Abstract
Adenovirus type 5 (Ad5) DNA packaging is initiated in a polar fashion from the left end of the genome. The packaging process is dependent on the cis-acting packaging domain located between nucleotides 230 and 380. Seven AT-rich repeats that direct packaging have been identified within this domain. A1, A2, A5, and A6 are the most important repeats functionally and share a bipartite sequence motif. Several lines of evidence suggest that there is a limiting trans-acting factor(s) that plays a role in packaging. Both cellular and viral proteins that interact with adenovirus packaging elements in vitro have been identified. In this study, we characterized a group of recombinant viruses that carry site-specific point mutations within a minimal packaging domain. The mutants were analyzed for growth properties in vivo and for the ability to bind cellular and viral proteins in vitro. Our results are consistent with a requirement of the viral IVa2 protein for DNA packaging via a direct interaction with packaging sequences. Our results also indicate that higher-order IVa2-containing complexes that form on adjacent packaging repeats in vitro are the complexes required for the packaging activity of these sites in vivo. Chromatin immunoprecipitation was used to study proteins that bind directly to the packaging sequences. These results demonstrate site-specific interaction of the viral IVa2 and L1 52/55K proteins with the Ad5 packaging domain in vivo. These results confirm and extend those previously reported and provide a framework on which to model the adenovirus assembly process.
Adenovirus (Ad) assembly has been studied by using pulse-chase kinetic analyses, through the characterization of temperature-sensitive virus mutants blocked at different stages of assembly at the restrictive temperature, by studies of virus mutants defective in the expression of proteins involved in the packaging process, and by studies of mutants defective in the packaging sequences (2-4, 6-8, 12, 25, 31, 32). The results suggest that adenovirus assembly follows an ordered series of assembly and processing events analogous to those found in the assembly of bacteriophages. Whether the viral genome is involved in nucleating the initiating steps of capsid morphogenesis or whether viral DNA is inserted into an empty, preformed capsid has not been resolved. It is clear, however, that the viral genome contains cis-acting sequences that mediate the packaging process (reviewed in reference 20). Adenovirus DNA packaging occurs in a polar manner from the left end of the genome (1, 28). The genomes of adenovirus type 3 (Ad3), Ad5, and Ad16, representatives of adenovirus subgroups B and C, harbor conserved packaging signals (6, 10, 19, 22). These conserved cis-acting packaging domains suggest similar mechanisms of selective and polar DNA packaging for all adenovirus subgroups.
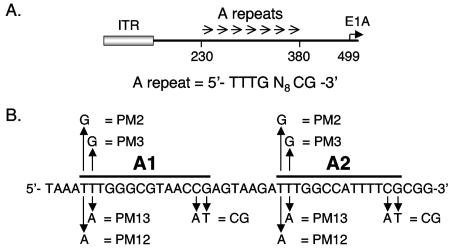
Ad5 DNA encapsidation is dependent on cis-acting sequences located between nucleotides (nt) 230 and 380 (Fig. 1A) (6, 7, 13, 25). Seven repeated sequences (termed A-repeats due to their AT-rich content) that contribute to viral DNA packaging are located within this domain. Although A-repeats are functionally redundant, they follow a hierarchy of importance: A1, A2, A5, and A6 are the most important repeats for packaging activity (6, 7, 25). A-repeats contain a bipartite consensus motif (5′-TTTG N8 CG-3′ [Fig. 1A]). Both the first and the second half-site of the consensus motif, as well as the 8-bp spacing between the half-sites, are critical for viral DNA packaging (25). Several lines of evidence suggest that a limiting trans-acting factor(s), which binds to the viral packaging sequences, plays a role in the packaging process (7, 25, 26). First, in vivo coinfection experiments show that viruses with a greater number of packaging repeats package more efficiently than viruses with fewer packaging repeats. Second, an isolated packaging domain on a multicopy plasmid represses packaging of a wild-type virus in vivo. Both results suggest competition for a limiting packaging factor(s) in vivo. Multimerized individual packaging repeats, termed minimal packaging domains, have been used to study Ad DNA packaging (5, 26). Minimal packaging sequences were shown to support packaging in vivo to various degrees when built into viruses lacking the authentic packaging domain. When minimal packaging sequences were used in binding assays in vitro as probes, three cellular transcription factors that bind to these sites were identified: chicken ovalbumin upstream promoter transcription factor (COUP-TF), OCT-1, and CCAAT displacement protein (CDP) (5, 26). It is very unlikely that COUP-TF and OCT-1 are required for Ad packaging, since viruses with mutated or synthetic packaging domains were identified that functioned efficiently in packaging in vivo but did not bind these transcription factors in vitro. However, a role for CDP in Ad packaging was suggested in earlier studies (5, 26).
FIG. 1.
Ad5 packaging sequences. (A) Schematic representation of the left end of the Ad5 genome. Nucleotide coordinates, relative to the left terminus, are indicated, and the inverted terminal repeat (ITR) is represented by a gray box. A-repeats 1 to 7 are represented by arrows between nt 230 and 380. The E1A transcription initiation site is located at nt 499. The packaging repeat consensus motif is shown under the diagram. (B) Sequences surrounding A-repeats 1 and 2 and site-specific point mutants. A-repeats 1 and 2 are indicated by bold lines above the sequence. Site-specific point mutations in these A-repeats are shown with arrows; the nucleotide(s) changed and the mutant designation are shown.
Two viral proteins, L1 52/55K and IVa2, have been found to play important roles in Ad packaging and virus assembly (8, 12, 31, 32), although their exact roles in this process remain unclear. A null mutant and a temperature-sensitive mutant of the L1 52/55K protein, respectively, form empty capsids or capsids that contain only the left end of the viral genome (8, 12). The Ad L1 52/55K protein is found within immature virus particles (8, 12) and forms a physical complex with the Ad IVa2 protein (9). In turn, the Ad IVa2 protein is essential for virus assembly and the formation of empty viral capsids. Ad IVa2 is found in both empty and mature virus particles (31, 32). The IVa2 protein binds to packaging A-repeats 1 and 2 as well as to A-repeats 4 and 5 in vitro (30). Collectively, these data demonstrate that the Ad L1 52/55K and IVa2 proteins play a key role(s) in the very early stages of the virus assembly process. IVa2 appears to be a multifunctional protein during Ad infection. In addition to its role in viral assembly, IVa2 has been implicated in the transcriptional control of the major late promoter (MLP) (29) by binding to the DE elements within the MLP (17).
