL, and gB, we searched for bioinformatics predictions of structural motifs and found that the ectodomain of HSV gH contains two predicted membrane α-helices, located at amino acid residues 377 to 397 and 513 to 531, respectively. We focused on the N-terminal one because it was positionally conserved in the gH from all examined herpesviruses, and we explored the possibility that it serves as an internal fusion peptide. Fusion peptides are key structural elements in viral fusion glycoproteins; they may be located at the N terminus, typically in glycoproteins that undergo a maturational cleavage or, alternatively, they may be present in a loop, contained in the ectodomain, which becomes exposed at fusion (12). Irrespectively of their location in the fusion glycoprotein, fusion peptides perform the key task of penetrating the target membranes, such that the fusion glycoproteins bridge the virion envelope and the cell membrane, and initiate fusion of the two lipid membranes (4, 16, 26, 36).
The characterization of the potential fusion peptide contained in the HSV gH ectodomain was performed by loss-of-function and gain-of-function approaches. Specifically, we report that the predicted membrane α-helix located at amino acid residues 377 to 397 of HSV gH ectodomain had the attribute of a membrane sequence; its partial or entire removal, or mutagenesis of critical residues abolished HSV infectivity and cell fusion activity. Its replacement with well-characterized fusion peptides from human immunodeficiency virus (HIV) gp41 or from vesicular stomatitis virus glycoprotein (VSV-G), but not with their antisense sequences, rescued the infectivity and the fusion activity of the deleted form of gH. Furthermore, while the interspecific form of gH in which the natural HSV gH α-helix was replaced with the positionally conserved α-helix from human cytomegalovirus (HCMV) exhibited trafficking defects, likely due to misfolding, the interpecific form of gH containing the predicted α-helix from varicella-zoster virus (VZV) gH exhibited almost normal values of infectivity and cell-cell fusion activity.
MATERIALS AND METHODS
Cells and virus.
BHK, COS, 143, and F6 cells were grown in Dulbecco modified Eagle medium supplemented with 5 to 10% fetal calf serum. F6 cells are a stably transformed Vero cell line that expresses HSV-1 gH under the control of the HSV-1 gD promoter (13). In the gH deletion mutant (ΔgH HSV) SCgHZ, the gH gene was replaced with lacZ gene; the virus was grown and titrated in the complementing F6 cells (13).
Plasmids.
Expression plasmids for HSV type 1 (HSV-1) gD, gB, gH, and gL were described earlier (2). EGFR2Δ (named erb-2) carries the extracellular domain and transmembrane (TM) sequences of rat HER-2/neu (nucleotides 25 to 2096) (GenBank accession number NM_017003) and is deleted of the tyrosine kinase domain (35). Plasmid pcagt7 contains the T7 RNA polymerase gene under control of the CAG promoter, and the pt7emcluc plasmid expresses the firefly luciferase under the T7 promoter (30, 33).
Constructs.
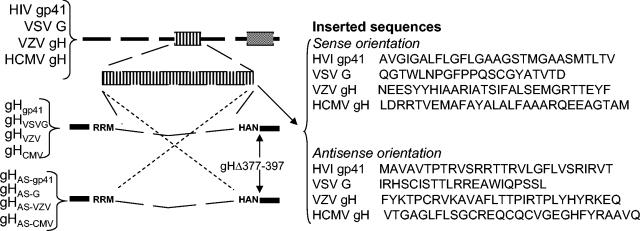
To generate pgD1-260t, an Asp718 restriction site was first engineered at amino acid 260 of a gD expression plasmid by site-directed mutagenesis (9); the Asp718-HindIII fragment was collapsed; the HindIII site was in the polylinker, downstream of the insert. To generate the chimeric gD-gH1 (gD1-260H354-416) construct, the gH sequence encoding amino acids 354 to 416 was PCR amplified from HSV-1 gH with the primers gH-354Asp718 (5-′-CGC CCA GGT ACC GTC GCG GGC ATA CGC-3′) and gH-416HindIII (5′-GGC TTT TTG GCC CAC TCG CGA AGC TTT GCC GGG TTG GC-3′). The amplicon was digested with Asp718 and HindIII and ligated into pgD1-260t (9), which was previously digested with the same enzymes. To generate gD-gH2 (gD1-260H505-551), we followed the same strategy described above for gD-gH1; the gH sequence encoding amino acids 505 to 551 was PCR amplified with the primers gH-505Asp718 (5′-CCT TCT GGT ACC CCG CGT GCT GAC CAC CCC-3′) and gH-551HindIII (5′-GGA CGT ACA CAA GCT TGA GGC TAT TCA GAA GGC T-3′), digested with Asp718 and HindIII, and ligated into pgD1-260t. gHσLeu, carrying L382P,GLL384-386WPP substitutions, was derived by site-directed mutagenesis with oligonucleotide 5′-CCT CTC TTC TGG CGC CCA ACG TGG CCG CCC GCG ACG TCG GGT TTT GC-3′. For the entire or partial deletion of the 377-to-397-amino-acid sequence of gH, a SphI site was first introduced at amino acid 377 in pMTS-gH (2) by means of the oligonucleotide 5′-GAG CAG GGC CCG CGC CGC ATG CTC TTC TGG CGC CTA-3′, thus generating gH377-SphI. To generate the gHΔ378-397 and gHΔ378-387 constructs, a second SphI site was introduced at residues 397 or 387, respectively, by means of the oligonucleotides 5′-GGT TTT GCT TTC GTG AAC GGC ATG CAC GCA AAC GGC GCG G-3′ or 5′-CGC CTA ACG GGG CTG CGC ATG CCG TCG GGT TTT GCT TTC G-3′. The SphI fragment was then collapsed. The mutagenesis inserted the following substitutions: PP376-377RM in both plasmids and T388P in gHΔ378-387. To construct gHVSV-G and gHAS-G, the sequence encoding amino acid residues 117 to 137 of VSV-G (QGTWLNPGFPPQSCGYATVTD) (41) was generated by annealing and extension of the partially overlapping oligonucleotides 5′-CCT CTA ACA TGC ATG CAA GGT ACC TGG CTG AAT CCA GGC TTC CCT CCT CAA AGT TGT GG-3′ and 5′-AGT ATT AGT AGC ATG CAA TCC GTC ACA GTT GCA TAT CCA CAA CTT TGA GGA GGG AAG C-3′. The forward oligonucleotide carried a silent Asp718 site for easiness of screening. The amplified fragment was digested with SphI and cloned into the SphI site of gHΔ378-397. The mutagenesis inserted two extra residues (CM) 3′ of the insert. Constructs carrying the insert in both the sense and the antisense directions were selected. The construction of gHgp41, gHAS-gp41, gHVZV, gHAS-VZV, gHCMV, and gHAS-CMV followed essentially the same strategy. The oligonucleotides used to amplify a fragment encoding for a 28-amino-acid peptide that included the fusion peptide and 12 downstream residues of HIV gp41 (residues 512 to 539 of HIV-1gp160) (AVGIGALFLGFLGAAGSTMGAASMTLTV) (3) were 5′-TAC TAA CAT GCA TGC TTG CAG TGG GAA TAG GAG CTT TGT TCC TTG GGT TCT TGG GAG CAG CAG GAA GC-3′ and 5′-CAG TAG ATG CAT GCT TAC CGT CAG CGT CAT TGA GGC TGC GCC CAT GGT GCT TCC TGC TGC TCC CAA GA-3′. The mutagenesis inserted an extra L at the 5′ end, and two extra residues (SM) at the 3′ end of the insert, respectively. The oligonucleotides used to amplify the predicted membrane α-helix of VZV gH plus four upstream and five downstream residues (residues 360 to 388, NEESYYHIAARIATSIFALSEMGRTTEYF) were 5′-CAA CAT GGC ATG CAC GAG GTA CCC TAT TAC CAT ATC GCC GCA AGA ATA GCC ACA TCA ATT TTT GCG-3′ and 5′-TCT AAC AGC ATG CAT TCT GTG GTA CGG CCC ATT TCC GAC AAC GCA AAA ATT GAT GTG GCT A-3′, respectively. The mutagenesis introduced the following substitutions (N360H, E362V, S363R, and YF-387,388-CM). The oligonucleotides used to amplify the predicted membrane α-helix of HCMV gH bracketed by two upstream and seven downstream residues (residues 334 to 362, LDRRTVEMAFAYALALFAAARQEEAGTAM) were 5′-GGT CGA TGT CGC ATG CTG GAC CGC CGC ACG GTA GAA ATG GCC TTC GCC TAC GCA TTG GCA CTG TTC G-3′ and 5′-TCG TGG GAT GGG CAT GCC GGT ACC GGC CTC TTC TTG TCG GGC TGC CGC GAA CAG TGC TAA CGC GTA G-3′, respectively. The mutagenesis introduced the EI-361,362-AM substitutions. All constructs were sequenced for accuracy.
