Abstract
BACKGROUND
Cervical cancer is a predominantly human papillomavirus (HPV)‐driven disease worldwide. However, its incidence is unexplainably low in western Asia, including Saudi Arabia. Using this paradigm, we investigated the role of HPV infection rate and host genetic predisposition in TP53 G72C single nucleotide polymorphism (SNP) presumed to affect cancer incidence.
METHODS
Patients treated between 1990 and 2012 were reviewed, and a series of 232 invasive cervical cancer cases were studied and compared with 313 matched controls without cancer. SNP was genotyped by way of direct sequencing. HPV linear array analysis was used to detect and genotype HPV in tumor samples.
RESULTS
The incidence of cervical cancer revealed bimodal peaks at 42.5 years, with a slighter rebound at 60.8 years. Among all cases, 77% were HPV‐positive and 16 HPV genotypes were detected—mostly genotypes 16 (75%) and 18 (9%)—with no difference by age, histology, or geographical region. Although the TP53 G72C genotype was not associated with overall cervical cancer risk, it was significantly associated with HPV positivity (odds ratio, 0.57; 95% confidence interval, 0.36‐0.90; P = .016). Furthermore, the variant C allele was significantly overtransmitted in the population (P < .0003).
CONCLUSION
Cervical cancer incidence displays bimodal curve peaking at a young age with secondary rebound at older age. The combination of relative low HPV infection and variant TP53 72C allele overtransmission provide a plausible explanation for the low incidence of cervical cancer in our population. Therefore, HPV screening and host SNP genotyping may provide more relevant biomarkers to gauge the risk of developing cervical cancer. Cancer 2017;123:2459–66. © 2017 The Authors. Cancer published by Wiley Periodicals, Inc. on behalf of American Cancer Society. This is an open access article under the terms of the Creative Commons Attribution NonCommercial License, which permits use, distribution and reproduction in any medium, provided the original work is properly cited and is not used for commercial purposes.
Keywords: cervical cancer, human papillomavirus, HPV genotyping, TP53 polymorphism, cancer predisposition, single nucleotide polymorphism
Short abstract
Cervical cancer incidence shows bimodal peaks at 43 years and relative rebound at 61 years of age. A reduced rate of human papillomavirus infection and overtransmission of the TP53 72C variant lower cancer incidence.
INTRODUCTION
Uterine cervical cancer (CxCa) is one of the most frequent malignancies affecting women worldwide.1 However, its incidence is not uniform in various populations having different socioeconomic and national human development levels; a large majority of the global burden occurs in less‐developed regions, and the lowest rates are in western Asia.1 Infection with human papillomavirus (HPV) stands out as being the main cause among all known risk factors, and high‐risk HPVs drive the major carcinogenic events in the pathogenesis of CxCa, with an estimated worldwide prevalence ranging between 85% and 99%.2, 3, 4 In contrast, CxCa incidence is low in a few countries, even though they lack national screening programs.5 In Saudi Arabia, CxCa is infrequent, accounting for only 2.4% of all new cases in women.6 The reason for this low incidence is not known.
Genetic inherited predisposition has largely been considered as an interfering factor modifying the risk of developing various types of malignancies, including CxCa.7 The hypothesis is that genetic polymorphisms in various classes of genes might foster HPV persistence and enhance progression to cancer.8 Moreover, emerging genome‐wide association studies have supported this assumption and detected susceptibility loci formerly not implicated in CxCa development.9 The pivotal gene encoding the tumor suppressor protein p53 (TP53) is the most widely evoked candidate gene with an alleged ability to affect the oncogenic potential of the viral HPV‐E6 protein.10 The p53 protein is a central regulator of cell cycle and DNA repair that orchestrates several pathways to preserve genomic integrity, which can be deregulated by HPV infection. A common nonsynonymous polymorphism (G/C, rs1042522) codes for arginine (Arg) or proline (Pro) at position 72 of the p53 amino acid sequence. The majority allele G (Arg form of p53) was associated with CxCa development and has been described as more susceptible than the Pro variant to HPV‐E6–mediated degradation.10
The low cervical cancer incidence observed in our population provides an ideal setting to study host genetic predisposition in conjunction with HPV infection. We hypothesized that low HPV presence in tumors and/or polymorphic variation in TP53 G72C contribute to reduced cancer risk. Because systematic information about CxCa in Saudi women is not available to help solve this dilemma,11 we investigated the prevalence of HPV infection and genotype in a series of CxCa patients and the possible association with TP53 single nucleotide polymorphism (SNP) rs1042522, which is presumed to predispose to cancer. The objective is to contribute to the understanding of this disease in our community, so health practitioners can deliver more insightful and personalized preventive medicine.
