Abstract
Virus-responsive signaling pathways that induce alpha/beta interferon production and engage intracellular immune defenses influence the outcome of many viral infections. The processes that trigger these defenses and their effect upon host permissiveness for specific viral pathogens are not well understood. We show that structured hepatitis C virus (HCV) genomic RNA activates interferon regulatory factor 3 (IRF3), thereby inducing interferon in cultured cells. This response is absent in cells selected for permissiveness for HCV RNA replication. Studies including genetic complementation revealed that permissiveness is due to mutational inactivation of RIG-I, an interferon-inducible cellular DExD/H box RNA helicase. Its helicase domain binds HCV RNA and transduces the activation signal for IRF3 by its caspase recruiting domain homolog. RIG-I is thus a pathogen receptor that regulates cellular permissiveness to HCV replication and, as an interferon-responsive gene, may play a key role in interferon-based therapies for the treatment of HCV infection.
Hepatitis C virus (HCV) is a major public health problem, infecting nearly 200 million people worldwide and causing hepatic fibrosis, end-stage cirrhosis, and hepatocellular carcinoma (16). A member of the Flaviviridae, HCV's positive-sense RNA genome contains highly structured 5′ and 3′ nontranslated regions (NTRs) flanking a large open reading frame (ORF) encoding a polyprotein that is processed into both structural (core-E2) and nonstructural (NS) proteins (Fig. 1A). The NS3-NS5B proteins support viral genome replication, which is also dependent upon conserved RNA sequences within the 5′NTR and 3′NTR that are highly structured and required for both protein translation and RNA replication (15, 20). HCV infection is treated with alpha interferon (IFN-α)-based therapy, but treatment is effective at best in only 50% of patients (10). The nearly unique ability of HCV to establish persistent infections in humans has been attributed, in part, to a variety of strategies to evade host immune and IFN-induced defenses (12). Epidemiological studies suggest that 25 to 50% of all persons resolve acute HCV infection without treatment (16), however, indicating that innate and/or adaptive immune responses are indeed capable of controlling the outcome of HCV infection. Processes that regulate innate intracellular antiviral responses may therefore serve as pivotal points of control, potentially limiting host permissiveness for HCV replication and favorably modulating subsequent adaptive immune responses.
FIG. 1.
The host response and cellular permissiveness to HCV RNA replication. (A) Diagram of the HCV genome (upper) and related replicon (lower). The polyprotein coding region and positions of individual protein products are shown. Arrows indicate positions of the RNA motifs described in the present study, including the IRES within the 5′NTR. (B) Huh7 cells were mock transfected or transfected with the indicated HCV replicon RNA and harvested 3, 8, 12, or 24 h later. ISG, replicon (HCV), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcript abundance was monitored by Northern blot analysis. (C) Huh7 cells were transfected with an IFN-β promoter-luc construct and 24 h later were mock treated (mock), infected with SenV, or transfected with the indicated RNA. Cells were harvested 20 h after virus infection or 8 h after RNA transfection and then assayed for firefly luciferase activity. Bars show the average values and standard deviation (SD) of firefly luciferase activity relative to a Renilla luciferase control from three experiments (relative luc activity). (D) In the left panel is shown RNA abundance in Huh7 cells that were mock transfected (mock) or transfected with luc-poly(A), HCV 5′NTR-luc, or encephalomyocarditis virus IRES-luc RNA were assessed by Northern blot with probes specific to the indicated ISGs or GAPDH. Cells were harvested at the time shown in hours above each lane, and transfected RNA was detected with a luciferase-specific probe (Luc). For the right panel, cells were mock transfected or transfected with purified L2198S HCV replicon RNA, luc-poly(A), or HCV 5′NTR-luc transcript and then harvested 12 h later. mRNA levels were quantified by real-time PCR. Bars show the average level ± the SD of IFN-β mRNA relative to GAPDH control from three experiments. (E) HCV RNA replication and cellular G418 resistance transduction efficiency were evaluated after transfection of Δ5B or HP replicon RNAs into Huh7 or Huh7.5 cells. On the left is shown the average fold increase ± the SD of HP replicon RNA relative to Δ5B replicon RNA at the indicated times. The replicon abundance was estimated from the luciferase activity expressed by a firefly luciferase sequence that was inserted in lieu of the Neo sequence in the replicon shown in Fig. 1A. This provides an accurate assessment of HCV RNA abundance (28). On the right, G418-resistant colony-forming efficiency of the indicated HCV replicon RNAs was determined by cell staining 3 weeks after transfection of Huh7 (panels 1 to 3) or Huh7.5 cells (panels 4 to 6) followed by G418 selection. The numbers at the bottom show the fold increase in numbers of Huh7.5 cell colonies compared to Huh7 colonies. The results shown are representative of three independent experiments.
Virus-induced production of IFN-α and IFN-β and the subsequent expression of IFN-stimulated genes (ISGs) are central to these antiviral defenses (22). This host response is initiated by cellular recognition of a pathogen-associated molecular pattern (PAMP) presented by the infection, in which a host protein receptor is engaged by the PAMP ligand and signals downstream components to activate intracellular immune defenses. In mammalian cells, replicating viral RNAs present features of nucleic acid sequence or structure that are recognized as distinct PAMPs by membrane-spanning Toll-like receptors (TLRs) or intracellular proteins coupled to signaling pathways that induce interferon production. Double-stranded RNA (dsRNA) and GU-rich single-stranded RNA (ssRNA) are recognized by TLR3 and TLR7/8, respectively (18). However, viral dsRNA can also initiate cellular responses through an intracellular, TLR3-independent mechanism (5, 32), thereby activating a set of latent transcription factors, including IFN-regulatory factor 3 (IRF3) and NF-κB that coordinately assemble onto the IFN-β promoter and induce IFN expression, ISG production and an intracellular antiviral state. Among these, IRF3 is essential for IFN production (21). Its activation occurs through redundant actions of TBK1 or IKKɛ protein kinases, which catalyze its carboxyl-terminal phosphorylation and result in nuclear translocation, DNA binding, and transcription-effector actions (6, 23). The IRF3-mediated induction of IFNs and ISGs, initiated by the recognition of an HCV-specific PAMP, is likely to impose intracellular restrictions to viral replication that limit host cell permissiveness (7, 25). To define how this host response influences HCV infection, we investigated the relationship between innate antiviral responses and cellular permissiveness for HCV RNA replication in a cell culture model.
MATERIALS AND METHODS
Cell culture and viruses.
Huh7.5 cells were a gift from C. Rice. Huh7 and Huh7.5 cells (3) were propagated in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum, 200 μM l-glutamine, and Sigma antibiotic-antimycotic solution. For treatment of cells, medium was removed and replaced with medium containing the indicated concentration of IFN-α2a (PBL Laboratories), anisomycin (Sigma) or interleukin-1 (R&D Systems). Sendai virus (SenV; Cantell strain) was obtained from Charles River Laboratory. Vesicular stomatitis virus (VSV) was a gift from M. Whitt. Virus infection of Huh7 or Huh7.5 cells was conducted as previously described (7, 9). For SenV infection, cells were exposed to 100 hemagglutinin units of SenV/ml of culture medium. For VSV infection, cells were exposed to a multiplicity of infection of 5.
