Abstract
Variation in life history contributes to reproductive success in different environments. Divergence of annual and perennial angiosperm species is an extreme example that has occurred frequently. Perennials survive for several years and restrict the duration of reproduction by cycling between vegetative growth and flowering, whereas annuals live for 1 year and flower once. We used the tribe Arabideae (Brassicaceae) to study the divergence of seasonal flowering behaviour among annual and perennial species. In perennial Brassicaceae, orthologues of FLOWERING LOCUS C (FLC), a floral inhibitor in Arabidopsis thaliana, are repressed by winter cold and reactivated in spring conferring seasonal flowering patterns, whereas in annuals, they are stably repressed by cold. We isolated FLC orthologues from three annual and two perennial Arabis species and found that the duplicated structure of the A. alpina locus is not required for perenniality. The expression patterns of the genes differed between annuals and perennials, as observed among Arabidopsis species, suggesting a broad relevance of these patterns within the Brassicaceae. Also analysis of plants derived from an interspecies cross of A. alpina and annual A. montbretiana demonstrated that cis‐regulatory changes in FLC orthologues contribute to their different transcriptional patterns. Sequence comparisons of FLC orthologues from annuals and perennials in the tribes Arabideae and Camelineae identified two regulatory regions in the first intron whose sequence variation correlates with divergence of the annual and perennial expression patterns. Thus, we propose that related cis‐acting changes in FLC orthologues occur independently in different tribes of the Brassicaceae during life history evolution.
Keywords: annual, Arabis, FLC, flowering time, PEP1, perennial
Short abstract
Introduction
Life history varies greatly among higher organisms and can diverge rapidly during evolution (Partridge & Harvey 1988). Two major life history strategies have been described, which are represented by semelparous organisms that reproduce only once in their lifetime and iteroparous organisms that undergo several reproductive periods (Silvertown & Charlesworth 2001). In plants, these life strategies are often referred to as monocarpic (semelparous) or polycarpic (iteroparous) and are defined by the rate of reproduction and probability of survival at each stage of the life cycle. Most monocarpic plants are annual, completing their life cycle within 1 year, while polycarpic plants are usually perennial, surviving for many years and entering the reproductive phase multiple times. Evolutionary transitions between perenniality and annuality have occurred often among higher plants. In most cases, annuality seems to be derived from perenniality (Hu et al. 2003; Grillo et al. 2009), but the reverse has also been described (Tank & Olmstead 2008).
Variation in life history is associated with adaptation to different environments. Annuals are usually found in open, dry habitats prone to seasonal drought but where seedlings have high survival rates if germination is appropriately timed. By contrast, perennials are more commonly found in mesic habitats where seedling mortality is high (Stebbins 1950; Silvertown & Charlesworth 2001). For example, perennial populations of Mimulus guttatus (Scrophulariaceae) have access to moisture throughout the year, while annual populations grow in areas that are arid during the summer months, and reciprocal transplant experiments indicate that each type is adapted to its respective environment (Hall & Willis 2006).
Theoretical models support the proposal that life history evolves to optimize the number of offspring produced while minimizing the costs of reproduction (Stearns 2000; Friedman & Rubin 2015). A deeper understanding of life history evolution and more specifically of the transition from perennial to annual requires knowledge of the developmental features that change during the transition and the associated underlying genetics. A critical feature is the initiation of reproduction through the induction of flowering. Perennials cycle multiple times through vegetative and reproductive states, which requires repeated decisions on the fate of individual meristems. These decisions involve, at least in part, flowering‐time genes whose roles are extended in perennials compared to annuals (Friedman & Rubin 2015). These mechanisms of flowering control have been studied in several perennial species and compared to that of the well‐established annual model species Arabidopsis thaliana (Albani & Coupland 2010; Wang et al. 2011; Andres & Coupland 2012; Iwata et al. 2012; Koskela et al. 2012; Bergonzi et al. 2013; Zhou et al. 2013).
PERPETUAL FLOWERING 1 (PEP1), the A. alpina orthologue of A. thaliana FLOWERING LOCUS C (FLC), has critical roles in the perennial life cycle. FLC is a MADS box transcription factor that delays flowering before vernalization (Michaels & Amasino 1999; Sheldon et al. 1999). PEP1, in addition to preventing flowering before vernalization, restricts flowering to a short episode after vernalization by causing reversion to vegetative growth after flowering (Wang et al. 2009; Castaings et al. 2014). This additional function of PEP1 is conferred by a distinct pattern of transcriptional regulation compared to FLC. The latter is stably repressed by vernalization allowing A. thaliana to flower continuously after vernalization. In contrast, PEP1 is only temporarily repressed by vernalization and rises in expression again when plants are returned to warm. This cyclical repression of flowering is proposed to contribute to the perennial life cycle by preserving meristems for vegetative growth upon the return to warm temperatures (Wang et al. 2009). Similar cycling patterns of expression of FLC orthologues were described in other perennial Brassicaceae species (Aikawa et al. 2010; Kemi et al. 2013). Repression of expression of both FLC and PEP1 during vernalization correlates with accumulation of trimethylation on lysine 27 of histone 3 (H3K27me3) starting from a region referred to as the nucleation region (Bastow et al. 2004; Wang et al. 2009; Angel et al. 2011; Yang et al. 2014).
At FLC, this modification remains after vernalization stably repressing gene expression, whereas in A. alpina, it disappears after the return of plants to warm, correlating with reactivation of PEP1 (Bastow et al. 2004; Sung & Amasino 2004; Finnegan & Dennis 2007; Wang et al. 2009). Repression of FLC is also regulated by noncoding RNAs expressed in cis during vernalization (Swiezewski et al. 2009; Heo & Sung 2011; Csorba et al. 2014). COOLAIR is an antisense RNA that is conserved at PEP1 (Swiezewski et al. 2009; Castaings et al. 2014; Csorba et al. 2014), whereas COLDAIR is a sense RNA expressed from the first intron and associated with a sequence important for repression of FLC transcription during vernalization called the vernalization response element (VRE; Sung et al. 2006; Heo & Sung 2011). PEP1 has a more complex structure than FLC, so that the first exon is duplicated in tandem along with the proximal promoter and part of the first intron giving rise to two overlapping transcripts (Albani et al. 2012). In perennial Arabidopsis lyrata, the FLC locus is tandemly duplicated, while in perennial Arabidopsis arenosa, it is partially triplicated (Nah & Chen 2010). This indicates that rearrangements at FLC orthologues might contribute to the rapid evolution of life history in the Brassicaceae (Alonso‐Blanco & Méndez‐Vigo 2014).
Understanding trait evolution requires systematic analysis of closely related species within a well‐developed phylogenetic framework. This approach has proven effective in, for example, identifying the mechanisms and direction of evolution of wing pigmentation patterns in the Drosophila genus (Prud'homme et al. 2006; Arnoult et al. 2013). The Brassicaceae provide similar advantages for studying the evolution of annual and perennial life history. Several genera in this family such as Arabidopsis, Brassica, Draba and Arabis contain both annual and perennial species. Notably, annual life history and perennial life history have diverged independently several times in the Arabis and Draba genera of the tribe Arabideae, which is the largest tribe of the Brassicaceae and comprises ~500 species (Couvreur et al. 2010; Al‐Shehbaz et al. 2011; Karl et al. 2012). Typically, perennial species‐rich groups are sister to annual species‐poor groups, and it has been suggested that life history changes are a major driving factor for diversification in the Arabideae (Karl & Koch 2013).
Comparative analyses of annual and perennial species within the Arabideae provide an opportunity to study the repeated divergence of these traits within relatively short evolutionary timescales. Arabis alpina has been used as a model genetic system to study perennial traits, its genome has been sequenced (Willing et al. 2015), and it exhibits a broad arctic–alpine distribution range originating from migrations out of Asia Minor (Koch et al. 2006; Ansell et al. 2011; Karl et al. 2012).
Here, we perform an intensive phylogenetic analysis of the main branches of the Arabideae represented by 16 taxa and relate this to the life history of individual species. These experiments demonstrate that A. montbretiana is an annual sister of perennial A. alpina and that their sister is the perennial Arabis nordmanniana. Consistent with their respective life histories, the FLC orthologue of A. montbretiana is stably repressed by vernalization in contrast to PEP1 of A. alpina. The close phylogenetic relationship of these two species allowed us to perform interspecies crosses and to demonstrate that cis‐acting variation contributes to the different patterns of expression of the FLC orthologues in these species. Detailed sequence comparisons of FLC orthologues isolated from species across the Arabideae and Camelineae tribes identified regions in the first intron that we propose contribute to the divergence of gene regulation between annual and perennial Brassicaceae species.
Materials and methods
Plant material
Plant material was obtained from Birol Mutlu, Turkey (A. montbretiana, BM7968, voucher HEID809801, RK043 in Karl & Koch 2013; Jipei Yue, Kunming, China (Scapiarabis setosifolia, RK025 in Karl & Koch 2013), Maarten Koornneef, MPIPZ Cologne, Germany (Arabis purpurea), Santiago Martin Bravo (Arabis nova subsp. iberica) and the Botanical Garden Heidelberg (Arabis alpina, RK001 and A‐Kili; Arabis aucheri RK041; Arabis auriculata, Arabis collina B‐2006‐0564 and JR045, Arabis hirsuta JR38, Arabis nordmanniana RK219, Arabis purpurea RK004, Arabis verna RK212, Aubrieta canescens subsp. macrostyla RK214, Draba aizoides Dra 002, Draba hispanica Dra 0026, Pseudoturritis turrita RK237 (all numbers referring to Karl & Koch 2013; Draba nemorosa B‐2002‐0157). As comparison, the A. alpina acc. Pajares (Wang et al. 2009) was used. Accession details are given in Table S1 (Supporting information).
Generation of an Arabis montbretiana × Arabis alpina F1 hybrid
To investigate the flowering behaviour of hybrids between annuals and perennials, A. montbretiana flowers were emasculated and cross‐pollinated with A. alpina acc. Pajares pollen. One viable plant was obtained through embryo rescue (adapted from Sauer & Friml 2008). Details are given in Supplementary Methods (Supporting information). For verification of the hybrid identity of the F1 plant, one of the phylogenetic markers used for the phylogenetic analysis (ITS) was amplified as described above and Sanger‐sequenced using the forward and reverse PCR primers. Sequences were analysed for double signals in positions being polymorphic in both parents.
Growth and vernalization conditions
Plants were cultivated on soil [Balster, Sinntal‐Altengronau, Germany (Type Mini‐Tray)] under a photoperiod of 16 h and a temperature of 20 °C during the day (minimum 18 °C at night) in three different glasshouse (GH1, GH2, GH3, Supplementary Methods, Supporting information). Plants were watered daily and fertilized regularly with Wuxal Super 8‐8‐6 and/or Wuxal Top K 21%, or a comparable fertilizer mix (Supplementary Methods, Supporting information).
For vernalization, two vernalization rooms were used (Viessmann, Allendorf, Germany, or Fitotron by Weiss Technik, UK) which all had a constant temperature of 4–5 °C and were run with short photoperiods of 8 h (light sources in Supplementary Methods V1, V2, Supporting information). During vernalization, plants were not fertilized and watered only when necessary.