Indirect immunofluorescence (IFA).
COS or BHK cells were grown on glass coverslips and transfected with the indicated plasmids by using Polyfect (Qiagen, Florence, Italy) according to the manufacturer's instructions. After 24 h the cells were either fixed with 4% paraformaldehyde for 10 min at room temperature followed, when requested, by permeabilization with 0.1%Triton X-100 in phosphate-buffered saline (PBS), or fixed with cold (−20°C) methanol. Samples were incubated with the following monoclonal antibodies (MAbs): MAb H170 to gD (Goodwin Institute, Plantation, Fla.); MAbs H12, 52S, and 53S to gH (32), followed by anti-mouse immunoglobulin G (IgG)-fluorescein isothiocyanate-conjugated antibody (Jackson Immunoresearch). Samples were observed with a Zeiss microscope, and micrographs were taken with a Kodak DC290 digital camera.
Western blot.
Confluent COS cells in 10-cm2 dishes were transfected with 750 ng of expression plasmids for wt-gD, pgD1-260t, gD-gH1, gD-gH2, or pcDNA3.1 vector. At 24 h after transfection the culture medium was harvested and concentrated to 1/10 of the original volume by using an Amicon Bioseparations Microcon YM-10 (Millipore Corp.). The cells were lysed directly in sodium dodecyl sulfate-containing buffer, and the proteins were separated by denaturing polyacrylamide gel electrophoresis and transferred to Hybond enhanced chemiluminescence nitrocellulose membrane (Amersham). The membranes were blocked overnight with 5% nonfat dry milk in PBS at 4°C and reacted with purified IgG from MAb H170, followed by the addition of anti-mouse IgG peroxidase-conjugated antibody (Sigma Aldrich, Milan, Italy). The blot was developed by enhanced chemiluminescence (Amersham Pharmacia, Milan, Italy), according to the manufacturer's instructions. For Western blotting of virions, the virions were pelleted by high-speed centrifugation from the medium of COS cells transfected with the appropriate plasmid (wt-gH, gHgp41, gHAS-gp41, gHVSV-G, gHAS-G, gHVZV, and gHAS-VZV) (75 μg of plasmid DNA/T-150 flask) and superinfected with a stock of complemented (gH−/+) SCgHZ (see description of complementation assay below). Virions were subjected to polyacrylamide gel electrophoresis, transferred to enhanced chemiluminescence membranes, and visualized by enhanced chemiluminescence with MAb H12 to gH and with MAb H170 to gD.
Immunoprecipitation.
COS cells were transfected with the indicated gH plasmid plus a gL plasmid. At 8 h after transfection, cells were metabolically labeled for 16 h with a mixture of [35S]methionine and [35S]cysteine (Amersham) at 25 μCi/ml of medium containing 1/20 of the usual concentration of unlabeled methionine and cysteine and 1% fetal bovine serum. Cells were solubilized in PBS* (1% sodium deoxycholate-1% Nonidet P-40 in PBS containing 0.1 mg each of TLCK (Nα-p-tosyl-l-lysine chloromethyl ketone) and TPCK (tolylsulfonyl phenylalanyl chloromethyl ketone) (Sigma)/ml and then centrifuged at 55,000 × g for 75 min. gH was immunoprecipitated from the lysate supernatants with MAb 53S, as described elsewhere (32). The immunocomplexes were harvested on protein A-Sepharose (Sigma), separated by polyacrylamide gel electrophoresis, and visualized by autoradiography in a Bio-Rad molecular imager.
CELISA and FACS.
Cell enzyme linked immunosorbent assay (CELISA) was performed as described previously (33). Briefly, subconfluent cultures of COS or BHK cells in 24-well plates were transfected with 250 ng of gL plasmid and plasmids encoding wt-gH or mutant gH in the amounts indicated in Table 1. The Erb-2 plasmid DNA was used to make the amounts of DNA equal. After 8 h, the cells were replated into 96-well plates and, after incubation for 18 h, were reacted with MAb 52S. Subsequently, cells were fixed with 4% formaldehyde in PBS, followed by biotinylated anti-mouse IgG conjugate, streptavidin-β-galactosidase conjugate, and ONPG (o-nitrophenyl-β-d-galactopyranoside) substrate. The optical density at 405 nm (OD405) was read. For fluorescence-activated cell sorting (FACS) analysis, cells in T-25 flasks were transfected as indicated above with one- or fivefold amounts of the indicated plasmid relative to the wt-gH plasmid, as detailed in Table 1. The total amount of transfected plasmid DNA per flask was made equal by addition Erb-2 plasmid DNA. At 24 h, unfixed cells were reacted with MAb 52S, followed by the addition of fluorescein isothiocyanate-labeled anti-mouse antibody.
TABLE 1.
