Supplemental Digital Content is available in the text.
Key Words: inflammatory bowel disease, crohn’s disease, ulcerative colitis, specific carbohydrate diet, nutrition
Abstract
Goal:
To determine the effect of the specific carbohydrate diet (SCD) on active inflammatory bowel disease (IBD).
Background:
IBD is a chronic idiopathic inflammatory intestinal disorder associated with fecal dysbiosis. Diet is a potential therapeutic option for IBD based on the hypothesis that changing the fecal dysbiosis could decrease intestinal inflammation.
Study:
Pediatric patients with mild to moderate IBD defined by pediatric Crohn’s disease activity index (PCDAI 10-45) or pediatric ulcerative colitis activity index (PUCAI 10-65) were enrolled into a prospective study of the SCD. Patients started SCD with follow-up evaluations at 2, 4, 8, and 12 weeks. PCDAI/PUCAI, laboratory studies were assessed.
Results:
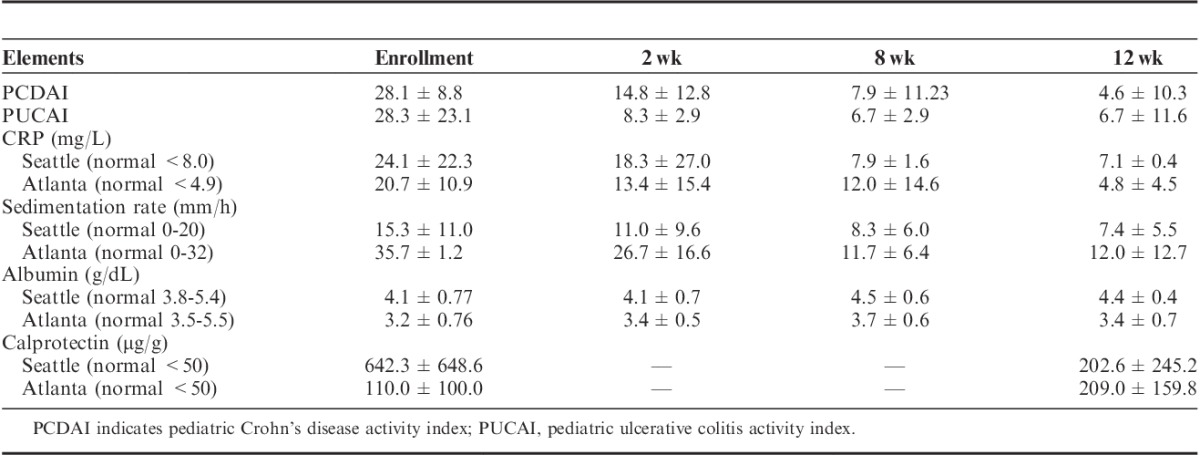
Twelve patients, ages 10 to 17 years, were enrolled. Mean PCDAI decreased from 28.1±8.8 to 4.6±10.3 at 12 weeks. Mean PUCAI decreased from 28.3±23.1 to 6.7±11.6 at 12 weeks. Dietary therapy was ineffective for 2 patients while 2 individuals were unable to maintain the diet. Mean C-reactive protein decreased from 24.1±22.3 to 7.1±0.4 mg/L at 12 weeks in Seattle Cohort (nL<8.0 mg/L) and decreased from 20.7±10.9 to 4.8±4.5 mg/L at 12 weeks in Atlanta Cohort (nL<4.9 mg/L). Stool microbiome analysis showed a distinctive dysbiosis for each individual in most prediet microbiomes with significant changes in microbial composition after dietary change.
Conclusions:
SCD therapy in IBD is associated with clinical and laboratory improvements as well as concomitant changes in the fecal microbiome. Further prospective studies are required to fully assess the safety and efficacy of dietary therapy in patients with IBD.
Inflammatory bowel disease (IBD) is thought to be the result of abnormal immune responses to the fecal microbiota and other environmental exposures in genetically susceptible individuals.1 Dysbiosis, the idea that the microbiome changes from a normal to an abnormal state which can be associated with disease, has been better defined in Crohn’s disease (CD) and ulcerative colitis (UC) patients in recent years. Some individuals have specific proteobacteria like Escherichia coli proliferation, whereas others can cluster in principle component analysis away from normal.2,3 Although an indisputable cause and effect of the fecal microbiome contributing to IBD symptoms and pathology has not been proven, many studies demonstrate strong associations, suggesting a causal contribution to disease state. The development of IBD has been closely associated with antibiotic exposure with a reported 84% relative risk increase in development of IBD in children with antibiotic use.4,5 In addition, antibiotics have been shown to be clinically efficacious in treating active IBD, and surgical diversion of the fecal stream in CD has been associated with clinical improvement in distal disease.6–9 In CD, there is also serum reactivity toward microbial antigens including antibodies to Saccharomyces cerevisiae (ASCA) and outer membrane protein C of E. coli (OmpC).10–12 Despite, the presumed role of the fecal microbiome in IBD pathogenesis, the majority of current therapies for IBD focus on suppressing the immune system and are not directed toward modulating the fecal microbiome.
Diet is known to have a significant impact on the fecal microbiome.13,14 Exclusive enteral nutrition (EEN) has been accepted internationally as first line induction therapy with equivalent efficacy to steroids with far fewer side effects and higher rates of mucosal healing.15 Other than EEN, diet as a primary or adjunct therapy for IBD is not a standard medical treatment for CD or UC. Yet, alternative and complementary therapies, which include diets, are used frequently by patients with IBD without medical direction or oversight.16 Many diets have anecdotally been reported to be efficacious but require further rigorous scientific evaluation.17 One of the more commonly used dietary therapies for IBD is the specific carbohydrate diet (SCD), developed by Dr Sydney Haas, a pediatrician, in the 1930s to treat patients with celiac disease.18 It was popularized in the late 20th century by Elaine Gottschall, whose daughter’s UC was successfully treated with SCD by Dr Haas.19 The SCD diet excludes all grains, sugars, except for honey, processed foods, and dairy, aside from specific fermented yogurt and some hard cheeses.