In this study, we have examined the binding of the Ad IVa2 and L1 52/55K proteins to wild-type and mutant packaging sequences in vitro and in vivo in comparison to the growth properties of corresponding mutant viruses. Our results demonstrate that the binding of IVa2 to the packaging sequences in vitro correlates directly with the packaging efficiency of the virus in vivo. Furthermore, it appears that the higher-order IVa2-containing complexes that form on adjacent packaging A-repeats in vitro are the complexes required for the packaging activity of these sites in vivo. Further, both the IVa2 and L1 52/55K proteins associate with the packaging domain in vivo in direct correlation with packaging function. Finally, our results indicate that the cellular protein CDP does not play a role in packaging. These data are consistent with the known role of the IVa2 and L1 52/55K proteins in the Ad assembly process, and they provide strong evidence for a causal link between the binding of these proteins to packaging sequences and the assembly of virus particles.
MATERIALS AND METHODS
Virus constructions.
Ad5 dl309 (14), the parent for all viruses described in this report, is phenotypically wild type for growth in vivo. Plasmid pE1A-194/814 contains the left-end Ad5 XbaI fragment (nt 1 to 1399) with a deletion between nt 194 and nt 814 and a unique XhoI restriction site at the junction of the deletion (26). A head-to-tail dimer of wild-type Ad5 A1 and A2 (A1/2-WT) (5′-TCGACTGTAGTAAATTTGGGCGTAACCGAGTAAGATTTGGCCATTTTCGCGGC-3′) was cloned into the pE1A-194/814 background at the deletion junction (A1 and A2 TTTG and CG consensus sequences are underlined). The recombinant plasmid was subsequently rebuilt into intact virus (Ad5 dl309) as described elsewhere (27). Comparable plasmids and viruses that contained individual point mutations within A-repeat consensus sequences were generated as described above. Viral lysates were titered by a plaque assay using N52.E6 cells. Purified virus particles were prepared by cesium chloride equilibrium centrifugation.
Cultured cells and infections.
Ad E1-positive N52.E6 cells (24) and N52.E6-Cre cells (which express Cre recombinase; a generous gift from G. Schiedner and S. Kochanek, ZMMK, University of Cologne, Cologne, Germany) were used to amplify and titer recombinant viruses and were maintained in Alpha Modification of Eagle medium containing 10% fetal bovine serum. Virus infections were performed at a multiplicity of infection of 3 PFU per cell for 1 h at 37°C. Cells were then washed twice with a Tris-buffered saline solution and overlaid with fresh medium. For the determination of virus yield, infected-cell lysates were prepared at 4, 24, 48, and 72 h postinfection, and the infectious virus yield was determined by plaque assays on N52.E6 cells. Packaging assays were performed as described elsewhere (19). Percent packaging efficiencies were calculated by dividing the relative percent packaging by the relative percent replication of two coinfecting viruses. This packaging ratio corrects for the level of replication of each input virus. The ratios are expressed as a percentage to indicate the relative packaging efficiency of a viral mutant compared to that of a coinfecting wild-type virus.
Extract preparation and gel mobility shift assays.
Nuclear extracts were prepared at 24 h after infection of N52.E6 cells, as described previously (17, 30), and were stored at −80°C. Two to five micrograms of nuclear extract was incubated with 1 μg of poly(dI-dC) and 20,000 to 50,000 cpm of 32P-labeled probe DNA (5 to 10 fmol of DNA) per in vitro binding reaction. The binding reaction was carried out in a total volume of 15 μl for 15 min at room temperature in 10 mM HEPES (pH 7.5)-20 mM KCl-3 mM MgCl2-10 mM EDTA-12% glycerol-1 mM dithiothreitol-300 μg of bovine serum albumin per ml. The complexes were resolved electrophoretically at 10 V/cm on a 4% 30:1 (acrylamide/bisacrylamide) polyacrylamide gel in 0.5× TBE (25 mM Tris [pH 8.3], 25 mM boric acid, 0.5 mM EDTA) at 4°C. For gel mobility supershift experiments, the anti-IVa2 monoclonal antibody 1A1 (16) was added 1/2 h before or 1/2 h after the addition of probe. Oligonucleotide probes were generated by 32P labeling using the Klenow fragment of DNA polymerase and [α-32P]dATP or [α-32P]dCTP. Aliquots of radiolabeled DNA fragments were subjected to trichloroacetic acid precipitation to measure the specific activity of each probe. The sequences of the top strands of the probes used are shown in Fig. 3A.
FIG. 3.
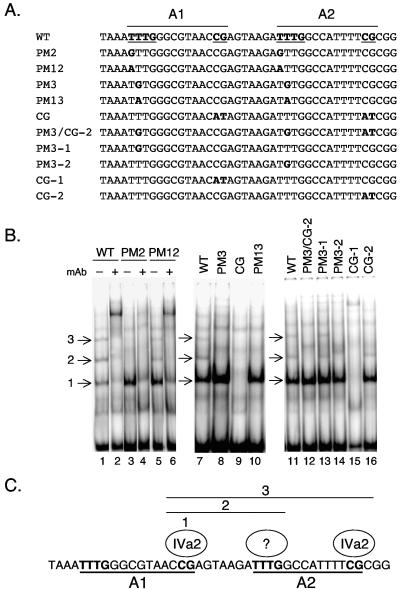
Binding of IVa2 to wild-type (WT) and mutant minimal packaging domains. (A) The top line shows the sequence of the wild-type A1-A2 region; packaging consensus sequences are boldfaced and underlined. Point mutations introduced into A-repeat 1 or 2 are shown below, with the mutated nucleotides boldfaced. (B) Gel mobility shift assays were performed using Ad-infected cell nuclear extracts and a dimeric A1+2-WT (lanes 1, 2, 7, and 11), PM2 (lanes 3 and 4), PM12 (lanes 5 and 6), PM3 (lane 8), PM-CG (lane 9), PM13 (lane 10), PM3/CG-2 (lane 12), PM3-1 (lane 13), PM3-2 (lane 14), PM-CG-1 (lane 15), or PM-CG-2 (lane 16) probe. Several binding activities specific for IVa2 are evident with the wild-type probe and are indicated by arrows (lane 1, complexes 1, 2, and 3). An anti-IVa2 monoclonal antibody (mAb) was added to the binding reaction mixtures in lanes 2, 4, and 6. (C) Schematic depiction of the basis for the formation of IVa2 complexes 1, 2, and 3 on A-repeats 1 and 2. IVa2 complex 1 forms over the CG motif of A-repeat 1. Complex 2 additionally contains an infection-specific protein(s) bound to the TTTG motif of A-repeat 2, and complex 3 likely contains an additional IVa2 protein bound to the CG motif of A-repeat 2.
Protein fractionation.
Uninfected HeLa cell nuclear extracts were prepared as described elsewhere (26), and 50 mg of total protein was subjected to heparin agarose chromatography with a bed volume of 1 ml/10 mg of loaded protein. The nuclear extract was added to buffer DB-100 (26) and washed with 5 bed volumes of loading buffer. Bound proteins were eluted by using a 0.1 to 1.0 M linear NaCl gradient in DB. Binding activities were identified by gel shift analysis using packaging sequences as a probe. Generally, 2 μl of each heparin agarose column fraction was used for gel mobility shift assays with CDP binding conditions as described previously (5).