Cultivation of plants used for expression analyses
Seeds of A. montbretiana, A. alpina acc. Pajares, A. auriculata and A. nordmanniana were either stratified on wet filter paper or sown directly on soil and stratified for 3–4 days at 4 °C in darkness. Four to six plants were grown in the glasshouse (GH1, Supplementary Methods, Supporting information) under the conditions described above. Vernalization treatment was performed 3 (annuals) or 6 weeks (perennials) after seed germination for 12 weeks (Viessmann, Allendorf, Germany, V1, Supplementary Methods, Supporting information). The ages at which plants were exposed to vernalization were selected because A. alpina acc. Pajares has a juvenile phase of 5 weeks during which flowering cannot be induced. The annuals on the other hand were found to have a shorter juvenile phase (for details on A. montbretiana and Arabis auriculata, see the Results and Supplementary Results sections, Supporting information). At the end of the vernalization period, plants were transferred back to the glasshouse conditions described above. Cuttings from the A. montbretiana × A. alpina hybrid were grown in the same way. For the experiment on the lines containing the introgressed AmFLC region, seeds were stratified for 4 days at 4 °C in darkness and then germinated on filter paper. Seedlings were transferred to soil and then cultivated under glasshouse conditions for 8 weeks (GH3, V2, Supplementary Methods, Supporting information). Plants were vernalized for 12 weeks at 5 °C. After vernalization, plants were returned to glasshouse conditions. Vernalization time‐courses were run in two replicates.
Validation of life history
All accessions except Scapiarabis setosifolia, Pseuditurritis turrita, Draba aizoides, Arabis purpurea RK004 and Arabis alpina RK001 were cultivated under glasshouse conditions (GH1, Supplementary Methods, Supporting information). After flowering, it was determined whether the plant underwent complete senescence (annual) or continued to grow vegetatively (perennial). These results were reproduced in the other two glasshouses used in this study.
Assessment of vernalization response
After germination, plants were grown either constantly under glasshouse conditions (GH1, GH2, Supplementary Methods, Supporting information) or exposed to vernalization 3 or 6 weeks after germination for 12 weeks (V1, Supplementary Methods, Supporting information). Flowering was scored as total leaf number (TLN) on the main shoot under control conditions or after vernalization. For A. montbretiana and A. auriculata, also the age at which the plant responds to vernalization was determined experimentally. For this experiment, A. montbretiana was grown under long‐day glasshouse conditions. Plants were shifted into vernalization at an age ranging from 1 to 6 weeks or left in long‐day conditions without vernalization. After 12 weeks, plants were returned to warm temperatures and long‐day conditions and TLN at flowering was scored. In a second experiment, A. montbretiana plants were grown in a plant growth chamber under 16 h long days (Grow Bank, CLF Climatic, Wertingen, Germany) at constant 22 °C and a light intensity of 150 μE. They were then vernalized for 12 weeks at 1–6 weeks after germination, as described above. A similar experiment was run for Arabis auriculata, which was shifted at two different ages (after 3 and 6 weeks of growth in GH1), and a control was maintained under long‐day glasshouse (GH1) conditions and not exposed to vernalization.
DNA extraction
For PCR and sequencing, genomic DNA was extracted with the DNeasy Plant Mini or Maxi Kit (Qiagen, Germany) following the manufacturer's protocol. DNA concentration and quality were checked by measuring OD 260 and OD 260/280 using a Nanodrop photometer (Thermo Fisher Scientific, USA). If the DNA concentration was too low, DNA was concentrated by evaporation using a Concentrator plus apparatus (Eppendorf, Hamburg, Germany). The quality of the DNA used for whole‐genome sequencing of A. montbretiana and A. nordmanniana was also checked on an agarose gel [0.5% agarose in TAE (40 mm Tris, 20 mm acetate, 1 mm EDTA)].
Amplification and sequencing of phylogenetic marker sequences, alignment and phylogenetic reconstruction
Four nuclear and two plastidic marker sequences were amplified and sequenced for use in phylogenetic reconstruction (for material, see Table S1, Supporting information). All of the selected phylogenetic marker sequences were successfully used for phylogenetic reconstruction in previous studies (White et al. 1990; Taberlet et al. 1991; Mummenhoff et al. 1997; Koch et al. 2000; Dobeš et al. 2004; Duarte et al. 2010). As orthologues of At2g13360 had only been used in a small study to demonstrate its value as a phylogenetic marker, its orthologue was identified in the genome assembly of A. alpina acc. Pajares (Willing et al. 2015). Synteny of the respective genomic regions around At2g13360 of A. thaliana and its orthologue in A. alpina was analysed using the program gata (Nix & Eisen 2005). When synteny was confirmed, primers were designed using the program primer3 (Rozen & Skaletsky 2000) amplifying exons 1 to 3 including all introns (product ~1 kb, primers in Table S2, Supporting information). Primer sequences, including the ones for the amplification of ITS (White et al. 1990; Mummenhoff et al. 1997), trnL (Taberlet et al. 1991), the trnL‐F IGS (Taberlet et al. 1991; Dobeš et al. 2004), CHS (Koch et al. 2000) and ADH (Koch et al. 2000), are given in Table S2 (Supporting information). Phylogenetic marker sequences were amplified by PCR from each species (see supplement for method, Supporting information). PCR products were separated on agarose gels which confirmed that only one PCR product was present, purified and sequenced directly or after cloning (see supplement for method, Supporting information) by Sanger sequencing. Electropherograms of the obtained sequences were controlled for quality using the program SeqMan (DNASTAR, Lasergene).
Both chloroplast markers and if necessary both parts of the ADH fragment were combined prior to aligning and were analysed as a unit. Sequences of the six utilized phylogenetic markers (ITS, trnL, trnL‐F, CHS, ADH and orthologue of At2g13360) were aligned with clustalx (Thompson et al. 1997) and subsequently adjusted by hand. For the markers CHS, ADH and orthologue of At2g13360, only the exons of the sequences were employed for the analyses (alignment in Table S3, Supporting information). After model testing (Table S4, Supporting information), Bayesian MCMC analyses (Yang & Rannala 1997) were performed with the mpi (message parsing interface) version of mrbayes version 3.1.2 (Ronquist & Huelsenbeck 2003). Additionally, a maximum parsimony analysis was run using both co‐orthologues of the nuclear phylogenetic markers of A. nordmanniana (alignment in Table S5 , Supporting information). Details are given in Supplementary Methods (Supporting information).
Contig assembly of A. montbretiana and A. nordmanniana and identification of FLC orthologues in five Arabideae taxa
For A. montbretiana and A. nordmanniana, Illumina libraries with an insert size of 400 bp were constructed, sequenced by Illumina sequencing (120 bp, paired ends), and contig assemblies based on 185 (A. montbretiana) and 415 million reads (A. nordmanniana) were generated using the clc genomics workbench version 4.7 or version 8.5, respectively, using standard settings. The contig assemblies were indexed as blast libraries and searched using PEP1 (Wang et al. 2009) as query. In the case of A. nordmanniana, the blast search resulted in several blast hits to FLC or flanking genes. When the contigs were assembled, two co‐orthologues of FLC were generated. The assembly of the individual contigs was confirmed by PCR and Sanger sequencing of the PCR fragments. The genome size of A. nordmanniana was estimated by kmer‐depth. The A. nordmanniana assembly was annotated using the maker pipeline (Cantarel et al. 2008). orthomcl (Li et al. 2003) was used to define orthologous groups. Details of the settings and methods used for the A. nordmanniana assembly and annotation are described in Supplementary Methods and Results (Supporting information).
Regions containing the FLC orthologues from A. auriculata, A. purpurea and A. nova subsp. iberica were each amplified by PCR (primers in Table S2, Supporting information) as three overlapping fragments. The first fragment spanned a region from the adjacent upstream gene (orthologue of At5g10150) and ended at the beginning of the first intron (amplification of intergenic region between orthologue of At5g10150 and FLC orthologue), and the second covered the region containing a part of exon 1, intron 1 and a part of exon 2 (amplification exon 1 to exon 2). The third PCR fragment contained a part of exon 2 and ended downstream of exon 7 (amplification exon 2 to exon 7). Various combinations of primers were used for Sanger sequencing the PCR products before or after cloning into the pGEM‐T vector (see Supplementary Methods, Supporting information). Primers were designed manually or using the program primer3 (Rozen & Skaletsky 2000). To follow the structural evolution of the FLC orthologues, sequences were aligned by mVISTA (Mayor et al. 2000; Frazer et al. 2004) and compared to A. thaliana FLC. Alignments were adjusted manually. Exons (by using cDNA data) and sequences with homology to known regulatory elements from FLC were identified and annotated. The annotations were used as a basis for generating locus schemes that were used for following the structural evolution of FLC orthologues. gata (Nix & Eisen 2005), mvista (Mayor et al. 2000; Frazer et al. 2004), yass (Noe & Kucherov 2005) and clustalw (Larkin et al. 2007; Goujon et al. 2010; McWilliam et al. 2013) were used for different aspects of sequence analysis (details in Supplementary Methods, Supporting information).
Expression analysis of PEP1, AmFLC, AauFLC and AnFLC‐A and AnFLC‐B
Leaf samples of A. alpina acc. Pajares, A. auriculata, A. montbretiana and A. nordmanniana were taken between ZT 4 and ZT 7 before (3 or 6 weeks in long‐day conditions depending on the experiment), at the end (after 12 weeks) and after vernalization (2, 3, 4 weeks in long day after vernalization depending on the experiment) in two biological replicates grown at different times (only one replicate was used for A. alpina because this confirmed data published by Wang et al. 2009). One young leaf from the main shoot was harvested at each time point, and samples were pooled from four to six plants, except for A. nordmanniana where one individual was monitored in each replicate.
Total RNA was extracted using the rneasy mini kit (Qiagen, Germany). The obtained RNA was treated with DNase (Ambion, Thermo Fisher Scientific, USA) and then transcribed into cDNA using the Superscript II Reverse Transcription Kit (Invitrogen, Thermo Fisher Scientific, USA). All kits were used according to the manufacturer's protocols.
All qPCRs were performed in a volume of 20 or 15 μL containing 2 μL cDNA, 2/1.5 μL 10× PCR buffer (Invitrogen, Thermo Fisher Scientific, USA), 1 μL Eva green (Biotium, USA), 2.5 mm MgCl2, 0.5 mm dNTPs, and 0.2 μL My Budget Taq polymerase (Bio‐Budget Technologies GmbH, Krefeld, Germany) or Taq DNA polymerase (Invitrogen, Thermo Fisher Scientific, USA). PCRs were run on a Bio‐Rad (California USA) Realtime PCR cycler or Roche iCycler using the primers published previously for A. alpina (Wang et al. 2009 and Bergonzi et al. 2013; primer names PEP1 (qPCR) and PP2A (qPCR) as reference gene; Table S2, Supporting information) in triplicates or quadruplicates. If necessary, primer sequences were adjusted to point mutations present in the priming sites in the different species (cDNA sequences of FLC orthologues in Fig. S1 (Supporting information); A. nordmanniana primers AnFLC‐A (qPCR), AnFLC‐B (qPCR) and AnRAN3 (qPCR) as reference gene; A. montbretiana primers AmFLC (qPCR) and PP2A (qPCR) as reference gene; A. auriculata primers AauFLC (qPCR), AauPP2A (qPCR) as reference gene; primer sequences in Table S2, Supporting information). As there is no mutation in the priming site for PP2A between A. alpina and A. montbretiana, both AmPP2A and AaPP2A were measured simultaneously. Data were analysed using the program excel 2013 (Microsoft Office) by the delta Ct method. The standard deviation was calculated separately for triplicates or quadruplicates of reference and target gene and then combined by error propagation. For easier display in the figures, the expression at 0 weeks of vernalization was set to 100% and expression at the other time point was recalculated relative to this time point. Efficiency of the PCR was determined by standard curves.