DNA methods.
pHCV 1bpt luc, pHCV HP luc, and pHCV Δ5B luc encode variants of the Con1 HCV 1b subgenomic RNA sequence (15) possessing the firefly luciferase gene under the control of the HCV 5′NTR/internal ribosome entry site (IRES) and were constructed by ligating a DNA fragment encoding firefly luciferase into the AscI/PmeI sites of the respective pHCV 1bpt, pHCV HP, or pHCV Δ5B constructs (7, 25). Plasmids pHCV 1b K2040 and pHCV 1b L2198S, respectively, encoding the Con1-derived K2040 and L2198S subgenomic HCV RNA replicons were previously described (25, 31). cDNA was prepared from mRNA isolated from Huh7 or Huh7.5 cells. Full-length cDNA products encoding IKKɛ, TBKI, RIP2, TRIF, or RIG-I were amplified from each by PCR with the specific primer sets shown in Table S1 in the supplemental material. Endotoxin-free plasmid DNA was prepared by using the endo-free Midiprep kit (Sigma). pCMV-IRF-3 5D was a gift from J. Hiscott. pIRF-3-EGFP was a gift from A. Garcia-Sastre. pEF Bos (encoding an amino-terminal FLAG epitope tag; vector control), pEF Bos-TBK1, and pEF Bos-IKKɛ were gifts from T. Maniatis. pEF Bos-TRIF was a gift from K. Fitzgerald. The wild-type RIG-I cDNA was cloned into the XbaI/ClaI restriction sites of pEF Bos to generate pEF Bos-RIG-I. pEF Bos-N-RIG and pEF Bos-C-RIG encode amino acids (aa) 1 to 284 and aa 218 to 925 of RIG-I (32). Full-length RIP2 was cloned into the HindIII/EcoRI restriction sites of pFLAG CMV-2 (Sigma) to yield pFlag RIP2. pEF Bos-RIG-I T55I and pEF Bos N-T55I were created from pEF-Bos-RIG-I and pEF Bos N-RIG, respectively, by a site-directed mutagenesis strategy using the QuikChange kit from Stratagene and the mutagenic primers T55Is (5′-GCCCAATGGAGGCTGCCATACTTTTTCTCAAGTTCC-3′) and T55Ia (5′-GGAACTTGAGAAAAAGTATGGCAGCCTCCATTGGGC-3′). pF3-EGFP was constructed by by ligating a HindIII/XbaI fragment from pEGFP (Clontech) into pF3-Luc (a gift from J. Hiscott). pDsRed was purchased from Clontech. pSP6 luc poly(A) was constructed by ligating a HindIII/XbaI fragment derived from pGL3-Basic (Promega) and encoding the firefly luciferase gene into pSP6 poly(A) (Promega). pCDNA3.1 luc poly(A) was then constructed by ligating a HindIII/EcoRI fragment from pSP6 luc poly(A) into pCDNA3.1 DNA transfections were performed by using Lipofectamine 2000 transfection reagent (Invitrogen).
Promoter-reporter luciferase assays were conducted as described previously (7) from 2 × 104 cells transfected with 50 ng of pIFN-β-luc, pISG56-luc or pISRE-luc encoding firefly luciferase and 12.5 ng of pCMV-luc encoding the Renilla luciferase gene (8). Luciferase activity was quantified by using a Bio-Rad luminometer.
RNA methods.
HCV replicon RNA was transcribed and purified from the respective linearized plasmids exactly as described previously (7). RNA was synthesized from plasmids by using the T7 Megascript kit (Ambion) and the manufacturer's protocol. Luciferase poly(A) RNA was transcribed from pCDNA3.1 luc poly(A). HCV 5′NTR-luc RNA was transcribed from pHCV 1bpt luc. Encephalomyocarditis virus IRES-luc RNA was transcribed from pIRES luc, which was constructed by ligating firefly luciferase into pIRES (Clontech) digested with SmaI. HCV ss1 RNA was transcribed from pCDNA3.1 HCV 1bpt NS3/4A (7) linearized with SgrAI. HCV ss2 RNA was transcribed from pCDNA3.1 HCV 1bpt NS5A (25). HCV 5′NTR and 3′NTR were transcribed directly from the PCR products T7 HCV 5′NTR and T7 HCV 3′NTR, respectively, which were amplified from pHCV 1bpt by using the primer set (encoding the T7 promoter) T7 HCV 5′NTRs (5′-GTAATACGACTCACTATAGGGCCAGCCCCCTGATGGGGGCGACA-3′) and pHCV 1bpt 520a (5′-TGCGTGCAATCCATCTTGTTC-3′) and the primer set T7 HCV 3′NTRs (5′-TAATACGACTCACTATAGGGAGATGAAGGTTGGGGTAAACACTC-3′) and HCV 3′NTRa (5′-ACATGATCTGCAGAGAGGCCA-3′), respectively. Amplified DNA was purified by agarose gel electrophoresis and extracted from the gel by using a QIAquick kit according to the manufacturer's protocol (Qiagen). RNA was purified by gel extraction, resuspended in water, and transfected by using the Transmessenger RNA transfection reagent (Qiagen) according to the manufacturer's protocol.
Northern blot analysis of RNA levels was performed exactly as described previously (7) with the indicated cDNA probes. Real-time PCR analyses were performed as described previously (25) with the oligonucleotide primer pairs listed in Table S2 in the supplemental material.
For small interfering RNA (siRNA) studies, Huh7 cells were transfected with 5 to 20 pmol of control siRNA (Ambion Silencer negative control #1) or the RIG-I si265 oligonucleotides si265s [5′-GAGGUGCAGUAUAUUCAGG(dTdT)-3′]) and si265a [5′-CCUGAAUAUACUGCACCUC(dTdT)-3′] by using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol.
Assessment of replicon RNA transduction efficiency and initial HCV RNA replication.
Transduction efficiency for HCV subgenomic RNA replicons in Huh7 and Huh7.5 cell lines was determined exactly as described previously (3). For determination of initial HCV RNA replication efficiency, 4 × 104 Huh7 or Huh7.5 cells were plated per well of a 24-well plate. After 24 h the cells were transfected with HCV replicon RNA transcribed from pHCV1b ΔNS5B luc or pHCV 1b HP luc. Then, 3 h later the cells were harvested for luciferase assay to determine relative transfection efficiencies or the cells were replated and cultured in six-well dishes to facilitate the evaluation of HCV RNA replication defined by luciferase activity. With this system the level of HCV RNA replication directly parallels luciferase activity values (28). The data were expressed as the increase in luciferase activity from cells transfected with HP luc RNA compared to cells transfected with the ΔNS5B luc RNA control and normalized for transfection efficiency derived from the initial 3-h time point.
Nucleotide sequence analysis.
Plasmids were verified by nucleotide sequence analysis (ABI Prism). For mRNA analysis cDNA was first produced from total cellular RNA isolated from Huh7 or Huh7.5 cells by using Omniscript (Qiagen) reverse transcriptase and an oligonucleotide dT primer (Ambion). A total of 10% of the reverse transcriptase reaction was used for PCR (ExTaq; Takara). Amplicons were sequenced directly, and complete mRNA sequences were assembled by using Vector NTI software (Informax).
Protein analysis.