Antibodies to Ad5 IVa2 and L1 52/55K proteins.
Full-length Ad5 IVa2 and L1 52/55K were expressed as glutathione S-transferase fusion proteins and were purified by glutathione-agarose chromatography by following conventional procedures. Affinity-purified fusion proteins were resolved on preparative sodium dodecyl sulfate (SDS)-polyacrylamide gels, extracted, and used for immunization of rabbits at Cocalico Biologicals (Reamstown, Pa.) according to procedures approved by the institutional animal care and use committee. Antibodies were developed in two separate animals for each antigen.
CHIP.
Chromatin immunoprecipitation (CHIP) experiments followed the overall approach described by Schepers et al. (23). N52.E6 cells were infected at 100 particles/cell by using the viruses described below. At 18 h postinfection, infected cells were processed for CHIP experiments. All buffers contained a cocktail of protease inhibitors. The serum-containing medium was removed from the cell monolayer, and serum-free medium was added. Formaldehyde was added directly to the culture medium to a final concentration of 1%, and the cells were incubated for 10 min at 37°C. Cross-linking reactions were terminated by the addition of glycine to a final concentration of 125 mM, and the cultures were incubated 5 min at room temperature. Cell monolayers were washed twice with an ice-cold phosphate-buffered saline solution, and the cells were detached by scraping into phosphate-buffered saline and then pelleted at 1,000 × g for 4 min at 4°C. The cell pellet was resuspended in SDS lysis buffer (50 mM Tris-HCl [pH 8.0], 10 mM EDTA, 1% SDS) and incubated on ice for 10 min. Chromatin was sheared to an average length of ∼500 bp by sonication using a Branson Sonifier 450 with three sets of 20- to 30-s pulses (by setting the output control at 5 and the duty cycle at constant) on ice. Samples were clarified by centrifugation at 25,000 × g and stored at −80°C. Chromatin preparations were normalized by quantitative real-time PCR (Q-PCR), with appropriate primer pairs for the viruses under study, using an aliquot of chromatin where the cross-links were reversed and the DNAs were deproteinized by proteinase K digestion at 65°C overnight, followed by phenol-chloroform extraction and purification using a MinElute PCR purification kit (QIAGEN).
Chromatin for immunoprecipitation was precleared in immunoprecipitation dilution buffer (16.7 mM Tris-HCl [pH 8.0], 167 mM NaCl, 1.2 mM EDTA, 0.01% SDS, and 1.1% Triton X-100) using protein A agarose (Upstate Biotechnology) for 1 h at 4°C with rotation. Samples were clarified and used for immunoprecipitation with excess polyclonal rabbit antibody (determined empirically) to either the IVa2 or the L1 52/55K protein. Samples were incubated overnight at 4°C with rotation. Protein A agarose was added, and samples were incubated at 4°C for 1 h with rotation. Protein A beads were washed five times with radioimmunoprecipitation assay buffer (50 mM Tris-HCl [pH 8.0], 750 mM NaCl, 5 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholic acid, 0.1% SDS), twice with LiCl wash buffer (10 mM Tris-HCl [pH 8.0], 0.25 M LiCl, 1 mM EDTA, 0.5% Triton X-100, 1% SDS, 1% sodium deoxycholic acid), and twice with TE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA). Protein A beads were resuspended in 100 μl of TE buffer, 150 μl of freshly prepared elution buffer (50 mM NaHCO3, 1% SDS) was added, and the samples were incubated for 15 min at room temperature with rotation. Samples were precipitated, supernatants were collected, and two additional elution steps were performed. Formaldehyde cross-linking was reversed by the addition of proteinase K to 500 μg/ml and incubation at 65°C overnight, followed by phenol-chloroform extraction and purification using a MinElute PCR purification kit (QIAGEN). Q-PCR was performed using appropriate primer pairs (indicated below) with a LightCycler-Fast Start DNA Master SYBR Green I kit (Roche) according to the manufacturer's instructions. Temperatures for primer annealing and MgCl2 concentrations were optimized experimentally.
RESULTS
Functional analysis of viruses with wild-type and mutant minimal packaging domains.
Four proteins that bind to adenovirus packaging sequences have been identified: cellular proteins CDP, OCT-1, and COUP-TF (5, 26) and the viral protein IVa2 (30). All of these proteins regulate transcriptional processes, and IVa2 additionally plays a role in Ad assembly. The packaging sequences of Ad overlap the transcriptional enhancer elements for the viral early genes. Our goal has been to probe the potential function of each of these proteins in Ad DNA packaging distinct from any involvement in early gene expression. We have approached this investigation in two ways. First, we take advantage of helper cell lines that complement the Ad E1 genes to avoid complications of mutations that reduce virus yield due to effects on E1 transcription, as opposed to effects on viral DNA packaging. Second, we assess packaging directly by performing coinfection experiments with wild-type and viable mutant viruses to directly measure viral DNA packaging efficiencies. Collectively, these assays allow us to distinguish the roles of different proteins in transcription versus packaging functions.