PATIENTS AND METHODS
Ethical Considerations
The study used cervical tumor tissues obtained during routine diagnostic procedures of CxCa patients. The samples were processed anonymously. Blood samples, where applicable, were obtained after informed consent was given. The Institutional Review Board and Research Centre Ethics Committee approved the study.
Clinical Specimens
CxCa patients treated between 1990 and 2012 in our tertiary hospital were reviewed, and 232 patients with histopathologically proven invasive tumors were included in the study. Sufficient pathological specimens to test for HPV infection were available for 213 patients, whereas blood samples were obtained from the remaining 19 patients to perform SNP genotyping. Patient eligibility criteria included cervical cancer without restriction on age or histological type (squamous cell carcinoma, adenocarcinoma). Pathological tumor samples were from regular biopsies or paraffin‐embedded tissues. Normal female volunteers without history of cancer were used as controls for SNP genotyping. They were recruited between employees or individuals undergoing physical examination for benign conditions. The control group consisted of 313 best matched women for age, ethnicity, and gravida. Blood samples (5 mL) were obtained upon providing approved informed consent for the genetic study.
DNA Extraction, SNP Sequencing, and Genotyping
DNA extraction was performed using a Puregene DNA Purification Kit (Gentra Systems, Big Lake, MN). The PCR thermal cycling conditions, primers used for amplification and genotyping of TP53 G72C Arg/Pro (rs1042522), have been described previously.12 The DYEnamic ET Dye Terminator Cycle Sequencing Kit (Amersham Biosciences, Little Chalfont, United Kingdom) was used to sequence the amplified fragment on the MegaBase 1000 sequencer (Applied Biosystems, Foster City, CA). Results were aligned to the reference sequence using SeqManII sequence analysis software (DNASTAR, Madison, WI) to determine the SNP genotype.
Detection and Genotyping of HPV
We used the Linear Array HPV Genotyping Test (LA HPV GT; Roche Diagnostics, Basel, Switzerland) according to the manufacturer's recommended methodology. The kit detects and genotypes the 37 most common anogenital HPVs (13 high‐risk: 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 68, and 24 low‐risk: 6, 11, 26, 40, 42, 53, 54, 55, 61, 62, 64, 66, 67, 69, 70, 71, 72, 73 [MM9], 81, 82 [MM4], 83 [MM7], 84 [MM8], 89 [CP6108], and IS39). The adequacy of samples is determined by the betaglobin gene as an internal control. HPV‐positive reactions show blue bands on the strip. Negative results show no HPV band after at least 2 repeated tests with confirmed adequacy of samples.
Statistical Analysis
Univariate analysis was performed to screen for factors that might be related to cancer (age, geographic region, parity, histology, cancer stage, and associated diseases). The SNP genetic association was assessed by the odds ratio (OR) and its confidence interval (CI). Significance level was analyzed using a chi‐square test, with the continuity uncorrected. One sample z‐test was used to detect differences in proportions when the referenced proportion was deemed constant. Each allele's transmission was assessed using a chi‐square test with 1 degree of freedom. A P value ≤ .05 was considered statistically significant. Deviations from Hardy‐Weinberg equilibrium (HWE) were tested to study the influences of genotype distribution by comparing the observed frequencies with the expected frequencies. Clustering of age‐related incidence of CxCa was verified by the analysis of mixture of 2 Gaussian distributions and was fitted using the EM algorithm in the R package (https://www.r-project.org) using the mixtools function. Statistical analysis softwares used were the SigmaPlot platform (version 12.5; SPSS Science, Chicago, IL), Case Control Studies (http://ihg.gsf.de/cgi-bin/hw/hwa1.pl; Helmholtz Center, Munich, Germany), and MedCalc (https://www.medcalc.org/calc/test_one_proportion.php; Ostend, Belgium).
