Large granular lymphocytic (LGL) leukaemia is a rare lymphoproliferative disorder characterized by the expansion of CD3+CD8+ cytotoxic T-lymphocytes (CTLs) or CD3− natural killer (NK) cells. LGL leukaemia can be subdivided into T-cell LGL (T-LGL) and chronic lymphoproliferative disorders of NK cells (CLPD-NK) depending on the type of cells affected. Patients with LGL leukaemia exhibit anaemia, neutropenia, splenomegaly and autoimmune disorders (Steinway et al, 2014). Somatic mutations in the signal transducer and activator of transcription 3 gene (STAT3) have been found in 40% of T-LGL and 30% of CLPD-NK patients. These STAT3 mutations result in constitutive activation of STAT3 protein. The discovery of STAT3 mutations in T-LGL and CLPD-NK has suggested a role for STAT3 mutations in the pathogenesis of these diseases (Koskela et al, 2012; Jerez et al, 2012). However, it remains still unclear whether STAT3 mutations play a causal role in the development of LGL leukaemia.
To investigate the effects of STAT3 mutations on haematopoietic cells, murine interleukin 3 (IL3) -dependent Ba/F3 cells were transduced with MSCV-IRES-GFP retrovirus expressing empty vector, wild-type STAT3 (STAT3WT) or LGL leukaemia-associated STAT3 mutants (STAT3D661V, STAT3Y640F), and the infected cells were sorted for GFP. Immunoblot analysis showed increased phosphorylation of STAT3 in Ba/F3 cells expressing STAT3D661V or STAT3Y640F mutant compared to vector or STAT3WT (Fig.S1A). However, expression of STAT3D661V or STAT3Y640F mutant did not provide any growth advantage to Ba/F3 cells when compared with vector or STAT3WT expression (Fig.S1B).
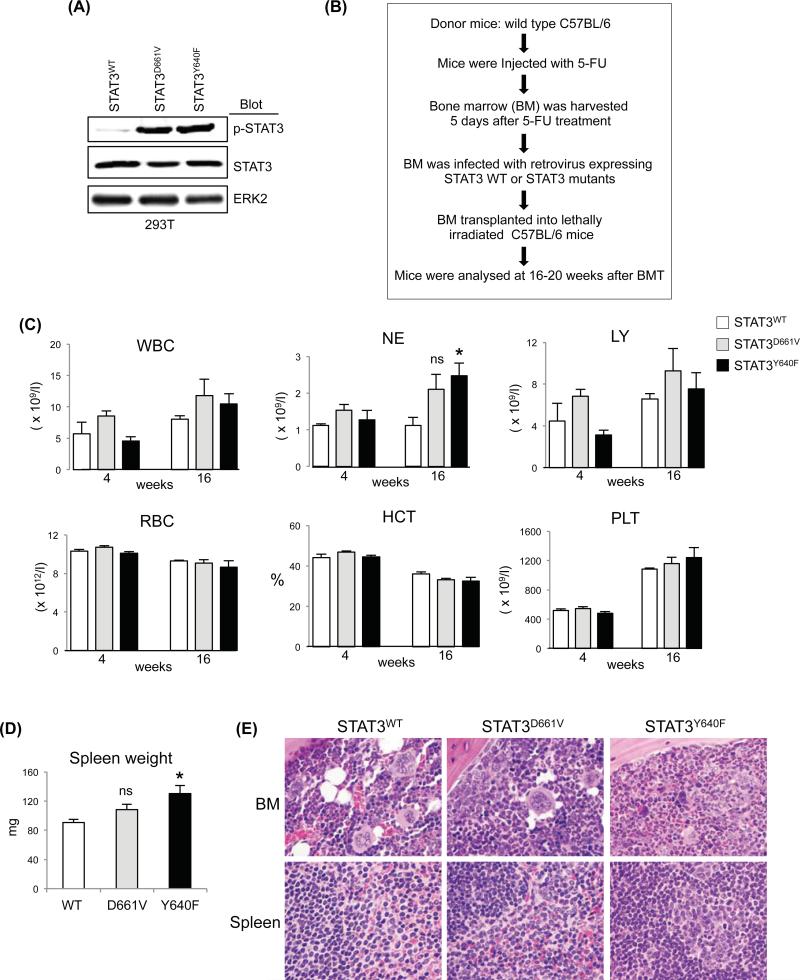
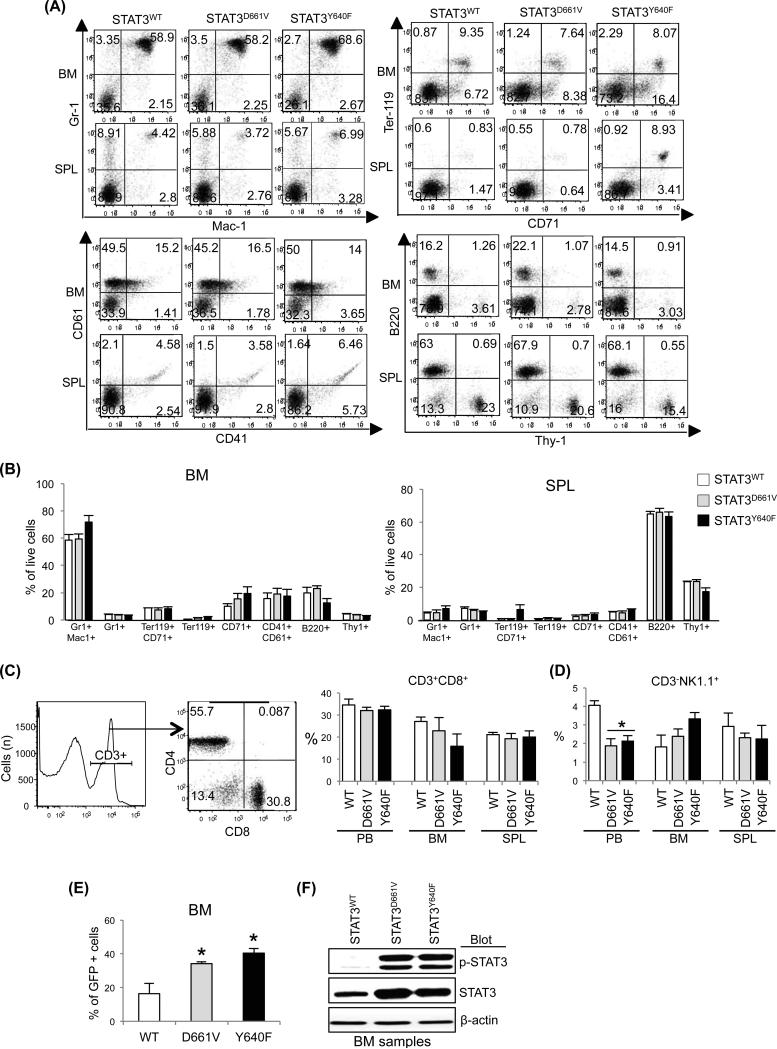
To assess the in vivo effects of STAT3 mutations on haematopoiesis, we performed retroviral bone marrow (BM) transduction and transplantation assays as outlined in Fig.1B. Expression of STAT3D661V or STAT3Y640F resulted in increased phosphorylation of STAT3 in 293T cells (Fig.1A). Transplanted animals expressing STAT3Y640F exhibited significantly increased neutrophil counts compared with mice expressing STAT3WT at 16 weeks after transplantation (Fig.1C). Also, there was trend of increased neutrophil counts in the transplanted animals expressing STAT3D661V (Fig.1C). No significant difference was observed in white blood cell, lymphocyte, red blood cell and platelet counts or haematocrit among transplanted animals