Extent of cell surface expression of wt-gH and chimeric gH in COS and BHK cells cotransfected with gH and gL plasmidsa
| Plasmid | Relative amt (fold) of gH plasmid | Cell surface expression (%)b in:
|
||
|---|---|---|---|---|
| COS cells
|
BHK cells (CELISA) | |||
| FACS | CELISA | |||
| wt-gH | 1 | 100 | 100 | 100 |
| gHgp41 | 1 | 45 | 48 | 70 |
| gHgp41 | 3 | NDc | 68 | 77 |
| gHgp41 | 5 | 73 | 72 | 99 |
| gHAS-gp41 | 1 | 53 | 54 | 98 |
| gHAS-gp41 | 3 | ND | 64 | 120 |
| gHAS-gp41 | 5 | ND | 73 | 180 |
| gHVSV-G | 1 | 30 | 58 | 70 |
| gHVSV-G | 3 | ND | 77 | 98 |
| gHVSV-G | 5 | 120 | 99 | 105 |
| gHAS-G | 1 | 38 | 50 | 110 |
| gHAS-G | 3 | ND | 77 | 150 |
| gHAS-G | 5 | ND | 99 | 190 |
| gHVZV | 1 | ND | 110 | ND |
| gHVZV | 3 | ND | 120 | ND |
| gHVZV | 5 | ND | 136 | ND |
| gHAS-VZV | 1 | ND | 102 | ND |
| gHAS-VZV | 3 | ND | 122 | ND |
| gHAS-VZV | 5 | ND | 138 | ND |
COS or BHK cells were transfected with plasmids encoding the indicated gH construct plus a gL-encoding plasmid. Amounts of gH plasmid indicated as one-, three-, or fivefold were 120, 360, or 600 ng/well in 24-well trays or 1 and 5 μg/T25 flask, respectively. The times of cell harvest were 24 h for COS cells and 48 h for BHK cells. Cells were reacted with MAb 52S for determination of cell surface expression of gH. In CELISA; each figure represents the average of triplicates.
Figures are expressed as percentages of the value obtained with wt-gH.
ND, not done.
Cell-cell fusion assay.
The luciferase-based cell-cell fusion assay was performed as detailed elsewhere (25, 33) by using the Luciferase Assay System from Promega (Florence, Italy). The total amount of transfected plasmid DNA was made equal by the addition Erb-2 plasmid DNA. All samples were run in triplicates.
Infectivity complementation assay.
The complementation assay was performed as detailed (9). Briefly, cells in T-25 or T-150 flasks were transfected with the appropriate gH plasmid. The total amount of plasmid DNA transfected per flask was made equal by the addition of Erb-2 plasmid DNA. After 4 h, the cells were infected with a gH−/+ stock of SCgHZ (7 PFU/cell). Unpenetrated virions were inactivated by two washes with PBS, followed by a 1-min rinse with 40 mM sodium citrate-10 mM KCl-135 mM NaCl (pH 3). The monolayers were then rinsed twice with PBS and overlaid with medium containing 1% fetal calf serum. Cells were incubated overnight at 37°C, and the progeny virus was titrated in F6 cells. When appropriate, extracellular virions were pelleted and analyzed by Western blotting.
RESULTS
Prediction of a positionally conserved membrane α-helix in the ectodomain of herpesviruses gH.
We searched for a number of predicted structural motifs in HSV gH, gL, and gB. The most striking outcome of the bioinformatics search was that provided by ENSEMBLE, a method suited for the location of all-alpha membrane domains, based on an ensemble of neural networks and hidden Markov models (24). ENSEMBLE predicted two membrane α-helices in HSV gH ectodomain, located at residues 377 to 397 and residues 513 to 531, respectively (Fig. 1A). The N-terminal one displayed a much higher probability score than the C-terminal one. More importantly, the predicted N-terminal α-helix was positionally conserved in all examined gHs, although with no significant sequence homology (Fig. 1B). To validate the prediction, well-known viral fusion glycoproteins, such as, for example, HIV gp41, VSV-G, hemagglutinin, and paramyxovirus SV5 F, were analyzed by means of ENSEMBLE; the method predicted high score probability membrane α-helices in the sequences that serve as fusion peptides (data not shown).
FIG. 1.
Prediction of membrane helices in gH sequences. (A) The two α-helices predicted in HSV-1 gH ectodomain by means of ENSEMBLE (24). Numbers in brackets indicate the first and last residue of the predicted helix. The reliability index (Rel.Index) evaluates the probability of correct prediction on a scale from 0 to 9. (B) Prediction of positionally conserved membrane α-helices in the gH of a number of herpesviruses. Numbers in brackets are as described for panel A. HHV, human herpesvirus; EBV, Epstein-Barr virus; GHV, gallid herpesvirus; EHV, equine herpesvirus; MCMV, murine cytomegalovirus; SHV, suid herpesvirus.
The predicted N-terminal membrane α-helix contained in HSV gH ectodomain has the attribute of a bona fide membrane segment.
In order to verify whether the predicted N- and C-terminal membrane α-helices (residues 377 to 397 and residues 513 to 531) contained in the ectodomain of HSV gH have the attribute of a membrane sequence, we determined whether they convert a soluble protein into a membrane-bound protein. To this end, we made use of a soluble form of gD, named gD1-260t (9), in which the sequences downstream of residue 260 were detected, thus removing the natural TM segment and cytoplasmic tail. The gH α-helix sequences bracketed by their respective upstream and downstream sequences were fused at the C terminus of gD1-260t, thus generating the constructs gD1-260H354-416, and gD1-260H505-551, herein named gD-gH1 and gD-gH2, respectively (Fig. 2). The upstream and downstream sequences of the predicted gH α-helices were included in the constructs, so as to provide to the chimeric gD-gH1 and gD-gH2 a membrane-proximal region and a cytoplasmic tail. The plasmids encoding gD-gH1 and gD-gH2 and, for comparison, gD1-260t and wt-gD were transfected in COS cells. The medium and the cell lysates were analyzed for the presence of gD by Western blotting with MAb H170, directed to a linear epitope of gD N terminus. As can be seen in Fig. 3A, gD1-260t was secreted in the medium and did not accumulate intracellularly. In contrast, the chimeric gD-gH1 accumulated intracellularly and was absent from the medium. This distribution could not be differentiated from that of membrane-bound wt-gD. gD-gH2 exhibited an intermediate phenotype between that of wt-gD and gD1-260, in that it was mostly secreted in the medium and partly retained within the cell. The intracellular distribution of gD-gH1 was further analyzed by IFA of the transfected COS cells. Figure 3b and c show that the intracellular distribution of the chimeric gD-gH1 did not substantially differ from that of wt-gD and was prominent at the cell surface. This distribution differed strikingly from that of gD1-260t, which was reticular and diffuse in the cytoplasm, and absent from the cell surface. Thus, the N-terminal predicted membrane α-helix transformed the soluble gD1-260t into a membrane-bound gD. The C-terminal α-helix was much weaker in retaining gD within the cell. We infer that the N-terminal predicted gH membrane α-helix has the attribute of a membrane segment. Because of this property and because the N-terminal membrane α-helix is positionally conserved in all examined gHs, subsequent studies focused on this α-helix.
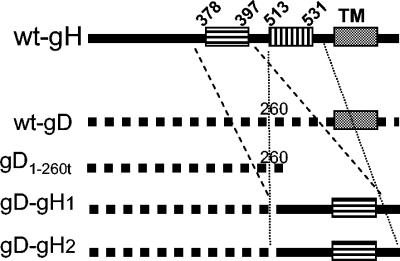
FIG. 2.
Schematic representation of the gD-gH1 and gD-gH2 constructs. The top line shows a wt-gH linear map with predicted α-helices and TM region. The second line shows a wt-gD linear map. An Asp718 site was inserted at residue 260 of gD; deletion of downstream sequences generated soluble gD1-260t (third line). Insertion of gH amino acid residues 354 to 416 and residues 505 to 551 at the C terminus of gD1-260t generated gD-gH1 and gD-gH2, respectively, is shown in the fourth and fifth lines.