Genotyping of the A. montbretiana × A. alpina F1 and resulting introgression lines
Based on the genomic information available (Willing et al. 2015; Kiefer et al. in preparation), length polymorphism markers distributed across the complete genomes were designed to distinguish A. alpina and A. montbretiana. To confirm the cross of A. montbretiana × A. alpina and initial genotyping of the subsequent generations, 40 length polymorphism markers were used (Table S8, Supporting information). For genotyping the introgression lines further, 84 markers were employed (Table S8, Supporting information). Length polymorphism markers were amplified in multiplexing PCRs containing two to three primer pairs (2 μL GoTaq buffer (Promega, Wisconsin, USA), 2 μL template, 1 μL MgCl2 (25 mm), 0.4 μL dNTPs (10 mm), 0.4 μL of each primer pair (each primer 10 μm), 0.05 μL GoTaq (Promega, Wisconsin, USA), 3.35 μL water), separated on 3% agarose gels (TAE) and scored as heterozygous or homozygous for either parent. Arabis alpina and A. montbretiana were used as control. All primers were designed using the program primer3 (Rozen & Skaletsky 2000). For the introgression lines, genotypes were plotted using the program flapjack 1.016.03.04 (Milne et al. 2010).
Estimates of evolutionary divergence of the first intron of FLC orthologues
All calculations on (average) evolutionary divergence of sequences of the first intron of the FLC orthologue from A. alpina acc Pajares and A. montbretiana (number of base substitutions per site) were performed in mega5 (Tamura et al. 2011) using the maximum composite likelihood model (Tamura et al. 2004). In the analysis referring to the alignment used for Fig. 4, the alignment was split at position 10 332 into the ‘5’ part’ (10 332 alignment positions; 2755 positions without missing data and gaps) and the ‘3’ part’ (10 333 to end >4167 alignment positions; 4018 positions without gaps).
Figure 4.

Distribution of SNPs (A) and indels (B) among PEP1 and AmFLC analysed by a sliding window approach (window size 100 bp, shift 20 bp). For the analysis of SNP distribution, all indels were excluded, while for the distribution analysis of indels, all indels were coded as 1 irrespective of their length; exons as well as sequence features homologous to regulatory elements known from FLC are annotated on top of the graph as a colour‐coded scheme. Nucleation region is the sequence homologous to the region showing elevated H3K27me3 before vernalization in Arabidopsis (Yang et al. 2014); COLDAIR represents the regions homologous to the COLDAIR RNA‐encoding region (Heo & Sung 2011); vernalization response element (VRE) indicates the region homologous to the VRE of A. thaliana identified in Sung et al. (2006); peak regions with windows containing significantly more SNPs than expected in a random distribution are marked by *.
Analysis of SNP and indel distribution between PEP1 and AmFLC
To define SNP evolutionary patterns among PEP1 and AmFLC, AlFLC1 and FLC, AmFLC and AuFLC as well as PEP1 and AnFLC‐A SNPs were scored for each alignment position in a Microsoft Excel table. Positions including gaps, missing data, ambiguous data or where more than two possibilities were present were excluded from the analysis. SNPs were then analysed by a sliding window approach. The window size was 100 bp, and the window was shifted by 20 bp or 1 bp. We used a binomial test to examine the over‐ or underrepresentation of SNPs within each window, comparing the actual number of observed SNPs with the distribution of SNPs, given an overall SNP occurrence rate. P‐values were converted into expected false discovery rates (FDR) to account for multiple testing. An FDR of <20% was considered significant. Insertions were coded as one mutation and also scored by a sliding window approach using the same window size and shift as above.
Results
Phylogenetic reconstruction based on annual and perennial representatives of the Arabideae
To study the evolution of life history in the Arabideae, a phylogenetic tree was constructed using selected annual and perennial species representing all known major lineages of the Arabideae (Karl et al. 2012; Karl & Koch 2013). The phylogenetic tree based on four nuclear and two plastidic markers was fully resolved, and all splits were well supported (Fig. 1).
Figure 1.

Left: Bayesian phylogenetic analysis of 17 representative annual and perennial species of the Arabideae based on a concatenated alignment of ITS, trnL, trnLF,CHS,ADH and At2g13360. Names of groups and/or larger genera of which only representatives were used in the phylogenetic reconstruction are given in boxes. Annuals are indicated by an encircled 1, posterior probability values are given at all nodes. Arabis alpina and Arabis montbretiana are confirmed to be sister species and together are sisters to the perennial A. nordmanniana. The reconstructed ancestral state for life history (Karl & Koch 2013) has been overlaid on the tree. Expression was studied in species indicated by *. Right: Structural evolution of FLC orthologues in the Arabideae. Throughout the branch leading to A. alpina, the structure of the locus becomes increasingly more complex. However, there is no relation of locus complexity and life cycle as A. nordmanniana loci orthologous to FLC have similar structures to that found in annual A. montbretiana. Colours are explained in the legend below the locus schemes. The sequence labelled as COLDAIR RNA encoding was defined by homology to A. thaliana (Heo & Sung 2011).
Results indicate that the species aggregate containing the perennial model A. alpina is sister to the annuals A. montbretiana and A. nova subsp. iberica. Moreover, perennial A. nordmanniana, which represents four to five perennial polycarpic species (Karl & Koch 2013), is sister to the clade containing A. alpina and A. montbretiana. Thus, the tree is consistent with previous studies (Karl et al. 2012; Karl & Koch 2013) but is based on additional phylogenetic markers and incorporates all annual and perennial species of interest in one analysis. In addition, life history was plotted on the phylogeny for each taxon used in the phylogenetic reconstruction (Figs 1 and S2, Supporting information).
This analysis is consistent with perenniality being the ancestral state and annuality having evolved independently in several lineages due to severe environmental change, as proposed previously (Karl & Koch 2013), but does not unequivocally exclude the possibility that annuality was ancestral in exceptional cases.
Structural evolution of FLC orthologues in the Arabideae. FLC orthologues in perennial Arabidopsis and Arabis species exhibit more complex gene structures when compared to annual Arabidopsis thaliana (Wang et al. 2009; Nah & Chen 2010; Albani et al. 2012; Kemi et al. 2013). Consistent with this observation, A. alpina PEP1 has a complex, duplicated gene structure (Wang et al. 2009; Albani et al. 2012). Therefore, a mechanistic link might exist between complexity of the FLC/PEP1 gene and life history. To further test this hypothesis, FLC orthologues were examined in a wider range of annual and perennial Arabis species. Six Arabis species (A. alpina, A. purpurea, A. montbretiana, A. nova subsp. iberica, A. nordmanniana and A. auriculata) were tested for their vernalization response to determine whether they are likely to harbour functional FLC alleles. Plants were exposed to vernalization treatment of 4 °C for 12 weeks at different times after germination and then returned to 20 °C under long photoperiods until flowering. Control plants were not exposed to vernalization but maintained under long photoperiods. Except for A. purpurea, all tested taxa showed either an obligate (A. alpina, A. nordmanniana) or a facultative (A. auriculata, A. montbretiana, A. nova subsp. iberica; Table S6, Supporting information) vernalization response and were therefore assumed to harbour at least one active FLC orthologue. Furthermore, A. montbretiana and A. auriculata responded to vernalization from soon after germination, and therefore did not exhibit a detectable juvenile phase (Fig. S3 , Supporting information). Three FLC orthologues were sequenced either directly after PCR amplification or after cloning (A. purpurea = ApFLC, A. nova subsp. iberica = AiFLC, A. auriculata = AauFLC). Another two FLC orthologues were identified in contig assemblies based on Illumina sequence data (A. montbretiana = AmFLC, A. nordmanniana = AnFLC‐A and AnFLC‐B; two genes because the analysed A. nordmanniana accession is tetraploid; for method and result of assemblies, see Supplementary Methods and Results, Supporting information). The structures of the loci were then compared to PEP1 and plotted on the phylogeny (Fig. 1, right panel). All analysed orthologues comprised seven exons, except PEP1 in which exon 1 is duplicated (Wang et al. 2009; Albani et al. 2012). Furthermore, no other duplicated regions were detected in the FLC orthologues as determined from the number of obtained PCR fragments or based on the results of blast searches performed on the contig assemblies. Therefore, AnFLC‐A and AnFLC‐B, AauFLC, AiFLC and AmFLC have simple gene structures, similar to FLC in A. thaliana, except that on the phylogeny separating A. auriculata and the other species, a sequence in the first intron showing homology to A. thaliana COLDAIR (Heo and Sung 2011) was duplicated. Therefore, among the analysed Arabis species, there is no correlation between FLC duplication or complexity and life history.
Differential expression of FLC orthologues isolated from four Arabis species correlates with life history
Transcription of PEP1 of A. alpina (Wang et al. 2009) as well as of the FLC orthologues of perennial Arabidopsis lyrata (Kemi et al. 2013) and Arabidopsis halleri (Aikawa et al. 2010) is reactivated after vernalization, while in A. thaliana, FLC is stably repressed upon return to warm temperatures (Bastow et al. 2004; Sung & Amasino 2004; Finnegan & Dennis 2007). To evaluate the extent to which FLC expression pattern is correlated with life history, we analysed the expression of the FLC orthologues of A. montbretiana, A. nordmanniana (allele specific for AnFLC‐A and AnFLC‐B) and A. auriculata using PEP1 as a control. In all four species, expression was detected before vernalization and was strongly reduced at the end of vernalization (Figs 2 and S4, Supporting information). Upon transfer to warm temperatures after vernalization, however, only the alleles from perennial species (AnFLC‐A and AnFLC‐B, PEP1) increased in expression, although to different extents, while the alleles from the annual species (AauFLC and AmFLC) were stably repressed (Figs 2 and S4; Data S1, Supporting information). These data demonstrate that the FLC orthologues follow a different expression pattern in annual and perennial Arabis taxa, consistent with data obtained from perennial and annual Arabidopsis species (Bastow et al. 2004; Sung & Amasino 2004; Finnegan & Dennis 2007; Aikawa et al. 2010; Kemi et al. 2013). Arabidopsis and Arabis belong to two different evolutionary lineages of the family (Couvreur et al. 2010), and therefore, our data demonstrate that a difference in repression of FLC orthologues between annual and perennial taxa after vernalization is found widely in the Brassicaceae.
Figure 2.
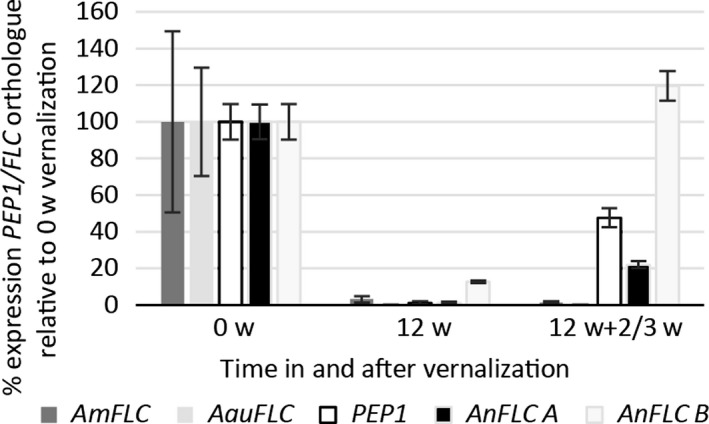
Relative expression of FLC orthologues in the perennials A. alpina (PEP1) and tetraploid A. nordmanniana (AnFLC‐A and AnFLC‐B) as well as in the annuals A. montbretiana (AmFLC) and A. auriculata (AauFLC). All examined orthologues are expressed before vernalization (0w = 0 weeks vernalization after growth at warm temperature for 3 or 6 weeks after germination) and repressed by vernalization (12 wv = 12 weeks vernalization at 4 °C). However, only in the perennial species are the FLC orthologues derepressed after vernalization because in the annual species, they stay stably repressed (12 wv + 2/3w = 12 weeks vernalization followed by 2 or 3 weeks at warm temperature). Expression values are relative to the respective orthologues of PP2A or RAN3 (A. nordmanniana) and relative to 0 weeks (0w) of vernalization. For each sample, leaves of four to six plants were pooled, error bars represent standard deviation of three or four technical qPCR replicates (n = 3–4). A biological replicate is given in the supplement (Fig. S4, Supporting information).
