Abstract
Most eukaryotic mRNAs are recruited to the ribosome by recognition of a 5’ m7GpppN cap. 30 years of genetic and biochemical evidence point to a role for interaction between the 5’ cap-interacting factors and the 3’ poly(A)-binding protein in bringing the ends of the mRNA into close proximity and promoting both translation and stability of the mRNA, in a form known as the closed loop. However, the results of recent RNA-protein interaction studies suggest that not all mRNAs have equal access to the closed loop factors. Furthermore, association with closed loop factors appears to be strongly biased towards mRNAs with short open reading frames, echoing the trend for higher translation of short mRNAs that has been observed in many eukaryotes. We recently reported that the ribosomal signaling scaffold protein RACK1 promotes the efficient translation of short mRNAs that strongly associate with the closed loop factors. Here, we discuss the implications of these observations with respect to translational control and suggest avenues through which the universality of the closed loop in eukaryotic translation could be revisited.
Keywords: Ribosome, RACK1, closed loop, translation
Background
In the early 1960s, it was discovered that ribosomes form polyribosomal aggregates that cluster together and act as the functional unit of protein synthesis (Warner et al. 1962; Gierer 1963; Wettstein et al. 1963). Realization of the cooperative nature of protein synthesis in which one mRNA is translated by many ribosomes simultaneously led to an immediate interest in the spatial topology of these complexes. Although the first images of polysomes suggested linear chains of ribosomes, some later reports found loop-like arrangements of ribosomes (Mathias et al. 1964; Ladhoff et al. 1981). Complementary biochemical experiments demonstrated that newly added mRNAs would not readily exchange with pre-existing mRNAs already engaged in protein synthesis (Philipps 1965; Adamson et al. 1969; Baglioni et al. 1969). Taken together, these experiments strongly implicated a mechanism in which the mRNA is arranged in a loop-like conformation and ribosomes go around the loop, thus repeatedly translating the same mRNA molecule (Baglioni et al. 1969).
Over the next few decades, functional data supporting the existence of a circular form of mRNA during translation amassed (Gallie 1998). First, the poly(A) tail was suggested to be a 3’ enhancer of translation initiation, indicating that it must somehow communicate with the mRNA 5’ end during initiation (Jacobson and Favreau 1983; Palatnik et al. 1984). Several groups later reported that the mRNA cap and poly(A) tail act synergistically to promote translation both in vivo and in vitro (Gallie 1991; Iizuka et al. 1994; Tarun and Sachs 1995), and this finding was explained as the cooperative function of factors interacting with the 5’ and 3’ ends during mRNA recruitment to ribosomes (Tarun and Sachs 1995; Preiss and Hentze 1998). These factors were soon shown to be the eIF4G component of the eIF4F cap-binding complex and the poly(A)-binding protein, PABP (Pab1 in yeast) (Tarun and Sachs 1996; Wells et al. 1998). Hence, the model that mRNAs circularize via the interaction of PABP with the poly(A) tail and eIF4F with the 5’ cap was born. This model for translation has become known as the ‘closed loop’ model (Jacobson 1996) and is commonly presented as the general model for eukaryotic translation in biology text books today.
The current model is mRNA-centric, where the ribosome neither recognizes nor modifies the closed loop state of the mRNA. However, our recent study suggests that a ribosome-associated protein, RACK1 (Asc1 in S. cerevisiae), is involved in either the recognition and/or function of the closed loop complex (Thompson et al. 2016). This finding suggests the presence of a mechanism for the ribosome to sense polysome topology, and raises questions about whether such a mechanism could be employed in translational control.
Questioning the universality of the closed loop
The key biochemical experiments supporting the function of the closed loop in translation demonstrated that the mRNA 5’ cap and 3’ poly(A) tail functionally interact during translation of a reporter mRNA (Gallie 1991; Iizuka et al. 1994; Tarun and Sachs 1995; Tarun et al. 1997; Bergamini et al. 2000; Michel et al. 2000; Svitkin et al. 2001). In these experiments, and subsequent, similar studies, four mRNA translation substrates were compared: an mRNA with both a 5’ cap and 3’ poly(A) tail, an mRNA with a cap but without a poly(A) tail, an mRNA with a poly(A) tail but without a cap, and an mRNA with neither cap nor poly(A) tail. In all experiments, adding either a cap or a poly(A) tail to the mRNA independently enhanced translation. Moreover, adding both a cap and poly(A) tail to the mRNA simultaneously conferred more translation than expected based on the independent levels of enhancement.1 This effect is referred to as cap-poly(A) ‘synergy’ and is taken as evidence for eIF4F and PABP functioning together as a complex connected to the 5’ and 3’ mRNA ends during translation. In parallel, genetic evidence supported an in vivo requirement for communication between the 5’- and 3’-interacting factors. Specifically, mutations that disrupt the interaction of eIF4G and Pab1 were shown to cause defects in translation and slowed growth rates in yeast (Tarun et al. 1997; Park et al. 2011).