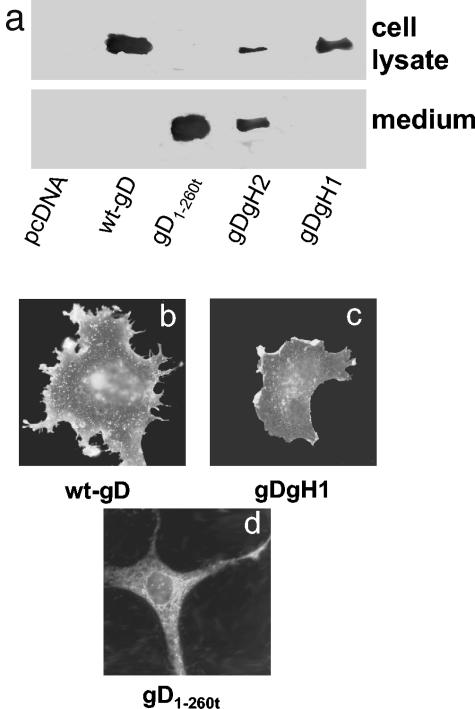
FIG. 3.
Distribution of gD-gH1, gD-gH2, gD1-260t, and wt-gD and their cell surface expression. (a) Western blot analysis of the medium and cell lysates from COS cells transfected with plamids encoding gD1-260t, gD-gH1, gD-gH2, wt-gD, or pcDNA3.1. Cells and media were harvested 24 h after transfection. Western blot was developed with MAb H170 to gD. Note that the medium of cells expressing gD1-260t contains a secreted gD, whereas the medium of cells expressing gD-gH1 contains no gD and that of gD-gH2 contains some secreted gD; the lysates from cells expressing gD-gH1 accumulate higher quantities of gD than cells expressing gD-gH2 and gD1-260t. (b to d) IFA localization of gD in methanol-fixed COS cells expressing wt-gD (b), gD-gH1 (c), and gD1-260t (d) as detected with MAb H170. In panels b and c, the prominent localization of gD is at the cell surface. In panel d, the prominent localization of gD is reticular and diffuse to the cytoplasm.
Deletion of the predicted membrane α-helix contained in HSV gH ectodomain or mutational disruption of the α-helix inhibits HSV infectivity and cell-cell fusion.
To attain a preliminary characterization of the membrane α-helix contained in HSV gH ectodomain, a loss-of-function approach was applied. Three gH mutants were generated and tested for ability to induce cell-cell fusion and to complement the infectivity of the ΔgH mutant SCgHZ (13). The gH mutants were gHΔ378-397 and gHΔ378-387, in which the α-helix sequence was deleted entirely or in part, and gHσLeu, carrying three L-to-P and one G-to-W (L382P and GLL384-386WPP) substitutions that disrupt the predicted α-helix (Fig. 4). For the cell fusion assay, the mutant forms of gH, or wt-gH, all cloned in MTS vector under the immediate-early cytomegalovirus promoter, were cotransfected with expression plasmids encoding gL, gB, and gD. In the luciferase-based assay, the target cells express the T7-promoter-driven luciferase gene and the effector cells express the quartet of gB, gD, gL, and gH plus a T7 RNA polymerase (1, 25, 33, 38). The extent of fusion was expressed as luciferase units. Figure 5A shows that all three gH mutants were totally unable to carry out cell-cell fusion, in contrast to wt-gH, which readily induced fusion in COS and BHK cells.
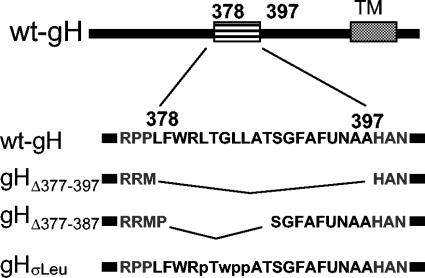
FIG. 4.
Schematic representation of the gH deletion and mutation constructs. The top line shows a wt-gH linear map with predicted α-helix and TM regions. The second line shows the sequence of the α-helix, residues 378 to 397, and flanking residues. In gHΔ378-397 the α-helix was collapsed, after insertion of two SphI sites at residues 377 and 397. In gHΔ378-387 residues 378 to 387 were collapsed after insertion of SphI sites at residues 377 and 387. gHσLeu carries the L382P, GLL384-386WPP substitutions.
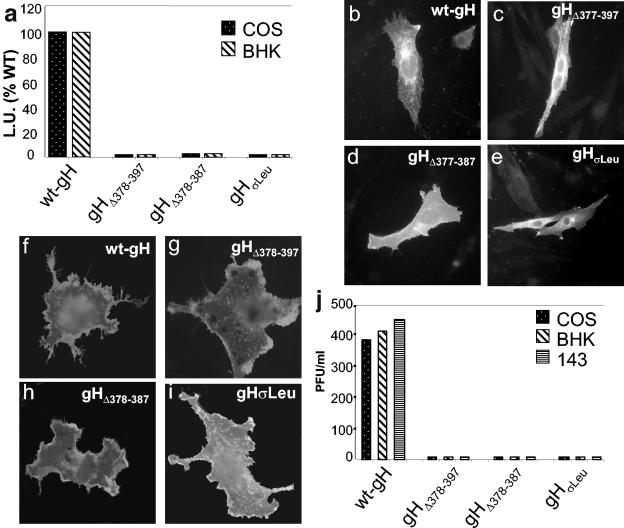
FIG. 5.
Cell fusion, cell surface expression, and infectivity complementation of gH mutants. (a) Quantification of luciferase-based cell fusion assay in effector cells transfected with plasmids encoding gL, gD, gB plus wt-gH, or the constructs gHΔ378-397, gHΔ378-387, and gHσLeu. The luciferase activity was expressed as luciferase units (L.U.). Each experiment was performed at least three times; the mean values are shown. (b to e) Paraformaldehyde-fixed permeabilized COS cells transfected with the indicated plasmid plus gL plasmid and stained with MAb 53S; reactivity denotes heterodimer formation with gL. (f to i) Paraformaldehyde-fixed COS cells transfected with the indicated plasmid plus gL, and stained with MAb 52S. (j) Infectivity complementation. The indicated cells were transfected with one of the gH plasmids and infected 4 h later with a gH−/+ stock of SCgHZ (7 PFU/cell). Progeny virus was titrated at 24 h in gH-expressing cells.