RESULTS
Patients and Clinical Data
The characteristics of the cervical cancer patients are summarized in Table 1. The median patient age at diagnosis of CxCa was 46 years (range, 28‐78 years). There were 224 Saudis and 8 expatriate women. Most (72%) of the patients originated geographically from central and eastern areas of Saudi Arabia followed by southern, western, and northern regions (Table 1). Most patients had several pregnancies with a median number of gravidity of 8 (range, 0‐19) and parity of 7 (range, 0‐16). Only 6 patients (3%) had prior screening, whereas 11 patients (5%) reported history of other unidentified cervical infections. Normal controls had similar socio‐economic distribution and comparable age (median, 45 years; range, 28‐78 years), ethnicity (94% Saudis), and gravida (median, 7; range, 0‐18). No history of Pap smear screening or previous sexually transmitted disease (STD) infection were reported.
Table 1.
Characteristics of Cervical Cancer Patients
| HPV Status | No. of Patients | Age, y, Median (Range) | Region of Saudi Arabia, n (%) | Gravidity, Median (Range) | Parity, Median (Range) | Histology, n (%) | Stage, n (%) | Associated Diseases, n (%) | Previous Screening, n (%) | Previous STD, n (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| HPV‐positive | 163 | 46 (29‐78) |
C: 74 (46) E: 43 (26) S: 16 (10) W: 15 (9) N: 15 (9) |
8 (0‐19) | 7 (0‐16) | AC: 31 (19) SCC: 132 (81) |
I: 42 (26) II: 78 (48) III: 36 (22) IV: 7 (4) |
Yes: 67 (41) No: 96 (59) |
Yes: 6 (4) No: 157 (96) |
Yes: 11 (7) No: 152 (93) |
| HPV‐negative | 50 | 44 (28‐74) |
C: 21 (42) E: 9 (18) S: 7 (14) W: 7 (14) N: 6 (12) |
6 (1‐13) | 6 (0‐12) |
AC: 9 (18) SCC: 41 (82) |
I: 17 (34) II: 21 (42) III: 9 (18) IV: 3 (6) |
Yes: 17 (34) No: 33 (66) |
Yes: 0 (0) No: 50 (100) |
Yes: 0 (0) No: 50 (100) |
| P a | .217 | .570 | .051 | .049 | .872 | .627 | .369 | .999 | .999 | |
| Unknown | 19 | 51 (30‐72) |
C: 13 (68) E: 6 (32) S: 0 (0) W: 0 (0) N: 0 (0) |
9 (5‐12) | 8 (5‐12) |
AC: 1 (5) SCC: 18 (95) |
I: 5 (26) II: 9 (48) III: 4 (21) IV: 1 (5) |
Yes: 10 (53) No: 9 (47) |
Yes: 0 (0) No: 19 (100) |
Yes: 0 (0) No: 19 (100) |
| Total | 232 | 46 (28‐78) |
C: 108 (47) E: 58 (25) S: 23 (10) W: 22 (9) N: 21 (9) |
8 (0‐19) | 7 (0‐16) |
AC: 41 (18) SCC: 191 (82) |
I: 64 (28) II: 108 (46) III: 49 (21) IV: 11 (5) |
Yes: 94 (41) No: 138 (59) |
Yes: 6 (3) No: 226 (97) |
Yes: 11 (5) No: 221 (95) |
AC, adenocarcinoma; C, central; E, eastern; N, northern; S, southern; SCC, squamous cell carcinoma; STD, sexually transmitted disease; W, western.
Significance level for the difference between HPV‐positive and HPV‐negative patients.
FIGO (International Federation of Gynecology and Obstetrics) cancer staging varied between IA and IV, with most patients (74%) having stage II/III disease (Table 1). Cancer histology revealed that 41 patients (18%) had adenocarcinoma, whereas 191 (82%) had squamous cell carcinoma of the cervix. The distribution of the 2 histopathological types of CxCa by patient age is shown in Figure 1A. The age distribution of CxCa evoked a bimodal curve separating the patients into 2 age groups: 25‐50 years and 51‐80 years. The first peak appeared at 41‐45 years, followed by a relative rebound peak at 56‐60 years. The analysis of mixture of 2 Gaussian distributions fitted using the EM algorithm in the R package showed distinct 2 curves (Fig. 1B) with means of 42.56 and 60.84 (standard deviation = 6.33 for both) and mixing proportions of 74% and 26% for the first and second clusters, respectively. There was no discernable difference between histological types (adenocarcinoma versus squamous cell carcinoma) between the 2 age clusters.
Figure 1.