However, the complex nature of the biochemical functions of the 5’ and 3’ ends of the mRNA, as well as those of the protein factors that associate with them, make straightforward interpretation of cap-poly(A) synergy findings difficult. Firstly, cap-poly(A) synergy may not be dependent on formation of the closed loop per se because PABP can promote translation – in trans – of a capped mRNA lacking a poly(A) tail (Munroe and Jacobson 1990; Borman et al. 2002). This observation makes sense in light of biochemical data showing that PABP enhances the affinity of the eIF4F complex for the mRNA cap even in the absence of a poly(A) tail (Borman et al. 2000; von Der Haar et al. 2000). Additionally, adding a cap and poly(A) tail to an mRNA may promote mRNA stability in a synergistic manner (because mRNA degradation can occur from either the 5’ or 3’ end) and it is unclear in some studies to what extent the synergistic effect on reporter expression can be assigned primarily to translational enhancement instead of RNA stability (Kozak 2004). Finally, PABP has additional functions in translation initiation that may not be connected to its interactions with eIF4F (Munroe and Jacobson 1990; Otero et al. 1999; Kahvejian et al. 2005). Nevertheless, the preponderance of biochemical evidence for interactions between the 5’- and 3’-bound factors, together with genetic evidence of their biological significance, make a compelling case for the existence of closed loop mRNAs in the cell.
What is the biochemical mechanism whereby the closed loop enhances translation? Several studies have pointed to a system of cooperative interactions between the 5’- and 3’-interacting factors that could explain this effect. PABP binding to eIF4G enhances the affinity of eIF4G for eIF4E. Likewise, eIF4G binding to eIF4E enhances eIF4E affinity for the mRNA 5’ cap (Ptushkina et al. 1998; Wei et al. 1998; Borman et al. 2000; von Der Haar et al. 2000). Thus, mRNAs in a closed loop are expected to have higher affinity for the eIF4F cap-binding complex than linear mRNAs unless there is sufficient free PABP to stabilize all eIF4F-cap complexes in trans. Intriguingly, two different approaches demonstrated that distinct mRNAs do differ in their affinities for the eIF4F cap-binding complex. The first challenged the translation of synthetic mRNAs with an excess of free cap in an extract system (Amrani et al. 2008), and the second assayed the stable association of yeast mRNAs with eIF4F components by immunoprecipitation and RNA sequencing (Costello et al. 2015). In both cases, a simple metric predicted which mRNAs would show high affinity for the cap-binding complex—a short open reading frame (Amrani et al. 2008; Thompson et al. 2016). Importantly, high levels of association with the closed loop factors is predictive of higher levels of translation (Fig. 1). Taken together, these data suggest that not all mRNAs form stable closed loops, but rather are selectively partitioned into the closed loop complex based on size.
Fig. 1.
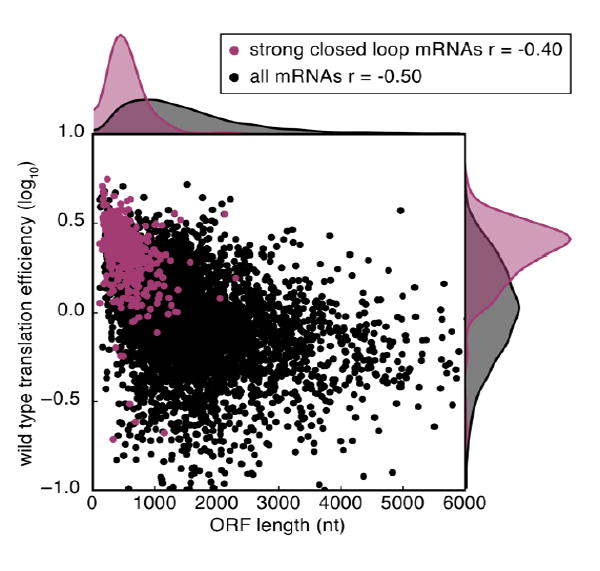
mRNAs that associate highly with the closed loop complex (Costello et al. 2015) have short open reading frames and are highly translated. Wild type translation efficiency data is taken from (Thompson et al. 2016). The Spearman correlation coefficient for each group is indicated (r=-0.50 for all mRNAs and r=-0.40 for strong closed loop mRNAs). The Mann-Whitney U test one-sided p-values for differences between strong closed loop mRNAs and other mRNAs is p<10-134 for ORF length and p<10-131 for wild type translation efficiency.