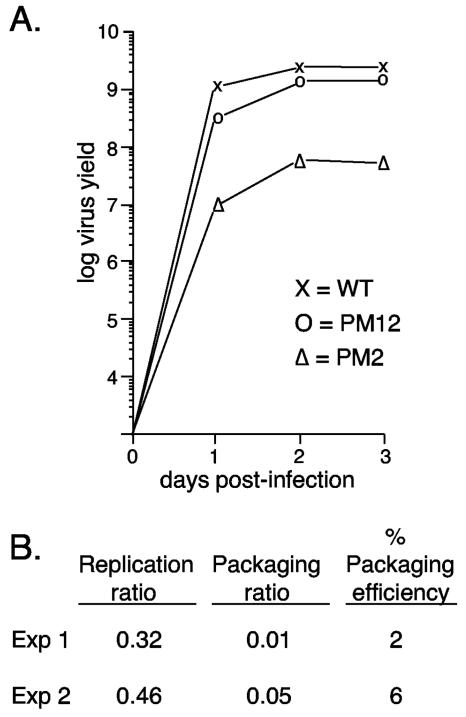
In order to assess the contribution of each observed binding activity to viral DNA packaging in vivo, we constructed and analyzed viruses where the authentic Ad5 packaging domain was replaced by two copies of a synthetic oligonucleotide containing either wild-type A-repeats 1 and 2 or mutated versions of A-repeats 1 and 2. These repeats were chosen for the analysis because a virus containing this minimal packaging domain grows as well as a virus with the authentic packaging domain (see below). Furthermore, all three cellular activities as well as the viral IVa2 protein were found to bind these minimal sites in vitro (5, 26, 30). The wild-type and mutant sites are diagramed in Fig. 1B. Site-specific point mutations were introduced individually into the first two T residues of the packaging consensus sequence (T→G for PM2 and -3; T→A for PM12 and -13), or the CG consensus motif was mutated to AT. All mutations were engineered in both A-repeats 1 and 2 and into both copies of the dimeric inserts in recombinant viruses. Mutant viruses PM13 and CG were not viable, as demonstrated by the inability to recover viral plaques after repeated attempts to propagate the mutants. The wild-type parent and mutants PM2, PM3, and PM12 all gave rise to viable viruses. The growth properties of these viruses were measured by using one-step growth curves in the E1-complementing cell line N52.E6 (Fig. 2). Insertion of the A1/2-WT dimer in place of the authentic packaging domain gave rise to a virus that grew as well as wild-type Ad5. Mutant virus PM12 grew nearly as well as A1/2-WT, whereas mutant PM2 showed ∼50-fold-reduced growth (Fig. 2) and mutant PM3 showed ∼1,000-fold-reduced growth potential (data not shown). Results of the packaging assays comparing the efficiency of packaging of the PM2 virus with that of a coinfecting wild-type virus show that the difference in growth is consistent with a packaging defect (Fig. 2B). PM3 could not be assayed for packaging by using this assay because of the low yield of virus. The ability of a dimeric copy of A-repeats 1 and 2 to substitute for the authentic packaging domain, as well as the requirement of the CG sequence for efficient packaging in vivo, is consistent with our previous analysis (25). The variable growth properties of viruses with mutations in the first 2 nt of the TTTG motif of the packaging consensus formed the basis for subsequent DNA binding studies to assess the functional relevance of cellular and viral proteins that bind to these sequences in vitro.
FIG. 2.
Single-step growth curve analysis of mutant viruses. (A) N52.E6 cells were infected with the different viruses indicated and subsequently washed. Total cellular lysates were prepared at 4, 24, 48, and 72 h postinfection. The yield of infectious virus was measured by plaque assays in N52.E6 cells. WT, wild type. (B) N52.E6 cells were coinfected with PM2 and wild-type Ad5, and packaging efficiency was determined. The replication ratio is the level of PM2 viral DNA divided by the level of total viral DNA in coinfected N52.E6 cells. The packaging ratio is the level of packaged PM2 DNA divided by the level of total viral DNA packaged. “% Packaging efficiency” is the percentage of PM2 packaged compared to that of the coinfecting wild-type virus, corrected for replication. Results of two independent packaging assays of PM2 are shown. Exp, experiment.
The Ad5 IVa2 protein forms multiple complexes with the packaging repeats in vitro that correlate with functional activity in vivo.
Zhang and Imperiale previously described the binding of the viral IVa2 protein to oligonucleotide probes that contain either A-repeats 1 and 2 or A-repeats 4 and 5 (30). Two IVa2-specific DNA-protein complexes were described in this analysis. The synthetic wild-type and mutant sites diagramed in Fig. 1B were used as radiolabeled probes in gel mobility shift assays with nuclear extracts prepared 24 h after infection of N52.E6 cells with wild-type Ad5 (Fig. 3B). The probes corresponded to a monomeric copy of the A1-A2 synthetic packaging sequence. With the wild-type A1-A2 probe, three complexes that contained the IVa2 protein, as measured by their ability to be supershifted by use of a monoclonal antibody directed against the IVa2 protein (Fig. 3B, lanes 1 and 2), were found. Only complex 1 was detected by using PM2 as a probe (Fig. 3B, lanes 3 and 4), whereas all three complexes were evident by using a probe carrying the PM12 mutations (lanes 5 and 6), although complexes 2 and 3 were slightly reduced compared to their levels with the wild-type probe. The same analysis was carried out using the other three mutants (PM3, CG, and PM13). Mutants PM3 and PM13 formed only IVa2 complex 1, whereas mutant CG was unable to form any IVa2-specific complex (Fig. 3B, lanes 8 to 10). Comparing these results to the growth properties of the corresponding viruses, we conclude from these analyses that viruses that grow to wild-type or nearly wild-type levels in vivo (A1/2-WT and A1/2-PM12) form all three IVa2-specific complexes in vitro. Mutant viruses that show significantly (PM2 and -3) or completely (PM13 and CG) defective growth either form only IVa2 complex 1 or form no IVa2 complexes at all. Thus, we correlate the formation of IVa2 complexes 2 and 3 in vitro with packaging activity in vivo.
In order to more accurately determine the basis for the formation of IVa2 complexes 1, 2, and 3, we performed mobility shift assays using probes that contained single, site-specific point mutations within the TTTG (PM3) or CG motifs of either A-repeat 1 or A-repeat 2. The location of each of these mutations is shown in Fig. 3A, and a representative mobility shift assay using a subset of these mutant sites is shown in Fig. 3B (lanes 12 to 16). Formation of IVa2 complex 1 required the CG motif of A-repeat 1; elimination of this site also eliminated the formation of IVa2 complexes 2 and 3. Formation of IVa2 complex 2 required the TTTG motif of A-repeat 2, while formation of IVa2 complex 3 required the CG motif of A-repeat 2. These results are summarized in Fig. 3C. We conclude that the CG motif of A-repeat 1 nucleates the formation of all three IVa2 complexes formed on A-repeats 1 and 2. The formation of complex 2 takes place by the binding of an additional infection-specific protein of unknown origin or perhaps IVa2, while the formation of complex 3 likely includes an additional IVa2 component at the CG motif of A-repeat 2. The complexes form in a sequential manner, starting with the binding of IVa2 to the CG motif in A-repeat 1 and then proceeding with additional binding to the TTTG motif of A-repeat 2 and subsequent binding to the CG motif of A-repeat 2. However, experiments using titrations of nuclear extracts in the in vitro binding reactions did not suggest that the different IVa2-containing complexes formed in a cooperative manner on these adjacent binding sites (data not shown). Supershift experiments using two polyclonal antibodies and a collection of monoclonal antibodies raised to L1 52/55K (11) yielded negative results (data not shown). This finding suggests that L152/55K is not part of the complexes observed in vitro and is consistent with the previous observation of Zhang and Imperiale (30).
Binding of the IVa2 and L1 52/55K proteins to the Ad5 packaging domain in vivo.