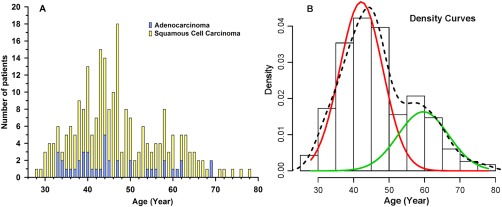
(A) Age distribution of adenocarcinoma and squamous cell carcinoma in 232 cervical cancer patients. (B) Density curves (histogram representing the percent of patients per unit age) of the incidence of cervical cancer by 5‐year age groups computed using the EM algorithm in the R package showing 2 clusters fitted with 2 curves with a first peak at younger age of 42.5 years (red curve) and a second peak at older age of 60.8 years (green curve). The dotted line represents the overall curve fitting.
Detection of HPV Infection and Genotyping
HPV tests were performed for 213 of the 232 patients for whom sufficient pathological materials were available. The results indicated that 163 patients (77%) were HPV‐positive, whereas 50 specimens (23%) were negative for HPV infection after at least 2 separate tests and an independent confirmation using the Xpert HPV Assay (Cepheid, Sunnyvale, CA). By histopathology, 78% of the adenocarcinomas and 79% of the squamous cell carcinomas were positive for HPV infection. The comparison between HPV‐positive and HPV‐negative patients showed no differences in age, geographic region, histology, stage, associated diseases, previous screening, or having other STDs in their medical history, except marginally significant (P = .05) differences for gravidity and parity (Table 1).
HPV genotyping detected 9 single genotypes and 10 double coinfections, with a total of 16 different HPV genotypes (Fig. 2). The majority of patients (90%) had a single infection. The most frequent HPV genotype by far was HPV16 (67.5%), followed by HPV31 (6.8%), HPV18 and HPV45 (5.5% each), HPV73 (1.8%), HPV59 (1.3%), and HPV6, HPV56, and HPV64 (0.6% each). Sixteen patients had double infections involving HPV16/18 (3.7%), HPV16/45 (1.3%), and HPV16/31, 16/39, 16/51, 16/70, 16/82(IS39), 33/52, 35/52, and 45/59 (0.6% each). By combining single and double infections, HPV16 remained the most common HPV genotype with an overall prevalence of 75%, followed by HPV18 (9%) and HPV31 and HPV45 (7% each). In addition, HPV16 and/or HPV18 were found in 62% (132/213) of patients altogether and represented 81% (132/163) of all HPV‐positive patients. The frequency of the 2 most prevalent genotypes (HPV16 and HPV18) was apparently not different in adenocarcinoma and squamous cell carcinoma (87% and 80%, respectively). In addition, there were no obvious variations in the occurrence of the main HPV genotypes across the age groups (Fig. 3), particularly when the 2 age clusters (25‐50 years and 51‐80 years) were compared.
Figure 2.
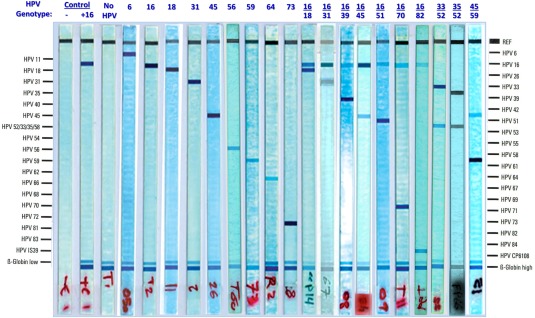
Illustrations of the Linear Array HPV Genotyping (Roche Diagnostics) results showing the different HPV genotypes, in single and double coinfections, detected in cervical cancer patients along with negative and positive controls.
Figure 3.
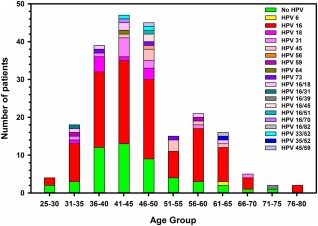
Age distribution by 5‐year groups of HPV detection and genotypes in 232 cervical cancer patients.