To rule out that the lack of fusion activity was dependent on a defect in heretodimer formation with gL, or in trafficking to the cell surface, cells transfected with each of the gH construct plus gL plasmid were stained by IFA with two MAbs to gH; MAb 53S is directed to a conformation-dependent epitope that requires gL for reactivity, and MAb 52S is directed to a conformation-dependent epitope that does not require gL for reactivity. The results of a typical experiment with permeabilized COS cells stained with MAb 53S show that all mutants exhibited gL-dependent reactivity (Fig. 5b to e). The results with paraformaldehyde-fixed unpermeabilized COS cells stained with MAb 52S show that all mutant forms of gH were able to reach the cell surface when coexpressed with gL (Fig. 5f to i). No cell surface expression was detected in the absence of gL (data not shown). Altogether, the characterization of the three gH mutants indicated that they did not substantially differ from wt-gH in terms of heterodimer formation with gL and trafficking to the plasma membrane. To gain biochemical evidence that the mutant forms of gH were not hampered in heterodimer formation with gL, we determined whether gL was coimmunoprecipitated with gH by MAb 53S. Cells transfected with the mutant gH plasmids and the gL plasmid were metabolically labeled with [35S]methionine and [35S]cysteine; gH was immunoprecipitated with MAb 53S. As can be seen in Fig. 6, gHΔ378-397, gHΔ378-387, and gHσLeu were all capable of coimmunoprecipitating gL.
FIG. 6.
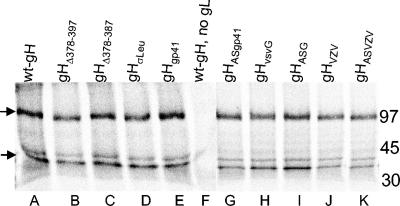
Coimmunoprecipitation of gH and gL by MAb 53S directed to gH. COS cells transfected with the indicated gH plasmid plus a gL plasmid were metabolically labeled with a mixture of [35S]methionine and [35S]cysteine for 16 h. gH was immunoprecipitated from the cell lysates with MAb 53S. gH coimmunoprecipitated gL in all samples. In lane F, in the absence of gL, MAb 53S failed to immunoprecipitate both gH and gL. On the right are indicated the relative migrations of molecular mass markers. Arrows on the left indicate the positions of gH and gL.
For the infectivity complementation assay, cells were transfected with the mutant forms of gH, or wt-gH, superinfected at 4 h with the ΔgH SCgHZ virus; the progeny virus was titrated in the complementing F6 cells at 24 h after transfection (13). When the ΔgH virus replicates in complementing cells, gH is supplied to virions in trans by transgenic gH, and the progeny virus is infectious for one cycle (gH−/+ stock). If gH is defective, the virus infectivity decreases or is abolished. Figure 5J shows that none of the mutant forms of gH was functional in infectivity complementation in any of the cell lines tested, COS, BHK, or 143. We conclude from these experiments that the α-helix represents a critical domain of HSV gH.
Substitution of the membrane α-helix contained in gH ectodomain with heterologous fusion peptides from HIV gp41 or VSV-G rescues HSV infectivity and cell-cell fusion activity of the gH with α-helix deleted.
To verify the hypothesis that the membrane α-helix in gH ectodomain serves as an internal fusion peptide, we applied a gain-of-function approach and replaced the α-helix with heterologous fusion peptides from HIV gp41 or from VSV-G. To this end, the appropriate sequences were cloned into the gH with the α-helix deleted (gHΔ378-397) in the sense or antisense direction (Fig. 7). The resulting constructs were named gHgp41, gHAS-gp41, gHVSV-G, or gHAS-G, respectively.
FIG. 7.
The top line shows a schematic linear map of HIV gp160, VSV-G, VZV gH, and HCMV gH; the HIV or VSV fusion peptides, or the predicted α-helix from VZV and HCMV gH, are marked as an enlarged box. Their sequences were cloned into the SphI site of gHΔ378-397, in sense or antisense directions. The right panel shows the sequence of the inserted fragments.
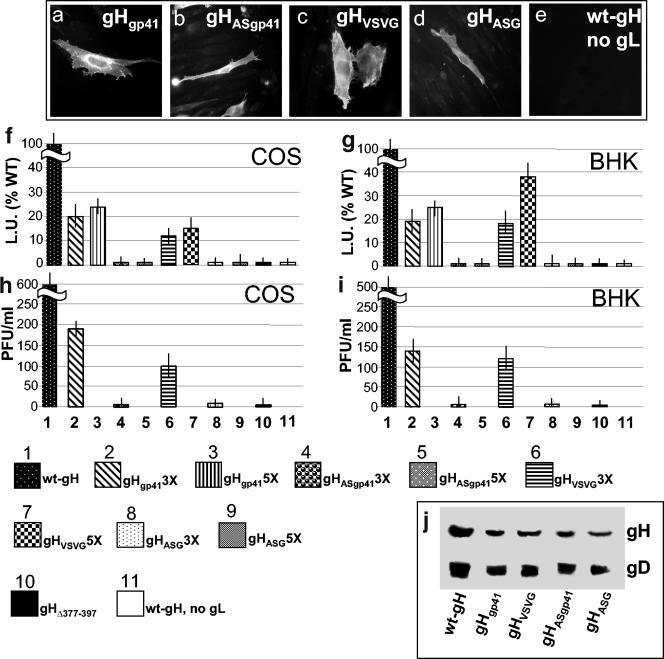
Preliminarily, we ascertained that the chimeric gHgp41 and gHVSV-G were able to form a heterodimer with gL (seen as immunofluorescence reactivity to MAb 53S and coimmunoprecipitation) and to traffic to the cell surface. The results shown in Fig. 8a to d indicate that all constructs reacted with MAb 53S. As expected, the reactivity of MAb 53S was indeed dependent upon the presence of gL (panel e), since cells expressing wt-gH in the absence of gL failed to stain. Figure 6 further shows that the chimeric forms of gH were capable of coimmunoprecipitating gL (lanes E, G, H, and I). Cell surface expression was quantified by CELISA, with MAb 52S, in COS and BHK cells. When the chimeric gH plasmids were transfected in the same amount as wt-gH plasmid (onefold amount), the cell surface expression of the chimeric forms of gH was reduced relative to wt-gH (Table 1). The reduction was overcome, at least in part, by increasing the amounts of transfected plasmids three- or fivefold (Table 1). FACS analysis conducted on some samples fully confirmed the extent of cell surface expression measured by CELISA (Table 1).
FIG. 8.
Expression, cell-cell fusion, and infectivity complementation of chimeric forms of gH. (a to e) BHK cells transfected with the indicated gH plasmid plus gL plasmid and reacted with MAb 53S. In panel e, gL plasmid was omitted. Positive reactivity indicates heterodimer formation with gL. (f and g) Quantification of luciferase-based cell-cell fusion assay in COS or BHK cells expressing the indicated gH plasmid plus gL, gD, and gB. The luciferase activity is expressed as luciferase units (L.U.). Figures represent percent values relative to wt-gH. Each experiment was performed at least three times. In lane 11, the transfection mixture contained wt-gH, gB, and gD plasmid and no gL plasmid. (h and i) Infectivity complementation, performed as described in legend to Fig. 5. Vertical bars represent the SD. (j) Quantification of gH and gD in complemented virions. Extracellular virions from the experiment in panel H were pelleted by high-speed centrifugation and analyzed by Western blotting with MAbs H12 to gH and H170 to gD. In the lane containing wt-gH a triple amount of virus was loaded.
The chimeric gH constructs were then tested for cell-cell fusion activity in the luciferase-based assay. The results illustrated in Fig. 8F and G show that gHgp41 rescued the cell fusion activity of the gHΔ378-397 to a 23% efficiency in either cell line. gHVSV-G rescued the fusion activity to a 37% efficiency in BHK cells. Importantly, the constructs carrying the antisense sequences exhibited no fusion activity.
