Abstract
A unique behavioral paradigm has been developed for Periplaneta americana that assesses the timing and success of memory consolidation leading to long-term memory of visual-olfactory associations. The brains of trained and control animals, removed at the critical consolidation period, were screened by two-directional suppression subtractive hybridization. Screens identified neurobiologically relevant as well as novel genes that are differentially expressed at the consolidation phase of memory. The differential expression of six transcripts was confirmed with real-time RT-PCR experiments. There are mitochondrial DNA encoded transcripts among the up-regulated ones (COX, ATPase6). One of the confirmed down-regulated transcripts is RNA polymerase II largest subunit. The mitochondrial genes are of particular interest because mitochondria represent autonomous DNA at synapses. These transcripts will be used as one of several tools in the identification of neuronal circuits, such as in the mushroom bodies, that are implicated in memory consolidation.
Experimental evidence has suggested that analogous learning and memory centers are shared by mammalian and insect taxa, and that these share similar dynamics, functional organizations, and molecular pathways (Alberini 1999, Strausfeld 2002, Cayre et al. 2002; Heisenberg 2003). For example, place memory, which in mammals is mediated by the hippocampus (O'Keefe and Conway 1978; Morris et al. 1982), has been shown in one insect taxa to require the integrity of the paired mushroom bodies (Mizunami et al. 1998b; Li and Strausfeld 1997; Strausfeld and Li 1999), neuropils that share architectural organization with the hippocampus and that integrate multimodal and contextual information (Mizunami et al. 1998a; Li and Strausfeld 1999). Behavioral responses and molecular mechanisms of sequential stages, or phases, of memory acquisition have been characterized in Drosophila melanogaster (Tully et al. 1994), Aplysia californica (Ghirardi et al. 1995), and the rat (Saitoh and Inokuchi 2000). The essential role of a single gene in learning was first demonstrated in a D. melanogaster mutant (Dudai et al. 1976; Byers et al. 1981). Long-term memory is differentiated from short-term in that it requires protein synthesis, and the transition from short- to long-term may be facilitated by intermediate-term memory. Memory consolidation occurs during the transition from intermediate-term to long-term memory, in which de novo protein synthesis is high and memory readily disrupted (Tully et al. 1994; Ghirardi et al. 1995). However, a behavioral assay that suggests a neural basis for such consolidation is lacking. The purpose of the present study is not to attempt to identify all transcription events associated with sensory association but instead to determine what is transcribed during the specific event of consolidation. We focus on this period for the very reason that subsequent in situ studies of consolidation-associated gene expression might better inform us about the identity of neuropils in which this stage of memory formation occurs.
Molecular mechanisms that accompany associative learning are crucial for understanding memory formation, as famously demonstrated first in gastropods (Kandel and Schwartz 1982; Alkon 1984), in which molecular profiling detected four newly synthesized proteins during an elementary form of learning (Castellucci et al. 1988). Recent studies utilizing molecular genetics can now identify differentially expressed transcripts, for example, between the amygdali of naive and fear conditioned mice (Stork et al. 2001), in brains of naive and experienced honey bees (Kucharski and Maleszka 2002), and in the hippocampi of naive and actively learning mice (Leil et al. 2003).
Here we identify the consolidation phase of memory in P. americana by exploiting a component of the insect's characteristic foraging behavior. This consists of antennal movements that are made toward an odor source. These movements, termed antennal projection responses (APR), can be precisely quantified to demonstrate associative learning of paired sensory stimuli (Kwon et al. 2004; Lent and Kwon 2004). We demonstrate that long-term memory is not established if sensory input is restricted unilaterally to one eye and the corresponding antenna while denying the contralateral antenna and eye sensory input. By using behavioral measures to compare memory dynamics of such restricted and nonrestricted (bilateral) sensory input, we identify a critical time period during which memory is consolidated. We utilized this unique behavioral strategy to identify concomitant up- and down-regulated gene transcripts during memory consolidation. We employed subtractive hybridization to enrich transcripts that are either overexpressed or suppressed in the brain of a cockroach trained under nonrestricted sensory conditions. We have identified neurobiologically relevant as well as novel differentially expressed P. americana genes at the consolidation phase of memory. Relationships with neurological phenotypes have been established among these genes such as Leigh syndrome and encephalopathy, NARP (neuropathy, ataxia, and retinitis pigmentosa) and bilateral striatal necrosis (Schon et al. 2001; Shoubridge 2001).
Results
Behavioral test for memory consolidation
APRs toward olfactory stimuli can be classically conditioned in P. americana (Fig. 1) using the visual-olfactory association spaced training paradigm developed by Lent and Kwon (2004). The method uses restrained cockroaches that are presented with an odor (US) paired with a spatially coincidental green LED visual cue (CS) and results in APRs that are made toward a learned visual cue. The CS alone does not elicit an APR in naive animals before training, but following five pairings of the US and CS an association of the green LED with the odor cue results in APRs toward the visual cue in the absence of the odor stimulus (Lent and Kwon 2004). Here we compare cockroaches trained in this fashion, but using two general conditions of the animals. In nonrestricted sensory conditions, both eyes and both antenna are unobscured. In restricted conditions, the antenna and eye on one side of the animal are shielded before and during training as to not expose them to the CS and US. This is done by ensheathing the antenna in a tube, closed at its tip, and covering the eye with an opaque shield. Unrestricted and restricted animals are then tested as just described. When tested between 5 min and 24 h after training, unrestricted animals (N = 27) show significantly higher APRs compared with those of control animals (N = 15). Unrestricted animals also evidence stable long-term memory, as evidenced by the high percentage of APRs to the CS in trained animal tested beyond the 24-h period (Mann-Whitney U test, U = 81, P < 0.01) (Fig. 2A; Lent and Kwon 2004).
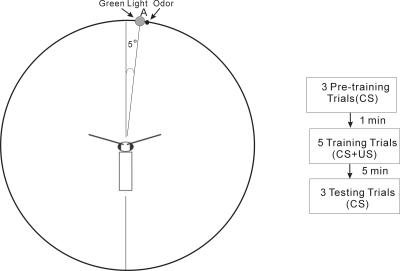
Figure 1.
Visual associative learning. Experimental setup and training protocol. Restrained cockroaches were positioned at the center of the arena. The distance from the head to the position of visual and olfactory cues was 15 cm. A green LED as a visual cue was positioned in parallel with an odor cue ∼5° from the midline of the head. Under restricted sensory conditions, one eye and the ipsilateral antenna were covered (naive half), restricting sensory information to the opposite side (trained half). Training protocol comprised three pretraining trials, five training trials, and three testing trials.
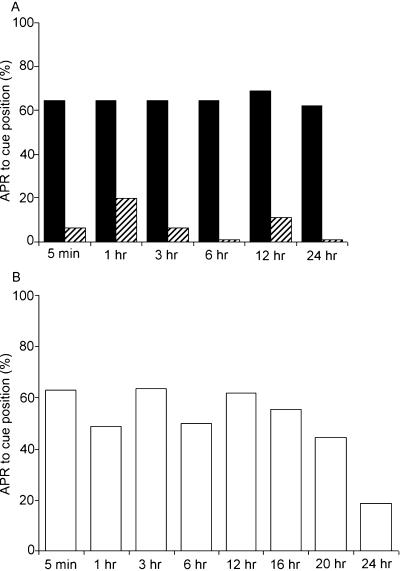
Figure 2.
(A) Antennal projection responses (APRs) of cockroaches trained in nonrestricted sensory conditions (black bars) were tested for up to 24 h and compared with control/untrained animals (hatched bars). A high percentage of APRs were retained in animals trained in nonrestricted sensory conditions from 5 min to 24 h with no significant difference between intervals. Trained animals were significantly different from control animals. (B) APRs of cockroaches trained in restricted sensory conditions were tested for up to 24 h. APRs of the trained half were elicited at 5 min and retained up to 20 h, but show a marked decrease to pretraining levels at 24 h.