In order to assess the binding of the viral IVa2 and L1 52/55K proteins to the Ad5 packaging domain in vivo, CHIP experiments using specific antisera directed against these products were performed. Viral chromatin was sheared by sonication to an average size of ∼500 bp, and the products of immunoprecipitation were quantified by using Q-PCR. This assay permits accurate quantification over a wide range of DNA concentrations. In the first set of experiments, chromatin was prepared 18 h after infection of N52.E6 cells with Ad5 mutant virus ts147 at the restrictive temperature of 39.5°C. At the nonpermissive temperature of 39.5°C, Ad5 ts147 shows a 5-log-unit reduction in infectious virus yield and does not make detectable virus particles, because the hexon capsid protein cannot be transported to the nucleus (15). Thus, the nuclei of cells infected with this mutant at the restrictive temperature would be expected to be a rich source of viral chromatin that is available for packaging but that has not been packaged because of the hexon defect. Specific antisera directed against the IVa2 and L1 52/55K proteins, as well as preimmune sera from the corresponding animals, were used for CHIP. Primer pairs corresponding to different intervals of the Ad5 genome were used in Q-PCR to assess the quantity and specificity of immunoprecipitated chromatin; standard curves were utilized for all primer pairs in order to quantitatively measure and compare DNA recovery with CHIP of different segments of the viral genome.
The results, shown in Table 1, are presented in three ways. First, the crossing points obtained by Q-PCR using different primer pairs are shown. Second, these data are converted to quantity of DNA recovered for each primer pair by utilizing their respective standard curves. Finally, the data are presented as fold enrichment to compare the amounts of DNA recovered by using immune versus preimmune serum. Significantly enhanced immunoprecipitation of the left end of the Ad5 genome was observed by using either the anti-IVa2 or the anti-L1 52/55K antiserum in comparison to matched preimmune serum (50- or 65-fold, respectively) when a primer pair corresponding to the left-end packaging domain was used in the CHIP analysis. Similar results were obtained using anti-IVa2 and anti-L1 52/55K antisera derived from separate animals. The specificity of the immune precipitation for the left end of Ad5 was confirmed by using primer pairs internal to the viral genome (∼nt 9000) (2.1- and 3.8-fold by using the anti-IVa2 and anti-L1 52/55K sera, respectively [Table 1]) and primer pairs corresponding to ∼nt 15,000 (data not shown). Thus, the Ad5 IVa2 and L1 52/55K proteins are bound to left-end sequences at late times after viral infection, consistent with their interaction with the packaging domain.
TABLE 1.
CHIP analysis of binding of IVa2 and L1 52/55K proteins to ts147 genomes in vivoa
| Antiserumb | Result with the indicated primer pair
|
|||||
|---|---|---|---|---|---|---|
| Crossing pointc
|
Amt of DNA recovered (ng)d
|
Fold enrichmente
|
||||
| Pack | 9000 | Pack | 9000 | Pack | 9000 | |
| Preimmune | 23.53 | 22.33 | 0.037 | 0.077 | ||
| α-IVa2 | 16.99 | 21.04 | 1.842 | 0.164 | 50 | 2.1 |
| Preimmune | 24.46 | 25.63 | 0.027 | 0.013 | ||
| α-L1 52/55K | 17.70 | 23.47 | 1.748 | 0.050 | 65 | 3.8 |
CHIP analysis used chromatin isolated from ts147-infected cells, anti-IVa2 and anti-L1 52/55K sera versus preimmune sera, and Q-PCR with primer pairs corresponding to the left-end packaging region (Pack) or ∼nt 9000 (9000).
α-IVa2, anti-IVa2, α-L1 52/55K, anti-L1 52/55K.
Results from Q-PCR analyses.
Quantification of recovery of specific DNA sequences was obtained from results of Q-PCR analyses by integration using a standard curve.
Fold increase in the amount of DNA recovered by using immune serum over that recovered with preimmune serum.
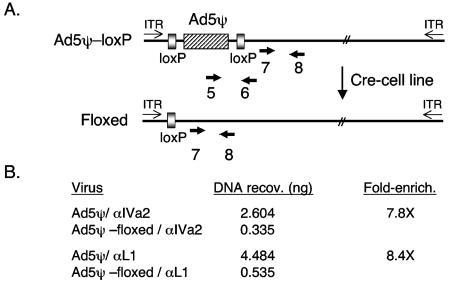
These results likely do not reflect the interaction of the IVa2 protein (and perhaps the L1 52/55K protein) with known binding sites in the MLP region for two reasons. First, the MLP is located at ∼nt 6000. Viral chromatin was sheared to ∼500-bp fragments that would physically separate the vast majority of the left-end sequences from those near the MLP. Second, the packaging-region primers used for Q-PCR are nearly 6 kbp from the MLP, whereas the nt-9000 primer pair is 3 kbp from the MLP. Yet the nt-9000 primers showed minimal binding of IVa2 and L1 52/55K in the CHIP assay. To further pinpoint where the IVa2 and L1 52/55K proteins were binding to the left end of the Ad5 genome, we used two additional assays. First, we compared the binding of the IVa2 and L1 52/55K proteins to left-end sequences by using viruses that contained or lacked a packaging domain. This was accomplished by using a virus that carries loxP sites flanking a minimal packaging domain (Fig. 4A). When this virus is grown in a cell line that expresses Cre recombinase (N52.E6-Cre), recombination between the loxP sites removes the packaging domain (termed floxing) in a large percentage of the nuclear viral genomes. The efficiency of floxing was determined by comparing the results of Q-PCR using input chromatin and primer pairs that hybridize only to unfloxed viral genomes versus those of Q-PCR using primer pairs that recognize both floxed and unfloxed genomes (Fig. 4A, primer pair 5+6 versus primer pair 7+8). The efficiency of floxing in these assays was 96 to 98%, depending on the experiment (data not shown). Using this experimental approach, we found 7.8- and 8.4-fold enrichment of left-end sequences by using IVa2- and L1 52/55K-specific antisera, respectively, with unfloxed viral genomes in comparison to floxed viral genomes (Fig. 4B). Thus, there was clear enrichment for left-end sequences in the CHIP assay when an intact packaging domain was present in the viral genomes. The difference in CHIP of floxed versus unfloxed genomes was not as great as the difference in the selectivity of ts147 genomes when immune serum was compared to preimmune serum (Table 1). This may represent the presence of additional packaging elements located outside the floxed interval (nt 185 to 1010) that are able to bind the IVa2 and L1 52/55K proteins. The contribution of sequences flanking A-repeats 1 to 7 to packaging has been described in previous reports (6, 7, 25), and we note the presence of perfect A-repeat consensus sequences between Ad5 nt 60 and 73 and between Ad5 nt 177 and 190 present in the floxed viral genomes.
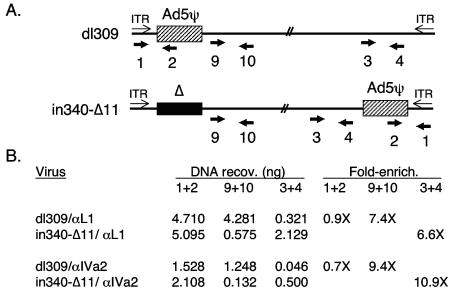
FIG. 4.