Given the potential for the fecal microbiome to be modulated by food exposures and therefore to contribute to IBD inflammation, we undertook a prospective open-label study examining the effects of the SCD on clinical disease activity, biological markers of inflammation, and fecal microbial composition in patients with active CD and UC.
MATERIALS AND METHODS
Study Setting and Participants
This is a multicenter, open-label study designed to determine tolerability, preliminary safety, and potential efficacy of the SCD in pediatric patients with IBD. The protocol was approved by the Institutional Review Board of Seattle Children’s Hospital and Children’s Center for Digestive Health Care. All patients/participants provided written informed consent or assent. The study was registered with ClinicalTrials.gov (number: NCT02213835). Study participants were recruited from Seattle Children’s Hospital and Children’s Center for Digestive Health Care outpatient gastroenterology clinics.
Patients with CD or UC ages 8 to 21 with mild or moderate disease activity as defined by pediatric Crohn’s disease activity index (PCDAI) score of 10 to 45 or pediatric ulcerative colitis index (PUCAI) of 10 to 60 were enrolled into this study. Before the study no change in medication(s) for IBD could occur for a minimum of 1 month for immunosuppressive medications and 2 months for biologics.
Study Intervention
Patients went on to the SCD as the sole intervention for the entire 12 weeks of the study. Patients received one-on-one education and counseling by a dietitian trained in the SCD during each visit. Before each visit patients completed a 3-day food intake record to help assure compliance with the diet. Dietary education included overview of the SCD; which foods are included and excluded. The dietitian counseled on weight loss prevention/management and provided several resources to help with meal planning, recipe books, meal, and snack recommendations. A staged approach was used introducing new SCD foods in a stepwise manner, working toward the complete SCD. Patients followed-up and were in contact with the dietitian, research assistant, and primary gastroenterologist for questions and problem intervention with the diet over the 12-week study.
Assessment of Participants
Study subject initial evaluation included history, physical examination, and laboratory tests including complete blood count with differential, C-reactive protein (CRP), erythrocyte sedimentation rate, albumin, vitamin D level, a stool study for Clostridium difficile, bacterial pathogen culture and ova and parasites was performed, as well as stool calprotectin and microbiome analysis.
Patients had clinical follow-up at 2, 4, 8, and 12 weeks. Standardized questionnaires, including the PCDAI or PUCAI were completed during each study visit.20,21 In addition, patients had a physical examination and standard blood work including complete blood count, sedimentation rate, CRP, albumin, and stool for microbiome analysis at each follow-up visit. Stool calprotectin was carried out at weeks 4 and 12. Vitamin D level was measured at baseline and again at week 12.
MICROBIOME
DNA Extraction
Total genomic DNA was extracted from stool using the PowerSoil DNA Isolation Kit (MoBio, Carlsbad, CA). The protocol was customized to include 2 incubation steps (65 and 95°C for 10 min each) after the addition of lysis buffer (solution C1). In addition, the provided garnet beads for mechanical disruption were substituted for 0.5 g of 0.1 mm diameter Zirconia/Silica beads (BioSpec Products, Bartlesville, OK). Final DNA yield was quantified using the Qubit 2.0 Fluorometer (Life Technologies, Carlsbad, CA) before next generation sequencing library construction.
Metagenomic Sequencing
Sequencing was performed on either the Illumina HiSeq 2000 or MiSeq platform. Sequencing libraries were constructed from genomic DNA using Illumina’s Nextera technology (Illumina Inc., San Diego, CA). Briefly, DNA preparations were simultaneously fragmented and tagged with adapter oligomers. A limited-cycle polymerase chain reaction amplified all tagged fragments and added: (1) index sequences (the dual indexing strategy uses two 8-base indices) to allow demultiplexing of sequence reads for pooled samples, and (2) sequencing primer sequences. Following polymerase chain reaction enrichment, libraries were denatured and hybridized via DNA/DNA binding of adapters to existing features on a glass flow cell compatible with the Illumina sequencers. Sequencing was performed using well-established, ultra-high throughput methods. The HiSeq 2000 produced ∼200 million pairs of 93 bp reads per lane, and we generated 25 to 30 million raw read pairs per sample using 7-8-plex pools. The single lane of the MiSeq generated 15 to 20 million raw pairs of 150 bp reads per sample.
Bioinformatics Analysis
Human DNA sequence was identified and removed using BMTagger19 (Rotmistrovsky, K. and Agarwala, R., BMTagger: Best Match Tagger for removing human reads from metagenomics datasets, 2011, unpublished) with the Hg-19 Homo sapiens reference genome. Duplicate reads were marked and removed using EstimateLibraryComplexity, part of the Picard tool package (http://picard.sourceforge.net/index.shtml). Sequence reads with ambiguous bases were trimmed from each end. Reads with Phred quality scores <6 over the first 80 (HiSeq) or 120 (MiSeq) bases of each read and reads shorter than 80 (HiSeq) or 120 (MiSeq) bases after trimming were removed. Sequences for all samples were limited to between 20 and 30 million reads to maintain comparable sample sizes. Taxonomic classification and relative species abundance of bacteria were obtained using MetaPhlAn2.22 Bacterial proportions assigned to taxonomic categories designated “unclassified” were not considered, and species with maximum abundance of 0.01% across all samples were removed to minimize sampling noise. Taxonomic proportions were rescaled to sum to unity. Multidimensional scaling of healthy control and Crohn’s patient microbiome samples from the Lewis et al14 study with bio2mds (https://CRAN.R-project.org/package=bios2mds) established a high-dimensional space into which longitudinal data from this study were projected using the method of Abdi.23 The Bray-Curtis distance between microbiome samples was calculated using vegan (https://CRAN.R-project.org/package=vegan). MetaPhlAn taxonomic profiles of 137 healthy adult gut microbiomes were obtained from participants in the NIH Human Microbiome Project (http://segatalab.cibio.unitn.it/tools/metaphlan2/). Metagenomics sequence for healthy controls and Crohn’s patients participating in a diet study were obtained from the NCBI Sequence Read Archive and analyzed using MetaPhlAn 2.14
Statistical Analysis
Descriptive statistics were prepared for all demographic, clinical, and laboratory measures including frequencies and percentages for categorical variables (eg, gender, receipt of medications) and means, SDs, and ranges for quantitative variables (eg, PCDAI, PUCAI). The comparison between proportions of elevated CRP values at baseline and 12 weeks was conducted using the McNemar test for paired binary data. Linear mixed-effects models, extensions of paired t tests that allow for inclusion of >2 repeated measures as well as additional factors and covariates, were used to examine changes from baseline in laboratory values. These models included site, time, and the interaction between site and time. All hypothesis testing is 2-sided, and P <0.05 are considered statistically significant. Analyses were conducted using SAS version 9.4 (SAS Institute Inc., Cary, NC).