Protein kinase assays and IRF3 dimerization analysis were performed exactly as described previously (11, 33). For evaluation of protein expression, cell extracts were prepared, and immunoblot analysis was conducted (7). Antibodies used for immunoblot analysis included anti-PKR monoclonal antibody 71/10 (A. Hovanessian), rabbit polyclonal anti-IRF3 (M. David), rabbit polyclonal anti-ISG56 (G. Sen), rabbit polyclonal anti-RIG-I (32), rabbit polyclonal anti-VSV (M. Whitt), rabbit polyclonal anti-SenV (I. Julkunen), monoclonal anti-TBK1 (Imgenex), monoclonal anti-IKKɛ (Imgenex), rabbit polyclonal anti-phospho-Ser51-eIF2α (Cell Signaling Technologies), monoclonal anti-eIF2α (Research Genetics), rabbit polyclonal anti-phospho-Thr180/Tyr182-P38 (Cell Signaling Technologies), rabbit polyclonal anti-P38 (Cell Signaling Technologies), monoclonal anti-Flag M2 (Sigma), and goat polyclonal anti-actin (Santa Cruz). Proteins were detected with a secondary antibody coupled to horseradish peroxidase and were visualized by chemiluminescence.
For immunofluorescence analysis of intracellular protein localization, 2 × 104 cells were cultured and treated on chamber slides then fixed and probed with polyclonal rabbit anti-IRF3 serum or monoclonal anti-Flag serum and fluorescein isothiocyanate-conjugated or rhodamine donkey anti-rabbit secondary antibodies exactly as described previously (7). After antibody staining, the nuclei were stained with DAPI (4′,6′-diamidino-2-phenylindole). Cells were visualized by fluorescence microscopy by using a Zeiss Axiovert digital imaging microscope in the UT Southwestern Pathogen Imaging Facility.
RNA-binding analysis of RIG-I.
For RNA-binding analysis, we used agarose beads (Sigma) conjugated to poly(C) or poly(I). The beads were washed several times in 50 mM Tris (pH 7.0)-200 mM NaCl and then resuspended in 50 mM Tris (pH 7.0)-50 mM NaCl. To make poly(I-C)-coated agarose beads, poly(I)-coated beads were resuspended in 2 volumes of 2 mg of poly(I) (Sigma)/ml prepared in 50 mM Tris (pH 7.0)-150 mM NaCl. The mixture was then rocked gently overnight at 4°C, collected by centrifugation at 1,000 × g, washed with 50 mM Tris (pH 7.0)-150 mM NaCl, resuspended in the same buffer as a 50% final slurry, and stored at 4°C for use. For poly(C) and poly(I-C) pull-down assays, poly(C)- or poly(I-C)-coated beads were equilibrated in binding buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1% NP-40) as a 10% slurry and then combined with an equal volume of whole-cell extract that was prediluted to contain 3 μg of protein. The cell extracts were supplemented with protease and phosphatase inhibitors and 25 U of RNase inhibitor/ml. The mixture was incubated with gentle agitation for 1 h at 4°C. For competition experiments, pull-down reactions were supplemented with 10 to 50 μg of the indicated competitor RNA/ml. Beads were centrifuged at 1,000 × g, rinsed three times with binding buffer, and then resuspended in 3 volumes of 1× sodium dodecyl sulfate-polyacrylamide gel electrophoresis sample buffer. Samples were incubated at 100°C for 5 min, centrifuged at 13,000 × g for 30 s, and loaded immediately onto sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels and processed for immunoblot analysis.
RESULTS
Structured HCV RNA presents a PAMP that induces antiviral responses.
Since HCV replication in cultured cells is insufficient for support of molecular virologic studies (20), we constructed a series of subgenomic HCV RNA replicons that encode different adaptive mutations and replicate autonomously, albeit with different efficiencies, when introduced into cultured human hepatoma (Huh7) cells (L2198S, HP, and K2040; Fig. 1A). We also studied a replication-defective HCV RNA (Δ5B) with a deletion in the RNA-dependent RNA polymerase. Regardless of its replication properties, each HCV replicon RNA induced expression of ISGs, including ISG6-16, ISG56, and ISG15 within 12 h of transfection into Huh7 cells (Fig. 1B). To determine whether structured regions of the HCV genome specifically present a dsRNA-like PAMP capable of stimulating this host response, we transfected cells with purified, in vitro-transcribed RNA representing the 5′NTR or 3′NTR of HCV. Cells were similarly transfected with RNAs corresponding to nucleotides 3423 to 3772 or nucleotides 6261 to 6702 of HCV, which, unlike the NTRs, do not contain well-documented RNA structures (termed ss1 and ss2, respectively; Fig. 1A) (27). Transfection of cells with the 5′NTR or 3′NTR RNAs induced IFN-β promoter activity, whereas only minimal increases were seen with the nonstructured ss1 or ss2 HCV (Fig. 1C). To confirm these results, we transfected Huh7 cells with RNA transcripts encoding firefly luciferase (luc), either with or without an upstream structured viral RNA segment. Northern blotting revealed that luciferase RNA appended with a poly(A) tail and lacking a structured upstream segment [luc-poly(A)] was a poor inducer of ISG expression (Fig. 1D, left panel). In contrast, inclusion of the HCV 5′NTR or the structured IRES of encephalomyocarditis virus resulted in ISG expression within 8 h of transfection. Luciferase activity assays confirmed efficient transfection of each RNA (data not shown), indicating that the luc sequence or its gene product were not responsible for ISG induction. The level of induction of IFN-β promoter activity by the 5′NTR-luc RNA was comparable to that induced by the entire subgenomic L2198S HCV replicon RNA 8 h after transfection (Fig. 1D, right panel). We conclude that structured regions of the HCV RNA, including the 5′NTR and 3′NTR present a specific PAMP capable of triggering a host response characterized by IFN-β and ISG expression.
Lesion(s) in the host response to the HCV RNA PAMP enhance cellular permissiveness to viral RNA replication.
Most HCV replicon RNAs containing wild-type sequence replicate poorly or not at all in Huh7 cells, and efficient replication requires specific cell culture-adaptive mutations (1). However, a subline of Huh7 cells, termed Huh7.5, exhibit a marked increase in permissiveness for HCV RNA replication (3). We compared the capacity of Huh7 and Huh7.5 cells to support replication after transfection of an HCV replicon RNA directing expression of luciferase. Replication of this RNA, as reflected in increases in luciferase activity relative to that produced by a replication-defective control RNA, was significantly greater in the Huh7.5 cells. Similarly, we observed a 74- to 128-fold increased efficiency in selection of G418-resistant cell colonies in the Huh7.5 cells after transfection of replicon RNAs expressing the selectable neo marker (Fig. 1E).
We examined dsRNA signaling pathways and the host response to virus infection and IFN treatment in these cell lines. Both demonstrated equivalent activation of protein kinase R (PKR) and p38 mitogen-activated protein after infection with VSV (Fig. S1 in the supplemental material). Thus, these dsRNA signaling pathways are intact in both cell lines. Similarly, IFN treatment induced robust expression of ISG56, a well-defined ISG and IRF3 target gene (19), in both Huh7 and Huh7.5 cells. However, unlike Huh7 cells, the Huh7.5 cells failed to induce ISG56 expression in response to SenV infection or transfection of the HCV 5′NTR-luc RNA (Fig. 2A). Moreover, infection with VSV or Sendai virus (SenV) failed to induce IFN-β promoter activity in the Huh7.5 cells but did so in Huh7 cells (Fig. 2B), despite the fact that the viruses replicated equally well in both cell lines (Fig. 2A and Fig. S1C in the supplemental material). Strikingly, transfection of the Huh7 cells with HCV RNA triggered ISG expression within 8 to 12 h, whereas this response was not observed in Huh7.5 cells despite equivalent transfection efficiency (Fig. 2C). Similar results were observed when cells were transfected with the synthetic dsRNA, poly(I-C) (data not shown). Thus, signaling pathways that confer responsiveness to virus infection or HCV RNA and that are intact in Huh7 cells are defective in Huh7.5 cells.