In contrast to the eIF4F cap-binding complex, PABP is found to bind most yeast mRNAs (Costello et al. 2015). Differences in the dynamics of mRNA dissociation could explain this observation. eIF4F dissociates very rapidly from capped RNA in vitro (koff ~3 sec-1) (O’Leary et al. 2013) likely leading to loss of mRNA upon dilution (e.g. during wash steps of immunoprecipitation). Although the rate of dissociation of PABP from poly(A) RNA was not determined, the presence of multiple RNA-binding domains within a single molecule of PABP is predicted to increase the lifetime of the complex. If the poly(A)-bound form of PABP substantially reduced the rate of dissociation of eIF4F from the cap (Fig. 2a), closed loop mRNAs would preferentially co-immunoprecipitate with eIF4E and eIF4G. Consistent with this model, interactions between eIF4G, PABP and poly(A) are mutually stabilizing in vitro (Le et al. 1997; Safaee et al. 2012).
Fig. 2.

Translational enhancement and regulation via the closed loop. a Closed loop mRNAs may be more highly translated than linear mRNAs due to higher de novo initiation rates and/or intrapolysomal ribosome recycling. The mutually reinforcing network of interactions between the cap, eIF4E, eIF4G, and PABP on closed loop mRNAs decreases the dissociation rate of the complex. b During activating conditions, eIF4E phosphorylation reduces the affinity of eIF4E for the cap, which could repress translation of linear mRNAs because eIF4E-cap binding is not stabilized by the eIF4G-PABP interaction. eIF4E binding to closed loop mRNAs is stabilized by protein-protein interactions. Closed loop mRNAs may benefit from reduced competition for limiting translation factors. c Under repressing conditions, 4E-BPs disrupt translation in a graded manner. Sub-saturating levels of 4E-BPs repress translation of linear mRNAs, but do not affect closed loop mRNAs because of the low dissociation rate of eIF4E from eIF4G-PABP in the closed loop complex. At saturating 4E-BP concentrations, 4E-BP concentration overcomes the high affinity of the closed loop complex for closed loop mRNAs leading to their translational repression.
The short open reading frame: A universal cis-acting enhancer element?
mRNAs with short ORFs associate preferentially with the closed loop factors and are some of the most highly translated in the cell (Arava et al. 2003; Costello et al. 2015; Thompson et al. 2016). In theory, it is possible that short mRNAs as a group have some other common attribute such as a cis-acting motif that enables them to effectively recruit the closed loop factors and maintain high levels of translation. However, such a motif has never been reported. Furthermore, short mRNAs are better translated in diverse eukaryotes and across unrelated functional categories of the encoded proteins (including exogenous reporters), suggesting a universal mechanism. In light of these observations, we suggest an alternative possibility—that the high translational activity of these mRNAs is largely endowed by the simple characteristic of their length. Although the mechanistic basis for translational privileging of short mRNAs is unclear, one suggestion is that diffusion will favor end collision in shorter molecules, which would give the closed loop more opportunities to form (Guo et al. 2015). This model predicts that total mRNA length matters, and short ORFs are better translated because they tend to occur within shorter mRNAs. Another possibility is that the ribosome somehow senses the length of the ORF during translation and alters the interaction of the mRNA with the closed loop factors, perhaps during intrapolysomal ribosome recycling.
Whatever the mechanism, a length-dependent mechanism for translational enhancement could have several advantages for the cell. First, mRNAs with short ORFs tend to encode highly expressed proteins with ‘housekeeping’ functions, such as histones, ribosomal proteins, and mitochondrial components (Eisenberg and Levanon 2003). The functions of this group are tightly coupled to growth and cell division (Warner 1999; Lempiäinen and Shore 2009; De Silva et al. 2015). Hence, a mechanism to ensure high expression of short ORFs by ‘default’ would prioritize production of growth-promoting proteins under optimal conditions. Conversely, a mechanism to repress translation of closed-loop-associated mRNAs would allow the cell to conserve its resources in response to stress or poor growth conditions. Length-dependent translational enhancement may provide the base upon which more nuanced sequence-specific regulatory circuits are built.
Can translation’s mRNA length bias be regulated?
We recently identified RACK1, a eukaryote-specific ribosomal protein of the small subunit, as a ribosomal factor that can modulate the mRNA length bias of translation (Thompson et al. 2016). Loss of the RACK1 protein decreased translation of short mRNAs relative to long mRNAs, suggesting a defect in the mechanism that normally acts to promote the heightened translation efficiency of short mRNAs. In the preceding section we have summarized the evidence that such a mechanism is provided by preferential formation or stabilization of the closed loop complex on short mRNAs. How might a ribosomal protein influence the strength of the mRNA length bias in translation efficiency?