Next, the chimeric gH constructs were assayed for infectivity complementation. Figure 8H and I shows that both gHgp41 and gHVSV-G were functional and exhibited efficiencies very similar to those attained in the cell fusion assay. Figure 8J further shows that there was no major defect in the amount of gH incorporated into complemented virions, ruling out that the reduced infectivity was the result of a reduced incorporation of gH in the virion envelope. Altogether, these results indicate that the replacement of the natural α-helix in HSV gH with heterologous fusion peptides partially rescued the fusion and infectivity activities of gH. The effect was specific since the insertion of the same sequences in antisense direction did not rescue any activity. The finding that replacement of the natural sequence with heterologous fusion peptides rescued the infectivity and fusion activity only partially was not surprising, since the insertion of heterologous sequences in a protein cannot achieve the optimal structure-function fitness that has been achieved during evolution.
Replacement of the natural HSV gH α-helix with the positionally conserved α-helix from VZV gH, but not HCMV gH, maintains the cell-cell fusion activity and HSV infectivity.
To preliminarily investigate whether the α-helix predicted in the gH from other herpesviruses has attributes of a fusion peptide, we generated interspecific forms of gH. The natural HSV gH α-helix was replaced with the one predicted in the ectodomain of VZV gH (amino acids residues 364 to 383) or HCMV gH (residues 336 to 355) (Fig. 1). The VZV and HCMV constructs carrying the sense and antisense (indicated by “AS”) sequences were named gHVZV and gHAS-VZV and gHCMV and gHAS-CMV, respectively (Fig. 7). Surprisingly, these interspecific forms of gH behaved in different manners. gHCMV and gHAS-CMV exhibited a marked defect in heterodimer formation with gL and in plasma membrane trafficking, as determined by lack of immunofluorescence with MAb 53S; reactivity to MAb 52S showed an intracytoplasmic reticular distribution, suggesting intracellular retention, possibly at the level of the endoplasmic reticulum (data not shown). It is very likely that this chimeric form of gH was misfolded.
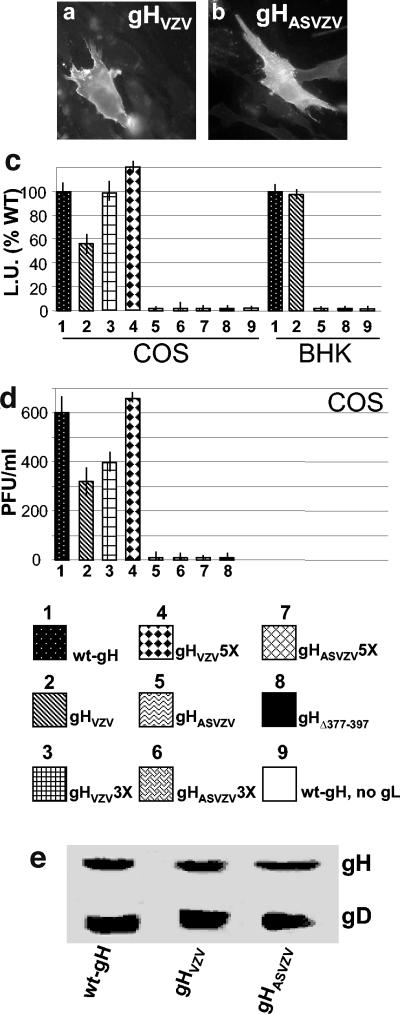
For the gHVZV and gHAS-VZV constructs, measurement of their reactivity to MAb 53S (Fig. 9a and b) and coimmunoprecipitation (Fig. 6, lanes J and K) indicated that they were capable of heterodimer formation with gL; measurement of their cell surface expression by CELISA showed no reduction relative to wt-gH (Table 1).
FIG. 9.
Expression (a and b), cell-cell fusion (c), and infectivity complementation (d) of interspecific forms of gH, gHVZV, and gHAS-VZV. Details are as in the legend to Fig. 8. Vertical bars represent the SD. (e) Quantification of gH and gD in complemented virions, as detailed in Fig. 8J.
In the cell fusion assay gHVZV reached nearly the same usion activity as wt-gH when transfected at three- and fivefold amounts in COS cells and was >90% efficient at a onefold amount in BHK cells. Of note, the antisense construct gHASVZV exhibited no fusion activity even in fivefold amounts (Fig. 9C).
In the infectivity complementation assay in COS cells gHVZV, but not its antisense construct, was only slightly hampered relative to wt-gH and reached wt-gH values when the amount of the transfected plasmid was increased (Fig. 9D). There was no defect in incorporation of the chimeric gHVZV in virions (Fig. 9E). Cumulatively, the results with gHCMV show that not any substitution of the natural HSV gH α-helix with exogenous α-helix results in a functional molecule able to interact with gL and to traffic to the plasma membrane. In contrast, the results with gHVZV clearly indicate that the predicted membrane α-helix contained in VZV gH ectodomain can functionally substitute the native HSV gH α-helix.
DISCUSSION
The results described here provide evidence that a predicted membrane α-helix contained within the ectodomain of HSV-1 gH (amino acid residues 377 to 397) has the attributes of an internal fusion peptide. First, the sequence behaves as a bona fide membrane segment. Second, we found that the predicted α-helix constitutes a critical domain of HSV-1 gH for the fusion that leads to virus entry into the cell and for cell-cell fusion. Its partial or entire deletion, or substitution of critical residues predicted to disrupt the α-helix, leads to the abolishment of HSV infectivity and syncytium formation in the cell-cell fusion assay. These results are consistent with the properties of an insertion mutant at residue 381, which was defective in cell fusion and infectivity complementation activity, although in that instance there was a defect in cell surface expression (14). Third, the predicted α-helix has attributes of an internal fusion peptide. Its replacement with the fusion peptide from HIV gp41 or from VSV-G partially rescued the infectivity and fusion activity of the HSV gH form with the α-helix deleted; in contrast, its replacement with the corresponding antisense sequences did not rescue any activity. Because HSV gH does not undergo cleavage, we favor the view that the α-helix contained in gH ectodomain serves as an internal fusion peptide that is probably exposed on a fusion loop rather than as a N-terminal fusion peptide. Finally, we found that potential fusion peptides are positionally conserved in the gH of all examined herpesviruses. The one present in VZV gH ectodomain (amino acid residues 364 to 383) can replace the natural HSV-1 gH α-helix with conservation of HSV infectivity and fusion activity. However, the natural HSV-1 gH α-helix cannot be exchanged with the α-helix predicted in HCMV gH, probably because of defects in folding and heterodimer formation with gL.
These findings form the basis of two fundamental conclusions with broad implications regarding the entry of HSV-1 and, in general, of herpesviruses into susceptible cells.
The key concept to emerge from our studies is that the presence of a membrane α-helix with the attributes of an internal fusion peptide points to gH as a most likely executor of HSV fusion. In this respect, gH shares key properties with classical fusion glycoproteins and appears to perform a function universally required in viral and cellular fusions. As a consequence, the molecular mechanism by which HSV-1 causes membrane fusion for entry of the virus into cells and for cell-cell fusion appears to be basically similar to that used by other viruses that utilize a fusion glycoprotein. Taking into account previous and current data, we propose that HSV entry involves the following steps. The interaction of gD with one of its receptors serves as the trigger of fusion, likely through the membrane-proximal proline-rich profusion domain. Ultimately, this leads to conformational modifications and activation of gH and insertion of its fusion peptide into the target membrane, a key event that culminates in fusion pore formation and the fusion of the viral and cellular membranes.