CHIP analysis of IVa2 and L1 52/55K protein binding to Ad genomes containing or lacking the packaging domain in vivo. (A) Schematic depiction of the parent virus containing a synthetic packaging domain (dimer of A-repeats 5 to 7 in place of nt 194 to 814 [26] flanked by loxP sites [designated Ad5ψ-loxP]). Infection of N52.E6-Cre cells results in the recombination (floxing) of the packaging sequences from the viral genome. Primer pair 7+8 is located outside of the floxed interval and is common to both viral genomes, while primer pair 5+6 is specific to the unfloxed genome and was used to determine the efficiency of recombination in viral DNAs present in the starting chromatin preparation. ITR, inverted terminal repeat. (B) CHIP analysis using chromatin isolated from N52.E6-Cre cells infected with Ad5-194/814+A5-72 or Ad5-194/814+A5-72+loxP (Ad5ψ-loxP), anti-IVa2 and anti-L1 52/55K sera, and Q-PCR with a primer pair (7+8) corresponding to the left-end packaging region. The concentration of DNA recovered (copies per nanogram) was determined by extrapolation of the crossing point from a standard curve for each specific primer pair. The data were evaluated and are presented as described for Table 1.
In the second approach, we took advantage of a virus, in340-Δ11, that lacks a packaging domain at the left end of the genome and, instead, carries a packaging domain at the right end of the genome (Fig. 5A). CHIP was performed using IVa2- and L1 52/55K-specific antisera, and the products were analyzed by Q-PCR using left-end and right-end primers, as well as packaging-domain primers common to both viruses (Fig. 5A, primer pairs 9+10, 3+4, and 1+2, respectively). The results of this analysis are presented in Fig. 5B and strongly support the notion that the IVa2 and L1 52/55K proteins specifically interact with the Ad5 packaging sequences. First, comparable CHIP of both proteins was observed when a primer pair that recognizes the packaging sequences contained in both viruses (primer pair 1+2) was used. In contrast, left-end sequences were enriched with the viral genome that contains a left-end packaging domain by using both IVa2- and L1 52/55K-specific antisera (primer pair 9+10; 7.4- and 9.4-fold enrichment, respectively). Similarly, right-end sequences were enriched with the viral genome that contains a right-end packaging domain (primer pair 3+4; 6.6- and 10.9-fold enrichment, respectively). Thus, two independent functional assays verify that the specificities of the interactions of the IVa2 and L1 52/55K proteins with the left end of the Ad5 genome are conferred by the packaging domain.
FIG. 5.
CHIP analysis of IVa2 and L1 52/55K protein binding to Ad genomes carrying left-end or right-end packaging domains in vivo. (A) Schematic depiction of the parent viruses that carry left-end (dl309) or right-end (in340-Δ11) packaging domains. Primer pair 1+2 is located within the packaging region and is common to both viral genomes. Primer pair 9+10 is specific to the left end of the genome outside the packaging domain, and primer pair 3+4 is common to both viruses and specific to the right end of the genome, at a location to the left of where the packaging domain is inserted into virus in340-Δ11. ITR, inverted terminal repeat. (B) CHIP analysis using chromatin isolated from N52.E6 cells infected with dl309 or in340-Δ11, anti-IVa2 and anti-L1 52/55K sera, and Q-PCR with primer pairs 1+2, 9+10, and 3+4. The data were evaluated and are presented as described for Table 1.
CDP binding is not required for Ad5 packaging.
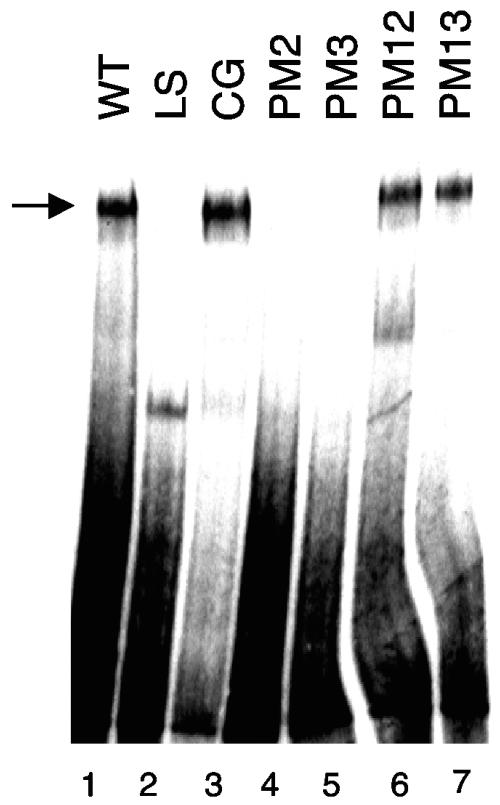
We previously described a strong correlation between the binding of CDP to Ad5 minimal packaging domains in vitro and the activity of these sites for packaging in vivo (5, 26). However, no definitive evidence for a role of CDP in Ad packaging was found. To investigate this question further, we analyzed the binding of CDP to minimal packaging domains containing point mutations in the packaging consensus sequence (Fig. 1B). CDP was partially purified from HeLa cell nuclear extracts and used in mobility shift assays (Fig. 6). CDP bound to the wild-type A1-A2 probe as expected (Fig. 6, lane 1); the identity of CDP was confirmed by using specific antisera directed against this protein (data not shown), as previously reported (5). A linker scanning mutation that disrupts the TTTG motif of the Ad5 packaging consensus sequence abolished CDP binding (Fig. 6, lane 2), whereas mutation of the CG motif did not interfere with CDP interaction (lane 3); these results are consistent with those previously reported with the same mutations in the context of A-repeats 5 and 6 (5). CDP did not bind to probes containing point mutations PM2 and PM3, whereas CDP bound efficiently to probes with the PM12 and PM13 mutations (Fig. 6, lanes 4 to 7). These results are consistent with the preference of CDP for binding AT-rich sequences (18), since the PM2 and PM3 mutations changed T residues to G, while the PM12 and PM13 mutations changed T residues to A. Since the PM13 mutation eliminated virus viability in vivo but, in contrast, did not reduce CDP binding in vitro, we conclude that binding of CDP to the Ad5 packaging domain is not required for viral DNA packaging. This conclusion is consistent with our inability to immunoprecipitate CDP with the packaging domain in CHIP experiments using four different antibodies that recognize CDP efficiently in the context of mobility shift assays (data not shown).
FIG. 6.
CDP binding to A-repeats does not correlate with packaging efficiency. Gel mobility shift assays were performed by using the CDP present in fractionated HeLa cell nuclear extracts and a dimeric A1/2-WT (lane 1), LS (lane 2), CG (lane 3), PM2 (lane 4), PM3 (lane 5), PM12 (lane 6), or PM13 (lane 7) probe. Arrow indicates CDP binding activity.