One possibility is that the ribosome recognizes the closed loop form of the cap-binding complex more readily than the non-closed-loop form and that the RACK1 protein contributes to this selectivity (Fig. 2a). Intriguingly, the exact mechanism by which the cap-binding complex is recognized by the ribosome is still shrouded in some mystery. In mammals, eIF4G interacts with the small ribosomal subunit via a bridging interaction with the eIF3 complex (Lamphear et al. 1995; Korneeva et al. 2005). However, in yeast no interaction between eIF4G and eIF3 has ever been demonstrated, and the domain of eIF4G that binds eIF3 in mammals is not present in yeast (Morino et al. 2000; Marintchev and Wagner 2005; Jivotovskaya et al. 2006). eIF5 has been suggested to bridge the interaction between the yeast small subunit and eIF4G (Asano et al. 2001), but mRNA recruitment is unaffected after eIF5 depletion in yeast arguing that the function of eIF5 must be redundant with at least one other factor (Jivotovskaya et al. 2006). The location of the RACK1 protein near the putative eIF4G docking site and its conservation throughout eukaryotes are consistent with a direct and important role in eIF4G-dependent mRNA recruitment. Moreover, RACK1 was found to co-purify with yeast eIF4G under stringent conditions, suggesting they may interact directly (Gavin et al. 2002; Gavin et al. 2006). To explain the selective effect of RACK1 on the translation of closed loop mRNAs, it is necessary to propose that the affinity of eIF4G-PABP for the ribosome is higher than that of eIF4G alone. Although it is unknown if and how PABP affects eIF4G’s interaction with the small subunit, there is extensive evidence for allosteric enhancement of binding affinities among the components of the closed loop (see above).
How could environmental signals specifically promote translation of mRNAs in the closed loop? One possibility is that these signals could alter the activity of proteins involved in closed loop formation. eIF4E, eIF4G, and PABP each have identified phosphorylation sites (Joshi et al. 1995; Le et al. 2000; Raught et al. 2000; Rush et al. 2005; Raught and Gingras 2007; Albuquerque et al. 2008; Dephoure et al. 2008; Holt et al. 2009; Swaney et al. 2013). Although these phosphorylation events are often correlated with growth-inducing signals, the roles of specific phosphorylation sites in promoting translation remain unclear (Scheper and Proud 2002; Jackson et al. 2010). Phosphorylation of eIF4E on Serine 209 increases in response to a variety of growth promoting stimuli, and expression of non-phosphorylatable eIF4E causes reduced growth in Drosophila and reduced tumorigenesis in mice (Lachance et al. 2002; Scheper and Proud 2002; Furic et al. 2010). It was thus unexpected to find that this phosphorylation event decreases the affinity of eIF4E for capped RNA ~5-fold (Scheper et al. 2002). However, in the context of the mutually stabilizing interactions of the closed loop, this modest reduction in cap affinity might be inconsequential. Thus, eIF4E S209 phosphorylation could make cellular translation more dependent on closed loop formation and indirectly enhance translation of growth-promoting genes by reducing competition from non-closed-loop mRNAs for limiting initiation factors (Fig. 2b). Intriguingly, ribosomal RACK1 enhances PKCβII-dependent phosphorylation of eIF4E on S209 in the context of chemotherapy-resistant hepatocellular carcinoma (Ruan et al. 2012). Thus, RACK1’s position on the ribosome might enable coordination of signaling events that promote or repress closed loop mRNA translation in response to changing growth conditions.
Another potential avenue for selective closed loop mRNA regulation is via repression by the eIF4E-binding proteins (4E-BPs), a class of proteins that have been studied extensively for their regulatory link to nutrient status (Richter and Sonenberg 2005). In their dephosphorylated state, 4E-BPs bind to eIF4E and sterically occlude binding of eIF4G. Upon nutrient or growth factor stimulation, phosphorylation of 4E-BPs causes their dissociation from eIF4E and allows eIF4E binding to eIF4G. Although all eIF4G-dependent initiation is potentially downregulated by 4E-BPs, closed loop mRNAs are likely to show distinct responses. First, decreased dissociation of eIF4E from eIF4G in the context of the closed loop should make this class of mRNA relatively resistant to translational repression by transient exposure to a dephosphorylated 4E-BP. Even if eIF4G dissociates from eIF4E, short mRNAs could maintain a higher local concentration of eIF4G due tethering by PABP and more frequent encounters between their 5’ and 3’ ends. Thus, in the presence of sub-saturating levels of dephosphorylated 4E-BPs, short mRNAs would be expected to spend more time bound to eIF4G and therefore translationally active. On the other hand, in the presence of saturating levels of 4E-BPs, these mRNAs would lose all the benefits of the closed loop, including enhanced affinity with the eIF4F complex and the ability to support ribosome recycling that would allow the same mRNA to be translated several times after one primary initiation event (Fig. 2c). Therefore, the magnitude of translational repression that 4E-BPs exert on closed loop mRNAs may be greater than the repression of a non-closed-loop mRNA.