TP53 G72C Genetic Polymorphic Variation
The candidate TP53 G72C SNP was genotyped in all 232 CxCa patients and the 313 female control volunteers without cancer. The genotype and allele frequencies were comparable between cancer patients and controls (Table 2). Interestingly, the C allele (Pro; 52%) was more common than the considered majority G allele (Arg; 48%) in this cohort of unrelated women (patients and controls). To further elucidate the genetic predisposing effects of the SNP in the presence of HPV infection, the association study was reanalyzed in cancer patients with HPV results (213 patients, 163 HPV‐positive patients compared with 50 HPV‐negative patients). This nested analysis revealed a significant association of TP53 G72C with CxCa in the presence of HPV infection. The variant TP53 72C allele (Pro) was slightly but significantly (OR, 0.57; 95% CI, 0.36‐0.90; P = .016) less frequent than the majority allele G (Arg) in HPV‐positive cervical cancers compared with HPV‐negative patients. The Cochran‐Armitage test for trend was statistically significant (common OR, 0.54; P = .011), indicating a dose‐dependent relationship of TP53 G72C genotypes in HPV‐positive patients. Testing for deviation from HWE in TP53 G72C genotypes (by comparing observed distributions with expected distributions) showed borderline significant deviation for HPV‐positive patients (P = .09) but not HPV‐negative patients (P = .72).
Table 2.
Genotype and Allele Frequencies of TP53 G72C Polymorphism and Its Genetic Association Between Cervical Carcinoma Patients Compared With Control Subjects Without Cancers and Between HPV‐Positive and HPV‐Negative Patients
| Genotype, Allele | Cases, n (%) | Controls, n (%) | OR (95% CI) | P | HWE Test, Expected Genotypes | |
|---|---|---|---|---|---|---|
| Cases | Controls | |||||
| Cancer (n = 232) | Volunteers (n = 313) | |||||
| G/G | 47 (20) | 64 (20) | 51.21 | 76.26 | ||
| G/C | 124 (53) | 181 (58) | 0.93 (0.60‐1.44) | .757 | 115.58 | 156.47 |
| C/C | 61 (26) | 68 (22) | 1.22 (0.73‐2.03) | .442 | 65.21 | 80.26 |
| G | 218 (47) | 309 (49) | — | — | — | — |
| C | 246 (53) | 317 (51) | 1.10 (0.86‐1.39) | .437 | — | — |
| P = .267 | P = .005 | |||||
| HPV+ (n = 163) | HPV− (n = 50) | |||||
| G/G | 40 (25) | 7 (14) | — | — | 45.37 | 7.61 |
| G/C | 92 (56) | 25 (50) | 0.64 (0.25‐1.61) | .344 | 81.25 | 23.79 |
| C/C | 31 (19) | 18 (36) | 0.30 (0.11‐0.81) | .014 | 36.37 | 18.61 |
| G | 172 (53) | 39 (39) | — | — | — | — |
| C | 154 (47) | 61 (61) | 0.57 (0.36‐0.90) | .016 | — | — |
| P = .091 | P = .719 | |||||
Abbreviations: C, cytosine; CI, confidence interval; G, guanine; HPV+, human papillomavirus–positive; HPV−, human papillomavirus–negative; HWE, Hardy‐Weinberg equilibrium; OR, odds ratio.
DISCUSSION
To the best of our knowledge, this study represents the largest investigation related to HPV infection and CxCa in Saudi Arabia,13, 14 particularly in connection with the association with a candidate cancer‐predisposing genetic polymorphic variation. Our results show that the ratio of histological types (18% adenocarcinoma vs 82% squamous cell carcinoma) is in agreement with published data15 with no apparent difference across age of patients, particularly when the 2 age clusters (25‐50 years and 51‐80 years) were compared (Figs. 1 and 3). A total of 16 different HPV genotypes in single or double coinfections were detected using the Linear Array HPV Genotyping test (Fig. 2). The majority of patients (90%) had single HPV infection. Patient characteristics revealed that besides marginally significant (P = .05) differences for gravidity and parity, where HPV‐positive women had slightly higher median numbers compared with HPV‐negative women, there were no differences in age, geographic region, histology, tumor stage, associated diseases, previous screening, or other STDs (Table 1).