RESULTS
Twelve individuals (9 from Seattle Children’s and 3 from Children’s Center for Digestive Health Care, Atlanta, Georgia) enrolled in the study. Mean age of participants was 12.8±2.2 years (range, 10 to 17 y). Six were male. Average disease duration before study was 1.3±1.6 years (range, 0 to 5 y). For patients with CD, macroscopic disease at time of diagnosis per Paris classification was ileocolonic (L3) in 4 patients, ileal (L1) in 4 patients, and colonic (L2) in 1 patient. For patients with UC disease was extensive in 1 patient and pancolitis was present in 2 patients. At the time of entrance into the study, patients were on methotrexate (n=1), azathioprine (n=2), mesalamine (n=4), adalimumab (n=1), ustekinumab (n=1), and no medication (n=5)24 (Table 1).
TABLE 1.
Patient Demographics and Clinical Characteristics of Patients Initiating the Specific Carbohydrate Diet

Clinical Results
No adverse events were reported. Two patients stopped the study, 1 from Seattle and 1 from Atlanta, at 2 and 8 weeks, respectively, because of difficulty maintaining the diet.
Two weeks after initiation of the SCD, 5 of the 12 patients were in clinical remission based upon PCDAI/PUCAI scoring, defined for both PCDAI/PUCAI as <10. At 8 weeks, 8 of the remaining 11 patients achieved clinical remission. At 12 weeks, 8 of 10 patients remained in remission (Table 2). Mean PCDAI at baseline was 28.1±8.8; at 2 weeks after initiation of the diet, 14.8±12.8; and at 12 weeks postinitiation of diet, 4.6±10.3. Mean PUCAI at baseline was 28.3±23.1; at 2 weeks after initiation of the diet, 8.3±2.9; and at 12 weeks postinitiation of the diet, 6.7±11.6. Dietary therapy was felt to be ineffective for 2 patients who were able to maintain the diet for the full 12 weeks (patients 9 and 11), whereas 2 individuals were unable to maintain the diet (patients 2 and 12).
TABLE 2.
Mean Clinical Disease Activity Index and Mean Laboratory Measures for Patients With Inflammatory Bowel Disease on the Specific Carbohydrate Diet (Mean±SD)
Anthropometric measurements of weight and body mass index (BMI) over the 3 month trial varied significantly for patients within the study. Although the 7/10 individuals completing the study had positive weight gain, 3 individuals had weight loss. The mean weight z-scores changed from baseline of −0.40±0.60 to −0.26±0.75. The mean BMI z-scores changed from −0.46±0.64 to −0.33±0.67.
Laboratory Results
All but 1 patient had improvement/normalization in their CRP at the week 2 follow-up. Of those individuals who were able to maintain the diet for the full study, baseline CRP was elevated in 7 of 10 patients, and by 12 weeks only 2 individuals had continued elevation of CRP (P=0.025) (Table 3). The mean CRP levels decreased from 24.1±22.3 mg/L at baseline to 18.3±27.0 mg/L at the 2-week visit in the Seattle Cohort (normal CRP<8.0 mg/L) and decreased from 20.7±10.9 mg/L at baseline to 13.4±15.4 mg/L at the 2-week visit in the Atlanta Cohort (normal CRP<4.9 mg/L). At 8 weeks and 12 weeks the mean CRP still remained below baseline level at 7.9±1.6 and 7.1±0.4 mg/L, in the Seattle cohort and 12.0±14.6 and 4.8 ±4.5 mg/L, respectively in the Atlanta cohort.
TABLE 3.
Percentage of Patients Able to Maintain the Specific Carbohydrate Diet With Active Disease Based Upon PCDAI/PUCAI and Abnormal Laboratory Studies
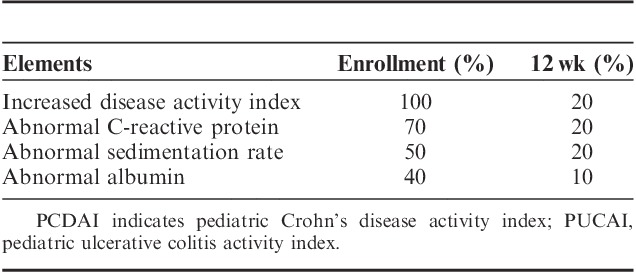
With respect to sedimentation rate, of those individuals who were able to maintain the diet for the full study, baseline sedimentation rate was elevated in 5 of 10 patients, and by 12 weeks only 2 individuals had continued elevation of sedimentation rate (Table 3). The mean sedimentation rate decreased from 15.3±11.0 mm/h at baseline to 11.0±9.6 mm/h at the 2-week visit in the Seattle cohort (normal sedimentation rate, 0 to 20 mm/h) and decreased from 35.7±1.2 mm/h at baseline to 26.7±16.6 mm/h at the 2-week visit in the Atlanta cohort (normal sedimentation rate, 0 to 32 mm/h). At 8 and 12 weeks the mean sedimentation rate still remained below baseline level at 8.3±6.0 and 7.4±5.5 mm/h in the Seattle Cohort and 11.7±6.4 and 12.0±12.7 mm/h, respectively, in the Atlanta Cohort. Although there were decreases of sedimentation rate in both the Seattle and Atlanta groups, only Atlanta changes in sedimentation rate are statistically significant (P=0.047 at 2 wk; P<0.001 at 8 and 12 wk).