DISCUSSION
Like many Ad-encoded proteins, the viral IVa2 product appears to be multifunctional. IVa2 was originally described as a DNA binding activity that bound to two sites (DE1 and DE2) within transcribed sequences of the major late promoter (17). Binding of IVa2 to these elements stimulates late gene transcription (29). The IVa2 protein subsequently was found to interact with the L1 52/55K protein in vitro and in vivo and to bind to adenovirus packaging repeats in vitro (9, 30). That the IVa2 product is involved in virus assembly is supported by several lines of research. First, a virus that carries tandem termination codons early in the IVa2 coding region has a defect in virus particle assembly in vivo (32). Second, the IVa2 protein is physically found within immature and mature Ad virions (8). Third, the L1 52/55K protein plays an important role in virus assembly, since a temperature-sensitive mutant at the restrictive temperature and a null mutant display similar phenotypes—either a short segment of viral DNA is packaged or no viral DNA is packaged into immature virus particles, respectively (8, 12). The interaction of IVa2 with the L1 52/55K protein links these two products in one or more very early steps in virus assembly and viral DNA packaging. Finally, the IVa2 protein binds to Ad packaging repeats in vitro (30).
The results presented in this study strengthen these observations in important ways. We find that the IVa2 and L1 52/55K proteins bind to the Ad5 packaging domain in vivo (Table 1; Fig. 4 and 5). This is the first demonstration that the L1 52/55K protein interacts with Ad packaging sequences, and our results provide confirmation in vivo of IVa2 binding to Ad packaging repeats detected using in vitro assays (30). A direct correlation was observed between the packaging activities of mutant viruses in vivo and the binding of higher-order IVa2-containing complexes to Ad5 packaging repeats in vitro (Fig. 3B, complexes 2 and 3). Zhang and Imperiale (30) observed two IVa2-specific complexes in their analyses, and we found three such complexes with our A1-A2 packaging probe. Through the analysis of individual mutants with mutations in each of the packaging repeat consensus sequences (Fig. 3), we conclude that IVa2-containing complexes 1 to 3 form by the sequential addition of IVa2 to the CG motif of A-repeat 1, the subsequent addition of an infection-specific protein on the adjacent TTTG of A-repeat 2 to form complex 2, and likely the binding of an additional IVa2 protein at the CG motif of A-repeat 2 to form complex 3. We believe that IVa2 is not the additional protein utilized in the formation of complex 2. In results not shown, we found that transfection of an expression plasmid encoding full-length IVa2 into N52.E6 cells resulted in the synthesis of IVa2 protein that could generate only complex 1 by using a packaging A1-A2 probe. Complexes 2 and 3 were not observed in these assays, consistent with a role for another infection-specific protein(s). Our data and those of Zhang and Imperiale (30) clearly support the conclusion that the CG motif of the Ad5 packaging consensus nucleates the formation of a primary IVa2 complex and that additional IVa2-containing complexes form subsequent to this event. Since our data directly correlate the formation of IVa2 complexes 2 and 3 with packaging activity in vivo (Fig. 2 and 3), it will be important to understand the basis for the formation of the these complexes.
Kedinger and colleagues (17) suggested that two types of IVa2-containing complexes form on the MLP. One contains a dimer of the IVa2 protein alone, while a second complex contains IVa2 with another, unknown protein of ∼40 kDa. Recently, Pardo-Mateos and Young (21) reported that a smaller form of the IVa2 protein, with a molecular size of ∼40 kDa, is synthesized by translational initiation at methionine 75 in the IVa2 coding region. We do not believe that complex 2 on the packaging domain is formed by a heterodimer of full-length IVa2 and the 40-kDa IVa2, since identical complexes were observed in gel shift experiments with nuclear extracts prepared from cells infected with a virus (21) that does not synthesize the 40-kDa IVa2 protein (data not shown). Complexes 2 and 3 do not appear to represent IVa2 complexes that also contain the L1 52/55K protein for several reasons. First, Zhang and Imperiale (30) found efficient formation of two IVa2-containing complexes on Ad packaging repeats by using extracts prepared from cells infected with an L1-null virus. Second, we did not detect a supershift of these complexes by using the same antisera against L1 52/55K proteins that were successfully used in the CHIP experiments. In addition, we did not detect any supershift complexes by using a collection of monoclonal antibodies to L1 52/55K. Although neither we nor Zhang and Imperiale (30) observed L1 52/55K binding in vitro, we did detect an association of L1 52/55K with the packaging domain in vivo. The methods for preparing chromatin are quite different from the methods for preparing nuclear extracts for gel mobility shift assays. Significantly, formaldehyde is added to the cells in the course of chromatin preparation. This can lead to protein-protein cross-linking as well as protein-DNA cross-linking. Immunoprecipitation of chromatin by antibodies to L1 52/55K could occur because L1 52/55K interaction with IVa2 is stabilized by cross-linking where the IVa2 protein binds directly to the DNA. This protein complex may not be stable in the nuclear extracts used for mobility shift assays.
We previously described the binding of the cellular transcription factors COUP-TF, OCT-1 and CDP to minimal Ad5 packaging domains in vitro (5, 26). We concluded that COUP-TF likely was not involved in Ad packaging, since this protein bound well in vitro to packaging repeat A6, which worked poorly for packaging in vivo, and bound poorly to packaging repeat A1, which functioned efficiently for packaging. Similarly, we ruled out the involvement of OCT-1 in Ad packaging, since this protein does not bind A-repeats 5 and 6 of Ad3, yet these sites function efficiently to direct packaging in place of the natural Ad5 packaging domain. It now also appears very unlikely that CDP is involved in Ad5 packaging, since CDP bound efficiently to A-repeats 1 and 2 carrying the PM13 mutation (Fig. 6), yet a virus containing this mutation was completely defective in growth. We speculate that the binding of transcription factors to Ad packaging elements may reflect a role for these activities in the function of the E1A enhancer region—either as activators (in the case of OCT-1) or as repressors (in the cases of COUP-TF and CDP) at early or late times after infection. In conclusion, our results support a functional role for the binding of the Ad IVa2 protein to packaging sequences with the selective encapsidation of the viral genome in vivo. Further studies are required to clarify the role of other infection-specific proteins in site-specific recognition of Ad packaging repeats and to understand the earliest steps in virus assembly that depend on these DNA-protein interactions.
Acknowledgments
We thank all the members of our lab for many helpful discussions and Paul Freimuth, Michael Hayman, Janet Hearing, and Eckard Wimmer for expert advice. We thank Jackie Dudley, Claude Kedinger, Alain Nepveu, and Thomas Shenk for the generous gifts of antibodies, Hamish Young for Ad IVa2 viral mutants, and Steffan Kochanek and Gudrun Schiedner for the generous gifts of the N52.E6 and N52.E6-Cre cell lines. We acknowledge the excellent technical support provided by Vanesa Noriega in the preparation of recombinant IVa2 and L1 52/55K proteins and the screening of polyclonal antisera.