FIG. 2.
Cells highly permissive to HCV RNA replication have a defective host response. (A) Huh7 and Huh7.5 cells were mock treated (mock), infected with SenV for 20 h (SenV), treated with 10 or 50 U of IFN-α2a/ml for 12 h, or transfected with luc-poly(A) or 5′NTR-luc RNA (HCV) for 8 h. Extracts were subjected to immunoblot analysis for ISG56, SenV, and β-actin abundance. (B) The activity of a transfected IFN-β promoter expression constructs was measured in cells that were mock infected or infected with SenV for 24 h or VSV for 10 h. The results shown are the average firefly luciferase activity ± the SD relative to a Renilla luciferase control. (C) Huh7 and Huh7.5 cells were mock transfected or transfected with 5′NTR-luc, L2198S, or HP HCV replicon RNA. Total cellular RNA was collected at 3, 8, or 12 h, and the level of replicon (HCV), 2′-5′ oligoadenylate synthetase 1 (OAS1), ISG6-16, ISG56, ISG15, and GAPDH RNA as determined by using specific probes. 5′NTR-luc RNA was monitored by using the Luc probe. (D) Huh7 and Huh7.5 cells were mock treated (mock), infected with SenV for 24 h, or transfected with luc-poly(A) (poly A) or HCV 5′NTR-luc RNA (HCV 5′NTR) for 8 h. Cells were processed and stained with anti-IRF3 rabbit serum and a rhodamine-conjugated secondary antibody (panels 1 to 8). Nuclei were visualized by staining with DAPI (panels 9 to 16). Magnification, ×40. The transfection efficiency was monitored by a parallel assessment of luciferase activity; the unit activity values are shown below the corresponding panels. (E) In the left panel, IRF3, ISG56, and actin abundance were evaluated by immunoblot analysis of extracts from untreated Huh7 and Huh7.5 cells or cells treated with 10 U of IFNα-2a/ml for 12 h (IFN) or transfected with HCV 5′NTR-luc RNA for 0, 4, or 8 h. The line at left denotes the position of the high-mass hyperphosphorylated IRF3 isoforms. On the right, proteins were separated on a nondenaturing gel in order to define the abundance of the inactive IRF3 monomer and the active IRF3 dimer complex (indicated at right) in cells that were mock infected or infected with SenV for 20 h.
The lack of IFN-β promoter activity and ISG56 expression in Huh7.5 cells could reflect a lesion in the IRF3 activation pathway. We therefore compared the subcellular distribution of endogenous IRF3 in these cells in response to virus infection or HCV RNA transfection. In resting Huh7 cells, IRF3 is distributed in a perinuclear and/or cytoplasmic context; it redistributes to the nucleus, a finding consistent with its activated state, after infection with SenV (Fig. 2D) (7). A similar redistribution of IRF3 occurs in Huh7 cells upon transfection with HCV 5′NTR-luc RNA, but not the luc-poly(A) control RNA, demonstrating that HCV RNA triggers IRF3 activation. In contrast, in Huh7.5 cells, there was no activation of IRF3 by SenV or HCV RNA, since IRF3 was retained in the cytoplasm under all conditions (Fig. 2D). In addition, transfection of the 5′NTR-luc RNA triggered accumulation of high-mass hyperphosphorylated, active isoforms of IRF3 within 4 h after transfection into Huh7 cells but not into Huh7.5 cells (Fig. 2E, left panel set). A similar difference was observed after SenV infection (data now shown). Consistent with this, SenV infection induced the formation of active, phosphorylation-dependent IRF3 dimers only in Huh7 cells (Fig. 2E, right panel). Thus, the Huh7.5 cells possess a lesion in essential transducer(s) of IRF3 phosphorylation that normally trigger its activation in response to virus infection or the HCV RNA PAMP.
Characterization of the Huh7.5 defect in IRF3 activation.
Further studies indicated that the lack of response in the Huh7.5 cells was most likely due to defects in signaling to IRF3 and not in IRF3 itself, since an ectopically expressed green fluorescent protein (GFP)-IRF3 protein accumulated in the nucleus of Huh7 cells, but not Huh7.5 cells, in response to SenV infection (Fig. S2A in the supplemental material). In addition, the lack of IRF3 nuclear accumulation in Huh7.5 cells was not due to disruption of nuclear transport, because the active, phosphomimetic IRF3 5D mutant was constitutively localized to the nucleus upon expression in Huh7 or Huh7.5 cells (Fig. S2B in the supplemental material). Ectopic expression of the TLR3 adaptor protein, TRIF (30), induced IFN-β promoter activation in both cell types, demonstrating that under these conditions the endogenous IRF3 kinases were intact and were themselves competent to signal IRF3-dependent promoter activation (Fig. S2C in the supplemental material). Consistent with this, there was an equivalent abundance of endogenous TBK1 and IKKɛ protein (Fig. S2D in the supplemental material) and TRIF mRNA (data not shown) in Huh7 and Huh7.5 cells, and these proteins were found to signal to IRF3 in a genetic complementation assay (described below). We found no mutations in the sequences of TBK1, IKKɛ, and TRIF cDNAs recovered from Huh7.5 cells (data not shown). Huh7 cells express little, if any, TLR3, and they do not respond to extracellular pIC (14), indicating that their response to intracellular HCV RNA is TLR3 independent. Together, these data suggest that the lesion in the IRF3 pathway in Huh7.5 cells is within a previously undefined intracellular signaling pathway that triggers IRF3 activation, either independent of or above the level of the TBK1 or IKKɛ kinases.
RIG-I complements the IRF3 activation defect.
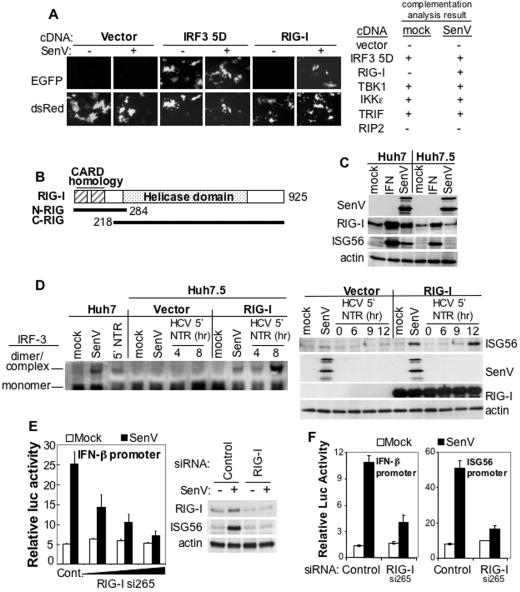
To identify a defective upstream signaling component that normally activates the IRF3 pathway, we subjected Huh7.5 cells to a genetic complementation assay searching for cDNA products capable of restoring IRF3 responsiveness to virus infection. Among other candidate cellular proteins selected because of prior evidence for roles in innate immune signaling, retinoic acid inducible gene I (i.e., RIG-I) was unique in restoring virus responsiveness to an IRF3-dependent promoter in Huh7.5 cells (Fig. 3A). Recently identified as an intracellular pattern recognition molecule that signals the downstream activation of IRF3 during virus infection (32), RIG-I is 925 aa in length and contains an amino-terminal region possessing tandem motifs with limited homology to the caspase activation and recruitment domain (CARD) (4) and a downstream DExD/H-box helicase domain that binds to RNA (Fig. 3B). RIG-I is a cytosolic protein that is expressed at a low basal level. We found that RIG-I mRNA (Fig. S3 in the supplemental material) and protein in Huh7 and Huh7.5 cells were increased in response to IFN treatment in a fashion similar to ISG56. In Huh7 cells the level of RIG-I and ISG56 also increased in response to SenV infection, but infection did not signal an increase in RIG-I expression in Huh7.5 cells, a finding consistent with an IRF3 activation defect (Fig. 3C). Importantly, ectopic expression of RIG-I led to accumulation of activated IRF3 dimers in Huh7.5 cells, restoration of ISG56 expression (Fig. 3D), and IRF3 nuclear translocation in response to either SenV infection or HCV 5′NTR-luc RNA transfection (Fig. S4 in the supplemental material).