Serum albumin improved or was maintained for all except 1 patient. Three of 4 patients who presented with hypoalbuminemia had normal of albumin levels by the end of the study (Table 3). The mean albumin levels increased from 4.1±0.77 g/dL at baseline to 4.4±0.44 g/dL at the 12-week visit in the Seattle cohort (normal albumin, 3.8 to 5.4 g/dL) and increased from 3.2±0.76 g/dL at baseline to 3.4±0.71 g/dL at the 12-week visit in the Atlanta Cohort (normal albumin, 3.5 to 5.5 g/dL) (Table 2). The mean calprotectin levels decreased from 642.3±648.6 μg/g at baseline to 304.9±436.3 μg/g at the 4 weeks and 202.6±245.2 μg/g at 12-week visit in the Seattle cohort (normal, <50 μg/g), whereas mean calprotectin levels went from 110.0±100.0 μg/g at baseline to 121.0±106.1 μg/g at 4 weeks and 209.0±159.8 μg/g at 12-week visit in the Atlanta cohort (normal calprotectin, <50 μg/g) (Table 2). Four individuals had increased calprotectin at 12 weeks as compared with their 4-week level. The increase in mean calprotectin in the Atlanta group was largely driven by a single patient whose baseline calprotectin rose over 300 points from baseline. One individual confirmed eating non-SCD foods for 2 weeks before calprotectin. The other individual who had clinically performed quite well and maintained on the diet after the study had a normal repeat calprotectin 1 month after the study. The changes in calprotectin for both the Seattle and Atlanta cohorts were not clinically significant (P=0.10, 0.67, respectively).
MICROBIOME
Metagenomic sequencing of DNA extracted from stool for 9 of the 12 patients identified 201 bacterial species in 28 longitudinal samples (Table S1A, Supplemental Digital Content 1, http://links.lww.com/JCG/A297). Species that increased, or decreased, in relative abundance from baseline by >0.5% at the 2-week follow-up are tabulated by patient in Table S1B, Supplemental Digital Content 2, http://links.lww.com/JCG/A297. Shannon diversity is shown in Table S1C, Supplemental Digital Content 3, http://links.lww.com/JCG/A297. Diversity increased from baseline at the 2-week timepoint for 4 patients, and did not change or slightly decreased in 4 others (Fig. 1). Only for patient 11 was a large decrease observed at 2 weeks, mostly due to an expansion of Faecalibacterium prausnitzii from 9.6% to 54%, but by 12 weeks diversity had increased above the baseline value. Median diversity for the 9 patients showed only a moderate increase from 2.48 to 2.65.
FIGURE 1.
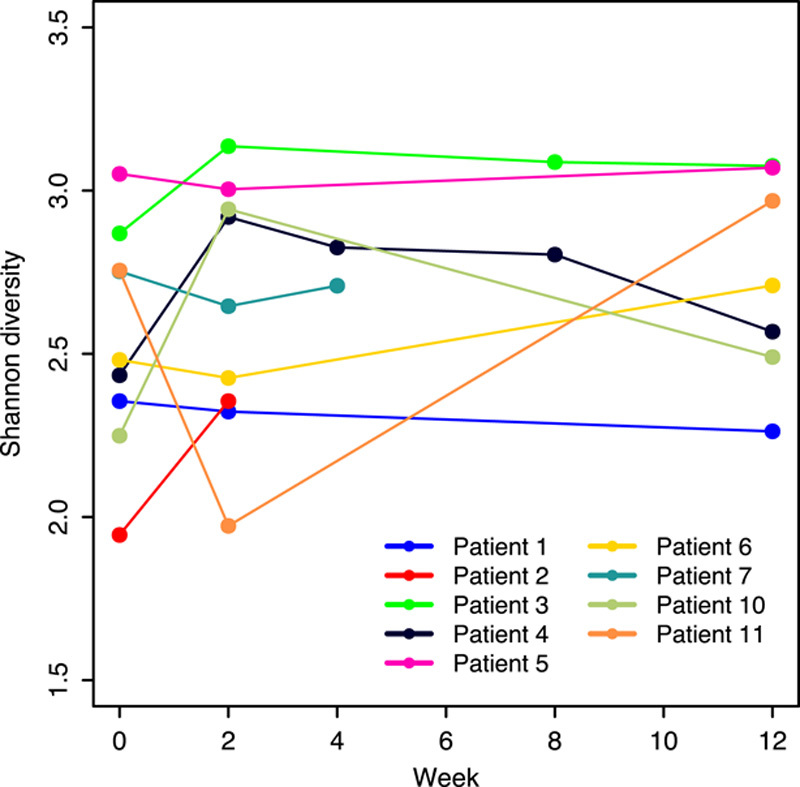
Within-subject (alpha) diversity of the gut microbiome. Shannon diversity derived from MetaPhlAn species abundance is shown for all patients. Missing timepoints reflect incomplete sequencing of samples and patients who did not complete the study.