expressing STAT3WT, STAT3D661V or STAT3Y640F (Fig.1C). Expression of STAT3Y640F resulted in modest but significant increase in spleen size/weight in the transplanted mice whereas STAT3D661V mice had slightly larger spleen than that of STAT3WT mice (Fig.1D). Histopathological analyses of the BM sections showed increased granulocytic precursors in STAT3Y640F mutant mice, whereas STAT3D661V mutant mice exhibited mildly increased myeloid cells compared with the BM of STAT3WT mice (Fig.1E). However, there were no increases in LGL cells in the BM of STAT3D661V or STAT3Y640F mutant mice. Spleen sections from both STAT3Y640F and STAT3D661V mice showed minimal increase in myeloid cells compared to STAT3WT mice (Fig.1E). Flow cytometric analysis showed modest increases in myeloid (Gr-1+/Mac-1+) cells in the BM of mice expressing STAT3Y640F compared with STAT3WT-expressing mice (Fig.2A, B and Fig.S2). However, we did not see any significant alterations in the erythroid (Ter119+/CD71+), megakaryocytic (CD41+/CD61+), B-cell (B220+) and T-cell (Thy-1+) precursors in the BM and spleens of STAT3D661V or STAT3Y640F mice compared with STAT3WT mice (Fig.2A, B and Fig.S2). As T-LGL is characterized by an expansion of CD3+CD8+ CTLs (Koskela et al, 2012), we also determined the CD3+CD8+ populations in the peripheral blood (PB), BM and spleens of the transplanted animals expressing STAT3WT, STAT3D661V or STAT3Y640F. We observed no significant difference in CD3+CD8+ populations in the PB, BM and spleens of STAT3D661V or STAT3Y640F mutant mice compared with STAT3WT mice (Fig.2C). We also did not observe any significant difference in CD3− natural killer cells (CD3−NK1.1+) in the BM and spleens of the STAT3D661V or STAT3Y640Fmutant mice compared with STAT3WT mice (Fig.2D). The frequency of CD3−NK1.1+ cells was rather decreased in the PB of STAT3D661V or STAT3Y640F mutant mice compared with STAT3WT mice (Fig.2D). The frequency of CD3+CD8+ and CD3-NK1.1+ cells in the thymus was comparable in STAT3WT, STAT3D661V and STAT3Y640F mice (Fig.S3A, B). We also assessed the engraftment of haematopoietic cells in the BM of the transplanted animals using GFP expression. As shown in Figure 2E, a significant proportion of cells (~40%) in the BM of transplanted mice receiving STAT3D661V or STAT3Y640F-transduced BM were GFP+, suggesting that donor BM cells were efficiently engrafted. Immunoblot analysis showed constitutive phosphorylation of STAT3 