Cockroaches trained in restricted sensory conditions (N = 18) also show significantly higher APRs compared with those of control animals when tested from 5 min up until 20 h after training (Mann-Whitney U test, U = 69, P < 0.05). In contrast, when tested after longer periods, for example, 24 h after training, there is no significant difference between naive and trained animals (Mann-Whitney U test, U = 121, P > 0.5) (Fig. 2B). Thus, under restricted training conditions there is a decay of memory between 20 and 24 h after training.
The divergence in the learned response between the nonrestricted and restricted sensory condition beginning ∼20 h after training and reaching an extreme at 24 h suggests an underlying difference in memory kinetics (Fig. 2., cf. A, black bars, and B). The 20-24-h period following training reveals the behavioral induction of long-term memory in animals that have undergone nonrestrictive training; conversely, this period shows the failure of this induction in animals that have undergone restricted sensory training resulting in the decay of the learned response (see Discussion). We refer to this 20-24-h time window of the nonrestricted sensory conditioning as the time when memory consolidation occurs, because this is the time window when the evident absence of stable long-term memory can be revealed earliest through behavioral experimentation (Fig. 2B). The term memory consolidation defines the initial phase of saturated long-term memory observed behaviorally. The next experimental step identifies molecular changes that are coincidental to the behaviorally observed memory consolidation.
Differential gene expression during memory consolidation
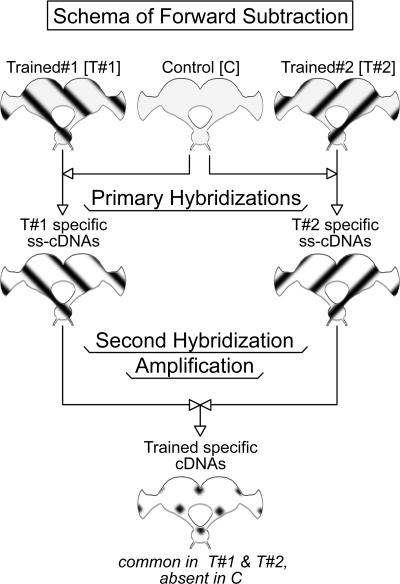
Gene expression analysis has been performed on the brains of cockroaches that demonstrated APRs 21-23 h after training under nonrestricted sensory conditions, that is, when consolidated memory emerges. Subtractive hybridization was used to screen for transcripts that are overexpressed and transcripts that are suppressed in the brains of trained P. americana. Two subtractive hybridizations were performed: one forward and one reverse subtraction. For each of the subtractive hybridizations, two primary hybridizations were performed with three dissected brains. Total RNA was prepared from these brains, and cDNA was synthetized on the RNA separately but side by side. In the forward subtraction, two trained brains were compared with one control brain (Fig. 3). The cDNA pools of each trained brain were ligated to different adaptors at the 5′ ends in the forward subtraction. During the primary hybridizations of the forward subtraction, each trained brain sample was mixed separately with an excess of the control sample, heat denatured, and allowed to anneal. The primary hybridizations leave trained sample-specific cDNAs single stranded (ss cDNA). The second hybridization of the forward subtraction is performed to anneal adaptor labeled trained specific ss cDNAs of two animals. The last step of the forward subtraction amplified 1/200th of the trained specific cDNA hybrids that were labeled with two different adaptors. About one-tenth of the amplified hybrids were cloned and analyzed. The schema in Figure 3 presents the protocol employed in the forward subtraction. In the reverse subtraction, two control brains were compared with one trained brain. The cDNA pools of each control brain were ligated to different adaptors at the 5′ ends in the reverse subtraction. Subsequent steps of the reverse subtraction were performed analogously to the forward subtraction. The forward and reverse subtractions were performed side by side throughout. Overexpressed genes are represented in the forward and suppressed genes are represented in the reverse subtracted cDNA libraries. Of the forward (abbreviated as T/C for Trained versus Control) and reverse (abbreviated as C/T for Control versus Trained) subtracted libraries, 288 randomly selected cDNA clones were purified and further analyzed. Of the 288 randomly selected clones, 216 were originated from the T/C, and 72 were from the C/T subtracted libraries. In the case of the C/T subtracted library, all insert bearing clones (72) were analyzed. For manageability clones from the T/C and C/T subtracted libraries were analyzed in three 96-well plates.
Figure 3.
Schema of forward subtraction to generate Trained-specific cDNAs. Patterns in the flowchart follow the progress of the subtraction. Opposing perpendicular patterns represent single-stranded cDNAs (ss-cDNA) that are specific to Trained#1 and T#2, respectively. Diamond pattern represents hybrids of T#1 and T#2 ss-cDNAs, i.e., cDNAs that are common in T#1 and T#2. These are Trained-specific cDNAs. For detailed description, see Results and Materials and Methods sections.
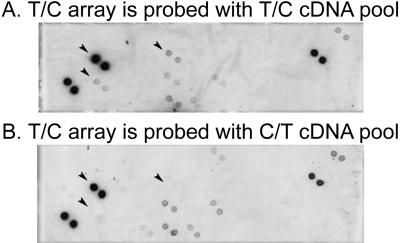
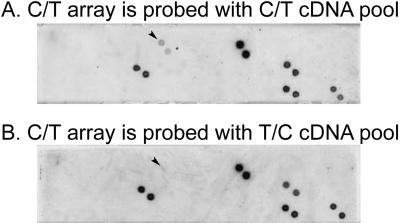
In order to eliminate cloning artifacts, differential screening was performed (Figs. 4, 5). Randomly selected clones were arrayed in duplicates on nylon membranes to make two identical arrays. One membrane was probed with the digoxigenin (DIG) labeled T/C cDNA pool, the other with the DIG-labeled C/T cDNA pool (subsets of the arrays are shown on Figs. 4, 5). Sixtysix of the 288 subtracted clones gave a hybridization signal with DIG-labeled T/C and/or C/T cDNA pools. Differential screening of T/C library clones resulted in 25 positive clones (subsets of the arrays are shown on Fig. 4). These 25 T/C clones exhibited a stronger hybridization signal with DIG-T/C cDNA pool than with the DIG-C/T cDNA pool. Differential screening of the C/T library resulted in 15 positive clones (subsets of the arrays are shown on Fig. 5). These 15 C/T clones exhibited stronger hybridization signal with DIG-C/T cDNA pool than with DIG-T/C cDNA pool. In summary, differential screenings verified that 40 clones originated from subtracted cDNA libraries are differentially expressed in trained or untrained P. americana.
Figure 4.
Differential screening of T/C library clones. For screening with the dot blot technique, 48 clones are arrayed here in duplicates on nylon membranes to make two identical arrays. The membranes are hybridized with DIG-labeled T/C (A) or DIG-labeled C/T (B) cDNA pools. Arrowheads indicate cDNA clones that correspond to genes with increased expression in the brain of the trained roach.
Figure 5.
Differential screening of C/T library clones. For screening with the dot blot technique, 36 clones are arrayed here in duplicates on nylon membranes to make two identical arrays. The membranes are hybridized with DIG-labeled C/T (A) or DIG-labeled T/C (B) cDNA pools. Arrowheads indicate cDNA clones that correspond to genes with increased expression in the brain of the trained roach.