Change in bacterial genera (>0.5%) at the 2-week timepoint are shown in Table S1D, Supplemental Digital Content 4, http://links.lww.com/JCG/A297. Dysbiosis in proteobacteria, defined as clonal expansion far above the distribution of abundances in a reference healthy population, was determined in this study by identifying taxa that exceeded the 95th quantile of 137 gut microbiomes of participants in the Human Microbiome Project.23 Genera at baseline and week 2 were compared with the 95th quantile for the most abundant genera (Fig. 2). Elevated abundance at baseline of Escherichia in patient 3 and Haemophilus in patient 2 was reduced to levels near or below Q95 at the 2-week timepoint. Klebsiella was also high in patient 2 but only marginally decreased, and Citrobacter increased. High levels of Clostridium and Streptococcus were reduced, but not below Q95. Median relative abundance of genera and proportions is shown in Table S1E, Supplemental Digital Content 5, http://links.lww.com/JCG/A297. Bacteroides and Parabacteroides had the largest decrease in median abundance and Eubacterium, Ruminoccocus, and Subdoligranulum had the largest increase (Table S1E, Supplemental Digital Content 6, http://links.lww.com/JCG/A297).
FIGURE 2.
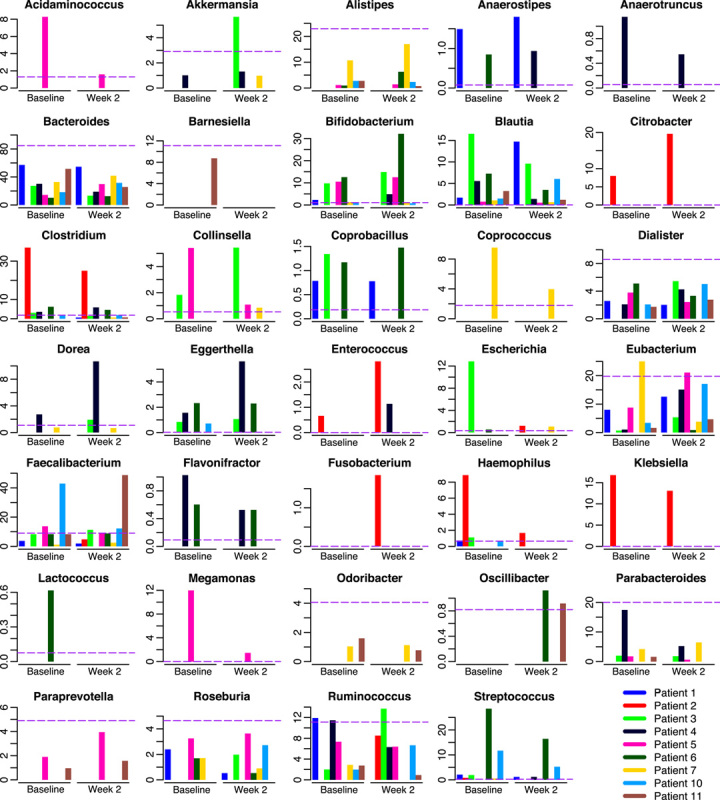
Correction of dysbiosis was observed in some patients at 2 weeks. Percent relative abundance of genera estimated by MetaPhlAn is shown for the baseline and 2-week timepoints. The 95th quantile of healthy adult gut microbiomes in the Human Microbiome Project (see Materials and methods section) is indicated by the horizontal dashed line.
Changes in phylum abundance are shown in Table S1F, Supplemental Digital Content 7, http://links.lww.com/JCG/A297. Proteobacteria decreased in all patients with the exception of patient 2 whose proteobacterial abundance was unusually high at 34% and patient 7 who had the lowest value at baseline of 0.3%. Bacteroides and Firmicutes inverted in abundance from 67% and 31% at baseline to 30 and 70% at 2 weeks, respectively, for Patient 11 resulting in the highest abundance of Firmicutes among all patients. There was no net change in phyla over all patients as a group.
DISCUSSION
The published experience of food-based dietary therapy for active CD and UC is limited. To date only a few case reports exist in which a whole food diet has been used as a potential treatment in IBD.25–27 The results of this prospective study show that diet in a small cohort of pediatric patients with active IBD was safe, and well tolerated. In addition, though a control group was not utilized, clinical and objective laboratory improvements were seen in the majority of patients with many patients achieving clinical remission and normalization of inflammatory markers. Although uncontrolled, the changes in the fecal microbiome suggests that diet in patients with IBD can alter the fecal microbiota with subsequent effect on clinical outcomes. Therefore, the therapeutic use of diet merits further study as a potential therapy for IBD.
The fecal microbiota of patients with CD is characterized by a decrease in commensal bacteria including members of Firmicutes and Bacteroides as well as a relative increase in proinflammatory bacteria such as Enterobacteriaceae.2,28 In addition, a decrease in butyrate producing bacteria which are important in intestinal health have been seen in patients with CD.29 Fluorescent in situ hybridization analysis has shown bacteria penetrating the mucus layer in 25% of colonic and 55% of ileal mucosal biopsies of patients with CD as compared with none in controls.30 The alterations in UC have been characterized by low phylotype diversity, depletion of commensal bacteria, with overrepresentation of Enterobacteriaceae and Enterococcus and underrepresentation of Ruminococcus and Bacteroides.3,31,32
In patients with IBD these alterations of the fecal microbiome, with unusual expansions of specific species, such as proteobacteria may be clinically detrimental. Clonal expansions of specific species with adaptation to the inflammatory milieu of the IBD intestinal tract may contribute to intestinal inflammation through invasion of an altered mucosal surface. Alteration of the fecal microbiome may also alter the inflammatory response through loss of microbiome diversity and subsequent loss of anti-inflammatory species. These ideas though plausible have not been associated with specific clinical outcomes and hence the implications of these alterations are still unknown. Other factors may also impact changes in the fecal microbiome including human genetic diversity, inflammatory burden, and use of concomitant medications. Further, diet is able to directly impact immunologic functioning as has been demonstrated by studies evaluating bile acid binding to the farnesoid X receptor.33,34 Given the genotypic diversity of patient with IBD as well as the microbial diversity, different mechanism of pathogenesis may be true for subsets of individuals with IBD, and the microbiome contribution to disease may vary. With this in mind, it will be important to understand how specific genotypes of IBD interact with the fecal microbial communities and how specific dietary therapies affect these interactions. We compared microbiomes of CD patients in this study by multidimensional scaling of baseline CD and controls from the Lewis et al14 study and baseline and 2 week samples from CD patients in this study (Fig. 3). Most of the 7 CD patients moved toward the centroid of the controls, suggesting that dietary therapy can improve dysbiosis in patients with IBD. In addition, no clear pattern of dysbiosis was observed in all patients. Instead most patients had evidence of a “unique” dysbiosis with an increase in organisms which are possibly proinflammatory in most prediet microbiomes. Significant changes in microbial composition were noted after dietary change. It is important to note that although microbial change were noted in patients stool microbiome, a clear cause and effect has not been proven, with the diet. In addition, an interesting counterpoint to the perceived effect of a specific microbiome on disease activity is seen with microbial changes after EEN. Although dysbiosis in the microbiome in patients on the SCD seem to correct, Crohn’s patients going onto EEN seem to have a worsening of their dysbiosis with a decrease of microbial diversity. Despite a variance in microbial composition both are associated with clinical and biochemical improvement.35,36 These divergent changes in the microbiome using different dietary interventions suggest that shifting the microbial communities maybe the most important factor for clinical improvement, or that the inflammatory changes found in IBD are multifactorial with microbial changes not being the sole trigger of the immune system. Another possible explanation is that the microbial changes found with dietary change are not the cause of the improvement seen with dietary intervention. Further studies are required to elucidate the functional role of the microbiome in disease activity.