in mice expressing STAT3D661V or STAT3Y640Fmutants (Fig.2F), confirming that the STAT3 mutants were efficiently expressed in the BM of the transplanted animals. Together, these data suggest that STAT3 mutations associated with LGL leukaemia results in constitutive activation of STAT3; however, expression of STAT3 mutant alone is not sufficient to induce LGL leukaemia in a mouse model. These data are in accordance with a previous report (Couronné et al, 2013) showing that expression of STAT3Y640F mutation primarily induces myeloid malignancy in a murine BM transplantation model.
Fig 1.
Analysis of the in vivo effects of STAT3 mutations in a mouse bone marrow transplantation model. (A) Retroviruses expressing STAT3WT, STAT3D661V or STAT3Y640F were produced in 293T cells. Immunoblot analysis show that expression of STAT3D661V or STAT3Y640F mutant resulted in increased phosphorylation of STAT3 in 293T cells. ERK2 was used as a loading control. (B) Experimental design on retroviral bone marrow (BM) transduction/transplantation assay. Wild-type C57BL/6 mice were injected with 5-fluorouracil (5-FU) 5 days prior to BM harvest. BM cells were transduced with retroviruses expressing STAT3WT, STAT3D661V and STAT3Y640F and the infected cells were transplanted into lethally irradiated C57BL/6 recipient mice (106 cells/recipient) by retro-orbital injection. Mice were analysed between 16-20 weeks after bone marrow transplantation (BMT). (C) Peripheral blood white blood cell (WBC), neutrophil (NE), lymphocyte (LY), red blood cell (RBC) and platelet (PLT) counts, and haematocrit (HCT) were assessed in the recipient mice expressing STAT3WT (n=4), STAT3D661V (n=4) and STAT3Y640F (n=4) at 4 and 16 weeks after BMT. (D) Spleen weights of transplanted animals expressing STAT3WT, STAT3D661V and STAT3Y640F are presented in bar graphs as mean ± SEM (* indicates p<0.05 by Student's t-test; ns indicates not significant). (E) Histopathological analyses of the haematoxylin and eosin (H&E) stained BM sections showed increased granulocytic precursors in transplanted animals expressing STAT3Y640F mutant, whereas BM of transplanted animals expressing STAT3D661V mutant exhibited mildly increased granulocytic cells compared with the BM of STAT3WT-expressing mice. Spleen sections from transplanted animals expressing STAT3D661V and STAT3Y640F showed minimal increase in myeloid cells compared to spleens of control STAT3WT-expressing mice.