Next we undertook sequence analysis of the 40 subtracted cDNA library clones. The inserts of these clones were sequenced and submitted to database searches. We encountered ribosomal RNA (rRNA) hits among the clones of both the T/C and C/T subtracted libraries. rRNA sequences were more prevalent among the candidate clones from the T/C subtracted library (15 out of 25 clones), than among the candidate clones subtracted from the C/T library (one out of 15 clones). We consider the rRNA hits as false positives. No other sequences common to both T/C and C/T subtracted libraries have been identified. There was one clone in the T/C and one in the C/T library with inserts that were not tagged by any adaptor sequence. Since these adaptor sequences are landmarks of a subtracted clone, these clones are cloning artifacts. An additional short clone of the C/T library corresponds to an oligonucleotide of the PCR-Select Subtractive Hybridization Kit. Two clones of the C/T library match unannotated low complexity sequences in the mouse genome. After omitting the rRNA clones, cloning artifacts, and repeats, we identified six clones of the T/C and seven clones of the C/T libraries for further analysis. These clones are listed in Tables 1 and 2. Identical clones were identified repeatedly and independently from both the T/C and from the C/T subtracted libraries. Those identical in the T/C library are D1, F5, and F7; those identical in the C/T library are 3B8 and 3B9. Identifying identical clones suggests that the screen has reached saturation.
Table 1.
Genes up-regulated at the consolidation phase of memory
| T/C Clone | Description, AC no. | Distribution | Genome |
|---|---|---|---|
| D1, F5, F7 | no homolog, AY622323 | ||
| E5 | Cytochrome C oxidase subunit I, AY622331 | mitochondrial | mitochondrial |
| G4 | no homolog, AY622324 | ||
| G5 | Cytochrome C oxidase subunit III, AY622322 | mitochondrial | mitochondrial |
| 3D6 | Cytochrome C oxidase subunit II, AY622333 | mitochondrial | mitochondrial |
| 3H5 | ATP Synthase A chain [ATPase6], AY622322 | mitochondrial | mitochondrial |
Table 2.
Genes down-regulated at the consolidation phase of memory
| C/T Clone | Description, AC no. | Distribution | Genome |
|---|---|---|---|
| A5 | peptide hormone ORF & 3′UTR, AY622312 | extracellular | nuclear |
| B10 | unknown, AY622325 | ||
| 3A11 | RNA pol II largest subunit ORF & 3′UTR, AY622330 | nuclear | nuclear |
| 3B4 | unknown, AY622326 | ||
| 3B8, 3B9 | Rieske Fe-S protein ORF & 3′UTR, AY622329 | mitochondrial | nuclear |
| 3C10 | 3′UTR, AY622327 | ||
| 3C12 | ENSANGP00000019854 ORF & 3′UTR, AY622328 | nuclear |
Categories of the isolated genes
Up-regulated genes
Genes that are candidates for being up-regulated in the brain of P. americana during memory consolidation are listed in Table 1. These include transcripts for three subunits of the multi-subunit cytochrome C oxidase enzyme (COX). Clones E5, 3D6, and G5 correspond to 242 aa, 148 aa, and 196 aa at the C-terminal of the ORFs for the roach cytochrome C oxidase subunits I, II, and III, respectively. The P. americana E5 clone shows 80%-90% identity to COX I of other insects in 240-aa overlap. E5 is most similar to the Blattella germanica COX I (cf. Martinez-Gonzalez and Hegardt 1994). The P. americana 3D6 clone is 97% identical to the published P. americana COX II cDNA (cf. Liu and Beckenbach 1992). Single nucleotide polymorphisms in the 434-bp-long 3D6 cDNA result in 3-aa substitution within the C-terminal 144 aa. The P. americana G5 cDNA clone is most similar to the D. melanogaster COX III subunit with 82% ungapped identity in 195-aa overlap. The P. americana COX I, II, and III protein sequences are very similar to their Homo sapiens homologs as well (cf. Anderson et al. 1981): 70% identical to human COX I, 60% identical to human COX II, and 71% identical to human COX III, respectively.
The transcript of ATPase6 subunit A, which is part of the proton transporting two-sector ATPase, is enriched in the brain of the trained roach as well. The P. americana 3H5 clone shows 80% ungapped identity to ATPase6 subunit A of a variety of Diptera in 66-aa overlap (cf., e.g., de Bruijn 1983; Beard et al. 1993). It is interesting that the predicted amino acid sequence of 3H5 is as similar to Pongo pigmaeus ATPase6 subunit A as it is to Apis mellifera ATPase6 subunit A (cf. Crozier and Crozier 1992; Xu and Arnason 1996).
The other four clones that are candidates for being up-regulated during memory consolidation are novel. D1, F5, and F7 clones carry identical 204-bp-long inserts. Clone G4 spans 395 bp. None of these novel sequences has an OPR, thus they likely correspond to untranslated regulatory regions. These inserts show no homology to any sequences deposited in online databases so far.
Down-regulated genes
Genes that are candidates for being suppressed in the brain of P. americana during memory consolidation are listed in Table 2. The transcript for one subunit of the cytochrome C reductase complex, the Rieske iron-sulfur protein, is suppressed in the brain of the trained roach. The P. americana 3B8 and 3B9 cDNA clones are identical and cover the C-terminal 70 aa of the Rieske iron-sulfur protein. The 3′ UTR of this clone is bordered by the CDS primer, which is employed by the SMART PCR cDNA synthesis kit as a modified oligo(dT) primer. The 70-aa-long P. americana 3B8 shows 92% identity to D. melanogaster (cf. Adams et al. 2000) and 81% to human Rieske protein (cf. Nishikimi et al. 1990).
The transcript of the RNA polymerase II largest subunit is also down-regulated during memory consolidation. The 3A11 clone aligns to ∼65 aa close to the C-terminal of the RNA polymerase II largest subunit protein. The P. americana 3A11 clone shows 76% identity to Glycine max (cf. Dietrich et al. 1990), 74% identity to human (cf. Wintzerith et al. 1992), and 67% to D. melanogaster RNA polymerase II largest subunit (cf. Jokerst et al. 1989). Eucaryotic RNA polymerases consist of 17-52 tandem heptapeptide repeats at the C-terminal region (for a review, see Corden 1990). The P. americana partial RNA polymerase II largest subunit of the 3A11 clone encodes seven heptapeptide repeats.
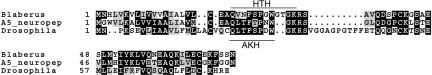
A homolog of the adipokinetic peptide hormone is down-regulated in the trained roach brain as well. The P. americana A5 clone encodes a full-length open reading frame (ORF) that shows 57% ungapped identity to the B. discoidalis hypertrehalosemic hormone (HTH) precursor (cf. Lewis et al. 1997) and 42% to the D. melanogaster adipokinetic hormone (AKH) precursor (cf. Noyes et al. 1995). SignalP predicts that the most likely signal peptide cleavage site is between positions 21 and 22: CEA-QL. This prediction is supported by comparing the precursors and mature peptide hormones of B. discoidalis HTH and D. melanogaster AKH to P. americana A5 polypeptide (Fig. 6). The P. americana A5 clone encodes a polypeptide that is 87.5% identical to the D. melanogaster mature AKH, although the signal and propeptide sequences are rather divergent. On the other hand, the mature B. discoidalis HTH and P. americana A5 are 58% identical. The predicted mature P. americana A5 is missing two internal amino acids compared with the mature B. discoidalis HTH. The signal and propeptide sequences in the B. discoidalis HTH are more similar to the A5 clone sequences than to the D. melanogaster AKH signal and propeptide sequences.
Figure 6.
Comparison of A5/neuropep with related peptides. The conceptual translation product of A5/neuropep is aligned with D. melanogaster adipokinetic hormone precursor (P17975) (Noyes and Schaffer 1990) and with B. discoidalis prepro-hypertrehalosemic hormone (U35277) (Lewis et al. 1997). Multiple sequence alignment was performed using PileUp (GCG, Wisconsin). The identical residues in the alignment were put on black background, and conserved subtitutions were put on gray (BoxShade server). Horizontal lines indicate the mature adipokinetic hormone (AKH) and hypertrehalosemic hormone (HTH).