This work was supported by Public Health grant AI41636 to P.H.
REFERENCES
- 1.Daniell, E. 1976. Genome structure of incomplete particles of adenovirus. J. Virol. 19:685-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.D'Halluin, J. C., G. R. Martin, G. Torpier, and P. A. Boulanger. 1978. Adenovirus type 2 assembly analyzed by reversible cross-linking of labile intermediates. J. Virol. 26:357-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.D'Halluin, J. C., M. Milleville, G. R. Martin, and P. Boulanger. 1980. Morphogenesis of human adenovirus type 2 studied with fiber- and fiber and penton base-defective temperature-sensitive mutants. J. Virol. 33:88-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edvardsson, B., S. Ustacelebi, J. Williams, and L. Philipson. 1978. Assembly intermediates among adenovirus type 5 temperature-sensitive mutants. J. Virol. 25:641-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Erturk, E., P. Ostapchuk, S. Wells, J. Yang, K. Gregg, A. Nepveu, J. Dudley, and P. Hearing. 2003. Binding of CCAAT displacement protein CDP to adenovirus packaging sequences. J. Virol. 77:6255-6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grable, M., and P. Hearing. 1990. Adenovirus type 5 packaging domain is composed of a repeated element that is functionally redundant. J. Virol. 64:2047-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grable, M., and P. Hearing. 1992. cis and trans requirements for the selective packaging of adenovirus type 5 DNA. J. Virol. 66:723-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gustin, K. E., and M. J. Imperiale. 1998. Encapsidation of viral DNA requires the adenovirus L1 52/55-kilodalton protein. J. Virol. 72:7860-7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gustin, K. E., P. Lutz, and M. J. Imperiale. 1996. Interaction of the adenovirus L1 52/55-kilodalton protein with the IVa2 gene product during infection. J. Virol. 70:6463-6467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hammarskjold, M. L., and G. Winberg. 1980. Encapsidation of adenovirus 16 DNA is directed by a small DNA sequence at the left end of the genome. Cell 20:787-795. [DOI] [PubMed] [Google Scholar]
- 11.Hasson, T. B., D. A. Ornelles, and T. Shenk. 1992. Adenovirus L1 52- and 55-kilodalton proteins are present within assembling virions and colocalize with nuclear structures distinct from replication centers. J. Virol. 66:6133-6142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hasson, T. B., P. D. Soloway, D. A. Ornelles, W. Doerfler, and T. Shenk. 1989. Adenovirus L1 52- and 55-kilodalton proteins are required for assembly of virions. J. Virol. 63:3612-3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hearing, P., R. J. Samulski, W. L. Wishart, and T. Shenk. 1987. Identification of a repeated sequence element required for efficient encapsidation of the adenovirus type 5 chromosome. J. Virol. 61:2555-2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones, N., and T. Shenk. 1979. Isolation of adenovirus type 5 host range deletion mutants defective for transformation of rat embryo cells. Cell 17:683-689. [DOI] [PubMed] [Google Scholar]
- 15.Kauffman, R. S., and H. S. Ginsberg. 1976. Characterization of a temperature-sensitive, hexon transport mutant of type 5 adenovirus. J. Virol. 19:643-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lutz, P., and C. Kedinger. 1996. Properties of the adenovirus IVa2 gene product, an effector of late-phase-dependent activation of the major late promoter. J. Virol. 70:1396-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mondesert, G., C. Tribouley, and C. Kedinger. 1992. Identification of a novel downstream binding protein implicated in late-phase-specific activation of the adenovirus major late promoter. Nucleic Acids Res. 20:3881-3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nepveu, A. 2001. Role of the multifunctional CDP/Cut/Cux homeodomain transcription factor in regulating differentiation, cell growth and development. Gene 270:1-15. [DOI] [PubMed] [Google Scholar]
- 19.Ostapchuk, P., and P. Hearing. 2001. Pseudopackaging of adenovirus type 5 genomes into capsids containing the hexon proteins of adenovirus serotypes B, D, or E. J. Virol. 75:45-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ostapchuk, P., and P. Hearing. 2003. Regulation of adenovirus packaging. Curr. Top. Microbiol. Immunol. 272:165-185. [DOI] [PubMed] [Google Scholar]
- 21.Pardo-Mateos, A., and C. Young. 2004. A 40 kDa isoform of the type 5 adenovirus IVa2 protein is sufficient for virus viability. Virology 324:151-164. [DOI] [PubMed] [Google Scholar]
- 22.Robinson, C. C., and C. Tibbetts. 1984. Polar encapsidation of adenovirus DNA: evolutionary variants reveal dispensable sequences near the left ends of Ad3 genomes. Virology 137:276-286. [DOI] [PubMed] [Google Scholar]
- 23.Schepers, A., M. Ritzi, K. Bousset, E. Kremmer, J. Yates, J. Harwood, J. Diffley, and W. Hammerschmidt. 2001. Human origin recognition complex binds to the region of the latent origin of DNA replication of Epstein-Barr virus. EMBO J. 20:4588-4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schiedner, G., S. Hertel, and S. Kochanek. 2000. Efficient transformation of primary human amniocytes by E1 functions of Ad5: generation of new cell lines for adenoviral vector production. Hum. Gene Ther. 11:2105-2116. [DOI] [PubMed] [Google Scholar]
- 25.Schmid, S. I., and P. Hearing. 1997. Bipartite structure and functional independence of adenovirus type 5 packaging elements. J. Virol. 71:3375-3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmid, S. I., and P. Hearing. 1998. Cellular components interact with adenovirus type 5 minimal DNA packaging domains. J. Virol. 72:6339-6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stow, N. D. 1981. Cloning of a DNA fragment from the left-hand terminus of the adenovirus type 2 genome and its use in site-directed mutagenesis. J. Virol. 37:171-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tibbetts, C. 1977. Viral DNA sequences from incomplete particles of human adenovirus type 7. Cell 12:243-249. [DOI] [PubMed] [Google Scholar]
- 29.Tribouley, C., P. Lutz, A. Staub, and C. Kedinger. 1994. The product of the adenovirus intermediate gene IVa2 is a transcriptional activator of the major late promoter. J. Virol. 68:4450-4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang, W., and M. J. Imperiale. 2000. Interaction of the adenovirus IVa2 protein with viral packaging sequences. J. Virol. 74:2687-2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang, W., and M. J. Imperiale. 2003. Requirement of the adenovirus IVa2 protein for virus assembly. J. Virol. 77:3586-3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang, W., J. A. Low, J. B. Christensen, and M. J. Imperiale. 2001. Role for the adenovirus IVa2 protein in packaging of viral DNA. J. Virol. 75:10446-10454. [DOI] [PMC free article] [PubMed] [Google Scholar]