Finally, regulation of closed loop mRNA translation could depend more generally on the concentration of ribosomes or certain translation factors in the cell. Lower concentrations of ribosomes may impair non-closed-loop translation more than closed loop translation, either because of the higher affinity of the cap-binding complex for closed loop mRNAs or due to the ability of closed loop mRNAs to reuse the same ribosomes during intrapolysomal ribosome recycling. Therefore, the lower relative protein synthetic capacity of cells in early G1 phase (Polymenis and Aramayo 2015) may favor closed loop translation. After a long period of growth, the higher protein synthetic capacity would be expected to decrease competition for limiting ribosomes and translation factors, thus allowing synthesis of other proteins, such as cyclins that are needed to initiate cell divison (Thomas 2000). Because nearly all cytosolic ribosomal proteins are encoded by closed loop mRNAs, such a translational hierarchy would promote rapid attainment of sufficient protein synthetic capacity before progressing through the cell cycle.
Although short mRNAs show relatively high ribosome density in diverse organisms and cell types, the relationship between mRNA length and closed loop association has not yet been established in metazoan eukaryotes. Regulation of the closed loop could complement the regulation of 5’ terminal oligopyrimidine (TOP) mRNAs in metazoans, which are highly translated in favorable growth conditions but rapidly repressed upon the onset of unfavorable growth conditions (Meyuhas and Hornstein 2000). It seems unlikely that the TOP motif or its associated signaling pathways (Wullschleger et al. 2006) directly explain the conserved translational privileging of short mRNAs throughout eukaryotes both because the TOP motif is not found in yeast, and because the group of short, closed-loop-associated mRNAs include many yeast mRNAs that are not homologous to TOP mRNAs (Costello et al. 2015). However, the fact that TOP mRNAs almost exclusively include ribosomal proteins and translation factors (Meyuhas and Kahan 2014) demonstrates the importance of tight regulation for this group of mRNAs, which is highly overlapping with short closed loop mRNAs in yeast (Costello et al., 2015; Thompson et al., 2016).
Perspective
Here we have presented evidence that mRNA length, and perhaps specifically the length of the mRNA open reading frame, can be sensed by the translation machinery to enhance translation of short mRNAs. Many questions remain unanswered, including an explanation for the primary event that determines preferential formation or stabilization of the closed loop on short mRNAs and the mechanism by which the closed loop state is communicated to the ribosome. Nevertheless, answering these questions will give us a deeper understanding of how intrinsic properties of eukaryotic mRNAs determine their translational activity and how the translation of a large group of growth-promoting mRNAs can be coordinately regulated according to the cellular growth state.
Acknowledgments
We thank members of the Gilbert lab for helpful discussions. This work was supported by the National Institutes of Health (GM094303) to WVG and in part by the NIH Pre-Doctoral Training Grant T32GM007287.
Footnotes
The literature is inconsistent regarding the magnitude of stimulation required for synergy. In many cases, stimulation by the combined action of the cap and poly(A) tail that is greater than the sum of their independent effects is described as “synergy”. We favor the null hypothesis that the combination of independent effects will produce stimulation equal to the product of their individual effects. Thus, only enhancement greater than the product constitutes synergy.
References
- Adamson SD, Howard GA, Herbert E. The ribosome cycle in a reconstituted cell-free system from reticulocytes. Cold Spring Harb Symp Quant Biol. 1969;34:547–554. doi: 10.1101/sqb.1969.034.01.062. [DOI] [PubMed] [Google Scholar]
- Albuquerque CP, Smolka MB, Payne SH, et al. A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol Cell Proteomics. 2008;7:1389–1396. doi: 10.1074/mcp.M700468-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amrani N, Ghosh S, Mangus DA, Jacobson A. Translation factors promote the formation of two states of the closed-loop mRNP. Nature. 2008;453:1276–1280. doi: 10.1038/nature06974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arava Y, Wang Y, Storey JD, et al. Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2003;100:3889–3894. doi: 10.1073/pnas.0635171100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano K, Shalev A, Phan L, et al. Multiple roles for the C-terminal domain of eIF5 in translation initiation complex assembly and GTPase activation. EMBO J. 2001;20:2326–2337. doi: 10.1093/emboj/20.9.2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baglioni C, Vesco C, Jacobs-Lorena M. The role of ribosomal subunits in mammalian cells. Cold Spring Harb Symp Quant Biol. 1969;34:555–565. doi: 10.1101/sqb.1969.034.01.063. [DOI] [PubMed] [Google Scholar]