The P. americana 3C12 clone is homologous to the ORF of the mosquito ENSANGP00000019854 sequence (cf. Holt et al. 2002). The P. americana 3C12 and the A. gambiae ENSANGP00000019854 predicted ORFs show 60% identity in a 62-aa overlap corresponding to the C-terminal region of ENSANGP00000019854. InterPro analysis identifies the “IPR001930 Peptidase_M1” domain within the ENSANGP00000019854 in a region that falls outside the region covered by the P. americana 3C12 clone. The signature sequence of M1 type metalloproteases is the HEXXH motif, which forms part of the metal binding site (Rawlings and Barrett 1995). This motif is absent in the A. gambiae ENSANGP00000019854. The HEXXH motif is present in characterized representatives of the Peptidase_M1 family (cf. InterPro), such as in the mouse glutamyl aminopeptidase (P16406) (Wu et al. 1990), in the S. cerevisiae Ala/Arg aminopeptidase (P37898, Caprioglio et al. 1993), or in the hypothetical D. melanogaster CG13420 (The FlyBase Consortium 2003). The mouse glutamyl aminopeptidase shows 26%, the yeast Ala/Arg aminopeptidase 31%, and the D. melanogaster CG13420 26% identity in 61-aa overlap to the P. americana C12 clone; that is, these Peptidase_M1 type proteins are more distant from P. americana C12, than A. gambiae ENSANGP00000019854, which does not have the Peptidase_M1 signature motif. The HEXXH motif is part of the metal binding site of the M1 type metalloproteases; therefore, ENSANGP00000019854 and 3C12 are probably not in the Peptidase_M1 family.
The P. americana B10, 3B4 and 3C10 clones are not homologous to sequences deposited in the public databases so far. None of these novel sequences has an ORF, thus they likely correspond to untranslated regulatory regions.
Relative quantification of up- and down-regulated transcripts
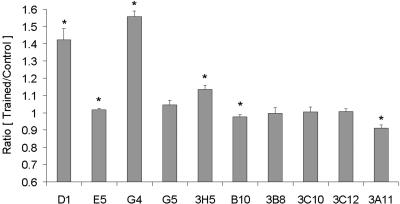
The relative expression levels of T/C (Table 1) and C/T (Table 2) transcripts were measured with real-time RT-PCR (Fig. 7). Statistical analysis confirmed that D1 (N = 26), E5 (N = 24), G4 (N = 40), and 3H5 (N = 38) of the T/C transcripts are significantly up-regulated in the brains of trained P. americana during memory consolidation (P < 0.05). The probability that there is no significant change in the up-regulation of G5 is 0.0725 (N = 35). Clones D1 and G4 are novel, E5 is COXI, G5 is COXII, and 3H5 is ATPase6. B10 (N = 19) and 3A11 (N = 6) of the C/T transcripts are significantly down-regulated in the brains of trained P. americana during memory consolidation (P < 0.05). For 3B8 (N = 21), 3C10 (N = 10), and 3C12 (N = 22), this test does not indicate significant deviation. Clone B10 is novel; 3A11 is RNA polymerase II largest subunit.
Figure 7.
Relative transcript levels are expressed as Trained versus Control ratios (gray bars). T/C and C/T clones were quantified with real-time RT-PCR. Mean ± SEM representation. Ratio equals 1 indicates no detectable change. *P < 0.05, two-tailed t-test.
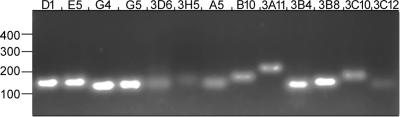
We accounted irreproducible results or experimental difficulties in the case of three transcripts, thus we omitted those from this comparison. The analysis of 3D6 of the T/C transcripts and of A5 of the C/T transcripts was inconclusive. Real-time RT-PCR provided quite divergent RT/C values for these transcripts for Trained-Control pair total RNA pools of different training days. 3B4 of the C/T transcripts has generated complex and apparently multifarious dissociation curves in the majority of the experiments when Trained-Control pair total RNA pools were compared for both trained and control samples. 3B4 is novel. Further sequence information should help to understand the complexity of amplification in the future. We did not see this variability in the preliminary experiments. The PCR amplification of all transcripts was analyzed on agarose gel with each primer pairs (Table 3) and one band was observed (Fig. 8). The template for this experiment was derived from brains of P. americana removed directly from the colony.
Table 3.
Primer sequences for real-time RT-PCR
| Transcript | Forward primer | Reverse primer | Expected size |
|---|---|---|---|
| D1, F5, F7 | GTTCAGCCAT TATCCTGCAC TATC | AGGGTTTATC GTGACATTAT GACC | 140 bp |
| E5 | CCATTGGAGG TTTAACAGGT GTCG | TCCTGCCATG ATAGCAAATA CTGC | 131 bp |
| G4 | GAGGACTGCA CATGCAGACT GG | AGCCCTTTAC GCCCTTTGTT CC | 118 bp |
| G5 | TGGAACAGGA TTCCACGGAC TG | CTGCTGCTTC AAACCCAAAA TGG | 119 bp |
| 3D6 | CATGAACAAT TCCGAGTCTT GGTG | TCCACAGATT TCGGAGCATT GAC | 119 bp |
| 3H5 | TTACCCTTAT TAGGAAATAC AGGTCC | TACGATTGAA TAGTAGCTAC TGCGG | 117 bp |
| A5 | TGGGTCTTGA AGGCTCTGGT TG | ATAGCTTGCA GGGGCCATCT TG | 121 bp |
| B10 | TCGGGATTTG CAGCAAGAGG TG | TGCGACCTGG ACCAGTTTTC TC | 148 bp |
| 3A11 | CACACCAAGT GCCTCCACAC G | CCGGCACAGC CGGTGAATAT G | 186 bp |
| 3B4 | CGAGGACATC GGGCTGCAAA TC | CCATTTTGGG TCCTCAATGT AAATG | 114 bp |
| 3B8, 3B9 | GTTGGTGATC ATCGGTGTGT GC | CCTTTCCGAA TGCGACCAGA GG | 126 bp |
| 3C10 | TACGCGGGCT GGATGTGTTG G | TCCGTTGTCG CGCAGATCAT TG | 152 bp |
| 3C12 | TTTGGCACAG CTGAGCAGAT CG | ATCGCAGGGA ATGTGAGAGT CG | 120 bp |
Figure 8.
PCR amplification of T/C and C/T transcripts (Tables 1, 2) with primer pairs described in Table 3 is analyzed on ethidium bromidestained agarose gel. Numbers at the left indicate size expressed as bp. All reactions resulted in one band of the expected size.