Fig 2.
Effects of STAT3 mutations on haematopoietic precursors. (A) Representative dot plots of flow cytometric analysis of myeloid (Gr-1+/Mac-1+), erythroid (Ter119+/CD71+), megakaryocytic (CD61+/CD41+), B-cell (B200+) and T-cell (Thy-1+) precursors in the bone marrow (BM) and spleens of transplanted animals expressing STAT3WT, STAT3D661V and STAT3Y640F at 16-20 weeks after bone marrow transplantation (BMT) are shown. (B) Percentages of myeloid (Gr-1+/Mac-1+), erythroid (Ter119+/CD71+), megakaryocytic (CD61+/CD41+), B-cell (B200+) and T-cell (Thy-1+) precursors in the BM and spleens (SPL) of transplanted animals expressing STAT3WT (n=4), STAT3D661V (n=4) and STAT3Y640F (n=4) are shown in bar graphs as mean ± SEM. (C) Flow cytometric analysis of CD3+CD8+ cells in the peripheral blood (PB), BM and spleens of transplanted animals expressing STAT3WT, STAT3D661V and STAT3Y640F at 16-20 weeks after BMT. (D) Percentages of CD3− natural killer cells (CD3−NK1.1+) in the peripheral blood (PB), BM and spleens of transplanted animals expressing STAT3WT, STAT3D661V and STAT3Y640F at 16-20 weeks after BMT are shown in histograms as mean ± SEM (n=4). (E) Flow cytometric analysis of GFP expression in the BM at 16-20 weeks after BMT. Data are shown in histograms as mean ± SEM (n=4; * indicates p<0.05 by Student's t-test). Note that significantly increased percentages of GFP+ cells in the BM of transplanted animals expressing STAT3D661V or STAT3Y640F compared with transplanted animals expressing STAT3WT. (F) Immunoblot analysis on total BM extracts shows constitutive phosphorylation of STAT3 in the BM of transplanted animals expressing STAT3D661V and STAT3Y640F mutants, suggesting that the STAT3 mutants were efficiently expressed and constitutively activated in the BM of the transplanted animals. β-actin was used as a loading control.
STAT3 can be activated by oncogenic tyrosine kinases (Vainchenker & Constantinescu 2013), by autocrine production of IL6 (Schuringa et al, 2000) or through somatic STAT3 mutations (Koskela et al, 2012; Jerez et al, 2012). Constitutive phosphorylation/activation of STAT3 has been observed in various human malignancies (Yu et al, 2014). The identification of STAT3 mutations in significant cases of T-LGL and CLPD-NK has suggested the use of STAT3 inhibitors as potential treatment for LGL leukaemia (Koskela et al, 2012; Jerez et al, 2012). If STAT3 mutations are not the driver of LGL leukaemia, therapies targeting STAT3 might not be very effective against LGL leukaemia. The absence of any LGL leukaemia phenotype in mice expressing STAT3 mutations implies that additional gene mutations or deregulation of other signalling molecules or pathways might be involved in association with STAT3 mutations in the pathogenesis of LGL leukaemia. Somatic mutations in STAT5B, PTPRT, BCL11B, SLIT2 and NRP1 genes affecting the STAT pathway and/or T-cell activation also have been found at a lower frequency in LGL leukaemia (Rajala et al, 2013; Andersson et al, 2013). Moreover, decreased SOCS3 expression and increased plasma levels of IL6 have been observed in patients with LGL leukaemia (Teramo et al, 2013). Future studies will identify co-existing mutations or genetic abnormalities in LGL leukaemia and determine the cooperative effects of such genetic abnormalities along with STAT3 mutations in the pathogenesis of LGL leukaemia.
Supplementary Material
Acknowledgements
This work was supported by the US National Institute of Health (NIH) grant (R01 HL095685) awarded to G.M. G.M. is a Scholar of the Leukemia & Lymphoma Society.
Footnotes
Authorship contributions
A.D. performed research, analysed data and wrote the manuscript; D.Y. performed research; R.E.H. conducted histopathological analysis and revised the manuscript; G.M. designed the research, analysed data, and wrote the manuscript.
Conflict of interest disclosure
The authors declare no conflict of interest.