- Bergamini G, Preiss T, Hentze MW. Picornavirus IRESes and the poly(A) tail jointly promote cap-independent translation in a mammalian cell-free system. RNA. 2000;6:1781–1790. doi: 10.1017/S1355838200001679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borman AM, Michel YM, Kean KM. Biochemical characterisation of cap-poly(A) synergy in rabbit reticulocyte lysates: the eIF4G-PABP interaction increases the functional affinity of eIF4E for the capped mRNA 5’-end. Nucleic Acids Res. 2000;28:4068–4075. doi: 10.1093/nar/28.21.4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borman AM, Michel YM, Malnou CE, Kean KM. Free Poly(A) Stimulates Capped mRNA Translation in Vitro through the eIF4G-Poly(A)-binding Protein Interaction. J Biol Chem. 2002;277:36818–36824. doi: 10.1074/jbc.M205065200. [DOI] [PubMed] [Google Scholar]
- Costello J, Castelli LM, Rowe W, et al. Global mRNA selection mechanisms for translation initiation. Genome Biol. 2015;16:10. doi: 10.1186/s13059-014-0559-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva D, Tu YT, Amunts A, et al. Mitochondrial ribosome assembly in health and disease. Cell Cycle. 2015;14:2226–2250. doi: 10.1080/15384101.2015.1053672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dephoure N, Zhou C, Villén J, et al. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci U S A. 2008;105:10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg E, Levanon EY. Human housekeeping genes are compact. Trends Genet. 2003;19:362–365. doi: 10.1016/S0168-9525(03)0140-9. [DOI] [PubMed] [Google Scholar]
- Furic L, Rong L, Larsson O, et al. eIF4E phosphorylation promotes tumorigenesis and is associated with prostate cancer progression. Proc Natl Acad Sci U S A. 2010;107:14134–14139. doi: 10.1073/pnas.1005320107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie DR. A tale of two termini. Gene. 1998;216:1–11. doi: 10.1016/S0378-1119(98)00318-7. [DOI] [PubMed] [Google Scholar]
- Gallie DR. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 1991;5:2108–2116. doi: 10.1101/gad.5.11.2108. [DOI] [PubMed] [Google Scholar]
- Gavin A-C, Aloy P, Grandi P, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- Gavin A-C, Bösche M, Krause R, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- Gierer A. Function of aggregated reticulocyte ribosomes in protein synthesis. J Mol Biol. 1963;6:148–157. doi: 10.1016/S0022-2836(63)80131-X. [DOI] [PubMed] [Google Scholar]
- Guo J, Lian X, Zhong J, et al. Length-dependent translation initiation benefits the functional proteome of human cells. Mol Biosyst. 2015;11:370–378. doi: 10.1039/c4mb00462k. [DOI] [PubMed] [Google Scholar]
- Holt LJ, Tuch BB, Villén J, et al. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science. 2009;325:1682–1686. doi: 10.1126/science.1172867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizuka N, Najita L, Franzusoff A, Sarnow P. Cap-dependent and cap-independent translation by internal initiation of mRNAs in cell extracts prepared from Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:7322–7330. doi: 10.1128/MCB.14.11.7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CUT, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson A. Poly(A) Metabolism and Translation: The Closed-loop Model. In: Hershey JWB, Mathews MB, Sonenberg N, editors. Translational Control. Cold Spring Harbor Laboratory Press; 1996. pp. 451–480. [Google Scholar]
- Jacobson A, Favreau M. Possible involvement of poly(A) in protein synthesis. Nucleic Acids Res. 1983;11:6353–6368. doi: 10.1093/nar/11.18.6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jivotovskaya AV, Valásek L, Hinnebusch AG, Nielsen KH. Eukaryotic translation initiation factor 3 (eIF3) and eIF2 can promote mRNA binding to 40S subunits independently of eIF4G in yeast. Mol Cell Biol. 2006;26:1355–1372. doi: 10.1128/MCB.26.4.1355-1372.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi B, Cai AL, Keiper BD, et al. Phosphorylation of eukaryotic protein synthesis initiation factor 4E at Ser-209. J Biol Chem. 1995;270:14597–14603. doi: 10.1074/jbc.270.24.14597. [DOI] [PubMed] [Google Scholar]
- Kahvejian A, Svitkin YV, Sukarieh R, et al. Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes Dev. 2005:104–113. doi: 10.1101/gad.1262905.(40S). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korneeva NL, First EA, Benoit CA, Rhoads RE. Interaction between the NH2-terminal domain of eIF4A and the central domain of eIF4G modulates RNA-stimulated ATPase activity. J Biol Chem. 2005;280:1872–1881. doi: 10.1074/jbc.M406168200. [DOI] [PubMed] [Google Scholar]