Discussion
The present visual olfactory conditioning paradigm was developed as a method for detecting learning and for monitor retention in cockroaches, a taxon that has proven, among investigated insects, the most suitable for intracellular recordings from higher centers that might support memory formation (Li and Strausfeld 1997, 1999; Mizunami et al. 1998a). The learning paradigm, which involves presenting bimodal sensory information such as recorded from mushroom body efferent neurons, provides crucial insight into the dynamics of short-term, intermediate-term, and long-term memory and memory consolidation in this species. Multiphasic stages of memory have been identified in A. californica (Ghirardi et al. 1995), D. melanogaster (Tully et al. 1994), and the rat (Saitoh and Inokuchi 2000). Comparing animals trained under either nonrestricted or restricted sensory conditions has suggested similar multiphasic stages of memory. In the nonrestricted sensory condition, the visual cue is detected unilaterally, whereas the olfactory cue is detected bilaterally by both antennae. Under the nonrestricted sensory condition, the learned APR, which is the movement of the antenna to the light cue on the same side of the animal as that stimulated, persists for at least 72 h (Lent and Kwon 2004), indicative of long-term memory. Restricted sensory conditioning is when the side opposite to that receiving the US and CS is physically denied any olfactory or visual input by sheathing the contralateral antenna and compound eye. Cock-roaches that have been trained to associate sensory information restricted to one side demonstrate APRs that are significantly different from the naive cockroach at 20 h, but this is no longer the case after 24 h. This suggests that there is a critical time period between 20 and 24 h after training, during which the processes of change from short-term and/or intermediate memory to long-term memory occur. This change, originally referred to as memory consolidation from behavioral experimentation (Müller and Pilzecker 1900), is thought to depend on the stabilization of modified circuits with the participation of proteins that are transcribed during this time (Tully et al. 1994).
Utilizing the kinetics of memory decay, indicated by comparing the results of restricted and unrestricted sensory conditioning, we identified the period at which we investigated gene expression possibly underlying such consolidation. Is this failure of transition to long-term memory due to inadequate gene expression levels? Comparing gene transcripts in the brain of trained and control roaches reveals genes that are differentially expressed during memory consolidation. Our study exemplifies the importance of performing differential screening with both forward and reverse subtracted cDNA libraries: Only 14% of the randomly selected subtractive cDNA library clones are differentially expressed using criteria of differential screening.
Up-regulated genes include mitochondrial genes that are overexpressed during memory consolidation. This increased expression would not be surprising if, during the consolidation phase, synaptic sites are undergoing structural change. Neuronal terminals are rich in mitochondria providing autonomous DNA to satisfy local needs for gene expression without lengthy transport in the neurites. We found that mitochondrial DNA encoded subunits of the 13-subunit Cytochrome C oxidase enzyme are up-regulated during memory consolidation. COX is the terminal enzyme of the respiratory chain complex. Mutations in COX I, II, and III subunits are associated with a variety of clinical phenotypes in man, including mental retardations such as Leigh syndrome and encephalopathy (Shoubridge 2001). COX has been used as an endogenous marker of neuronal activity to monitor pathological conditions. Markedly reduced COX activities have been observed in autopsied brains of humans with presenile dementia of the Alzheimer type, with Huntington's and Parkinson's diseases, and with other neurological diseases (Wong-Riley 1989). In epilepsy-prone mice, increased COX I expression is associated with the epileptogenesis as a consequence of mitochondrial hyperactivity (Nakagawa et al. 2002). COX activity has also been linked to experience-related neuronal activity in the honeybee brain (Armengaud et al. 2000). Antennal input deprivation leads to reduced COX activity staining in three layers of the vertical lobe of the mushroom body. Furthermore, chemical stimulation with potassium ions to cause global depolarization in neurons resulted in significantly increased COX activity in the antennal lobe of the bee (Armengaud et al. 2000). Altered COX activity has been encountered in task-relevant regions of the cattlefish brain after learning (Agin et al. 2001). Whether the above changes in COX activity are caused by changes in the regulation of the catalytic activity of the COX enzyme and/or its mRNA is still to be investigated. In any event, the literature supports the relevance of the COX subunit genes in neuronal functions both during the manifestation of cognitive behavior and under pathological conditions.
Subunit A, a mitochondrial DNA encoded subunit of ATPase 6 of the respiratory chain, was identified in the T/C subtractive cDNA library. Mutations in ATPase 6 subunit A result in fatal disorders, such as NARP (neuropathy, ataxia, and retinitis pigmentosa), Leigh syndrome, and bilateral striatal necrosis (Schon et al. 2001). ATPase 6 subunit A has been identified as a plasticity gene in the cat visual cortex (Yang et al. 2001). Thus the mitochondrial ATPase 6 subunit A is reported to have a neuronal phenotype as well.
Genes are also down-regulated during the consolidation phase, although none of the so-far-identified ones were mitochondrial in origin. Down-regulated genes include RNA polymerase II largest subunit. Protein coding genes are transcribed by RNA polymerase II. RNA polymerase II largest subunit mutants elicit external developmental defects and various levels of lethality in D. melanogaster (The FlyBase Consortium 2003). Why RNA polymerase II largest subunit is being suppressed during memory consolidation is not clear.
Some of the memory consolidation genes that were confirmed to be differentially expressed by real-time RT-PCR are novel. These transcripts are D1 and G4 among the up-regulated transcripts and B10 among the down-regulated transcripts. Further sequence analysis of longer clones should allow us to learn a molecular function of these transcripts.
There are several transcripts that were not found to be differentially expressed using real-time RT-PCR, although differential expression of these subtracted library clones were confirmed by the differential screen. For identifying differences, subtractive hybridization is exceptionally sensitive due to the last step of the employed schema when only the differentially expressed transcripts are amplified. Real-time RT-PCR is less sensitive for the same purpose because it amplifies the two samples to be compared separately, and the product concentrations at the early log linear phase are compared. Both subtractive hybridization and RT-PCR somewhat compensate for individual variance. In real-time RT-PCR experiments, the template was generated from a pool of five brains, in subtractive hybridization the second hybridization step was included to reduce individual differences.
The identified genes described above are differentially expressed at the time when long-term consolidated memory occurs. An interesting feature of our findings is the overrepresentation of mitochondrial DNA-encoded proteins among those genes that were up-regulated. A recent finding that 11 of the 282 human mental retardation genes are encoded by the mitochondrial genome underscores the importance of this cellular compartment in brain plasticity (Inlow and Restifo 2004). Future analyses are aimed at identifying the neurons, and hence the circuits in which they participate, in which differential gene expression occurs during memory consolidation.
Materials and Methods
Behavioral paradigm
Experiments were performed on adult male P. americana. Cock-roaches were purchased from Carolina Biological Supply (North Carolina, USA), placed in a colony, and allowed 1-2 wk to adapt to their new environment prior to experimentation. Animals were provided with a food mixture of catfood and peanut butter granola and water and kept as described by Lent and Kwon (2004).
The APR of P. americana was classically conditioned in a visual olfactory learning paradigm under nonrestricted sensory conditions as described by Lent and Kwon (2004). Briefly, after 20-24 h starvation, animals were anesthetized with carbon dioxide and restrained. One hour later the cockroach was placed in the middle of a uniformly round arena. The wall contained a single green LED in the right hemifield positioned 5° from the midline (for the restricted sensory condition, an additional LED was positioned in the left hemifield 5° from the midline). During training a food odor source (US) was spatially coincident with the green LED (CS). The nonrestricted training protocol comprised three pretaining trials, five training trials, and three test trials (Fig. 1; for details, see Lent and Kwon 2004). Cockroaches that elicited APRs toward the CS stimulus (green LED) after the training session were considered trained. In order to characterize the consolidation phase of memory formation, cockroaches were trained under restricted conditions as well. Under restricted sensory condition, the eye and the antenna are covered on one side with wax and a sealed plastic tube, respectively. This effectively restricts olfactory and visual sensory information to one half, and the CS and US are delivered to this side, unilaterally. The pretraining, training, and testing in the restricted sensory condition were identical to the nonrestricted except one half of the head, randomly determined, was covered as described. Nonparametric statistics were used for behavioral analysis as described by Lent and Kwon (2004). The Mann-Whitney U test was used to test differences between two groups. Values shown represent the responses “0” or “1” in percentages; therefore, standard deviations are not depicted.
For the subsequent genetic screen, all animals were trained under nonrestricted sensory condition. Control roaches were always kept side by side with the trained ones and were exposed to the same treatment—isolation from the colony, starvation, carbon dioxide treatment, period of time spent restrained—but the training itself was omitted. Between 21 and 23 h after training, the whole brains, including optic lobes, of both trained and control animals were dissected.