What remains to be deciphered is the role played by gB and gL. We propose that their role is to assist gH in the execution of fusion. Thus, gB may undergo conformational modifications that enable gH to switch from the fusion-inactive to the fusion-active conformation. As far as gL is concerned, because gL is the chaperone of gH, required for its folding and trafficking to the plasma membrane, and because the gL contacts to gH map to three regions, one of which includes residues 300 to 473 (7, 40), it is tempting to speculate that the role of gL is to mask the hydrophobic α-helix and thus enhance the water solubility and solvent interface of gH ectodomain and avoid hydrophobic aggregation.
The second conclusion to emerge from these studies is that a predicted membrane α-helix is positionally conserved in the gH of all examined herpesviruses (Fig. 1B). The one from VZV gH functionally replaces the native HSV-1 gH α-helix, arguing in favor of a conserved function. Taking into account that the trio of gH, gL, and gB is conserved throughout the Herpesviridae family and is essential for virus infectivity in all examined herpesviruses (see www.stdgen.lanl.gov), it seems likely that gH serves as the fusion glycoprotein also for other herpesviruses and is part of a conserved mechanism of fusion. This possibility is supported by the observation that virus-free cell-cell fusion assays for VZV, human cytomegalovirus, human herpesvirus 6, human herpesvirus 8, and Epstein-Barr virus all require gH (11, 18, 28, 37). An exception is the cell-cell fusion of VZV, which can alternatively be mediated by coexpression of gB and gE (11). The recent finding that peptides with the sequence of a predicted coiled-coil region in HCMV gH inhibit virus infectivity and the inherent implication that HCMV gH carries coiled-coil regions is in agreement with and strongly supports the proposed role of gH as a classical fusion glycoprotein (23).
Current findings render feasible the rational design of inhibitors of herpesvirus fusion that potentially could be broadly applicable to all human herpesviruses. Inhibitors of fusion are among the latest generation of anti-HIV and anti-influenza drugs (20, 29). The nucleoside analogues currently used for treatment of a variety of diseases caused by human herpesviruses other than HSV-1 and HSV-2 are either ineffective or unsuitable for long-term treatment. Inhibitors of fusion would increase the armamentarium available for the treatment of infections caused by these viruses, as well as by HSV-1 or HSV-2 strains resistant to nucleoside analogues.
Acknowledgments
We thank T. Minson, H. Browne, G. Cohen, and R. Eisenberg for reagents; Laura Menotti for invaluable suggestions; and Elisabetta Romagnoli for assistance.
This study was supported by the FIRB autonomous and coordinated project, Cofin-MIUR, CNR-Functional Genomics, and the University of Bologna Fondo Pallotti.
REFERENCES
- 1.Avitabile, E., G. Lombardi, and G. Campadelli-Fiume. 2003. Herpes simplex virus glycoprotein K, but not its syncytial allele, inhibits cell-cell fusion mediated by the four fusogenic glycoproteins, gD, gB, gH, and gL. J. Virol. 77:6836-6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avitabile, E., G. Lombardi, T. Gianni, M. Capri, and G. Campadelli-Fiume. 2004. Coexpression of UL20p and gK inhibits cell-cell fusion mediated by herpes simplex virus glycoproteins gD, gH-gL, and wt-gB or an endocytosis-defective gB mutant, and downmodulates their cell surface expression. J. Virol. 78:8015-8025. [DOI] [PMC free article] [PubMed]
- 3.Bergeron, L., N. Sullivan, and J. Sodroski. 1992. Target cell-specific determinants of membrane fusion within the human immunodeficiency virus type 1 gp120 third variable region and gp41 amino terminus. J. Virol. 66:2389-2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bressanelli, S., K. Stiasny, S. L. Allison, E. A. Stura, S. Duquerroy, J. Lescar, F. X. Heinz, and F. A. Rey. 2004. Structure of a flavivirus envelope glycoprotein in its low-pH-induced membrane fusion conformation. EMBO J. 23:728-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bzik, D. J., B. A. Fox, N. A. DeLuca, and S. Person. 1984. Nucleotide sequence of a region of the herpes simplex virus type 1 gB glycoprotein gene: mutations affecting rate of virus entry and cell fusion. Virology 137:185-190. [DOI] [PubMed] [Google Scholar]
- 6.Cai, W. H., B. Gu, and S. Person. 1988. Role of glycoprotein B of herpes simplex virus type 1 in viral entry and cell fusion. J. Virol. 62:2596-2604. (Erratum, 62:4438.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cairns, T. M., R. S. Milne, M. Ponce-de-Leon, D. K. Tobin, G. H. Cohen, and R. J. Eisenberg. 2003. Structure-function analysis of herpes simplex virus type 1 gD and gH-gL: clues from gDgH chimeras. J. Virol. 77:6731-6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campadelli-Fiume, G., F. Cocchi, L. Menotti, and M. Lopez. 2000. The novel receptors that mediate the entry of herpes simplex viruses and animal alphaherpesviruses into cells. Rev. Med. Virol. 10:305-319. [DOI] [PubMed] [Google Scholar]
- 9.Cocchi, F., D. Fusco, L. Menotti, T. Gianni, R. J. Eisenberg, G. H. Cohen, and G. Campadelli-Fiume. 2004. The soluble ectodomain of herpes simplex virus gD contains a membrane-proximal pro-fusion domain and suffices to mediate virus entry. Proc. Natl. Acad. Sci. USA 101:7445-7450. [DOI] [PMC free article] [PubMed]
- 10.Cocchi, F., L. Menotti, P. Mirandola, M. Lopez, and G. Campadelli-Fiume. 1998. The ectodomain of a novel member of the immunoglobulin superfamily related to the poliovirus receptor has the attributes of a bona fide receptor for herpes simplex viruses 1 and 2 in human cells. J. Virol. 72:9992-10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cole, N. L., and C. Grose. 2003. Membrane fusion mediated by herpesvirus glycoproteins: the paradigm of varicella-zoster virus. Rev. Med. Virol. 13:207-222. [DOI] [PubMed] [Google Scholar]
- 12.Colman, P. M., and M. C. Lawrence. 2003. The structural biology of type I viral membrane fusion. Nat. Rev. Mol. Cell. Biol. 4:309-319. [DOI] [PubMed] [Google Scholar]
- 13.Forrester, A., H. Farrell, G. Wilkinson, J. Kaye, N. Davis Poynter, and T. Minson. 1992. Construction and properties of a mutant of herpes simplex virus type 1 with glycoprotein H coding sequences deleted. J. Virol. 66:341-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galdiero, M., A. Whiteley, B. Bruun, S. Bell, T. Minson, and H. Browne. 1997. Site-directed and linker insertion mutagenesis of herpes simplex virus type 1 glycoprotein H. J. Virol. 71:2163-2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geraghty, R. J., C. Krummenacher, G. H. Cohen, R. J. Eisenberg, and P. G. Spear. 1998. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science 280:1618-1620. [DOI] [PubMed] [Google Scholar]