- Kozak M. How strong is the case for regulation of the initiation step of translation by elements at the 3’ end of eukaryotic mRNAs? Gene. 2004;343:41–54. doi: 10.1016/j.gene.2004.08.011. [DOI] [PubMed] [Google Scholar]
- Lachance PED, Miron M, Raught B, et al. Phosphorylation of eukaryotic translation initiation factor 4E is critical for growth. Mol Cell Biol. 2002;22:1656–1663. doi: 10.1128/MCB.22.6.1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladhoff AM, Uerlings I, Rosenthal S. Electron microscopic evidence of circular molecules in 9-S globin mRNA from rabbit reticulocytes. Mol Biol Rep. 1981;7:101–106. doi: 10.1007/BF00778739. [DOI] [PubMed] [Google Scholar]
- Lamphear BJ, Kirchweger R, Skern T, Rhoads RE. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. Implications for cap-dependent and cap-independent translational initiation. J Biol Chem. 1995;270:21975–21983. doi: 10.1074/jbc.270.37.21975. [DOI] [PubMed] [Google Scholar]
- Le H, Browning KS, Gallie DR. The phosphorylation state of poly(A)-binding protein specifies its binding to poly(A) RNA and its interaction with eukaryotic initiation factor (eIF) 4F, eIFiso4F, and eIF4B. J Biol Chem. 2000;275:17452–17462. doi: 10.1074/jbc.M001186200. [DOI] [PubMed] [Google Scholar]
- Le H, Tanguay RL, Balasta ML, et al. Translation Initiation Factors eIF-iso4G and eIF-4B Interact with the Poly (A)-binding Protein and Increase Its RNA Binding Activity. J Biol Chem. 1997;272:16247–16255. doi: 10.1074/jbc.272.26.16247. [DOI] [PubMed] [Google Scholar]
- Lempiäinen H, Shore D. Growth control and ribosome biogenesis. Curr Opin Cell Biol. 2009;21:855–863. doi: 10.1016/j.ceb.2009.09.002. [DOI] [PubMed] [Google Scholar]
- Marintchev A, Wagner G. eIF4G and CBP80 share a common origin and similar domain organization: Implications for the structure and function of eIF4G. Biochemistry. 2005;44:12265–12272. doi: 10.1021/bi051271v. [DOI] [PubMed] [Google Scholar]
- Mathias AP, Williamson R, Huxley HE, Page S. Occurrence and function of polysomes in rabbit reticulocytes. J Mol Biol. 1964;9:154–167. doi: 10.1016/s0022-2836(64)80097-8. [DOI] [PubMed] [Google Scholar]
- Meyuhas O, Hornstein E. Translational Control of TOP mRNAs. In: Sonenberg N, Hershey JWB, Mathews MB, editors. Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press; 2000. pp. 671–693. [Google Scholar]
- Meyuhas O, Kahan T. The race to decipher the top secrets of TOP mRNAs. Biochim Biophys Acta. 2014;1849:801–811. doi: 10.1016/j.bbagrm.2014.08.015. [DOI] [PubMed] [Google Scholar]
- Michel YM, Poncet D, Piron M, et al. Cap-Poly(A) synergy in mammalian cell-free extracts. Investigation of the requirements for poly(A)-mediated stimulation of translation initiation. J Biol Chem. 2000;275:32268–32276. doi: 10.1074/jbc.M004304200. [DOI] [PubMed] [Google Scholar]
- Morino S, Imataka H, Svitkin YV, et al. Eukaryotic translation initiation factor 4E (eIF4E) binding site and the middle one-third of eIF4GI constitute the core domain for cap-dependent translation, and the C-terminal one-third functions as a modulatory region. Mol Cell Biol. 2000;20:468–477. doi: 10.1128/mcb.20.2.468-477.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munroe D, Jacobson A. mRNA poly(A) tail, a 3’ enhancer of translational initiation. Mol Cell Biol. 1990;10:3441–3455. doi: 10.1128/MCB.10.7.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Leary SE, Petrov A, Chen J, Puglisi JD. Dynamic recognition of the mRNA cap by Saccharomyces cerevisiae eIF4E. Structure. 2013;21:2197–2207. doi: 10.1016/j.str.2013.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otero LJ, Ashe MP, Sachs AB. The yeast poly(A)-binding protein Pab1p stimulates in vitro poly(A)-dependent and cap-dependent translation by distinct mechanisms. EMBO J. 1999;18:3153–3163. doi: 10.1093/emboj/18.11.3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palatnik CM, Wilkins C, Jacobson A. Translational control during early Dictyostelium development: possible involvement of poly(A) sequences. Cell. 1984;36:1017–1025. doi: 10.1016/0092-8674(84)90051-5. [DOI] [PubMed] [Google Scholar]
- Park E-H, Walker SE, Lee JM, et al. Multiple elements in the eIF4G1 N-terminus promote assembly of eIF4G1 PABP mRNPs in vivo. EMBO J. 2011;30:302–316. doi: 10.1038/emboj.2010.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philipps GR. Haemoglobin synthesis and polysomes in intact reticulocytes. Nature. 1965;205:567–70. doi: 10.1038/205567a0. [DOI] [PubMed] [Google Scholar]