Construction of hybrids between A. montbretiana and A. alpina and analysis of the basis of the divergence in AmFLC and PEP1 mRNA patterns
To study the regulation of life history as well as the inheritance of PEP1 and AmFLC expression patterns, A. alpina acc. Pajares and A. montbretiana were crossed. The flowers of A. montbretiana were emasculated and cross‐pollinated with A. alpina acc. Pajares pollen, but no viable seeds were formed. Therefore, an embryo rescue protocol was applied (Sauer & Friml 2008). By this approach, one plant was successfully grown from approximately 100 cultured immature seeds and its hybrid identity was confirmed by sequencing and length polymorphism markers (Tables S7 and S8, Supporting information). The hybrid was vernalized for 12 weeks to ensure that it flowered. When returned to warm temperatures and long‐day (LD) conditions, the plant flowered abundantly from all shoots (Fig. 3D–F). Axillary shoots were produced from all leaf axils generating a plant that constantly increased in size and flowered perpetually from all branches. Siliques were formed, but all seeds aborted early in development and no viable seeds could be obtained. Therefore, the hybrid could not be reliably scored as annual or perennial because it did not complete the life cycle by forming seeds.
Figure 3.

Arabis montbretiana (A), A. alpina (B) and A. montbretiana × A. alpina (C) at 50 days after germination. D–F A. montbretiana × A. alpina at (D) the onset of flowering at 99 days after germination, (E) fully flowering but not setting viable seeds after self‐pollination 129 days after germination, (F) perpetually growing and flowering 427 days after germination. (G) Relative expression of AmFLC and PEP1 through a vernalization cycle in two cuttings obtained from the hybrid in D–F. Both FLC orthologues are expressed before vernalization (0w = 0 weeks) and repressed by vernalization (12w = 12 weeks vernalization). After vernalization and return to warm temperatures (12d = 12 days and 19d = 19 days in warm), AmFLC stays stably repressed while PEP1 rises again in expression. Thus, in the interspecies hybrid, both FLC orthologues follow the expression pattern observed in the parents indicating that the differential expression is cis‐mediated. Expression is expressed relative to PP2A and relative to 0 weeks of vernalization as percentage. Error bars represent the propagated error of the standard deviation calculated for two biological replicates with four technical replicates (n = 3–4) each. H‐I Relative expression of PEP1 and/or AmFLC through a vernalization cycle in three introgression lines obtained from back crossing the hybrid in D–F to A. alpina. Expression is relative to PP2A and then expressed as % relative to expression at 0‐w vernalization. Error bars represent the standard deviation derived from three or four technical (n = 3–4) replicates. Different introgression lines represent biological replicates. H Analysis of three independent heterozygous plants (5034, 5002, 5078). The orthologues show different behaviours after vernalization and maintain the same or similar expression pattern that they show in the parental species (Fig. 2). I Analysis of three different AmFLC homozygous plants (5012, 5057, 5006). AmFLC displays the same stable expression pattern after vernalization as in the A. montbretiana control.
The basis of the differential expression patterns of PEP1 and AmFLC is unknown. The generated hybrid, however, offers the opportunity to test whether the patterns of AmFLC and PEP1 transcription differed due to cis‐acting variation by analysing their expression after vernalization in the same genetic background (Wittkopp et al. 2004). AmFLC and PEP1 mRNA levels were evaluated in two cuttings before, at the end of and subsequent to a 12‐week‐vernalization cycle using gene‐specific primers designed based on polymorphisms detected between the two genes. Both PEP1 and AmFLC mRNAs were expressed before vernalization and repressed during vernalization (Fig. 3G). However, upon transferring the plants to warm LDs at the end of vernalization, AmFLC mRNA remained at very low levels, while abundance of PEP1 mRNA increased (Fig. 3G). In conclusion, in the interspecies hybrid, each gene exhibits the expression pattern characteristic of the parent from which it originated. This result suggests that cis‐acting sequence variation at PEP1 and AmFLC confers the observed difference in expression pattern in the hybrid. To verify this hypothesis, AmFLC introgression lines that were largely homozygous for A. alpina but retained a genomic segment including AmFLC were constructed by backcrossing to A. alpina three times and selecting for inheritance of AmFLC (Fig. S5, Supporting information). Three plants heterozygous and three plants homozygous for the AmFLC genomic segment were exposed to vernalization along with A. montbretiana as a control. AmFLC was stably repressed after vernalization in the homozygous AmFLC introgression line (Fig. 3I) and in the heterozygotes was much more strongly repressed than PEP1 (Fig. 3H). These results again indicate that both AmFLC and PEP1 responded differently to vernalization when in the same genetic background. Taken together, the analyses of the hybrid and of the introgression lines demonstrate that cis‐acting variation contributes to the differential behaviour of AmFLC and PEP1 after vernalization.
SNPs between PEP1 and AmFLC are distributed nonrandomly
The different expression patterns of PEP1 and AmFLC involve variation in cis‐acting elements. To identify polymorphisms that might contribute to the different expression patterns, the nucleotide sequences of the genomic loci were compared. Comparison of the AmFLC genomic DNA sequence with the cDNA showed that the protein is encoded by a single gene spanning 7 exons (Fig. S6A, B, Supporting information), as is FLC in A. thaliana. Furthermore, alignment of the AmFLC cDNA to that of PEP1 demonstrated that in A. alpina acc. Pajares, the splice site at the PEP1 intron 2–exon 3 junction is shifted, producing an additional 27 bp in the mRNA and therefore a predicted 9 amino acid insertion in the protein. In addition, five nonsynonymous substitutions are present within the coding sequence (Fig. S1, Supporting information).
In total, 176 SNPs and 57 indels were detected between AmFLC and PEP1 by comparing the genic regions spanning ~280 bp upstream of the start codon (of exon 1A in PEP1; Fig. S6C, D, Supporting information) that include sequences homologous to the functional promoter of FLC (Sheldon et al. 2002), all exons and introns as well as ~400 bp downstream of the stop codon. This number of SNPs and indels meant that no simple polymorphism could be identified as impairing a cis‐acting element conferring the differential expression of AmFLC and PEP1 after vernalization. Therefore, the distribution of SNPs and indels between AmFLC and PEP1 was examined by a sliding window approach to test whether these occurred at higher frequencies in specific genic regions (Fig. 4A, B, Supporting information). Six clusters of SNPs found in this analysis differed significantly (P ˂ 0.1) from the expected random distribution (Fig. S7, Supporting information). Five of these clusters were located in the 5′ part of the first intron leading to a higher evolutionary distance of this segment of the intron between A. alpina and A. montbretiana than for the 3′ part (0.044 vs. 0.017).
To obtain insight into the possible functional relevance of the clustered polymorphisms, the loci were annotated by comparison with FLC, in which several cis‐regulatory regions have been identified in intron 1 (Fig. 4, Tables S9, S10, S11, S12 and Figs S8, S9, S10, S11, Supporting information). A region showing homology to the nucleation region (Angel et al. 2011; Yang et al. 2014) (Fig. S11, Supporting information) as well as duplicated regions showing homology to the VRE (Sung et al. 2006) (Fig. S10, Supporting information) and the sequence encoding COLDAIR RNA (Heo & Sung 2011) were identified. The duplicated regions showing homology to the VRE and the COLDAIR RNA‐encoding sequence showed 79% (VRE and COLDAIR) identity to each other in A. alpina and 73% (VRE) and 77% (COLDAIR) in A. montbretiana (Table S12, Supporting information). The six statistically significant peaks in SNP frequency detected in the sliding window analysis coincided with the nucleation region, the region between the nucleation region and the more 5′ located segment showing homology to the VRE and the COLDAIR RNA‐encoding sequence (COLDAIR‐like 1) and the sequence separating COLDAIR‐like 1 from the second segment showing homology to the VRE (Fig. 4A).
Overall, the higher conservation of the 3′ part of intron 1 is consistent with it being required for transcription of FLC (Sheldon et al. 2002). By contrast, the higher variation in the 5′ part of intron 1, which includes sequences showing homology to the nucleation region and to the COLDAIR RNA‐encoding sequence of FLC, suggests that divergence within these segments could be responsible for the species‐specific expression patterns of PEP1 and AmFLC.
SNP distribution patterns detected by comparing annual and perennial species pairs in different tribes of the Brassicaceae
FLC orthologues are also differentially expressed after vernalization in other closely related annual and perennial species, so that, for example, FLC is stably repressed after vernalization in A. thaliana (Sheldon et al. 2000) while the expression of its orthologue from A. lyrata (AlFLC1) increases again after vernalization (Kemi et al. 2013). Therefore, to broaden the analysis of FLC orthologues, the genomic DNA sequences of FLC, AlFLC1, AmFLC, PEP1, AnFLC‐A and AauFLC were aligned, gap columns were excluded, and SNP distribution was scored using a sliding window approach. The analysis of FLC and AlFLC1 resulted in a SNP distribution pattern similar to the one described for the comparison of AmFLC and PEP1 (Fig. 5A). Several peaks in SNP frequency containing more SNPs than the expected random distribution were detected in the comparison of each species pair (Fig. 5A). Several of these peaks in SNP frequency are specific to only one of the species comparisons, but two were detected in the same regions in both comparisons. One of the common SNP frequency peaks was detected in a region homologous to the nucleation region (Yang et al. 2014), and the second was present upstream of the VRE (Sung et al. 2006) (Figs 5A, S19 and S20, Supporting information). Sequence differences in these two regions might contribute to the different expression patterns of FLC orthologues detected in annual and perennial species, because they are highly polymorphic in two independent occurrences of divergence of annual and perennial patterns.
Figure 5.
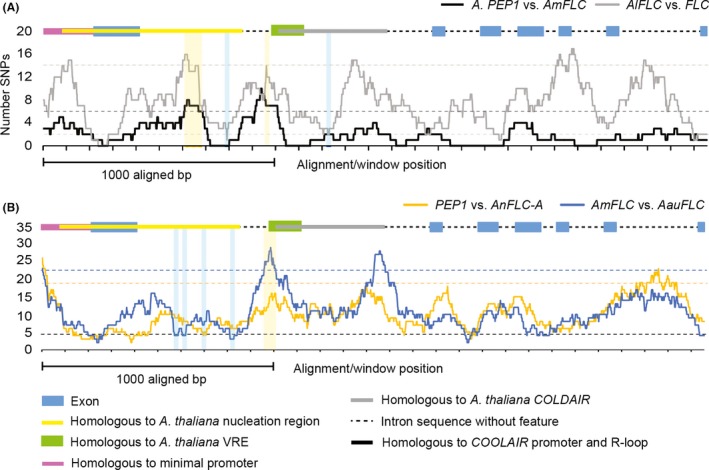
Comparison of SNP distribution in (A) PEP1 vs. AmFLC and AlFLC1 vs. FLC and (B) AmFLC vs. AauFLC and PEP1 vs. AnFLC‐A by a sliding window analysis (window size 100 bp, shift 1 bp). For the analysis of SNP distribution, all indels were excluded; exons as well as sequence features homologous to regulatory elements known from FLC are annotated on top of the graph as a colour‐coded scheme. Exclusion of gap columns leads to deletion of elements which are duplicated only in PEP1, AmFLC and AnFLC‐A, and therefore, the schematic overview of the locus is shorter than the locus schemes in Fig. 4. Nucleation region is the sequence homologous to the region showing elevated H3K27me3 before vernalization in Arabidopsis (Yang et al. 2014); COLDAIR represents the regions homologous to the COLDAIR RNA‐encoding region (Heo & Sung 2011); vernalization response element (VRE) indicates the region homologous to the VRE of A. thaliana identified in (Sung et al. 2006); dashed lines represent thresholds above or below which more or fewer SNPs than expected by chance occur: (A) grey dashed line at 14 SNPs threshold for more and at two SNPs for fewer SNPs than expected by chance in AlFLC1 vs. FLC; line at six SNPs threshold for more SNPs than expected by chance for PEP1 vs. AmFLC; no window with significantly fewer SNPs, (B) blue dashed line significantly more SNPs than expected by chance for AmFLC vs. AauFLC, orange dashed line significantly more SNPs than expected by chance for PEP1 vs. AnFLC‐A, black dashed line fewer SNPs than expected for both curves. Peak regions with windows containing significantly more SNPs than expected in a random distribution are shaded in yellow; regions containing significantly fewer SNPs than expected in a random distribution are shaded in blue.