FIG. 3.
RIG-I is an essential transducer of IRF3 activation. (A) Huh7.5 cells were subjected to complementation analysis with cotransfected plasmids encoding the IRF3-dependent F3-EGFP reporter construct and the constitutively expressed dsRed protein with a vector control plasmid or a plasmid from a panel of constructs encoding proteins implicated in antiviral signaling, including IRF3 5D (positive control), RIG-I (32), TBK1, IKKɛ (6, 23), TRIF (30), and RIP2 (13). At 24 h posttransfection the cells were mock infected (−) or infected with SenV (+). On the left, cells were transfected with RIG-I or control constructs and visualized by direct fluorescence imaging of EGFP and dsRED after mock or SenV infection. A summary of results from this particular analysis is shown on the right; “−” and “+” indicate the absence or presence of EGFP expression. (B) RIG-I structural features. The regions comprising the N-RIG and C-RIG constructs are shown. The RIG-I expression constructs used in the present study contained an amino-terminal FLAG epitope tag fused in frame to the RIG-I coding region. (C) The abundance of SenV proteins, endogenous RIG-I, ISG56, and β-actin was determined by immunoblot analysis of Huh7 and Huh7.5 cells that were mock treated, treated with 10 U of IFN-α2a/ml, or infected with SenV for 20 h. (D) On the left, IRF3 dimer and monomer abundance in nontransfected Huh7 cells or Huh7.5 cells transfected with either empty vector or a vector expressing RIG-I. Cells were mock treated, infected with SenV for 20 h, or transfected with HCV 5′NTR-luc RNA for 4 or 8 h prior to harvest and electrophoresis of extracts through a nondenaturing gel. The positions of the IRF3 monomer and dimer complex are indicated. On the right is shown ISG56, SenV, ectopically expressed RIG-I, and β-actin abundance in extracts from Huh7.5 cells transfected with empty vector or vector encoding RIG-I. At 24 h after transfection, cells were mock treated, infected with SenV for 20 h, or transfected with HCV 5′NTR-luc for 0, 6, 9, or 12 h prior to harvesting for immunoblot analysis. Ectopically expressed RIG-I was detected by using anti-FLAG M2 antibody. (E and F) Huh7 cells transfected with IFN-β-luc or ISG56-luc promoter expression constructs were transfected with a control siRNA (control) or a specific siRNA directed to the first exon of RIG-I (RIG-I si265), after which cells were mock infected or infected with SenV for 20 h, followed by quantification of relative luc activity. Bars show the average ± the SD values from triplicate experiments. In panel E, the left panel shows cells transfected with increasing amounts of RIG-I si265 siRNA. The right panel set shows endogenous RIG-I, ISG56, and actin protein abundance in extracts of mock-infected (−) or SenV-infected (+) cells transfected with control or RIG-I si265 siRNA.
To confirm that RIG-I signals IRF3 activation, we demonstrated that siRNA silencing of RIG-I suppressed SenV induction of IFN-β promoter activity in Huh7 cells. Suppression was dose dependent, not observed with a control siRNA (Fig. 3E), and associated with a block in SenV induction of ISG56 (Fig. 3F). We conclude from these results that RIG-I is essential for triggering IRF3 activation and ISG expression in response to virus infection or the HCV RNA PAMP in human hepatocytes and that RIG-I complements the IRF3 signaling defect in Huh7.5 cells, restoring responsiveness to the IRF3 pathway.
Unique domains of RIG-I bind HCV RNA and direct signaling to IRF3 in hepatocytes.
To determine which domains of RIG-1 confer host cell responsiveness to the viral PAMPs, we assessed the ability of the amino-terminal CARD homology domain and the carboxyl-terminal helicase domain (see Fig. 3B) to signal IFN-β promoter activity in response to SenV infection. When expressed in Huh7 cells, the CARD-homology domain (N-RIG) constitutively activated the IRF3 pathway, whereas expression of the carboxyl-terminal helicase domain (C-RIG) resulted in dominant-negative inhibition of virus-induced IRF3 activation (Fig. S5 in the supplemental material). This finding is consistent with those of another recent study (32). We verified that the helicase function of RIG-I is essential for IRF3 signaling, since RIG-I with a mutation (K270A) that disrupts the conserved Walker-A motif and eliminates helicase activity (32) failed to restore SenV or HCV RNA activation of IRF3 in Huh7.5 cells (R. Sumpter and M. Gale, Jr., unpublished observations). These results provide evidence that the CARD-homology domain of RIG-I mediates RNA signaling to the IRF3 pathway in human hepatocytes and indicate a regulatory role for the helicase domain in this process.
RNA-binding activity maps to the helicase domain of RIG-I (32). We demonstrated that ectopically expressed RIG-I was recovered from cell extracts in a pIC-agarose pull-down assay (Fig. 4A), a finding consistent with a role for dsRNA binding in RIG-I signaling. The specificity of this assay for dsRNA binding proteins was confirmed by the recovery of endogenous PKR, a known dsRNA binding protein (12), but not endogenous actin. RIG-I dsRNA binding activity was specifically competed for by the addition of increasing concentrations of soluble pIC and HCV 5′NTR or 3′NTR RNAs but not by the unstructured ss2 RNA (Fig. 4B). Thus, RIG-I is a dsRNA binding protein that recognizes and binds RNA PAMP(s) residing within structured regions of the HCV genome.
FIG. 4.