Suppression subtractive hybridization
To prepare cDNA for subtractive hybridization total RNA was prepared from each individual brain separately with TRIzol (GIBCO). Briefly, brains were dissected in DEPC-treated phosphate buffered saline and then transferred into TRIzol reagent. The manufacturer's standard protocol resulted in 2-4 μg total RNA per roach brain.
Suppression subtractive hybridization was performed using the PCR-Select cDNA subtraction system (Clontech) following the manufacturer's protocol. Briefly, full-length cDNA was prepared using the SMART PCR cDNA synthesis kit (Clontech). Reverse transcription was performed on ∼0.1 μg total RNA of each individual roach brain. The cDNA pools were cut with RsaI and ligated to different adaptor oligonucleotides provided by the manufacturer. RsaI digests were phenol purified using Phase-lock (Eppendorf) and ethanol precipitated. Ligation efficiency analysis was performed with PCR using two primers designed from a known P. americana cDNA sequence (elongation factor-1 α [EF1], GenBank accession no. U90054). For each roach brain sample, the PCR product using one EF1-specific primer (180-200 bp of U90054) and an adaptor-specific primer (provided with the kit) was about the same intensity as the PCR product amplified using two EF1-specific primers (180-200 bp and 556-575 bp of U90054). Custom primers were obtained from Invitrogen. Two-directional subtractive hybridization was performed using the brains of trained and control animals. In the forward subtraction, two different Trained brains were compared to one Control brain. Thus, the T/C subtractive cDNA pool was generated. The T/C pool was cloned into pT-Adv vector to generate a T/C library (Advantage PCR cloning kit, Clontech). This library represents genes with increased expression in the brain of the trained roach. In the reverse subtraction, two different Control brains were compared to one Trained brain. The C/T cDNA pool was created in this way. The C/T pool was cloned into pT-Adv vector to make a C/T library. The C/T library represents genes with suppressed expression in the brain of the trained roach.
Differential screening of the subtracted clones
Randomly selected clones of the T/C and the C/T libraries were tested with differential screening. We took advantage of a 96-well plate workstation, thus the number of clones that were analyzed is a multiple of 96. Briefly, plasmid arrays were made with 288 (3 × 96) randomly selected clones and probed with both subtracted cDNA pools. Plasmid minipreps were prepared using Qiagen 96 Turbo Miniprep kit on a Biomek FX automated workstation. Clones were applied in duplicates to nylon membranes to make two identical arrays. To remove adaptor oligonucleotides from the hybridization probes, subtracted cDNA pools were digested with RsaI restriction endonuclease and purified with MinElute (Qiagen); 450 ng of each subtracted cDNA pool was labeled with DIG (DIG-labeling, Boehringer). The DIG-labeled T/C and C/T cDNA probes were made with random priming and were added to the hybridization mixture at 20 ng/mL concentration. DNA arrays were prehybridized for at least 60 min at 72°C in ExpressHyb hybridization solution supplemented with Blocking solution for differential screening (Clontech). The Blocking solution contains sheared salmon sperm DNA and oligonucleotides corresponding to the 3′ terminal half of the adaptors and their complementary oligonucleotides. One membrane was probed with the DIG-labeled T/C cDNA pool, the other with the DIG-labeled C/T cDNA pool. Hybridization and washing steps were performed as described in the PCR-Select Differential Screening protocol (Clontech). A peroxidase conjugated anti-DIG antibody (Boehringer) and Supersignal west femto chemiluminescent substrate (Pierce) combination was used to visualize the hybridization signal on the cDNA arrays.
Sequencing and sequence analysis of candidate clones
Plasmid DNAs were prepared using Qiagen miniprep kit. DNA was sequenced with either Applied Biosystems PRISM 377 DNA or 3730xl DNA Sequencers. Initial sequence analysis was performed in GCG (Wisconsin Package version 10.3, Accelrys, Inc.). P. americana cDNA sequences were submitted to database search assisted by fasta (Pearson and Lipman 1988; Pearson 1990) and OPRs were submitted to the InterPro database (Mulder et al. 2003) at the European Bioinformatics Institute (www.ebi.ac.uk). Sequence data were also matched against specialized databases such as FlyBase for D. melanogaster sequences (The FlyBase Consortium 2003), SilkBase for Bombyx mori sequences (Shimada et al. 1999-2000), the A. gambiae genome at the National Center for Biotechnology (Holt et al. 2002; www.ncbi.nlm.nih.gov), and the Honeybee EST project (Whitfield et al. 2002). Database searches were performed through September 24, 2004. BoxShade server was utilized to shade PileUp multiple alignments (www.ch.embnet.org). Signal peptide and cleavage site prediction was performed with SignalP (Nielsen et al. 1997) at the Center for Biological Sequence Analysis (www.cbs.dtu.dk).
Quantitative real-time RT-PCR
Twenty-one hours after training, both trained and control animals were decapitated, and the heads were stored submerged in RNAlater (Ambion). The brains of trained and control animals that were compared were always obtained pair-wise from the same-day experiment. The whole brains, including optic lobes, of both trained and control animals were dissected under RNAlater. The retina was always peeled away. Five brains were pooled to perform each total RNA extractions. The trained and its corresponding control pool were processed side by side during the entire RNA isolation procedure. Total RNA was isolated with RNeasy Mini (Qiagen). This was followed by DNase I treatment and the subsequent application of DNase inactivation reagent (Ambion). In the final step, total RNA was concentrated with RNAqueous-Micro (Ambion). RNA concentration was determined spectrophotometrically with NanoDrop (Ambion). Five brains yielded 2 to 4 μg total RNA. Two to four microgram total RNA was reverse transcribed with SuperScript III first strand synthesis system (Invitrogen) using oligo(dT)20 primer. Aliquots of the reverse transcription reactions were used as templates in the real-time PCR experiments. The trained and the corresponding control total RNA were always transcribed at the same concentration side by side.
Quantitative real-time PCR (qPCR) was performed on ABI 7000 sequence detector. Transcript-specific primers for qPCR are listed in Table 3. For the PCR reactions, multiples of 20 μL mastermixes were prepared using 12.5 μL iTaq SYBR Green supermix with ROX (BioRad) and 7.5μL diluted cDNA corresponding to 400-3.125 ng total RNA. A 20-μL aliquot of the mastermix was supplemented with 2.5μL of 10 μM forward and 2.5μL of 10 μM reverse primers in each well of the 96-well optical plates. Cycling conditions include initial denaturation (2 min at 95°C), amplification, and quantification program repeated 45 times (10 sec at 95°C, 1 min at 60°C with a single fluorescent measurement at the end of each elongation step) and dissociation protocol (60°C to 95°C by 1°C increments followed by a 30-sec hold and fluorescent measurement). Real-time PCR efficiencies were calculated from the slopes of the “standard curves” of the ABI Prism 7000 SDS program according to the equation E = 10(—1/slope) (Rasmussen 2001). For the relative quantification of each transcript, the efficiencies were determined as the average of three to five independent experiments. In each experiment four to 10 template concentrations were used to perform the linear regression. The relative expression ratio (R) of the transcript is calculated based on the crossing point deviation (ΔCP) measured in the control and trained total RNA pools and based on the PCR efficiency of a transcript (Pfaffl 2001). The relative expression ratio in the trained versus control RNA pools was calculated with the following equation: RT/C = (Etranscript)ΔCP(control-trained). For each of the transcripts, six to 40 crossing point deviations (ΔCP) were obtained from two to three independent experiments. Each experiment was performed with at least four template concentrations. Statistics were carried out using Statistica 5.5 for Windows, and results were regarded as `not significant' if P > 0.05.