As a further control to test the functional significance of the two regions described above, we also examined whether they are highly polymorphic between related species that do not differ in life history. To this end, SNP frequencies were analysed in comparisons of the FLC orthologues of the closely related annual taxa A. montbretiana and A. auriculata as well as the closely related perennials A. alpina and A. nordmanniana (Fig. 5B). The peaks in SNP frequency obtained in these analyses were also compared to those detected in Fig. 5A. The region adjacent to the 5′ end of the VRE, which was highly polymorphic in the comparisons of the annual and perennial species (Fig. 5A), was also detected as variable in comparisons of FLC orthologues of the two annual species but not in comparing the two perennial species (Figs 5B and S20, Supporting information). Therefore, this sequence could be functionally conserved in perennials and diverge independently in annuals, so that it contains a high frequency of SNPs in comparisons of annuals and perennials or of annuals. By contrast, the polymorphic segment detected in the nucleation region by comparing annuals and perennials (Fig. 5A) showed fewer SNPs than expected in a random distribution when the orthologues of either the two genes from perennials (AnFLC‐A and PEP1) or the two genes from annuals (AmFLC and AauFLC) were compared (Figs 5B, S22 and S23, Supporting information). Variation in this segment of the nucleation region at the 5′ end of intron 1 could therefore be involved specifically in the divergence of annual and perennial patterns of regulation of FLC orthologues. Our sequence analysis of FLC orthologues from the Arabideae and Camelineae therefore identifies this region in the nucleation region and another at the 5′ end of the VRE as likely to be involved in the divergence of annual and perennial patterns of expression of FLC orthologues (Figs 6, S19 and S20, Supporting information).
Figure 6.

Summary of the regulatory regions of FLC and their variation among annual and perennial taxa. Top: Schematic representation of the FLC locus of A. thaliana including the minimal promoter, exon 1, intron 1 and exon 2 indicating the positions of SNPs, binding sites and regulatory elements that were identified in other studies as well as regions containing putative regulatory elements identified in this study. Lower diagrams depict segments of three reporter gene deletions reported by Sheldon et al. (2002) that showed unstable or stable repression after vernalization. The lowest diagram shows a deletion mutant in which FLC was not stably repressed after vernalization and defines the VRE (Sung et al. 2006).
Detection of common regulatory elements
The regions harbouring significantly fewer SNPs than expected might define regions that are important for FLC regulation in both annuals and perennials. These regions were therefore examined in all four alignments shown in Fig. 5A, B. No regions harbouring fewer SNPs than expected were detected in comparisons of AmFLC and PEP1, perhaps because many of the sequence windows in this comparison were 100% identical. However, the analysis of FLC and AlFLC1 identified six regions harbouring fewer SNPs than expected, and three of these regions were located in intron 1. The first was located at the end of the nucleation region, the second in the region homologous to the COLDAIR RNA‐encoding sequence and the third at the very end of the first intron (Figs 5A and S21, Supporting information). The comparison of AmFLC and AauFLC also detected the segment at the end of the nucleation region as harbouring fewer SNPs than expected (Fig. 5B). Interestingly, comparison of sequences of FLC and AlFLC1 in the segment homologous to the nucleation region and the COLDAIR RNA‐encoding sequence revealed that each segment includes the motif GGATTTGT and both of these motifs are perfectly conserved in all six species analysed (Fig. S21, Supporting information). Therefore, this sequence element could have a common function in FLC regulation across all taxa (Fig. 6).
Discussion
To study the divergence of life history, a phylogeny of 17 annual and perennial species from the tribe Arabideae was constructed using a combination of six different phylogenetic marker systems. This highly resolved and supported phylogeny was congruent to one that previously fully resolved the backbone of the tribe (Karl & Koch 2013), but the phylogeny described here also included D. nemorosa and A. nova subsp. iberica. These species were grouped together with their previously described relatives, which are other Draba species and A. montbretiana, respectively (Jordon‐Thaden et al. 2010; Karl et al. 2012). The phylogeny of the Arabideae is therefore robust and provides a framework for studying the evolution of annual and perennial life history, which have diverged within several of the established clades.
Evolution of FLC structure
Remarkable sequence variation at FLC genes occurs within and between Brassicaceae species, and increasing complexity of these loci has been suggested to be linked to perenniality in A. alpina and in Arabidopsis species (Alonso‐Blanco & Méndez‐Vigo 2014). Indeed, through the phylogenetic branch leading from annual A. auriculata to A. alpina, the orthologous FLC loci increased in complexity. AauFLC showed a similar simple structure to FLC of A. thaliana, while PEP1 from perennial A. alpina acc. Pajares was the most complex locus incorporating a partial duplication that includes exon 1 and generates two overlapping transcripts (Albani et al. 2012). Furthermore, AmFLC and AiFLC from annual A. montbretiana and A. nova subsp. iberica showed simple structures similar to A. thaliana FLC, further strengthening this hypothesis. However, in perennial tetraploid A. nordmanniana, single‐copy AnFLC genes were found that showed simple exonic structures similar to FLC, although they contained a duplication of part of intron 1 related to COLDAIR (Heo and Sung 2011) that was also present in AmFLC and AiFLC. Therefore, at least in the Arabideae, there does not seem to be a strict association between gene or exon duplication at the FLC locus and perenniality.
Correlation between annualism and FLC expression pattern
In perennials, flowering is strongly repressed at particular times during the life cycle, ensuring episodes of vegetative and reproductive development, whereas annuals usually flower more rapidly and do not return to vegetative development. In the Brassicaceae, the vernalization response pathway was proposed to play several roles in conferring these episodes of vegetative and reproductive development in the context of the perennial life cycle (Wang et al. 2009; Aikawa et al. 2010; Kemi et al. 2013). Furthermore, annual and perennial species in the Brassicaceae also differ in the age at which they acquire competence to flower in response to vernalization (Wang et al. 2011; Bergonzi et al. 2013; Zhou et al. 2013). Consistent with this conclusion, A. montbretiana and A. auriculata behaved in a similar way to annual A. thaliana and flowered in response to vernalization treatments given within 1 week of germination. Thus, the annuals A. montbretiana, A. auriculata and A. thaliana show similar flowering responses and differ from perennial A. alpina in that floral induction through vernalization is less restrained by age.
In A. thaliana, FLC expression is stably repressed during vernalization, so that upon return to warm temperatures, its expression stays low ensuring that the plant is stably induced to flower (Michaels & Amasino 1999; Bastow et al. 2004). By contrast, in several perennial Brassicaceae species, levels of expression of orthologues of FLC cycle in response to temperature, so that they fall in cold and rise again in subsequent periods of warm (Wang et al. 2009; Aikawa et al. 2010; Kemi et al. 2013). Thus, the cycling pattern of FLC expression correlates with episodes of vegetative growth and flowering in the perennials. The functional significance of these patterns was demonstrated genetically in A. alpina (Wang et al. 2009; Albani et al. 2012). The FLC orthologues of annuals A. montbretiana and A. auriculata were also found to be stably repressed after vernalization, similar to the pattern of expression of FLC found in A. thaliana and contrasting with the perennial pattern of PEP1 regulation in A. alpina and A. nordmanniana. In tetraploid A. nordmanniana, both FLC orthologues were found to be upregulated to different extents after vernalization, which might be a cis‐mediated effect caused by any of the numerous SNPs that differ between the two alleles. Thus, consistent with their flowering behaviour, annual and perennial species differ in the patterns of expression of FLC orthologues. This stable repression in response to vernalization must have evolved independently more than once during the diversification of the Arabideae and in the Arabidopsis genus.
Evolution of FLC regulation among annual and perennial Brassicaceae species
Differences in gene expression arise during evolution through cis‐acting changes in the gene of interest or by alterations in the activity of trans‐acting factors that contribute to the regulation of its expression (Emerson & Li 2010). These possibilities can be distinguished by the construction of hybrids, in which both orthologues of the gene of interest are present but the genetic background encoding trans‐acting factors is identical (Wittkopp et al. 2004; de Meaux et al. 2005). If the orthologues show different expression patterns in the parental species and these are retained in the hybrid, then cis‐acting changes must contribute to the differential expression. In the hybrid of A. montbretiana and A. alpina as well as in introgression lines derived from the hybrid, PEP1 and AmFLC were expressed in different patterns after vernalization, as observed in the parental species. Therefore, the differential expression must involve cis‐acting changes. Scans of SNP frequency were performed between FLC orthologues to search for putative regulatory elements that have diverged between annual and perennial species and to identify conserved sequences that may have a common function in FLC regulation in annual and perennial taxa.
Expression of FLC in warm temperatures
Transgenic approaches based on deletion analysis of a FLC::GUS transgene demonstrated that 272 bp of promoter as well as ~200 bp at the 5′ end and ~525 bp at the 3′ end of intron 1 are sufficient for transcription of FLC before vernalization (Sheldon et al. 2002). Our sequence comparisons included 295 bp of FLC sequence upstream of the ATG, and the comparisons of FLC and AlFLC1 or PEP1 and AnFLC‐A identified one region containing significantly more SNPs than expected. No known motifs were present in this region. However, the 272‐bp minimal promoter of FLC contained three putative bZIP recognition motifs (Sheldon et al. 2002) of which only one is highly conserved in all six taxa included in the analysis and is therefore likely to be functionally important. A later study reported a binding motif that is involved in recognition of the FLC promoter by SUF4, a component of the FRIGIDA complex that increases FLC transcription in A. thaliana (Choi et al. 2011). However, the binding motif, which is located upstream of the minimal promoter (Sheldon et al. 2002), was mutated or absent in A. alpina and entirely absent in A. montbretiana. Therefore, either regulation of FLC by the FRIGIDA complex does not occur in the Arabis genus or the binding site for the SUF4 complex is not conserved.