FIGURE 3.
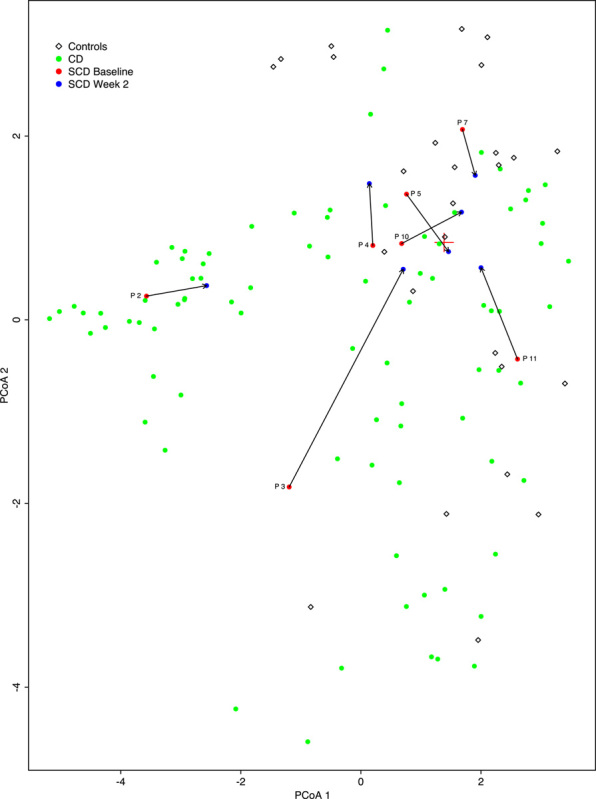
The microbiomes of Crohn’s disease (CD) patients showed progress toward controls. Multidimensional scaling (MDS) analysis of CD patients and controls in the Lewis and colleagues study was based on species proportions determined by MetaPhlAn and the Bray-Curtis distance measure. The baseline and week 2 samples from the specific carbohydrate diet (SCD) study were projected into the MDS plot (see Materials and methods section). The red “+” symbol represents the centroid of the controls. Arrows illustrate the change from baseline for 7 CD patients in our study.
Despite these limitations, accumulating evidence from animal studies shows that diet could have a significant impact on the fecal microbiome of individuals with IBD.13,14 In a murine model of mice deficient for immune genes relevant to host-microbial interactions (MyD88−/−, NOD2 −/−, ob/ob, and Rag1−/−) and >200 outbred mice, diet reproducibly altered the gut microbiota despite differences in host genotype.13 In humans, long-term dietary patterns have been shown to be associated with specific bacterial enterotypes, particularly protein and animal fats (Bacteroides) versus carbohydrates (Prevotella). Changing diet has also been shown to rapidly and reproducibly alter the fecal microbiome in humans within a day.37 Increasing animal protein intake seems to increase the abundance of bile tolerant microorganisms such as Bilophilia and Bacteroides and decrease the levels of Firmicutes which metabolize dietary plant polysaccharides.38 In addition, a western diet rich in fats and simple sugars was compared with regular chow in CEABAC10 mice, a genetically modified mouse predisposed to developing IBD. The diet was shown to induce dysbiosis with an increase in E. coli, as well as altered host barrier function favoring adherent invasive E. coli.39 In addition, common dietary emulsifiers, carboxymethylcellulose, maltodextrin, and polysorbate-80 (P80), have been shown in mouse models to promote a robust colitis in genetically predisposed mice. The emulsifiers were also noted to change the composition of the microbiota increasing the overall inflammatory potential of the fecal microbiota. In addition, these emulsifiers increased mucolytic bacteria causing erosion of the mucus layer and shortening the distance between fecal microbiota and intestinal epithelial cells by over 50%.40–42 Given the impact of diet on the fecal microbiome and the constructs of the SCD in which high fat, high sugar foods and all food additives, such as food emulsifiers and preservatives, are removed, a potential mechanism of action exists for clinical benefit from dietary interventions.