In agreement with results reported in a nearby country having comparable socioeconomic status,16 our results indicated that the prevalence of HPV infection of any genotypes in Saudi CxCa patients (77%) is below the estimated worldwide range (85%‐99%).3, 4, 17 Statistical analysis revealed that this rate is significantly lower than the lowest (85%) global estimation (1 sample z‐test; 95% CI, 71% ‐ 82%; P = .001). These results are in line with the finding that the prevalence of other STDs, including human immunodeficiency virus, in Saudi Arabia is low compared with global rates.18 The conservative society, sexual mores, and widely practiced male circumcision may reduce exposure to STD infections and shift the parameters that affect the frequency and outcome of related diseases, including CxCa. However, the assumption of low incidence of primary HPV infection in cervical swaps has not been substantiated in our population.14 Therefore, it can be speculated that the lower HPV infection rate in our cervical cancer patients could indicate a higher rate of HPV clearance, likely due to reduced susceptibility. In addition, age‐specific occurrence of cervical cancer and HPV distribution in our patients revealed a bimodal curve (Fig. 1B), with a first peak occurring at young ages (mean, 42.56 years) followed by a relative rebound at older ages (mean, 60.84 years). This bimodal occurrence was evoked in other population positioning peaks at younger age of 30‐34 years and slightly older age of 65‐69 years.19 These high points of cervical cancer incidence are shifted 5‐15 years after an analogous bimodal age‐specific HPV‐infection prevalence described at 25 and 45 years of age,20 which may represent the time of cervical malignant transformation to take place with some specificity of each population remnant of possible genetic and cultural differences. HPV16 was by far the most frequent genotype, including coinfections, with an overall prevalence of 75% compared with 54% worldwide.17 The next most common genotypes were HPV18 (9%), HPV31 (7%), and HPV45 (7%). In addition, HPV16 and/or HPV18 were present in 62% of all CxCa and formed 81% of HPV‐positive patients. The latter prevalence rate is slightly higher than that reported in Europe (74.5%), North America (76.5%), and the world altogether (70.9%).17
Studying the genetic relationship between CxCa and the most commonly suspected cancer predisposing polymorphism TP53 G72C revealed no significant association, whereas CxCa patients and control women without cancer had displayed similar genotype frequencies (Table 2). Following the first description of the potential usefulness of the Arg genotype as a marker of risk to develop uterine cervix neoplasia,10 there were many studies that examined the association between CxCa and TP53 G72C SNP; however, the results in various populations were inconsistent.21 Nevertheless, there is substantial collective evidence to support a sound association specifically when HPV status is considered. Thus, whereas Sousa et al22 could not confirm the association in a number of European countries other than Italy and the United Kingdom, 2 other large meta‐analyses have validated the significant association between the homozygous G allele (Arg) and CxCa.23, 24
Of particular importance, the nested association analysis within CxCa patients (Table 2) showed that the variant TP53 72C allele was significantly less frequent than the majority allele G in HPV‐positive CxCa patients compared with HPV‐negative CxCa patients (OR, 0.57; 95% CI, 0.36‐0.90; P = .016). This means that the variant allele (C, Pro) is associated with reduced odds for HPV infection and, therefore, is protective against cervical cancer. In other words, the majority risk allele (G, Arg) was more frequent in HPV‐positive patients compared with HPV‐negative patients (53% vs 39%, respectively). HWE revealed a borderline significant deviation (P = .091), indicating a possible coselection of risk allele and HPV. Furthermore, there was a statistically significant dose‐dependent relationship for TP53 G72C genotypes (Cochran‐Armitage trend test, P = .011), indicating an ordering in the effect of the G allele in HPV‐positive patients. In fact, it is this allele that is deemed more prone to degradation by high‐risk HPV‐E6 oncoprotein.10 Consistent with our results, Hu et al21 also reported that the TP53 G72 (Arg) was significantly overtransmitted in Caucasian CxCa subjects of a family‐based association study where HPV was determined, especially in subjects who were infected with HPV16/18.21 In validation of our results, a systematic meta‐analysis including 49 studies showed that HPV‐positive patients had significantly elevated odds of progression from squamous intraepithelial lesions to invasive CxCa when associated with the p53 Arg allele (OR, 1.37; 95% CI, 1.15‐1.62; P < .001) that was not seen in HPV‐negative individuals.25 The authors further stated that the Arg allele of p53 codon 72 is associated with the progression from squamous intraepithelial lesions to CxCa in the presence of HPV positivity only. Our results are in agreement with this conclusion and provide an independent confirmation cohort.