Repression of FLC by vernalization
The same segments of intron 1 required for expression of FLC prior to vernalization were also described as being important for its repression by vernalization (Sheldon et al. 2002). Later studies demonstrated that the 5′ end of intron 1 overlaps with a region referred to as the nucleation region, at which H3K27 accumulates most rapidly and to high levels during vernalization (Yang et al. 2014). The same region contains two RY elements which act as binding sites of the transcriptional repressor VAL1 that nucleates silencing at the FLC locus by interacting with the apoptosis‐ and splicing‐associated protein (ASAP) complex and maybe Polycomb Repressive Complex 1 (PRC1) (Qüesta et al. 2016; Yuan et al. 2016). One of the RY elements was perfectly conserved across all six taxa in our analysis, consistent with a highly conserved function, while the second one was mutated in A. alpina, but only in one of the two tandem copies of the first exon. Sequence variation in the nucleation region was proposed to cause differences in the kinetics of vernalization because four SNPs in the A. thaliana accession Lov‐1 were identified in this region, including two in the 5′ end of intron 1, and shown to be associated with requirement for an extended vernalization period to cause silencing of FLC after vernalization (Coustham et al. 2012). The first SNP in intron 1 was conserved among the Lov‐1 accession and all taxa included in this study, and only the FLC allele of A. thaliana Columbia was different, suggesting that among A. thaliana accessions, the Lov‐1 SNP is ancestral. The second SNP in intron 1 was variable across taxa. In this position, Lov‐1, PEP1 and AmFLC were the same, while AnFLC‐A, AlFLC1, AauFLC and FLC from A. thaliana Columbia shared the same base in this position. No regions with significantly more or fewer SNPs than expected in a random distribution were found to overlap with the two SNPs in intron 1 detected in the study of the Lov‐1 accession (Qüesta et al. 2016). However, the more 5′ VAL1 binding site was located within the region harbouring fewer SNPs than expected in annuals, while the second VAL1 binding site was located in the overlap of the regions harbouring fewer SNPs than expected in annuals or perennials (Fig. 6). The antisense RNA COOLAIR which plays an initial role in downregulation of FLC in response to cold (Swiezewski et al. 2009) is conserved in A. alpina (Castaings et al. 2014).
Maintenance of FLC repression after vernalization
Deletion analysis identified four regions within the first intron of FLC that are required for stable repression of transcription after vernalization (Sheldon et al. 2002). Another study identified the vernalization response element (VRE) whose deletion leads to unstable repression of FLC (Sung et al. 2006). Subsequently, parts of the VRE were reported to act as the promoter of a noncoding sense RNA named COLDAIR that interacts with components of the PRC2 complex in vitro (Heo and Sung 2011). The COLDAIR RNA‐encoding sequence also included one of the candidate regions for conferring stable repression of FLC after vernalization identified by Sheldon et al. (2002). However, there are some contradictions between the reporter construct data (Sheldon et al. 2002) and the analysis of the VRE mutant (Sung et al. 2006) that are summarized in Fig. 6. The comparison of AlFLC1 and FLC identified two regions within the first intron that contained fewer SNPs than expected in a random distribution. One of the regions was also found in the comparison of AauFLC and AmFLC, whereas in PEP1 and AmFLC comparisons, no fragments harbouring fewer SNPs than expected were found. Identification of the same region in two sequence comparisons in different evolutionary lineages, Camelineae and Arabideae, indicates that this region may be of general importance in the regulation of FLC and its orthologues. Interestingly, both conserved regions in the A. thaliana comparison with A. lyrata contained the same perfectly conserved sequence motif (GGATTTGT), and these were also present in all four Arabis species. The deletion analysis of Sheldon et al. (2002) is consistent with a role for this highly conserved ‘GGATTTGT’ motif in stable repression of FLC. In this study, deletion of the region including both ‘GGATTTGT’ motifs caused unstable repression after vernalization, while constructs where only one copy of the motif was deleted were still stably repressed after vernalization. Thus, these motifs might act redundantly in conferring stable repression of FLC by vernalization. If these motifs do contribute to stable repression of FLC in annual A. thaliana, then perennial taxa such as A. alpina must also contain some of the motifs contributing to stable repression in annuals.
Reactivation of FLC orthologues after vernalization
No cis‐regulatory elements have been described that are involved in the reactivation of FLC orthologues in perennials after vernalization. Unstable repression of FLC occurs upon deletion of the VRE (Sung et al. 2006), and therefore, a loss of function of the VRE in the perennial lineages could confer reactivation after vernalization. However, this is apparently in contradiction to the hypothesis that perenniality is the ancestral state. Nevertheless, parts of the VRE are indeed deleted in A. halleri and A. lyrata, and in A. alpina, a 232‐bp insertion is found at the 5′ end of the VRE. On the other hand, although expression of AnFLC‐A and AnFLC‐B was unstable after vernalization, its VRE sequence contained no insertions or deletions compared to that of FLC. The SNP scans and statistical tests identified regions that harbour significantly more or fewer SNPs than expected by chance. The combination of SNP scans comparing annual and perennial taxa and SNP scans within pairs of closely related annual and perennial taxa identified a segment within intron 1 at the 3′ end of the nucleation region that was particularly divergent within the annual–perennial pairs but more strongly conserved among annuals in one segment and among perennials in another segment. Therefore, this region might contain at least two regulatory elements that vary during evolution and one of which could be related to reactivation of FLC after vernalization (Fig. 6). The identified region could be tested by transgenic approaches to directly examine its functional significance.
A possible hypothesis from these studies is that perennials contain two types of regulatory elements that confer stable repression or reactivation of FLC orthologues after vernalization. The annual expression pattern could then evolve several times independently by recurrent deactivation of a perennial element required for reactivation. Thus, mutation would provide a flexibility in regulation of FLC orthologues that could rapidly evolve along with shifts from perennial to annual life history. The phylogenetic framework and sequence analysis presented here provide a basis for further testing of this hypothesis.
C.K. and G.C. designed research and wrote manuscript; G.C. and M.K. provided plant material; C.K. and S.B. performed vernalization shift experiment; C.K. helped in DNA extraction, genotyping, expression analysis, estimates on evolutionary divergence, SNP distribution analysis, sequencing of FLC orthologues, A. nordmanniana assembly, amplification of phylogenetic marker sequences, generation and verification of interspecies hybrid, A. montbretiana contig assembly and AmFLC contig detection and analysis; R.K. and C.K. involved in alignment and phylogenetic analysis; E.S. and C.K. involved in generation of length polymorphism markers; C.K. involved in A. montbretiana and A. nordmanniana contig assembly and FLC contig detection; E.S. involved in preparation of A. montbretiana assembly for submission, A. nordmanniana read preparation, genome annotation, k‐mer analysis, ortho‐MCL analysis; A.T. helped in statistical test on SNP distribution.
Data accessibility
Whole‐genome shotgun projects: deposited at GenBank under the Accession nos LNCG00000000 (A. nordmanniana) and LNCH00000000 (A. montbretiana).
DNA sequences: KC814706;JQ919880;GU181934;GU181999;JQ919835;JQ919813;JQ919857;KC814698;JQ919879;HM046191;HM046243;HM046216;KC814699;JQ919881;GU182084;GU181952;GU182017;KC814700;JQ919882;KC814707;JQ919884;HQ646785;HQ646724;FJ187937;FJ188234;FJ188083;KC814701;JQ919885;HM046193;HM046245;HM046218;KC814708;JQ919886;KF547353;KF547826;KF547624;KC814702;JQ919887;JQ919839;JQ919817;JQ919861;KC814709;JQ919888;KC814703;JQ919889;GU181938;GU182003;GU182089;GU181957;GU182022;KC814713;JQ919899;JQ919840;JQ919818;JQ919862;KC814704;JQ919890;HQ646642;HQ646763;HQ646702;KC814705;JQ919891;KF547181;GU202868;GU202800;KC814710;JQ919893;JQ919844;JQ919822;JQ919866;KC814711;JQ919894;JQ919845;JQ919823;JQ919867;KC814712;JQ919896;KU525030;KU525030;KU525030;KU525030;KU525030;KU525030;KU321504;KU321505;KU321506;KU321507;KU321508;KU321509;KU321510;KU321511;KU321512;KU321513;KU321514;KU321515;KU321516;KU321517;KU321518;KU321519;KU321520;KU321353;KU321354;KU321355;KU321356;KU321357;KU321358;KU321359;KU321360;KU321361.
Supporting information
Table S1 Details of accessions used in this study stating country of origin, provider of seeds and sequences used in the phylogenetic analysis and their genebank accession numbers; for the pairs of grey shaded accessions marker sequences were used in combination to obtain a full set for the phylogenetic reconstructions.
Table S2 Primers used in this study for amplification of phylogenetic marker sequences, qPCR and amplification of the mRNA of FLC orthologues as well as the genomic loci.
Table S3 Alignment of marker sequences used for phylogenetic analysis; the alignment is given in nexus format and partitions are indicated at the end of the file.
Table S4 Results of Model Testing and Bayesian analyses for the individual single marker dataset and the combined dataset
Table S5 Alignment of marker sequences (ADH, CHS, At2g13360) used for the maximum parsimony analysis including both co‐orthologues of A. nordmanniana for each marker. Introns were not included in the analysis and therefore deleted.
Table S6 Vernalization response of selected Arabideae; plants were grown either in long day for several months or in long day for 6 weeks, transferred into vernalization for 12 weeks and then returned to warm temperatures and long day conditions.
Table S7 Polymorphic sites in Arabis alpina Pajares and Arabis montbretiana and the polymorphisms found in the hybrid individual in the ITS confirming the hybrid identity of the plant.
Table S8 Primers used for genotyping the F1 plant and introgression lines carrying a genomic fragment including AmFLC.
Table S9 Number of SNPs and indel polymorphisms detected in different features of PEP1 versus AmFLC, Am = A. montbretiana, Aa = A. alpina, At = A. thaliana (exons, regulatory sequences known from FLC (proximal promoter (Sheldon et al. 2002); nucleation region is the sequence homologous to the region showing elevated H3K27me3 before vernalization in Arabidopsis (Yang et al. 2014); COLDAIR is the regions homologous to the COLDAIR RNA encoding region (Heo & Sung 2011); Vernalization Responsive Element (VRE) indicates the region homologous to the VRE of A. thaliana identified in (Sung et al. 2006)).
Table S10 Alignment of the PEP1, AmFLC, AlFLC1 and FLC loci comprising a partial sequence of the next upstream gene or its orthologues from A. thaliana (At5 g10150), all intergenic sequence between At5 g10150 and its orthologues and FLC and orthologues as well as all sequences orthologues/homologous to exons and introns of FLC (including indels) and approximately 500 bp of sequence downstream of the stop codon.
Table S11 Summary of the comparison of AtFLC, AmFLC, AlFLC1 and AaPEP1.
Table S12 Comparison of VRE/VRE‐like and COLDAIR/COLDAIR‐like sequences found in the first intron of PEP1 and AmFLC; intron 1 in A. alpina refers to the sequence homologous to A. montbretiana intron 1.
Table S13 Associated p‐values for the respective SH tests based on the different single marker datasets.
Table S14 Alignment of both FLC orthologues from A. nordmanniana (AnFLC A and B) to PEP1 and AmFLC; a stretch of N was inserted between the two contigs joined to form AnFLC A. The alignment includes a partial sequence of the orthologue of At5 g10150, exons 1 (A and B in A. alpina) to 7 including all intronic sequences (partial intron 6 in AnFLC A) as well as ~280‐bp sequence downstream of the stop codon (includes the COOLAIR promoter (Castaings et al. 2014; Csorba et al. 2014; Swiezewski et al. 2009).
Fig. S1 Alignment of cDNA sequences of AmFLC, AauFLC and PEP1 used for designing primers for qPCR.
Fig. S2 Maximum Parsimony (MP) phylogenetic reconstruction calculated in MEGA5 (Tamura et al. 2011) based on the orthologues of ADH, CHS and At2g13360.
Fig. S3 Vernalization response of (A) A. montbretiana and (B) A. auriculata measured in leaf number.
Fig. S4 Biological replicates of expression data shown in Fig. 2.
Fig. S5 Genotypes of introgression lines (A. montbretiana in A. alpina acc. Pajares background) used in the expression analysis of AmFLC and PEP1 for confirming the results obtained from the two cuttings of the original F1 plant (A. montbretiana x A. alpina acc. Pajares).