With clinical studies showing a positive impact of the SCD on symptoms and laboratory values,25,26,43,44 animal studies linking diet to the development of IBD in the setting of genetic predisposition,39,40,42 and broad patient interest in diet as therapy, the integration of diet into the treatment paradigm of IBD is likely to evolve over the coming years. Although the results of the study suggest an overall benefit for patients on the SCD with the majority of patients going into clinical remission and many showing an alteration of the microbiome, there are limitations to this study. As an open-labeled study, recruited patients and parents had a strong personal belief that SCD would improve symptoms. It cannot be excluded that participant bias could account for some of the effect seen in the PCDAI/PUCAI. The degree of mucosal healing in this study was not assessed with ileocolonoscopy, but objective evaluation of inflammation with both CRP and fecal calprotectin did support the improvements seen in clinical disease activity. Another limitation to dietary therapy is the difficulty in ascertaining compliance with the diet. Although we had study participants meet with a dietitian at each visit to review the diet, there are no objective measures of dietary compliance. Also, variations in treatment from the different study sites, interpretation of dietary protocols and local resources, could potentially impact the results of the study. Finally the small sample size for this study limits the precision of estimated effects of the SCD within our IBD patients. The study itself also highlights some of the difficulty of dietary therapy. Two individuals were unable to finish the study given difficulty maintaining the diet. In addition, although most individuals gained weight, some did not. Although there was an overall improvement in weight and BMI z-scores for patients on the SCD, assuring proper weight gain and nutritional adequacy is essential in assuring overall successful integration of diet as a primary therapy for IBD. Further study will be required to evaluate the long-term effect of the diet on growth. Despite these limitations, this study suggests a further link between the fecal microbiota and IBD and suggests that further study of diet as a potential therapeutic option using controlled clinical trials is warranted. This study also emphasizes the need for patients on dietary therapy to be followed by physicians and dieticians educated in nutritional therapy.
Supplementary Material
Supplemental Digital Content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's Website, www.jcge.com.
Footnotes
Supported by grants from the Keating Foundation, Woodward Crohn’s and Colitis Foundation and Seattle Children’s Center for Clinical and Translational Research Academic Enrichment Fund. This publication was also supported by the National Center for Advancing Translational Sciences of the National Institutes of Health under Award Number UL1TR000423. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
D.L.S. has written a patient handbook on nutrition in IBD, Nutrition in Immune Balance. The other authors declare that they have nothing to disclose.
REFERENCES
- 1.Kostic AD, Xavier RJ, Gevers D. The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology. 2014;146:1489–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frank DN, St, Amand AL, Feldman RA, et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104:13780–13785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rajilic-Stojanovic M, Shanahan F, Guarner F, et al. Phylogenetic analysis of dysbiosis in ulcerative colitis during remission. Inflamm Bowel Dis. 2013;19:481–488. [DOI] [PubMed] [Google Scholar]
- 4.Shaw SY, Blanchard JF, Bernstein CN. Association between the use of antibiotics and new diagnoses of Crohn’s disease and ulcerative colitis. Am J Gastroenterol. 2011;106:2133–2142. [DOI] [PubMed] [Google Scholar]
- 5.Virta L, Auvinen A, Helenius H, et al. Association of repeated exposure to antibiotics with the development of pediatric Crohn’s disease—a nationwide, register-based finnish case-control study. Am J Epidemiol. 2012;175:775–784. [DOI] [PubMed] [Google Scholar]
- 6.D’Haens GR, Geboes K, Peeters M, et al. Early lesions of recurrent Crohn’s disease caused by infusion of intestinal contents in excluded ileum. Gastroenterology. 1998;114:262–267. [DOI] [PubMed] [Google Scholar]
- 7.Rutgeerts P, Goboes K, Peeters M, et al. Effect of faecal stream diversion on recurrence of Crohn’s disease in the neoterminal ileum. Lancet. 1991;338:771–774. [DOI] [PubMed] [Google Scholar]
- 8.Khan KJ, Ullman TA, Ford AC, et al. Antibiotic therapy in inflammatory bowel disease: a systematic review and meta-analysis. Am J Gastroenterol. 2011;106:661–673. [DOI] [PubMed] [Google Scholar]
- 9.Turner D, Levine A, Kolho KL, et al. Combination of oral antibiotics may be effective in severe pediatric ulcerative colitis: a preliminary report. J Crohns Colitis. 2014;8:1464–1470. [DOI] [PubMed] [Google Scholar]
- 10.Zholudev A, Zurakowski D, Young W, et al. Serologic testing with ANCA, ASCA, and anti-OmpC in children and young adults with Crohn’s disease and ulcerative colitis: diagnostic value and correlation with disease phenotype. Am J Gastroenterol. 2004;99:2235–2241. [DOI] [PubMed] [Google Scholar]
- 11.Sartor RB. Therapeutic manipulation of the enteric microflora in inflammatory bowel diseases: antibiotics, probiotics, and prebiotics. Gastroenterology. 2004;126:1620–1633. [DOI] [PubMed] [Google Scholar]
- 12.Winslet MC, Andrews H, Allan RN, et al. Fecal diversion in the management of Crohn’s disease of the colon. Dis Colon Rectum. 1993;36:757–762. [DOI] [PubMed] [Google Scholar]
- 13.Carmody RN, Gerber GK, Luevano JM, Jr, et al. Diet dominates host genotype in shaping the murine gut microbiota. Cell Host Microbe. 2015;17:72–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lewis JD, Chen EZ, Baldassano RN, et al. Inflammation, antibiotics, and diet as environmental stressors of the gut microbiome in pediatric Crohn’s disease. Cell Host Microbe. 2015;18:489–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Critch J, Day AS, Otley A, et al. Use of enteral nutrition for the control of intestinal inflammation in pediatric Crohn disease. J Pediatr Gastroenterol Nutr. 2012;54:298–305. [DOI] [PubMed] [Google Scholar]