Finally, it is interesting to note that the assumed “protective” variant TP53 72C allele was the most common allele found in our patients (C, 52% vs G, 48%; patients and controls combined, n = 545) and appears to be overtransmitted in our population including both sexes, as we have previously reported and deposited at the National Center for Biotechnology Information website (https://www.ncbi.nlm.nih.gov/projects/SNP/snp_ss.cgi? ss=ss105111344). This is different from other populations known to have higher incidence of cervical cancer. For example, the TP53 G72 risk allele was shown to be overtransmitted in Caucasian, African American, and Mexican women.21, 26 Hu et al21 had reported 71% G allele and 29% C allele in Caucasian women (n = 514) and 65% G allele and 35% C allele in African American women (n = 62). In comparison with these published data, statistical analysis revealed significant overtransmission of the C allele in our patients compared with Caucasian (P < .0001) and African American (P = .00034) women. It is probable that the absence of significant association in this study between patients and controls without cancer is due to this overtransmitted TP53 72C allele in our population. The reappearance of the significant association became evident in nested analysis comparing HPV‐positive patients with HPV‐negative patients because the TP53 G72C predisposing effect is restricted to HPV infection. This is probably why TP53 G72C tended to deviate from HWE in HPV‐positive patients compared with HPV‐negative patients due to coselection of the TP53 G72 risk allele with HPV infection. Thus, it is plausible to speculate that the preponderance of the variant TP53 72C allele contributes to some extent to the low incidence of CxCa, in addition to the conservative societal characteristics of the patient population. These results support the coconsideration of HPV screening with host genetic predisposition to guide decision making toward a more effective and personalized preventive medicine.
In conclusion, cervical cancer incidence has a bimodal curve of occurrence. The relatively low incidence of HPV (77%) and the preponderance of the variant TP53 72C allele may contribute to the reduced incidence of CxCa observed in our population. Thus, to further gauge the risk of developing CxCa, HPV infection screening associated with host SNP genotyping could provide more relevant candidate susceptibility biomarkers for effective preventive and personalized medicine.
FUNDING SUPPORT
This study was supported by the National Science, Technology and Innovation Plan (NSTIP‐KACST) grant 12‐MED2945‐20 (RAC# 2060 029, 2130 025). The funder had no role in the design, data collection, analysis, writing, or decision to publish.
CONFLICT OF INTEREST DISCLOSURES
The authors made no disclosures.
AUTHOR CONTRIBUTIONS
Conceived and designed the study: Ghazi A. Alsbeih. Performed experiments: Najla M. Al‐Harbi, Sara S. Bin Judia. Analyzed the data: Ghazi A. Alsbeih, Mohamed M. Shoukri. Contributed reagents, materials and analysis tools: Hatim A. Khoja, Asma M. Tulbah. Wrote the manuscript: Ghazi A. Alsbeih.
We thank Medhat El‐Sebaie, Ismail Albadawi, Khalid Balaraj, Rana Mahmood, Fahad Almajhdi, Nasser Al‐Rajhi, Hadeel Almanea, Khaled Al‐Hadyan, Muneera Al‐Buhairi, Sara Elewisy, Reham Al‐Ahmadi, and Jenelyn Castro for facilitating samples and clinical data collection. We also thank Monies Dorota for help with DNA sequencing.
REFERENCES
- 1. Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 10. http://globocan.iarc.fr. Accessed April 29, 2012.
- 2. zur Hausen H. Human papillomaviruses in the pathogenesis of anogenital cancer. Virology. 1991;184:9‐13. [DOI] [PubMed] [Google Scholar]
- 3. Walboomers JM, Jacobs MV, Manos MM, et al. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol. 1999;189:12‐19. [DOI] [PubMed] [Google Scholar]
- 4. de Sanjose S, Quint WG, Alemany L, et al. Human papillomavirus genotype attribution in invasive cervical cancer: a retrospective cross‐sectional worldwide study. Lancet Oncol. 2010;11:1048‐1056. [DOI] [PubMed] [Google Scholar]
- 5. Nahvijou A, Daroudi R, Tahmasebi M, et al. Cost‐effectiveness of different cervical screening strategies in Islamic Republic of Iran: a middle‐income country with a low incidence rate of cervical cancer. PLoS One. 2016;11:e0156705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bazarbashi S, Al‐Zahrani A, eds. Cancer Incidence and Survival Report 2007. Riyadh, Kingdom of Saudi Arabia: Ministry of Health, Saudi Cancer Registry; 2011. [Google Scholar]