Characterization of RIG-I function. (A) Extracts from Huh7 cells that were transfected with empty vector or a vector expressing RIG-I were mixed with pIC-coupled agarose beads. The input proteins and those recovered as pull-down products (PD) were evaluated by immunoblot analysis with anti-FLAG (to detect plasmid-encoded RIG-I), anti-PKR, or anti-β-actin antibodies. (B) Extracts from Huh7 cells transfected with the RIG-I expression vector were mixed with pIC-coupled agarose beads, and the binding reactions were supplemented with 10 or 50 μg of soluble pIC, HCV ss1, 5′NTR, or 3′NTR RNA/ml. Input proteins and pull-down products (PD) were evaluated by immunoblot analysis with anti-FLAG and anti-β-actin antibodies. Competitive inhibition of pIC binding to RIG-I was quantified by densitometry and expressed as the percentage of the initial RIG-I pull-down signal remaining in each respective lane. (C) Comparison of the nucleotide sequences of RIG-I mRNA present in Huh7 and Huh7.5 cells demonstrates a C-to-T substitution at nucleotide position 164 in the RIG-I ORF in Huh7.5 cells. This changes codon 55 from threonine to isoleucine (T55I) as shown in the structural diagram. (D) In the left panel, Huh7 cells were cotransfected with IFN-β-luc expression plasmid and 50 ng of vector only (vector) or IFN-β-luc expression plasmid with (from left to right) increasing amounts (25, 50, and 100 ng) of expression vector encoding RIG-I, the full-length T55I mutant (T55I), N-RIG, N-RIG containing the T55I mutation (N-T55I), or full-length T55I mutant with 50 ng of RIG-I expression vector (T55I + RIG-I). Cells were mock infected or infected with SenV for 20 h, harvested, and subjected to luciferase assay. Bars show the average value ± the SD of each relative luciferase activity. In the right panel, Huh7.5 cells were transfected with vector control or plasmids encoding IRF3-5D (control), RIG-I, N-RIG, full-length T55I mutant (T55I), or N-T55I. After 24 h cells were mock infected (−) or infected with SenV (+) for 20 h and then harvested, and extracts were subjected to immunoblot analysis for ISG56, SenV, RIG-I, N-RIG, N-T55I, and β-actin. RIG-I proteins were detected with anti-FLAG antibody. (E) Huh7 cells were transfected with empty vector or vectors encoding RIG-I, C-RIG, or the full-length T55I mutant (T55I). Cells were harvested at 24 h, and extracts were mixed with agarose beads coupled to poly(C) or pIC. Proteins in the pull-down fraction (PD) and input were subjected to immunoblot analysis. RIG-I proteins were detected as in panel B.
RIG-I is defective in cells highly permissive for HCV RNA replication.
The distinct domain features of RIG-I indicate that mutations in either domain could potentially confer defects in dsRNA signaling to IRF3 during virus infection. To determine whether this could explain the phenotype of Huh7.5 cells, we sequenced the RIG-I cDNA derived from mRNA isolated from Huh7 and Huh7.5 cells. We found a single nucleotide substitution that rendered an amino acid change at codon 55 (T55I) in the RIG-I ORF and located within the first CARD-homology domain of a single sequence that was repeatedly recovered from Huh7.5 cells (Fig. 4C). When introduced into wild-type RIG-I, the T55I mutation caused a loss of signaling to IRF3. In an IFN-β promoter assay, ectopic expression of the T55I mutant had a dominant-negative effect upon endogenous RIG-I signaling in Huh7 cells, causing a reduction in promoter activity triggered by SenV infection (Fig. 4D, left panel). When introduced into a construct expressing only the amino-terminal CARD-homology domain, the T55I mutation resulted in a loss of constitutive activation of the IRF3-dependent IFN-β promoter (Fig. 4D). However, the dominant-negative activity of T55I required its expression as the full-length protein, because the endogenous response to SenV infection remained intact in cells expressing N-RIG T55I. Immunoblot analysis demonstrated that the T55I mutation disrupted RIG-dependent SenV-induced ISG56 expression in Huh7.5 cells (Fig. 4D, right panel set).
The T55I mutation had no effect upon the ability of RIG-I to bind dsRNA, since the RIG-I carboxyl-terminal helicase domain (C-RIG; aa 218 to 925) was sufficient for dsRNA binding (Fig. 4E). These experiments further confirmed the specificity of RIG-I interaction with dsRNA, since there was no binding to single-stranded poly(C)-agarose beads. Thus, the loss of function caused by the T55I mutation is due to disruption of one or more signaling events directed by the CARD-homology domain and not to alteration of the RNA binding activity of the protein.
RIG-I regulates permissiveness to HCV RNA replication in Huh7.5 cells.
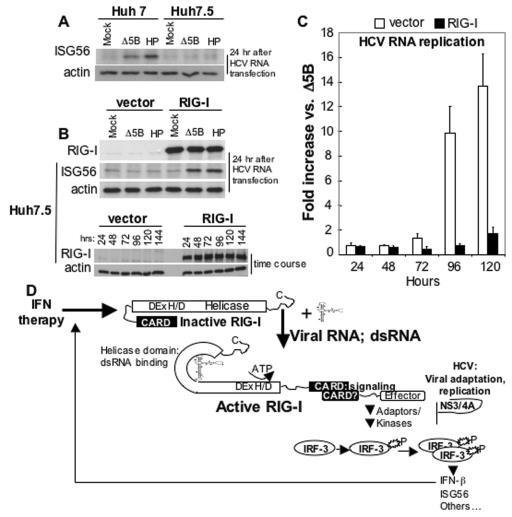
To define the role of RIG-I in cellular permissiveness to HCV RNA replication, we measured HCV RNA replicon amplification over a 120-h time course in Huh7.5 cells transfected either with an empty vector or one expressing RIG-I in the absence of G418 selection and using the replicon-encoded luciferase expression as a read out of HCV RNA levels. This approach allowed a direct assessment of the influence of RIG-I upon viral RNA replication over a short time course, without confounding effects due to the selection of cell culture-adaptive mutations in the replicon RNA (1, 28). As expected, transfection of HCV replicon RNA induced a host response leading to ISG56 expression in Huh7 cells, but not in Huh7.5 cells (Fig. 5A). The ectopic expression of RIG-I complemented the defect in Huh7.5 cells, restoring ISG56 expression after transfection of replicon RNA. RIG-I expression was maintained in the transfected Huh7.5 cells throughout an extended 144-h time course (Fig. 5B, upper and lower panel sets, respectively). To assess viral RNA replication in Huh7.5 cells complemented with ectopic RIG-I, we compared the activity of the luciferase protein encoded by replication-competent (HP) versus replication-defective (Δ5B) replicon RNA in cells in parallel as a direct marker of HCV RNA abundance. As shown in Fig. 5C, the HP replicon levels remained low and did not amplify beyond an approximately twofold increase in Huh7.5 cells expressing the functional RIG-I, whereas in those transfected with the empty vector, the HP RNA-encoded luciferase was continuously amplified over the same 120-h time course. At the end of this period the HP replicon had amplified by >13-fold over the level of the Δ5B replication-defective control RNA. Thus, RIG-I complementation of IRF3 signaling and restoration of the host response in Huh7.5 cells results in a phenotypic switch from hyperpermissiveness for HCV RNA replication to a relatively nonpermissive phenotype characterized by recognition of HCV RNA and suppression of viral RNA amplification.
FIG. 5.
Effect of RIG-I on cellular permissiveness for HCV RNA replication. (A) Huh7 and Huh7.5 cells were mock transfected or transfected with Δ5B or HP HCV replicon RNA. After 24 h, the cells were harvested and extracts were subjected to immunoblot for ISG56 and β-actin. (B) Huh7.5 cells were transfected with vector alone or a vector encoding RIG-I. In the upper panel the cells were secondarily transfected, 24 h later, with Δ5B or the HP replicon RNA. After 24 h, cells were harvested, and extracts were subjected to immunoblot analysis for β-actin, ISG56, and the ectopically expressed RIG-I protein. The lower panel shows an immunoblot analysis of β-actin and ectopic RIG-I protein abundance over a 144-h time course posttransfection. (C) Huh7.5 cells were transfected with empty vector (vector) or vector expressing RIG-I. At 24 h, parallel cultures were secondarily transfected with Δ5B or HP HCV replicon RNA containing the firefly luciferase sequence in lieu of Neo, as described in the legend for Fig. 1E. The level of HCV replicon-encoded luciferase activity was measured as a specific marker of HCV RNA abundance (28). The bars show the average fold increase in luciferase expression ± the SD compared to that expressed by the replication-defective Δ5B replicon control. (D) Model of viral RNA binding and IRF3 signaling by RIG-I. Details are given in the text.