Fig. S6 (A) Dot plot of the AmFLC genomic region (last exon upstream gene until the end of the COOLAIR promoter) vs. AmFLC cDNA; 7 exons can be detected, no exon has been duplicated (B, C, D) Comparison of the PEP1, AmFLC and FLC sequences by GATA plots.
Fig. S7 Statistical test on the SNP distribution in the alignment of AmFLC and PEP1.
Fig. S8 Alignment of the proximal promoter region including UTR of FLC, AmFLC and PEP1; the proximal promoter is defined as the region homologous to what was described as FLC minimal promoter (Sheldon et al. 2002).
Fig. S9 Alignment of COLDAIR and A. montbretiana and A. alpina COLDAIR ‐like 1 and COLDAIR ‐like 2; yellow positions indicate ancestral positions maintained in COLDAIR ‐like 1 while pink positions indicate conservation in COLDAIR ‐like 2; more ancestral positions are maintained in COLDAIR ‐like 1; COLDAIR‐likes are defined as regions showing homology to the Arabidopsis COLDAIR sequence (Heo & Sung 2011).
Fig. S10 Alignment of VRE and A. montbretiana and A. alpina VRE ‐like 1 and VRE ‐like 2; VRE‐like sequences are defined as sequences showing homology to the Vernalization Response Element of FLC in Arabidopsis thaliana (Sung et al. 2006).
Fig. S11 Alignment of the A. montbretiana and A. alpina region homologous to the nucleation region in A. thaliana defined by the ChIP primers from (Yang et al. 2014).
Fig. S12 Arabis montbretiana life cycle monitored under greenhouse conditions.
Fig. S13 Alignment of PEP1 exon 1A and 1B and AmFLC exon 1 along with adjacent intron sequences; the 416 bp used to differentiated copy 1A and copy 1B are indicated by $ while the SNPs differentiating the duplicated segment of intron 1 in PEP1 are marked by *
Fig. S14 Kmer abundance distribution.
Fig. S15 Ends of concatenated contigs which were found to show homology to FLC and which were shown to be part of the same molecule (AnFLC B) based on PCR and subsequent Sanger sequencing; All contigs showed overlapping ends.
Fig. S16 Statistical test on the SNP distribution in the alignment of (A) AmFLC and PEP1 and (B‐C) FLC and AlFLC1. The test measures whether the observed SNP number in a window is different from a random distribution (A and B more SNPs than expected, C fewer SNPs than expected); the left panel shows the number of SNPs per window, the right panel shows the –log10 of the associated p‐values; the dashed line is the threshold above which statistical significance was reached; values were corrected for FDR.
Fig. S17 Statistical test on the SNP distribution in the alignment of (A) AnFLC‐A and PEP1 and (B) AauFLC and AmFLC.
Fig. S18 Statistical test on the SNP distribution in the alignment of (A) AnFLC‐A and PEP1 and (B) AauFLC and AmFLC.
Fig. S19 Sequence alignment of six Brassicaceae species in the segment showing more SNPs than expected by chance in the nucleation region from Fig. 5A.
Fig. S20 Sequence alignment of six Brassicaceae species in the segment showing more SNPs than expected by chance upstream/in the region homologous to the VRE from Fig. 5A.
Fig. S21 Sequence alignment of six Brassicaceae species in the segment showing fewer SNPs than expected in a random distribution in the comparison of AlFLC1 and FLC showing homology to (A) the end of the nucleation region and (B) COLDAIR from Fig. 5A; the conserved GGATTTGT‐motif is indicated a red square.
Fig. S22 Sequence alignment of six Brassicaceae species in the segment showing fewer SNPs than expected by chance within the region showing homology to the nucleation region in the annual couple (AauFLC‐AmFLC) from Fig. 5B.
Fig. S23 Sequence alignment of six Brassicaceae species in the segment showing fewer SNPs than expected by chance within the region showing homology to the nucleation region in the perennial couple (PEP1‐AnFLC‐A) from Fig. 5B
Data S1 Ct values and evaluation of qPCR data
Acknowledgments
We are grateful to Renhou Wang, UCSD, USA, for making contact to Birol Mutlu, Inönü University, Turkey, whom we would like to thank for sharing some of his A. montbretiana (BM5948) seeds with us, and to Santiago Martín‐Bravo, Pablo de Olavide University, Spain, for providing us with seeds of A. nova subsp. iberica. We are grateful to Wen‐Biao Jiao for sharing the A. montbretiana genome annotation prior to publication. We would like to thank Maria Albani and Eva Madrid Herrero for providing some of the primers used in this study, Elisa Ansorena for performing some of the expression analyses and Michaela Lehnen for her help with the genotyping. Furthermore, we thank Korbinian Schneeberger, Ulla Kemi, Luise H. Brand, Samson Simon, Stefan Wötzel and Wilma van Esse for valuable discussions and comments on the manuscript. This work was supported by the Deutsche Forschungsgemeinschaft through SFB680 and the Cluster of Excellence on Plant Sciences (CEPLAS), and the European Research Council through HyLife. The laboratory of G.C. receives a core grant from the Max Planck Society.
References
- Aikawa S, Kobayashi MJ, Satake A, Shimizu KK, Kudoh H (2010) Robust control of the seasonal expression of the Arabidopsis FLC gene in a fluctuating environment. Proceedings of the National Academy of Sciences, 107, 11632–11637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albani M, Coupland G (2010) Comparative analysis of flowering in annual and perennial plants. Current Topics in Developmental Biology, 91, 323–348. [DOI] [PubMed] [Google Scholar]
- Albani M, Castaings L, Wötzel S et al (2012) PEP1 of Arabis alpina is encoded by two overlapping genes that contribute to natural genetic variation in perennial flowering. PLoS Genetics, 8, e1003130. doi: 10.1371/journal.pgen.1003130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso‐Blanco C, Méndez‐Vigo B (2014) Genetic architecture of naturally occurring quantitative traits in plants: an updated synthesis. Current Opinion in Plant Biology, 18, 37–43. [DOI] [PubMed] [Google Scholar]
- Al‐Shehbaz IA, German DA, Karl R, Jordon‐Thaden I, Koch MA (2011) Nomenclatural adjustments in the tribe Arabideae (Brassicaceae). Plant Diversity and Evolution, 129, 71–76. [Google Scholar]
- Andres F, Coupland G (2012) The genetic basis of flowering responses to seasonal cues. Nature Reviews Genetics, 13, 627–639. [DOI] [PubMed] [Google Scholar]
- Angel A, Song J, Dean C, Howard M (2011) A Polycomb‐based switch underlying quantitative epigenetic memory. Nature, 476, 105–108. [DOI] [PubMed] [Google Scholar]
- Ansell SW, Stenøien HK, Grundmann M et al (2011) The importance of Anatolian mountains as the cradle of global diversity in Arabis alpina, a key arctic‐alpine species. Annals of Botany, 108, 241–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnoult L, Su KF, Manoel D et al (2013) Emergence and diversification of fly pigmentation through evolution of a gene regulatory module. Science, 339, 1423–1426. [DOI] [PubMed] [Google Scholar]
- Bastow R, Mylne J, Lister C, Lippman Z, Martienssen R, Dean C (2004) Vernalization requires epigenetic silencing of FLC by histone methylation. Nature, 427, 164–167. [DOI] [PubMed] [Google Scholar]
- Bergonzi S, Albani MC, Loren V et al (2013) Mechanisms of age‐dependent response to winter temperature in perennial flowering of Arabis alpina . Science, 340, 1094–1097. [DOI] [PubMed] [Google Scholar]
- Cantarel BL, Korf I, Robb SMC et al (2008) MAKER: an easy‐to‐use annotation pipeline designed for emerging model organism genomes. Genome Research, 18, 188–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaings L, Bergonzi S, Albani MC, Kemi U, Savolainen O, Coupland G (2014) Evolutionary conservation of cold‐induced antisense RNAs of FLOWERING LOCUS C in Arabidopsis thaliana perennial relatives. Nature Communications, 5, 4457. doi:10.1038/ncomms5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K, Kim J, Hwang H‐J et al (2011) The FRIGIDA complex activates transcription of FLC, a strong flowering response in Arabidopsis, by recruiting chromatin modification factors. Plant Cell, 23, 289–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coustham V, Li P, Strange A, Lister C, Song J, Dean C (2012) Quantitative modulation of polycomb silencing underlies natural variation in vernalization. Science, 337, 584–587. [DOI] [PubMed] [Google Scholar]
- Couvreur TL, Franzke A, Al‐Shehbaz IA, Bakker FT, Koch MA, Mummenhoff K (2010) Molecular phylogenetics, temporal diversification, and principles of evolution in the mustard family (Brassicaceae). Molecular Biology and Evolution, 27, 55–71. [DOI] [PubMed] [Google Scholar]
- Csorba T, Questa JI, Sun Q, Dean C (2014) Antisense COOLAIR mediates the coordinated switching of chromatin states at FLC during vernalization. Proceedings of the National Academy of Sciences of the United States America, 111, 16160–16165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobeš C, Mitchell‐Olds T, Koch M (2004) Extensive chloroplast haplotype variation indicates Pleistocene hybridization and radiation of North American Arabis drummondii, A. divaricarpa, and A. holboellii (Brassicaceae). Molecular Ecology, 13, 349–370. [DOI] [PubMed] [Google Scholar]
- Duarte JM, Wall PK, Edger PP et al (2010) Identification of shared single copy nuclear genes in Arabidopsis, Populus, Vitis and Oryza and their phylogenetic utility across various taxonomic levels. BMC Evolutionary Biology, 10, 61. doi: 10.1186/1471‐2148‐10‐61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson JJ, Li W‐H (2010) The genetic basis of evolutionary change in gene expression levels. Philosophical Transactions of the Royal Society B: Biological Sciences, 365, 2581–2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan E, Dennis E (2007) Vernalization‐induced trimethylation of histone H3 lysine 27 at FLC is not maintained in mitotically quiescent cells. Current Biology, 17, 1978–1983. [DOI] [PubMed] [Google Scholar]
- Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I (2004) VISTA: computational tools for comparative genomics. Nucleic Acids Research, 32, W273–W279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J, Rubin MJ (2015) All in good time: understanding annual and perennial strategies in plants. American Journal of Botany, 102, 497–499. [DOI] [PubMed] [Google Scholar]
- Goujon M, McWilliam H, Li W et al (2010) A new bioinformatics analysis tools framework at EMBL‐EBI. Nucleic Acids Research, 38, W695–W699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillo MA, Li C, Fowlkes AM et al (2009) Genetic architecture for the adaptive origin of annual wild rice, Oryza nivara . Evolution, 63, 870–883. [DOI] [PubMed] [Google Scholar]
- Hall M, Willis J (2006) Divergent selection on flowering time contributes to local adaptation in Mimulus guttatus populations. Evolution, 60, 2466–2477. [PubMed] [Google Scholar]
- Heo JB, Sung S (2011) Vernalization‐mediated epigenetic silencing by a long intronic noncoding RNA. Science, 331, 76–79. [DOI] [PubMed] [Google Scholar]
- Hu FY, Tao DY, Sacks E et al (2003) Convergent evolution of perenniality in rice and sorghum. Proceedings of the National Academy of Sciences, 100, 4050–4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata H, Gaston A, Remay A et al (2012) The TFL1 homologue KSN is a regulator of continuous flowering in rose and strawberry. Plant Journal, 69, 116–125. [DOI] [PubMed] [Google Scholar]
- Jordon‐Thaden I, Hase I, Al‐Shehbaz I, Koch MA (2010) Molecular phylogeny and systematics of the genus Draba (Brassicaceae) and identification of its most closely related genera. Molecular Phylogenetics and Evolution, 55, 524–540. [DOI] [PubMed] [Google Scholar]
- Karl R, Koch M (2013) A world‐wide perspective on crucifer speciation and evolution: phylogenetics, biogeography and trait evolution in tribe Arabideae. Annals of Botany, 112, 983–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karl R, Kiefer C, Ansell S, Koch M (2012) Systematics and evolution of arctic‐alpine Arabis alpina L. (Brassicaceae) and its closest relatives in the eastern Mediterranean. American Journal of Botany, 99, 778–794. [DOI] [PubMed] [Google Scholar]