- 16.Gibbons, D. L., M. C. Vaney, A. Roussel, A. Vigouroux, B. Reilly, J. Lepault, M. Kielian, and F. A. Rey. 2004. Conformational change and protein-protein interactions of the fusion protein of Semliki Forest virus. Nature 427:320-325. [DOI] [PubMed] [Google Scholar]
- 17.Gompels, U., and A. Minson. 1986. The properties and sequence of glycoprotein H of herpes simplex virus type 1. Virology 153:230-247. [DOI] [PubMed] [Google Scholar]
- 18.Huber, M. T., and T. Compton. 1998. The human cytomegalovirus UL74 gene encodes the third component of the glycoprotein H-glycoprotein L-containing envelope complex. J. Virol. 72:8191-8197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hutchinson, L., H. Browne, V. Wargent, N. Davis-Poynter, S. Primorac, K. Goldsmith, A. C. Minson, and D. C. Johnson. 1992. A novel herpes simplex virus glycoprotein, gL, forms a complex with glycoprotein H (gH) and affects normal folding and surface expression of gH. J. Virol. 66:2240-2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.LaBonte, J., J. Lebbos, and P. Kirkpatrick. 2003. Enfuvirtide. Nat. Rev. Drug Discov. 2:345-346. [DOI] [PubMed] [Google Scholar]
- 21.Ligas, M. W., and D. C. Johnson. 1988. A herpes simplex virus mutant in which glycoprotein D sequences are replaced by beta-galactosidase sequences binds to but is unable to penetrate into cells. J. Virol. 62:1486-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lopez, M., F. Cocchi, L. Menotti, E. Avitabile, P. Dubreuil, and G. Campadelli-Fiume. 2000. Nectin2a (PRR2a or HveB) and nectin2d are low-efficiency mediators for entry of herpes simplex virus mutants carrying the Leu25Pro substitution in glycoprotein D. J. Virol. 74:1267-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lopper, M., and T. Compton. 2004. Coiled-coil domains in glycoproteins B and H are involved in human cytomegalovirus membrane fusion. J. Virol. 78:8333-8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martelli, P. L., P. Fariselli, and R. Casadio. 2003. An ENSEMBLE machine learning approach for the prediction of all-alpha membrane proteins. Bioinformatics 19(Suppl. 1):i205-i211. [DOI] [PubMed] [Google Scholar]
- 25.Milne, R. S., S. L. Hanna, A. H. Rux, S. H. Willis, G. H. Cohen, and R. J. Eisenberg. 2003. Function of herpes simplex virus type 1 gD mutants with different receptor-binding affinities in virus entry and fusion. J. Virol. 77:8962-8972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Modis, Y., S. Ogata, D. Clements, and S. C. Harrison. 2004. Structure of the dengue virus envelope protein after membrane fusion. Nature 427:313-319. [DOI] [PubMed] [Google Scholar]
- 27.Montgomery, R. I., M. S. Warner, B. J. Lum, and P. G. Spear. 1996. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell 87:427-436. [DOI] [PubMed] [Google Scholar]
- 28.Mori, Y., X. Yang, P. Akkapaiboon, T. Okuno, and K. Yamanishi. 2003. Human herpesvirus 6 variant A glycoprotein H-glycoprotein L-glycoprotein Q complex associates with human CD46. J. Virol. 77:4992-4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nicholson, K. G., J. M. Wood, and M. Zambon. 2003. Influenza. Lancet 362:1733-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okuma, K., M. Nakamura, S. Nakano, Y. Niho, and Y. Matsuura. 1999. Host range of human T-cell leukemia virus type I analyzed by a cell fusion-dependent reporter gene activation assay. Virology 254:235-244. [DOI] [PubMed] [Google Scholar]
- 31.Pellett, P. E., K. G. Kousoulas, L. Pereira, and B. Roizman. 1985. Anatomy of the herpes simplex virus 1 strain F glycoprotein B gene: primary sequence and predicted protein structure of the wild type and of monoclonal antibody-resistant mutants. J. Virol. 53:243-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peng, T., M. Ponce de Leon, M. J. Novotny, H. Jiang, J. D. Lambris, G. Dubin, P. G. Spear, G. H. Cohen, and R. J. Eisenberg. 1998. Structural and antigenic analysis of a truncated form of the herpes simplex virus glycoprotein gH-gL complex. J. Virol. 72:6092-6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pertel, P. E., A. Fridberg, M. L. Parish, and P. G. Spear. 2001. Cell fusion induced by herpes simplex virus glycoproteins gB, gD, and gH-gL requires a gD receptor but not necessarily heparan sulfate. Virology 279:313-324. [DOI] [PubMed] [Google Scholar]
- 34.Roop, C., L. Hutchinson, and D. C. Johnson. 1993. A mutant herpes simplex virus type 1 unable to express glycoprotein L cannot enter cells, and its particles lack glycoprotein H. J. Virol. 67:2285-2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rovero, S., A. Amici, E. D. Carlo, R. Bei, P. Nanni, E. Quaglino, P. Porcedda, K. Boggio, A. Smorlesi, P. L. Lollini, L. Landuzzi, M. P. Colombo, M. Giovarelli, P. Musiani, and G. Forni. 2000. DNA vaccination against rat her-2/Neu p185 more effectively inhibits carcinogenesis than transplantable carcinomas in transgenic BALB/c mice. J. Immunol. 165:5133-5142. [DOI] [PubMed] [Google Scholar]
- 36.Smith, A. E., and A. Helenius. 2004. How viruses enter animal cells. Science 304:237-242. [DOI] [PubMed] [Google Scholar]
- 37.Spear, P. G., and R. Longnecker. 2003. Herpesvirus entry: an update. J. Virol. 77:10179-10185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Turner, A., B. Bruun, T. Minson, and H. Browne. 1998. Glycoproteins gB, gD, and gHgL of herpes simplex virus type 1 are necessary and sufficient to mediate membrane fusion in a COS cell transfection system. J. Virol. 72:873-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Warner, M. S., R. J. Geraghty, W. M. Martinez, R. I. Montgomery, J. C. Whitbeck, R. Xu, R. J. Eisenberg, G. H. Cohen, and P. G. Spear. 1998. A cell surface protein with herpesvirus entry activity (HveB) confers susceptibility to infection by mutants of herpes simplex virus type 1, herpes simplex virus type 2, and pseudorabies virus. Virology 246:179-189. [DOI] [PubMed] [Google Scholar]
- 40.Westra, D. F., H. B. Kuiperij, G. W. Welling, A. J. Scheffer, T. H. The, and S. Welling-Wester. 1999. Domains of glycoprotein H of herpes simplex virus type 1 involved in complex formation with glycoprotein L. Virology 261:96-105. [DOI] [PubMed] [Google Scholar]
- 41.Zhang, L., and H. P. Ghosh. 1994. Characterization of the putative fusogenic domain in vesicular stomatitis virus glycoprotein G. J. Virol. 68:2186-2193. [DOI] [PMC free article] [PubMed] [Google Scholar]