Acknowledgments
This work was supported by grants from the Human Frontiers Science project (RG01432000B) and from the National Science Foundation (NSF IBN0411958) to N.J.S. D.D.L. holds a National Science Foundation Graduate Research Fellowship Award. We thank Brian Coullahan, Kevin Kiesler, and Ryan Sprissler at the Genomic Analysis and Technology Core facility (ARL, University of Arizona) for sequencing, 96-well format sample processing, and advice in real-time RT-PCR; Ellen Walworth for her advice on differential screens; and Myria Kyriacou for technical assistance.
Article published online before print. Article and publication date are at http://www.learnmem.org/cgi/doi/10.1101/lm.87905.
References
- Adams, M.D., Celniker, S.E., Holt, R.A., Evans, C.A., Gocayne, J.D., Amanatides, P.G., Scherer, S.E., Li, P.W., Hoskins, R.A., Galle, R.F., et al. 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185-2195. [DOI] [PubMed] [Google Scholar]
- Agin, V., Chicher, R., and Chichery, M.P. 2001. Effects of learning on cytochrome oxidase activity in cuttlefish brain. Neuroreport 12: 113-116. [DOI] [PubMed] [Google Scholar]
- Alberini, C.M. 1999. Genes to remember. J. Exp. Biol. 202: 2887-2891. [DOI] [PubMed] [Google Scholar]
- Alkon, D.L. 1984. Calcium-mediated reduction of ionic currents: A biophysical memory trace. Science 226: 1037-1045. [DOI] [PubMed] [Google Scholar]
- Anderson, S., Bankier, A.T., Barrell, B.G., de Bruijn, M.H., Coulson, A.R., Drouin, J., Eperon, I.C., Nierlich, D.P., Roe, B.A., Sanger, F., et al. 1981. Sequence and organization of the human mitochondrial genome. Nature 290: 457-465. [DOI] [PubMed] [Google Scholar]
- Armengaud, C., Causse, N., Aït-Oubah, J., Ginolhac, A., and Gauthier, M. 2000. Functional cytochrome oxidase histochemistry in the honeybee brain. Brain Res. 859: 390-393. [DOI] [PubMed] [Google Scholar]
- Beard, C.B., Hamm, D.M., and Collins, F.H. 1993. The mitochondrial genome of the mosquito Anopheles gambiae: DNA sequence, genome organization, and comparisons with mitochondrial sequences of other insects. Insect Mol. Biol. 2: 103-124. [DOI] [PubMed] [Google Scholar]
- Byers, D., Davis, R.L., and Kiger Jr., J.A. 1981. Defect in cyclic AMP phosphodiesterase due to the dunce mutation of learning in Drosophila melanogaster. Nature 289: 79-81. [DOI] [PubMed] [Google Scholar]
- Caprioglio, D.R., Padilla, C., and Werner-Washburne, M. 1993. Isolation and characterization of AAP1: A gene encoding an alanine/arginine aminopeptidase in yeast. J. Biol. Chem. 268: 14310-14315. [PubMed] [Google Scholar]
- Castellucci, V.F., Kennedy, T.E., Kandel, E.R., and Goelet, P. 1988. A quantitative analysis of 2-D gels identifies proteins in which labeling is increased following long-term sensitization in Aplysia. Neuron 1: 321-328. [DOI] [PubMed] [Google Scholar]
- Cayre, M., Malaterre, J., Scotto-Lomassese, S., Strambi, C., and Strambi, A. 2002. The common properties of neurogenesis in the adult brain: From invertebrates to vertebrates. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 132: 1-15. [DOI] [PubMed] [Google Scholar]
- Corden, J.L. 1990. Tails of RNA polymerase II. Trends Biochem. Sci. 15: 383-387. [DOI] [PubMed] [Google Scholar]
- Crozier, R.H. and Crozier, Y.C. 1992. The cytochrome b and ATPase genes of honeybee mitochondrial DNA. Mol. Biol. Evol. 9: 474-482. [DOI] [PubMed] [Google Scholar]
- de Bruijn, M.H. 1983. Drosophila melanogaster mitochondrial DNA, a novel organization and genetic code. Nature 304: 234-241. [DOI] [PubMed] [Google Scholar]
- Dietrich, M.A., Prenger, J., and Guilfoyle, T.J. 1990. Analysis of the genes encoding the largest subunit of RNA polymerase II in Arabidopsis and soybean. Plant Mol. Biol. 15: 207-223. [DOI] [PubMed] [Google Scholar]
- Dudai, Y., Jan, Y.N., Byers, D., Quinn, W.G., and Benzer, S. 1976. dunce, a mutant of Drosophila deficient in learning. Proc. Natl. Acad. Sci. 73: 1684-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The FlyBase Consortium. 2003. The FlyBase database of the Drosophila genome projects and community literature. Nucleic Acids Res. 31: 172-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghirardi, M., Montarolo, P.G., and Kandel, E.R. 1995. A novel intermediate stage in the transition between short- and long-term facilitation in the sensory to motor neuron synapse of Aplysia. Neuron 14: 413-420. [DOI] [PubMed] [Google Scholar]
- Heisenberg, M. 2003. Mushroom body memoir: From maps to models. Nat. Rev. Neurosci. 4: 266-275. [DOI] [PubMed] [Google Scholar]
- Holt, R.A., Subramanian, G.M., Halpern, A., Sutton, G.G., Charlab, R., Nusskern, D.R., Wincker, P., Clark, A.G., Ribeiro, J.M., Wides, R., et al. 2002. The genome sequence of the malaria mosquito Anopheles gambiae. Science 298: 129-149. [DOI] [PubMed] [Google Scholar]
- Inlow, J.K. and Restifo, L.L. 2004. Molecular and comparative genetics of mental retardation. Genetics 166: 835-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jokerst, R.S., Weeks, J.R., Zehring, W.A., and Greenleaf, A.L. 1989. Analysis of the gene encoding the largest subunit of RNA polymerase II in Drosophila. Mol. Gen. Genet. 215: 266-275. [DOI] [PubMed] [Google Scholar]
- Kandel, E.R. and Schwartz, J.H. 1982. Molecular biology of learning: Modulation of transmitter release. Science 218: 433-443. [DOI] [PubMed] [Google Scholar]
- Kucharski, R. and Maleszka, R. 2002. Molecular profiling of behavioural development: Differential expression of mRNAs for inositol 1,4,5-trisphosphate 3-kinase isoforms in naive and experienced honeybees (Apis mellifera). Brain Res. Mol. Brain Res. 99: 92-101. [DOI] [PubMed] [Google Scholar]
- Kwon, H.-W., Lent, D.D., and Strausfeld, N.J. 2004. Spatial learning in the restrained American cockroach, Periplaneta americana. J. Exp. Biol. 207: 377-383. [DOI] [PubMed] [Google Scholar]
- Leil, T.A., Ossadtchi, A., Nichols, T.E., Leahy, R.M., and Smith, D.J. 2003. Genes regulated by learning in the hippocampus. J. Neurosci. Res. 71: 763-768. [DOI] [PubMed] [Google Scholar]
- Lent, D.D. and Kwon, H.-W. 2004. Antennal movements reveal associative learning in the American cockroach, Periplaneta americana. J. Exp. Biol. 207: 369-375. [DOI] [PubMed] [Google Scholar]
- Lewis, D.K., Jezierski, M.K., Keeley, L.L., and Bradfield, J.Y. 1997. Hypertrehalosemic hormone in a cockroach: Molecular cloning and expression. Mol. Cell. Endocrinol. 130: 101-108. [DOI] [PubMed] [Google Scholar]
- Li, Y. and Strausfeld, N.J. 1997. Morphology and sensory modality of mushroom body extrinsic neurons in the brain of the cockroach, Periplaneta americana. J. Comp. Neurol. 387: 631-650. [PubMed] [Google Scholar]