- 16.Wong AP, Clark AL, Garnett EA, et al. Use of complementary medicine in pediatric patients with inflammatory bowel disease: results from a multicenter survey. J Pediatr Gastroenterol Nutr. 2009;48:55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hou JK, Lee D, Lewis J. Diet and inflammatory bowel disease: review of patient-targeted recommendations. Clin Gastroenterol Hepatol. 2014;12:1592–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haas SV, Haas MP. The treatment of celiac disease with the specific carbohydrate diet; report on 191 additional cases. Am J Gastroenterol. 1955;23:344–360. [PubMed] [Google Scholar]
- 19.Gottschall E. Breaking the Viscious Cycle, 2nd ed Baltimore, Ontario, Canada: Kirkton Press Limited; 1994. [Google Scholar]
- 20.Hyams J, Markowitz J, Otley A, et al. Evaluation of the pediatric Crohn disease activity index: a prospective multicenter experience. J Pediatr Gastroenterol Nutr. 2005;41:416–421. [DOI] [PubMed] [Google Scholar]
- 21.Turner D, Otley AR, Mack D, et al. Development, validation, and evaluation of a pediatric ulcerative colitis activity index: a prospective multicenter study. Gastroenterology. 2007;133:423–432. [DOI] [PubMed] [Google Scholar]
- 22.Segata N, Waldron L, Ballarini A, et al. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat Methods. 2012;9:811–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adbi H.Salkind NJ. Metric multidimensional scaling. Encyclopedia of Measurement and Statistics. Thousand Oaks, CA: Sage; 2007:598–605. [Google Scholar]
- 24.Levine A, Griffiths A, Markowitz J, et al. Pediatric modification of the Montreal classification for inflammatory bowel disease: the Paris classification. Inflamm Bowel Dis. 2011;17:1314–1321. [DOI] [PubMed] [Google Scholar]
- 25.Suskind DL, Wahbeh G, Gregory N, et al. Nutritional therapy in pediatric Crohn disease: the specific carbohydrate diet. J Pediatr Gastroenterol Nutr. 2014;58:87–91. [DOI] [PubMed] [Google Scholar]
- 26.Cohen SA, Gold BD, Oliva S, et al. Clinical and mucosal improvement with specific carbohydrate diet in pediatric Crohn disease. J Pediatr Gastroenterol Nutr. 2014;59:516–521. [DOI] [PubMed] [Google Scholar]
- 27.Burgis JC, Nguyen K, Park KT, et al. Response to strict and liberalized specific carbohydrate diet in pediatric Crohn’s disease. World J Gastroenterol. 2016;22:2111–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fujimoto T, Imaeda H, Takahashi K, et al. Decreased abundance of Faecalibacterium prausnitzii in the gut microbiota of Crohn’s disease. J Gastroenterol Hepatol. 2013;28:613–619. [DOI] [PubMed] [Google Scholar]
- 29.Wang W, Chen L, Zhou R, et al. Increased proportions of bifidobacterium and the lactobacillus group and loss of butyrate-producing bacteria in inflammatory bowel disease. J Clin Microbiol. 2014;52:398–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kleessen B, Kroesen AJ, Buhr HJ, et al. Mucosal and invading bacteria in patients with inflammatory bowel disease compared with controls. Scand J Gastroenterol. 2002;37:1034–1041. [DOI] [PubMed] [Google Scholar]
- 31.Nemoto H, Kataoka K, Ishikawa H, et al. Reduced diversity and imbalance of fecal microbiota in patients with ulcerative colitis. Dig Dis Sci. 2012;57:2955–2964. [DOI] [PubMed] [Google Scholar]
- 32.Angelberger S, Reinisch W, Makristathis A, et al. Temporal bacterial community dynamics vary among ulcerative colitis patients after fecal microbiota transplantation. Am J Gastroenterol. 2013;108:1620–1630. [DOI] [PubMed] [Google Scholar]
- 33.Gadaleta RM, van Erpecum KJ, Oldenburg B, et al. Farnesoid X receptor activation inhibits inflammation and preserves the intestinal barrier in inflammatory bowel disease. Gut. 2011;60:463–472. [DOI] [PubMed] [Google Scholar]
- 34.Nolan JD, Johnston IM, Walters JR. Altered enterohepatic circulation of bile acids in Crohn’s disease and their clinical significance: a new perspective. Expert Rev Gastroenterol Hepatol. 2013;7:49–56. [DOI] [PubMed] [Google Scholar]
- 35.Gerasimidis K, Bertz M, Hanske L, et al. Decline in presumptively protective gut bacterial species and metabolites are paradoxically associated with disease improvement in pediatric Crohn’s disease during enteral nutrition. Inflamm Bowel Dis. 2014;20:861–871. [DOI] [PubMed] [Google Scholar]
- 36.Quince C, Ijaz UZ, Loman N, et al. Extensive modulation of the fecal metagenome in children with Crohn’s disease during exclusive enteral nutrition. Am J Gastroenterol. 2015;110:1718–1729. Quiz 1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu GD, Chen J, Hoffmann C, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.David LA, Maurice CF, Carmody RN, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martinez-Medina M, Denizot J, Dreux N, et al. Western diet induces dysbiosis with increased E coli in CEABAC10 mice, alters host barrier function favouring AIEC colonisation. Gut. 2014;63:116–124. [DOI] [PubMed] [Google Scholar]
- 40.Chassaing B, Koren O, Goodrich JK, et al. Dietary emulsifiers impact the mouse gut microbiota promoting colitis and metabolic syndrome. Nature. 2015;519:92–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nickerson KP, Chanin R, McDonald C. Deregulation of intestinal anti-microbial defense by the dietary additive, maltodextrin. Gut Microbes. 2015;6:78–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nickerson KP, McDonald C. Crohn’s disease-associated adherent-invasive Escherichia coli adhesion is enhanced by exposure to the ubiquitous dietary polysaccharide maltodextrin. PLoS One. 2012;7:e52132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kakodkar S, Farooqui AJ, Mikolaitis SL, et al. The specific carbohydrate diet for inflammatory bowel disease: a case series. J Acad Nutr Diet. 2015;115:1226–1232. [DOI] [PubMed] [Google Scholar]
- 44.Obih C, Wahbeh G, Lee D, et al. Specific carbohydrate diet for pediatric inflammatory bowel disease in clinical practice within an academic IBD center. Nutrition. 2016;32:418–425. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Digital Content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's Website, www.jcge.com.