- Polymenis M, Aramayo R. Translate to divide: control of the cell cycle by protein synthesis. Microb Cell. 2015;2:94–104. doi: 10.15698/mic2015.04.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preiss T, Hentze MW. Dual function of the messenger RNA cap structure in poly(A)-tail-promoted translation in yeast. Nature. 1998;392:516–20. doi: 10.1038/33192. [DOI] [PubMed] [Google Scholar]
- Ptushkina M, von der Haar T, Vasilescu S, et al. Cooperative modulation by eIF4G of eIF4E-binding to the mRNA 5’ cap in yeast involves a site partially shared by p20. EMBO J. 1998;17:4798–4808. doi: 10.1093/emboj/17.16.4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raught B, Gingras A-C. Signaling to Translation Initiation. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational control in biology and medicine. Cold Spring Harbor Laboratory Press; 2007. pp. 369–400. [Google Scholar]
- Raught B, Gingras A-C, Gygi SP, et al. Serum-stimulated, rapamycin-sensitive phosphorylation sites in the eukaryotic translation initiation factor 4GI. EMBO J. 2000;19:434–444. doi: 10.1093/emboj/19.3.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter JD, Sonenberg N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature. 2005;433:477–480. doi: 10.1038/nature03205. [DOI] [PubMed] [Google Scholar]
- Ruan Y, Sun L, Hao Y, et al. Ribosomal RACK1 promotes chemoresistance and growth in human hepatocellular carcinoma. J Clin Invest. 2012;122:2554–2566. doi: 10.1172/JCI58488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rush J, Moritz A, Lee KA, et al. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat Biotechnol. 2005;23:94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- Safaee N, Kozlov G, Noronha AM, et al. Interdomain allostery promotes assembly of the poly (A) mRNA complex with PABP and eIF4G. Mol Cell. 2012;48:375–386. doi: 10.1016/j.molcel.2012.09.001. [DOI] [PubMed] [Google Scholar]
- Scheper GC, Proud CG. Does phosphorylation of the cap-binding protein eIF4E play a role in translation initiation? Eur J Biochem. 2002;269:5350–5359. doi: 10.1046/j.1432-1033.2002.03291.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheper GC, van Kollenburg B, Hu J, et al. Phosphorylation of Eukaryotic Initiation Factor 4E Markedly Reduces Its Affinity for Capped mRNA. J Biol Chem. 2002;277:3303–3309. doi: 10.1074/jbc.M103607200. [DOI] [PubMed] [Google Scholar]
- Svitkin YV, Imataka H, Khaleghpour K, et al. Poly(A)-binding protein interaction with elF4G stimulates picornavirus IRES-dependent translation. RNA. 2001;7:1743–1752. [PMC free article] [PubMed] [Google Scholar]
- Swaney DL, Beltrao P, Starita L, et al. Global analysis of phosphorylation and ubiquitylation cross-talk in protein degradation. Nat Methods. 2013;10:676–682. doi: 10.1038/nmeth.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarun SZ, Sachs AB. A common function for mRNA 5’ and 3’ ends in translation initiation in yeast. Genes Dev. 1995;9:2997–3007. doi: 10.1101/gad.9.23.2997. [DOI] [PubMed] [Google Scholar]
- Tarun SZ, Sachs AB. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- Tarun SZ, Wells SE, Deardorff JA, Sachs AB. Translation initiation factor eIF4G mediates in vitro poly(A) tail-dependent translation. Proc Natl Acad Sci U S A. 1997;94:9046–9051. doi: 10.1073/pnas.94.17.9046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas G. An encore for ribosome biogenesis in the control of cell proliferation. Nat Cell Biol. 2000;2:E71–72. doi: 10.1038/35010581. [DOI] [PubMed] [Google Scholar]
- Thompson MK, Rojas-Duran MF, Gangaramani P, Gilbert WV. The ribosomal protein Asc1/RACK1 is required for efficient translation of short mRNAs. Elife. 2016;5:1–22. doi: 10.7554/eLife.11154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Der Haar T, Ball PD, Mccarthy JEG. Stabilization of Eukaryotic Initiation Factor 4E Binding to the mRNA 5’-Cap by Domains of eIF4G. J Biol Chem. 2000;275:30551–30555. doi: 10.1074/jbc.M004565200. [DOI] [PubMed] [Google Scholar]
- Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- Warner JR, Rich A, Hall CE. Electron Microscope Studies of Ribosomal Clusters Synthesizing Hemoglobin. Science. 1962;138:1399–13403. doi: 10.1126/science.138.3548.1399. [DOI] [PubMed] [Google Scholar]
- Wei C-C, Balasta ML, Ren J, Goss DJ. Wheat Germ Poly(A) Binding Protein Enhances the Binding Affinity of Eukaryotic Initiation Factor 4F and (iso)4F for Cap Analogues. Biochemistry. 1998;37:1910–1916. doi: 10.1021/bi9724570. [DOI] [PubMed] [Google Scholar]
- Wells SE, Hillner PE, Vale RD, Sachs AB. Circularization of mRNA by Eukaryotic Translation Initiation Factors. Mol Cell. 1998;2:135–140. doi: 10.1016/S1097-2765(00)80122-7. [DOI] [PubMed] [Google Scholar]
- Wettstein F, Staehelin T, Noll H. Ribosomal aggregate engaged in protein synthesis: characterization of the ergosome. Nature. 1963;197:430–435. doi: 10.1038/197430a0. [DOI] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]