DISCUSSION
RIG-I is a signal transducer and HCV RNA PAMP receptor.
Our results define the cellular RNA helicase, RIG-I, as a transducer of signals that activate innate antiviral defenses limiting HCV RNA replication. In cultured hepatocytes, RIG-I plays an essential role as a TLR-independent PAMP receptor that specifically binds dsRNA and structured regions within the HCV genome to signal IRF3 activation, IFN production, and ISG expression. This response is an important component of the acute hepatic antiviral defenses that are triggered in vivo within days of exposure to HCV (2) and is therefore likely to contribute to the resolution of acute infection (26). Our data also demonstrate that RIG-I is essential for triggering IRF3 activation in response to SenV or VSV infection in hepatocytes (Fig. 2B and 3E), a finding consistent with its role as a viral PAMP receptor. We speculate that secondary or tertiary RNA structures, often present in viral RNAs (24), present a specific PAMP that is engaged by RIG-I. RIG-I specifically bound the structured 5′NTR and 3′NTR segments of the HCV genome but not the predicted nonstructured viral RNAs (Fig. 4B and E). The RIG-I-based cellular response exploits the need for HCV to conserve unique structured terminal RNA segments that are essential to viral replication. This interaction signals the induction of IRF3 responsive genes and other ISGs, including ISG56, that have been shown to suppress HCV RNA replication (7, 29). The binding of RIG-I to viral RNA thus induces critical antiviral effectors impacting cellular permissiveness for HCV through processes that limit viral RNA replication.
CARD signaling activates downstream IRF3.
CARD-containing proteins mediate signaling events in response to a variety of intracellular and extracellular pathogen-related stimuli that direct NF-κB activation via TLR-independent mechanisms (4). RIG-I and other CARD-containing DExD/H box helicase proteins thus appear to represent a family of proteins that activate a TLR3-independent response against a variety of viral pathogens (32). The RIG-I CARD-homology domain is capable of inducing IRF3 activation. Sequence differences, which distinguish the RIG-I homolog from other CARD proteins (4, 32), may provide for recruitment of unique signaling components that activate IRF3 through processes that depart from the canonical CARD-CARD homotypic interaction pathways. The kinase TBK1, which does not contain a CARD, is essential for dsRNA or virus-induced IRF3 activation (17), suggesting that it is required for RIG signaling to IRF3. TBK1 does not directly associate with RIG-I (E. Foy and M. Gale, Jr., unpublished observations). The most likely scenario for RIG-I-mediated IRF3 activation involves interaction with downstream adaptor(s) or CARD-containing proteins that ultimately results in TBK1/IRF3 activation. In this regard, the location of the T55I mutation in the first CARD domain of the Huh7.5 RIG-I mutant is of particular interest. It uncoupled signal transduction from viral RNA binding. Huh7.5 cells are derived from and are isogenic to Huh7 cells (3). Among other potential lesions in antiviral processes within Huh7-derived cells, the hyperpermissive phenotype of Huh7.5 cells can be attributed to the RIG-I T55I mutation that abolishes PAMP signaling to IRF3. The IRF3 response contributes to control of HCV RNA replication (7). Thus, activation of IRF3 is a critical determinant of cellular permissiveness for HCV RNA replication, although defects in other, RIG-I-independent innate antiviral defense pathways could contribute to the overall level of cellular permissiveness for HCV.
A new model for virus activation of IFN-α/β and ISGs.
We propose a model in which within the infected hepatocyte RIG-I binding to the HCV RNA PAMP induces events that are regulated by its helicase domain and alter its conformation sufficiently to recruit or interact with signaling partners that direct the downstream phosphorylation of IRF3 and expression of antiviral effector genes (Fig. 5D). This response is likely to be triggered by the HCV RNA PAMP immediately upon introduction of the viral genome into the host cell cytoplasm (see Fig. 5A). Moreover, recently published observations demonstrate that HCV RNA replication can trigger IRF3 activation (25), implying that RIG-I engages HCV during the process of viral replication. Since RIG-I is strongly IFN inducible (Fig. S3 in the supplemental material), one of the effects of IFN therapy is an enhanced sensing of virus replication, which initiates a cascade of antiviral events. The importance of the RIG-I pathway for cellular permissiveness to viral replication is underscored by the variety of ways that viruses antagonize downstream IRF3 actions (12). Indeed, HCV has the potential to block virus triggering of IRF3 phosphorylation through the actions of its NS3/4A serine protease (7). The results from recent work now indicate that this regulation is attributed to blockade of RIG-I-dependent signaling imposed by the protease function of NS3/4A (7a), and it is likely that upon its accumulation NS3/4A antagonizes RIG-I signaling in the infected cell. Therapeutic approaches to control RIG-I and NS3/4A function may provide novel strategies to limit HCV infection by modulating cellular permissiveness for virus replication.