- ———. 1999. Multimodal efferent and recurrent neurons in the medial lobes of cockroach mushroom bodies. J. Comp. Neurol. 409: 647-663. [DOI] [PubMed] [Google Scholar]
- Liu, H. and Beckenbach, A.T. 1992. Evolution of the mitochondrial cytochrome oxidase II gene among 10 orders of insects. Mol. Phylogenet. Evol. 1: 41-52. [DOI] [PubMed] [Google Scholar]
- Martinez-Gonzalez, J. and Hegardt, F.G. 1994. Cytochrome c oxidase subunit I from the cockroach Blattella germanica: Cloning, developmental pattern and tissue expression. Insect Biochem. Mol. Biol. 24: 619-626. [DOI] [PubMed] [Google Scholar]
- Mizunami, M., Okada, R., Li, Y., and Strausfeld, N.J. 1998a. Mushroom bodies of the cockroach: Activity and identities of neurons recorded in freely moving animals. J. Comp. Neurol. 28: 501-519. [PubMed] [Google Scholar]
- Mizunami, M., Weibrecht, J.M., and Strausfeld, N.J. 1998b. Mushroom bodies of the cockroach: Their participation in place memory. J. Comp. Neurol. 402: 520-537. [PubMed] [Google Scholar]
- Morris, R.G., Garrud, P., Rawlins, J.N., and O'Keefe, J. 1982. Place navigation impaired in rats with hippocampal lesions. Nature 297: 681-683. [DOI] [PubMed] [Google Scholar]
- Mulder, N.J., Apweiler, R., Attwood, T.K., Bairoch, A., Barrell, D., Bateman, A., Binns, D., Biswas, M., Bradley, P., Bork, P., et al. 2003. The InterPro Database, 2003 brings increased coverage and new features. Nucleic Acids Res. 31: 315-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, G.E. and Pilzecker, A. 1900. Experimentelle Beitrage zur Lehre von Gedächtnis. Z. Psychol. 1: 1-300. [Google Scholar]
- Nakagawa, Y., Mishima, T., Murashima, Y.L., and Nakano, K. 2002. Increased expression of mitochondrial respiratory enzymes in the brain of epilepsy-prone, naive EL mice. Brain Res. Mol. Brain Res. 101: 59-61. [DOI] [PubMed] [Google Scholar]
- Nielsen, H., Engelbrecht, J., Brunak, S., and von Heijne, G. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10: 1-6. [DOI] [PubMed] [Google Scholar]
- Nishikimi, M., Hosokawa, Y., Toda, H., Suzuki, H., and Ozawa, T. 1990. The primary structure of human Rieske iron-sulfur protein of mitochondrial cytochrome bc1 complex deduced from cDNA analysis. Biochem. Int. 20: 155-160. [PubMed] [Google Scholar]
- Noyes, B.E. and Schaffer, M.H. 1990. The structurally similar neuropeptides adipokinetic hormone I and II are derived from similar, very small mRNAs. J. Biol. Chem. 265: 483-489. [PubMed] [Google Scholar]
- Noyes, B.E., Katz, F.N., and Schaffer, M.H. 1995. Identification and expression of the Drosophila adipokinetic hormone gene. Mol. Cell. Endocrinol. 109: 133-141. [DOI] [PubMed] [Google Scholar]
- O'Keefe, J. and Conway, D.H. 1978. Hippocampal place units in the freely moving rat: Why they fire where they fire. Exp. Brain Res. 31: 573-590. [DOI] [PubMed] [Google Scholar]
- Pearson, W.R. 1990. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 183: 63-98. [DOI] [PubMed] [Google Scholar]
- Pearson, W.R. and Lipman, D.J. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. 85: 2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl, M.W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29: 2002-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen, R. 2001. Quantification on the LightCycler. In Rapid cycle real-time PCR methods and applications (eds. S. Meuer et al.), pp. 21-34. Springer Press, Heidelberg, Germany.
- Rawlings, N.D. and Barrett, A.J. 1995. Evolutionary families of metallopeptidases. Meth. Enzymol. 248: 183-228. [DOI] [PubMed] [Google Scholar]
- Saitoh, Y. and Inokuchi, K. 2000. A triphasic curve characterizes the retention of lever-pressing behavior rewarded by lateral hypothalamic stimulation during the immediate-post-trial period in rats: Implications for a transient-intermediate stage between short- and long-term memory. Neurosci Res. 37: 211-219. [DOI] [PubMed] [Google Scholar]
- Schon, E.A., Santra, S., Pallotti, F., and Girvin, M.E. 2001. Pathogenesis of primary defects in mitochondrial ATP synthesis. Semin. Cell Dev. Biol. 12: 441-448. [DOI] [PubMed] [Google Scholar]
- Shimada, T., Mita, K., and Kobayashi, M. 1999-2000. SilkBase: Bombyx mori EST database. http://www.ab.a.u-tokyo.ac.jp/silkbase.
- Shoubridge, E.A. 2001. Cytochrome c oxidase deficiency. Am. J. Med. Genet. 106: 46-52. [DOI] [PubMed] [Google Scholar]
- Stork, O., Stork, S., Pape, H.C., and Obata, K. 2001. Identification of genes expressed in the amygdala during the formation of fear memory. Learn. Mem. 8: 209-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strausfeld, N.J. 2002. Organization of the honey bee mushroom body: Representation of the calyx within the vertical and gamma lobes. J. Comp. Neurol. 450: 4-33. [DOI] [PubMed] [Google Scholar]
- Strausfeld, N.J. and Li, Y. 1999. Organization of olfactory and multimodal afferent neurons supplying the calyx and pedunculus of the cockroach mushroom bodies. J. Comp. Neurol. 409: 603-625. [PubMed] [Google Scholar]
- Tully, T., Préat, T., Boynton, S.C., and Del Vecchio, M. 1994. Genetic dissection of consolidated memory in Drosophila. Cell 79: 35-47. [DOI] [PubMed] [Google Scholar]
- Whitfield, C.W., Band, M.R., Bonaldo, M.F., Kumar, C.G., Liu, L., Pardinas, J.R., Robertson, H.M., Soares, M.B., and Robinson, G.E. 2002. Annotated expressed sequence tags and cDNA microarrays for studies of brain and behavior in the honey bee. Genome Res. 12: 555-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintzerith, M., Acker, J., Vicaire, S., Vigneron, M., and Kedinger, C. 1992. Complete sequence of the human RNA polymerase II largest subunit. Nucleic Acids Res. 20: 910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong-Riley, M.T. 1989. Cytochrome oxidase: An endogenous metabolic marker of neuronal activity. Trends Neurosci. 12: 94-101. [DOI] [PubMed] [Google Scholar]
- Wu, Q., Lahti, J.M., Air, G.M., Burrows, P.D., and Cooper, M.D. 1990. Molecular cloning of the murine BP-1/6C3 antigen: A member of the zinc-dependent metallopeptidase family. Proc. Natl. Acad. Sci. 87: 993-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, X. and Arnason, U. 1996. The mitochondrial DNA molecule of Sumatran orangutan and a molecular proposal for two (Bornean and Sumatran) species of orangutan. J. Mol. Evol. 43: 431-437. [DOI] [PubMed] [Google Scholar]
- Yang, C., Silver, B., Ellis, S.R., and Mower, G.D. 2001. Bidirectional regulation of mitochondrial gene expression during developmental neuroplasticity of visual cortex. Biochem. Biophys. Res. Commun. 287: 1070-1074. [DOI] [PubMed] [Google Scholar]