References
- Andersson EI, Rajala H, Eldfors S, Ellonen P, Olson T, Jerez A, Clemente MJ, Kallioniemi O, Porkka K, Heckman C, Loughran TP, Jr., Maciejewski JP, Mustjoki S. Novel somatic mutations in large granular lymphocytic leukemia affecting the STAT-pathway and T-cell activation. Blood Cancer J. 2013;3:e168. doi: 10.1038/bcj.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couronné L, Scourzic L, Pilati C, Della Valle V, Duffourd Y, Solary E, Vainchenker W, Merlio JP, Beylot-Barry M, Damm F, Stern MH, Gaulard P, Lamant L, Delabesse E, Merle-Beral H, Nguyen-Khac F, Fontenay M, Tilly H, Bastard C, Zucman-Rossi J, Bernard OA, Mercher T. STAT3 mutations identified in human hematologic neoplasms induce myeloid malignancies in a mouse bone marrow transplantation model. Haematologica. 2013;98:1748–1752. doi: 10.3324/haematol.2013.085068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerez A, Clemente MJ, Makishima H, Koskela H, Leblanc F, Peng Ng K, Olson T, Przychodzen B, Afable M, Gomez-Segui I, Guinta K, Durkin L, His ED, McGraw K, Zhang D, Wlodarski MW, Porkka K, Sekeres MA, List A, Mustjoki S, Loughran TP, Maciejewski JP. STAT3 mutations unify the pathogenesis of chronic lymphoproliferative disorders of NK cells and T-cell large granular lymphocyte leukemia. Blood. 2012;120:3048–3057. doi: 10.1182/blood-2012-06-435297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskela HL, Eldfors S, Ellonen P, van Adrichem AJ, Kuusanmäki H, Andersso EI, Lagström S, Clemente MJ, Olson T, Jalkanen SE, Majumder MM, Almusa H, Edgren H, Lepistö M, Mattila P, Guinta K, Koistinen P, Kuittinen T, Penttinen K, Parsons A, Knowles J, Saarela J, Wennerberg K, Kallioniemi O, Porkka K, Loughran TP, Jr, Heckman CA, Maciejewski JP, Mustjoki S. Somatic STAT3 mutations in large granular lymphocytic leukemia. N Engl J Med. 2012;366:1905–1913. doi: 10.1056/NEJMoa1114885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajala HL, Eldfors S, Kuusanmäki H, van Adrichem AJ, Olson T, Lagström S, Andersson EI, Jerez A, Clemente MJ, Yan Y, Zhang D, Awwad A, Ellonen P, Kallioniemi O, Wennerberg K, Porkka K, Maciejewski JP, Loughran TP, Jr, Heckman C, Mustjoki S. Discovery of somatic STAT5b mutations in large granular lymphocytic leukemia. Blood. 2013;121:4541–4550. doi: 10.1182/blood-2012-12-474577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuringa JJ, Wierenga AT, Kruijer W, Vellenga E. Constitutive Stat3, Tyr705, and Ser727 phosphorylation in acute myeloid leukemia cells caused by the autocrine secretion of interleukin-6. Blood. 2000;95:3765–3770. [PubMed] [Google Scholar]
- Steinway NS, Leblanc F, Loughran TP., Jr. The pathogenesis and treatment of large granular lymphocyte leukemia. Blood Rev. 2014;28:87–94. doi: 10.1016/j.blre.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teramo A, Gattazzo C, Passeri F, Lico A, Tasca G, Cabrelle A, Martini V, Frezzato F, Trimarco V, Ave E, Boscaro E, Piazza F, Facco M, Trentin L, Semenzato G, Zambello R. Intrinsic and extrinsic mechanisms contribute to maintain the JAK/STAT pathway aberrantly activated in T-type large granular lymphocyte leukemia. Blood. 2013;121:3843–3854. doi: 10.1182/blood-2012-07-441378. [DOI] [PubMed] [Google Scholar]
- Vainchenker W, Constantinescu SN. JAK/STAT signaling in hematological malignancies. Oncogene. 2013;32:2601–2613. doi: 10.1038/onc.2012.347. [DOI] [PubMed] [Google Scholar]
- Yu H, Lee H, Herrmann A, Buettner R, Jove R. Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nat Rev Cancer. 2014;14:736–746. doi: 10.1038/nrc3818. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.