Supplementary Material
Acknowledgments
This study was supported by NIH grants AI48235 and AI060389 (M.G.), U19-AI40035 (S.M.L.), T32-GM08203 (R.S.), and T32 AI 07520 (E.F.); a Burroughs Wellcome Fund Investigator in Pathogenesis of Infectious Disease award (M.G.); the Ellison Medical Foundation New Scholars in Global Infectious Disease Research program (ID-NS-0032; M.G.); and a gift from Mr. and Mrs. R. Batcheldor. K.L. is the John Mitchell Hemophilia of Georgia Liver Scholar of the American Liver Foundation. M.G. is the Nancy C. and Jeffrey A. Marcus Scholar in Medical Research in Honor of Bill S. Vowell.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Bartenschlager, R., and V. Lohmann. 2001. Novel cell culture systems for the hepatitis C virus. Antivir. Res. 52:1-17. [DOI] [PubMed] [Google Scholar]
- 2.Bigger, C. B., K. M. Brasky, and R. E. Lanford. 2001. DNA microarray analysis of chimpanzee liver during acute resolving hepatitis C virus infection. J. Virol. 75:7059-7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blight, K. J., J. A. McKeating, and C. M. Rice. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 76:13001-13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouchier-Hayes, L., and S. J. Martin. 2002. CARD games in apoptosis and immunity. EMBO Rep. 3:616-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edelmann, K. H., S. Richardson-Burns, L. Alexopoulou, K. L. Tyler, R. A. Flavell, and M. B. Oldstone. 2004. Does Toll-like receptor 3 play a biological role in virus infections? Virology 322:231-238. [DOI] [PubMed] [Google Scholar]
- 6.Fitzgerald, K. A., S. M. McWhirter, K. L. Faia, D. C. Rowe, E. Latz, D. T. Golenbock, A. J. Coyle, S. M. Liao, and T. Maniatis. 2003. IKKɛ and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4:491-496. [DOI] [PubMed] [Google Scholar]
- 7.Foy, E., K. Li, C. Wang, R. Sumpter, M. Ikeda, S. M. Lemon, and M. Gale, Jr. 2003. Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science 300:1145-1148. [DOI] [PubMed] [Google Scholar]
- 7a.Foy, E., K. Li, R. Sumpter, Y.-M. Loo, C. Johnson, C. Wang, P. Fish, M. Yoneyama, S. Lemon, and M. Gale, Jr. 2005. Control of antiviral defenses through hepatitis C virus disruption of RIG-I signaling. Proc. Natl. Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed]
- 8.Fredericksen, B., G. Akkaraju, E. Foy, C. Wang, J. Pflugheber, Z. J. Chen, and M. Gale, Jr. 2001. Activation of the interferon-β promoter during hepatitis C virus RNA replication. Viral Immunol. 15:29-40. [DOI] [PubMed] [Google Scholar]
- 9.Fredericksen, B. L., M. Smith, M. G. Katze, P. Y. Shi, and M. Gale, Jr. 2004. The host response to West Nile Virus infection limits viral spread through the activation of the interferon regulatory factor 3 pathway. J. Virol. 78:7737-7747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fried, M. W., M. L. Shiffman, K. R. Reddy, C. Smith, G. Marinos, F. L. Goncales, Jr., D. Haussinger, M. Diago, G. Carosi, D. Dhumeaux, A. Craxi, A. Lin, J. Hoffman, and J. Yu. 2002. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N. Engl. J. Med. 347:975-982. [DOI] [PubMed] [Google Scholar]
- 11.Gale, M., Jr., and M. G. Katze. 1997. What happens inside lentivirus or influenza virus infected cells: insights into regulation of cellular and viral protein synthesis. Companion Methods Enzymol. 11:383-401. [DOI] [PubMed] [Google Scholar]
- 12.Katze, M. G., Y. He, and M. Gale, Jr. 2002. Viruses and interferon: a fight for supremacy. Nat. Rev. Immunol. 2:675-677. [DOI] [PubMed] [Google Scholar]
- 13.Kobayashi, K., N. Inohara, L. D. Hernandez, J. E. Galan, G. Nunez, C. A. Janeway, R. Medzhitov, and R. A. Flavell. 2002. RICK/Rip2/CARDIAK mediates signalling for receptors of the innate and adaptive immune systems. Nature 416:194-199. [DOI] [PubMed] [Google Scholar]
- 14.Lanford, R. E., B. Guerra, H. Lee, D. R. Averett, B. Pfeiffer, D. Chavez, L. Notvall, and C. Bigger. 2003. Antiviral effect and virus-host interactions in response to alpha interferon, gamma interferon, poly(I)-poly(C), tumor necrosis factor alpha, and ribavirin in hepatitis C virus subgenomic replicons. J. Virol. 77:1092-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lohmann, V., F. Korner, J.-O. Kock, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 16.McHutchison, J. G. 2004. Understanding hepatitis C. Am. J. Manag. Care. 10:S21-S29. [PubMed] [Google Scholar]
- 17.McWhirter, S. M., K. A. Fitzgerald, J. Rosains, D. C. Rowe, D. T. Golenbock, and T. Maniatis. 2004. IFN-regulatory factor 3-dependent gene expression is defective in Tbk1-deficient mouse embryonic fibroblasts. Proc. Natl. Acad. Sci. USA 101:233-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Neill, L. A. 2004. Immunology: after the Toll rush. Science 303:1481-1482. [DOI] [PubMed] [Google Scholar]
- 19.Peters, K. L., H. L. Smith, G. R. Stark, and G. C. Sen. 2002. IRF-3-dependent, NFκB- and JNK-independent activation of the 561 and IFN-beta genes in response to double-stranded RNA. Proc. Natl. Acad. Sci. USA 99:6322-6327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reed, K. E., and C. M. Rice. 1998. Molecular characterization of hepatitis C virus, p. 1-37. In H. W. Reesink (ed.), Hepatitis C virus, vol. 1. Karger, Basel, Switzerland. [DOI] [PubMed]
- 21.Sato, M., H. Suemori, N. Hata, M. Asagiri, K. Ogasawara, K. Nakao, T. Nakaya, M. Katsuki, S. Noguchi, N. Tanaka, and T. Taniguchi. 2000. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity 13:539-548. [DOI] [PubMed] [Google Scholar]
- 22.Sen, G. C. 2001. Viruses and interferons. Annu. Rev. Microbiol. 55:255-281. [DOI] [PubMed] [Google Scholar]
- 23.Sharma, S., R. Benjamin, B. tenOever, N. Grandvaux, G.-P. Zhou, R. Lin, and J. Hiscott. 2003. Triggering the interferon antiviral response through a novel IKK-related pathway. Science 300:1148-1151. [DOI] [PubMed] [Google Scholar]
- 24.Simmonds, P., A. Tuplin, and D. J. Evans. Detection of genome-scale ordered RNA structure (GORS) in genomes of positive-stranded RNA viruses: implications for virus evolution and host persistence. RNA 10:1337-1356. [DOI] [PMC free article] [PubMed]
- 25.Sumpter, R., C. Wang, E. Foy, Y.-M. Loo, and M. Gale, Jr. 2004. Viral evolution and interferon resistance of hepatitis C virus RNA replication in a cell culture model. J. Virol. 78:11591-11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thimme, R., J. Bukh, H. C. Spangenberg, S. Wieland, J. Pemberton, C. Steiger, S. Govindarajan, R. H. Purcell, and F. V. Chisari. 2002. Viral and immunological determinants of hepatitis C virus clearance, persistence, and disease. Proc. Natl. Acad. Sci. USA 99:15661-15668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tuplin, A., J. Wood, D. J. Evans, A. H. Patel, and P. Simmonds. 2002. Thermodynamic and phylogenetic prediction of RNA secondary structures in the coding region of hepatitis C virus. RNA 8:824-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vrolijk, J. M., A. Kaul, B. E. Hansen, V. Lohmann, B. L. Haagmans, S. W. Schalm, and R. Bartenschlager. 2003. A replicon-based bioassay for the measurement of interferons in patients with chronic hepatitis C. J. Virol. Methods 110:201-209. [DOI] [PubMed] [Google Scholar]
- 29.Wang, C., J. Pflugheber, R. Sumpter, D. Sodora, D. Hui, G. C. Sen, and M. Gale, Jr. 2002. Alpha interferon induces distinct translational control programs to suppress hepatitis C virus RNA replication. J. Virol. 77:3898-3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamamoto, M., S. Sato, K. Mori, K. Hoshino, O. Takeuchi, K. Takeda, and S. Akira. 2002. Cutting edge: a novel Toll/IL-1 receptor domain-containing adapter that preferentially activates the IFN-beta promoter in the Toll-like receptor signaling. J. Immunol. 169:6668-6672. [DOI] [PubMed] [Google Scholar]
- 31.Ye, J., C. Wang, R. Sumpter, Jr., M. S. Brown, J. L. Goldstein, and M. Gale, Jr. 2003. Disruption of hepatitis C virus RNA replication through inhibition of host protein geranylgeranylation. Proc. Natl. Acad. Sci. USA 100:15865-15870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoneyama, M., M. Kikuchi, T. Natsukawa, N. Shinobu, T. Imaizumi, M. Miyagishi, K. Taira, S. Akira, and T. Fujita. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5:730-737. [DOI] [PubMed] [Google Scholar]
- 33.Yoneyama, M., W. Suhara, and T. Fujita. 2002. Control of IRF-3 activation by phosphorylation. J. Interferon Cytokine Res. 22:73-76. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.