- 7. Hildesheim A, Wang SS. Host and viral genetics and risk of cervical cancer: a review. Virus Res. 2002;89:229. [DOI] [PubMed] [Google Scholar]
- 8. Ferenczy A, Franco E. Persistent human papillomavirus infection and cervical neoplasia. Lancet Oncol. 2002;3:11‐16. [DOI] [PubMed] [Google Scholar]
- 9. Chen D, Gyllensten U. Lessons and implications from association studies and post‐GWAS analyses of cervical cancer. Trends Genet. 2015;31:41‐54. [DOI] [PubMed] [Google Scholar]
- 10. Storey A, Thomas M, Kalita A, et al. Role of a p53 polymorphism in the development of human papillomavirus‐associated cancer. Nature. 1998;393:229‐234. [DOI] [PubMed] [Google Scholar]
- 11. Alsbeih G, Al‐Harbi N, El‐Sebaie M, Al‐Badawi I. HPV prevalence and genetic predisposition to cervical cancer in Saudi Arabia. Infect Agent Cancer. 2013;8:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Al‐Hadyan KS, Al‐Harbi NM, Al‐Qahtani SS, Alsbeih GA. Involvement of single‐nucleotide polymorphisms in predisposition to head and neck cancer in Saudi Arabia. Genet Test Mol Biomarkers. 2012;16:95‐101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vaccarella S, Bruni L, Seoud M. Burden of human papillomavirus infections and related diseases in the extended Middle East and North Africa region. Vaccine. 2013;31(suppl 6):G32‐G44. [DOI] [PubMed] [Google Scholar]
- 14. Alhamlan FS, Al‐Qahtani AA, Al‐Ahdal MN. Current studies on human papillomavirus in Saudi Arabia. J Infect Dev Ctries. 2015;9:571‐576. [DOI] [PubMed] [Google Scholar]
- 15. Seoud M, Tjalma WA, Ronsse V. Cervical adenocarcinoma: moving towards better prevention. Vaccine. 2011;29:9148‐9158. [DOI] [PubMed] [Google Scholar]
- 16. Khorasanizadeh F, Hassanloo J, Khaksar N, et al. Epidemiology of cervical cancer and human papilloma virus infection among Iranian women—analyses of national data and systematic review of the literature. Gynecol Oncol. 2013;128:277‐281. [DOI] [PubMed] [Google Scholar]
- 17. World Health Organization/Institut Català d’Oncologia . Human Papillomavirus and Related Cancers in the World. Barcelona, Spain: HPV Information Centre; 2010. [Google Scholar]
- 18. Filemban SM, Yasein YA, Abdalla MH, Al‐Hakeem R, Al‐Tawfiq JA, Memish ZA. Prevalence and behavioral risk factors for STIs/HIV among attendees of the Ministry of Health hospitals in Saudi Arabia. J Infect Dev Ctries. 2015;9:402‐408. [DOI] [PubMed] [Google Scholar]
- 19. Harper DM, Vierthaler SL. Next generation cancer protection: the bivalent HPV vaccine for females. ISRN Obstet Gynecol. 2011;2011:457204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bruni L, Diaz M, Castellsague X, Ferrer E, Bosch FX, de Sanjose S. Cervical human papillomavirus prevalence in 5 continents: meta‐analysis of 1 million women with normal cytological findings. J Infect Dis. 2010;202:1789‐1799. [DOI] [PubMed] [Google Scholar]
- 21. Hu X, Zhang Z, Ma D, et al. TP53, MDM2, NQO1, and susceptibility to cervical cancer. Cancer Epidemiol Biomarkers Prev. 2010;19:755‐761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sousa H, Santos AM, Pinto D, Medeiros R. Is the p53 codon 72 polymorphism a key biomarker for cervical cancer development?. A meta‐analysis review within European populations. Int J Mol Med. 2007;20:731‐741. [PubMed] [Google Scholar]
- 23. Koushik A, Platt RW, Franco EL. p53 codon 72 polymorphism and cervical neoplasia: a meta‐analysis review. Cancer Epidemiol Biomarkers Prev. 2004;13:11‐22. [DOI] [PubMed] [Google Scholar]
- 24. Jee SH, Won SY, Yun JE, Lee JE, Park JS, Ji SS. Polymorphism p53 codon‐72 and invasive cervical cancer: a meta‐analysis. Int J Gynaecol Obstet. 2004;85:301‐308. [DOI] [PubMed] [Google Scholar]
- 25. Habbous S, Pang V, Eng L, et al. p53 Arg72Pro polymorphism, HPV status and initiation, progression, and development of cervical cancer: a systematic review and meta‐analysis. Clin Cancer Res. 2012;18:6407‐6415. [DOI] [PubMed] [Google Scholar]
- 26. Pina‐Sanchez P, Hernandez‐Hernandez DM, Taja‐Chayeb L, et al. Polymorphism in exon 4 of TP53 gene associated to HPV 16 and 18 in Mexican women with cervical cancer. Med Oncol. 2011;28:1507‐1513. [DOI] [PubMed] [Google Scholar]