- Kemi U, Niittyvuopio A, Toivainen T et al (2013) Role of vernalization and of duplicated FLOWERING LOCUS C in the perennial Arabidopsis lyrata . New Phytologist, 197, 325–335. [DOI] [PubMed] [Google Scholar]
- Koch M, Haubold B, Mitchell‐Olds T (2000) Comparative analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis and related genera (Brassicaceae). Molecular Biology and Evolution, 17, 1483–1498. [DOI] [PubMed] [Google Scholar]
- Koch MA, Kiefer C, Ehrich D, Vogel J, Brochmann C, Mummenhoff K (2006) Three times out of Asia Minor: the phylogeography of Arabis alpina L. (Brassicaceae). Molecular Ecology, 15, 825–839. [DOI] [PubMed] [Google Scholar]
- Koskela E, Mouhu K, Albani M et al (2012) Mutation in TERMINAL FLOWER1 reverses the photoperiodic requirement for flowering in the wild strawberry Fragaria vesca . Plant Physiology, 159, 1043–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M, Blackshields G, Brown N et al (2007) Clustal W and Clustal X version 2.0. Bioinformatics, 23, 2947–2948. [DOI] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Roos DS (2003) OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Research, 13, 2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor C, Brudno M, Schwartz J et al (2000) VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics, 16, 1046–1047. [DOI] [PubMed] [Google Scholar]
- McWilliam H, Li W, Uludag M et al (2013) Analysis Tool Web Services from the EMBL‐EBI. Nucleic Acids Research, 41, W597–W600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Meaux J, Goebel U, Pop A, Mitchell‐Olds T (2005) Allele‐specific assay reveals functional variation in the Arabidopsis thaliana chalcone synthase promoter region that is compatible with neutral evolution. Plant Cell, 17, 676–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM (1999) FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell, 11, 949–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne I, Shaw P, Stephen G et al (2010) Flapjack – graphical genotype visualization. Bioinformatics, 26, 3133–3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mummenhoff K, Franzke A, Koch MA (1997) Molecular phylogenetics of Thlaspi s. l. (Brassicaceae) based on chloroplast DNA restriction site variation and sequences of the internal transcribed spacers of nuclear ribosomal DNA. Canadian Journal of Botany, 75, 469–482. [Google Scholar]
- Nah G, Chen G (2010) Tandem duplication of the FLC locus and the origin of a new gene in Arabidopsis related species and their functional implications in allopolyploids. New Phytologist, 186, 228–238. [DOI] [PubMed] [Google Scholar]
- Nix D, Eisen M (2005) GATA: a graphic alignment tool for comparative sequence analysis. BMC Bioinformatics, 6. doi:10.1186/1471‐2105‐6‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noe L, Kucherov G (2005) YASS: enhancing the sensitivity of DNA similarity search. Nucleic Acids Research, 33, W540–W543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge L, Harvey PH (1988) The ecological context of life history evolution. Science, 241, 1449–1455. [DOI] [PubMed] [Google Scholar]
- Prud'homme B, Gompel N, Rokas A et al (2006) Repeated morphological evolution through cis‐regulatory changes in a pleiotropic gene. Nature, 440, 1050–1053. [DOI] [PubMed] [Google Scholar]
- Qüesta JI, Song J, Geraldo N, An H, Dean C (2016) Arabidopsis transcriptional repressor VAL1 triggers polycomb silencing at FLC during vernalization. Science, 353, 485–488. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck J (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics, 19, 1572–1574. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers In: Bioinformatics Methods and Protocols: Methods in Molecular Biology (eds Krawatz S, Misener S.), pp. 365–386. Humana Press, Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- Sauer M, Friml J (2008) In vitro culture of Arabidopsis embryos In: Methods in Molecular Biology vol. 427: Plant Embryogenesis (eds Suárez MF, Bozhkov PV.), pp. 71–76. Humana Press, Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- Sheldon CC, Burn JE, Perez PP et al (1999) The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell, 11, 445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon CC, Rouse DT, Finnegan EJ, Peacock WJ, Dennis ES (2000) The molecular basis of vernalization: the central role of FLOWERING LOCUS C (FLC). PNAS, 97, 3753–3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon CC, Conn AB, Dennis ES, Peacock WJ (2002) Different regulatory regions are required for the vernalization‐induced repression of FLOWERING LOCUS C and for the epigenetic maintenance of repression. Plant Cell, 14, 2527–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvertown J, Charlesworth D (2001) Introduction to Plant Population Biology. Blackwell Science Ltd, Oxford, UK. [Google Scholar]
- Stearns S (2000) Life history evolution: successes, limitations, and prospects. Naturwissenschaften, 87, 476–486. [DOI] [PubMed] [Google Scholar]
- Stebbins GL (1950) Variation and Evolution in Plants. Columbia University Press, New York. [Google Scholar]
- Sung S, Amasino RM (2004) Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature, 427, 159–164. [DOI] [PubMed] [Google Scholar]
- Sung S, He Y, Eshoo T et al (2006) Epigenetic maintenance of the vernalized state in Arabidopsis thaliana requires LIKE HETEROCHROMATIN PROTEIN 1 . Nature Genetics, 38, 706–710. [DOI] [PubMed] [Google Scholar]
- Swiezewski S, Liu F, Magusin A, Dean C (2009) Cold‐induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature, 462, 799–802. [DOI] [PubMed] [Google Scholar]
- Taberlet P, Gielly L, Pautou G, Bouvet J (1991) Universal primers for amplification of three non‐coding regions of chloroplast DNA. Plant Molecular Biology, 17, 1105–1109. [DOI] [PubMed] [Google Scholar]
- Tamura K, Nei M, Kumar S (2004) Prospects for inferring very large phylogenies by using the neighbor‐joining method. Proceedings of the National Academy of Sciences (USA), 101, 11030–11035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution, 28, 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tank DC, Olmstead RG (2008) From annuals to perennials: phylogeny of subtribe Castillejinae (Orobanchaceae). American Journal of Botany, 95, 608–625. [DOI] [PubMed] [Google Scholar]
- Thompson J, Gibson T, Plewniak F, Jeanmougin F, Higgins D (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research, 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Farrona S, Vincent C et al (2009) PEP1 regulates perennial flowering in Arabis alpina . Nature, 459, 423–427. [DOI] [PubMed] [Google Scholar]
- Wang R, Albani MC, Vincent C et al (2011) Aa TFL1 confers an age‐dependent response to vernalization in perennial Arabis alpina . Plant Cell, 23, 1307–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White T, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics In: PCR Protocols: A Guide to Methods and Applications (eds Innis M, Gelfand D, Sninsky J, White T.), pp. 315–322. Academic Press, Orlando, Florida. [Google Scholar]
- Willing E‐M, Rawat V, Mandáková T et al (2015) Genome expansion of Arabis alpina linked with retrotransposition and reduced symmetric DNA methylation. Nature Plants, 1, Article number: 14023. doi: 10.1038/nplants.2014.23 [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, Haerum BK, Clark AG (2004) Evolutionary divergence of cis and trans gene regulation. Nature, 430, 85–88. [DOI] [PubMed] [Google Scholar]
- Yang Z, Rannala B (1997) Bayesian phylogenetic Monte Carlo method inference using DNA sequences: a Markov Chain. Molecular Biology and Evolution, 14, 717–724. [DOI] [PubMed] [Google Scholar]
- Yang H, Howard M, Dean C (2014) Antagonistic Roles for H3K36me3 and H3K27me3 in the Cold‐Induced Epigenetic Switch at Arabidopsis FLC . Current Biology, 24, 1793–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W, Luo X, Li Z et al (2016) A cis cold memory element and a trans epigenome reader mediate Polycomb silencing of FLC by vernalization in Arabidopsis . Nature Genetics, 48, 1527–1535. [DOI] [PubMed] [Google Scholar]
- Zhou C‐M, Zhang T‐Q, Wang X et al (2013) Molecular basis of age‐dependent vernalization in Cardamine flexuosa . Science, 340, 1097–1100. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Details of accessions used in this study stating country of origin, provider of seeds and sequences used in the phylogenetic analysis and their genebank accession numbers; for the pairs of grey shaded accessions marker sequences were used in combination to obtain a full set for the phylogenetic reconstructions.
Table S2 Primers used in this study for amplification of phylogenetic marker sequences, qPCR and amplification of the mRNA of FLC orthologues as well as the genomic loci.
Table S3 Alignment of marker sequences used for phylogenetic analysis; the alignment is given in nexus format and partitions are indicated at the end of the file.
Table S4 Results of Model Testing and Bayesian analyses for the individual single marker dataset and the combined dataset
Table S5 Alignment of marker sequences (ADH, CHS, At2g13360) used for the maximum parsimony analysis including both co‐orthologues of A. nordmanniana for each marker. Introns were not included in the analysis and therefore deleted.
Table S6 Vernalization response of selected Arabideae; plants were grown either in long day for several months or in long day for 6 weeks, transferred into vernalization for 12 weeks and then returned to warm temperatures and long day conditions.
Table S7 Polymorphic sites in Arabis alpina Pajares and Arabis montbretiana and the polymorphisms found in the hybrid individual in the ITS confirming the hybrid identity of the plant.
Table S8 Primers used for genotyping the F1 plant and introgression lines carrying a genomic fragment including AmFLC.
Table S9 Number of SNPs and indel polymorphisms detected in different features of PEP1 versus AmFLC, Am = A. montbretiana, Aa = A. alpina, At = A. thaliana (exons, regulatory sequences known from FLC (proximal promoter (Sheldon et al. 2002); nucleation region is the sequence homologous to the region showing elevated H3K27me3 before vernalization in Arabidopsis (Yang et al. 2014); COLDAIR is the regions homologous to the COLDAIR RNA encoding region (Heo & Sung 2011); Vernalization Responsive Element (VRE) indicates the region homologous to the VRE of A. thaliana identified in (Sung et al. 2006)).
Table S10 Alignment of the PEP1, AmFLC, AlFLC1 and FLC loci comprising a partial sequence of the next upstream gene or its orthologues from A. thaliana (At5 g10150), all intergenic sequence between At5 g10150 and its orthologues and FLC and orthologues as well as all sequences orthologues/homologous to exons and introns of FLC (including indels) and approximately 500 bp of sequence downstream of the stop codon.
Table S11 Summary of the comparison of AtFLC, AmFLC, AlFLC1 and AaPEP1.
Table S12 Comparison of VRE/VRE‐like and COLDAIR/COLDAIR‐like sequences found in the first intron of PEP1 and AmFLC; intron 1 in A. alpina refers to the sequence homologous to A. montbretiana intron 1.
Table S13 Associated p‐values for the respective SH tests based on the different single marker datasets.
Table S14 Alignment of both FLC orthologues from A. nordmanniana (AnFLC A and B) to PEP1 and AmFLC; a stretch of N was inserted between the two contigs joined to form AnFLC A. The alignment includes a partial sequence of the orthologue of At5 g10150, exons 1 (A and B in A. alpina) to 7 including all intronic sequences (partial intron 6 in AnFLC A) as well as ~280‐bp sequence downstream of the stop codon (includes the COOLAIR promoter (Castaings et al. 2014; Csorba et al. 2014; Swiezewski et al. 2009).
Fig. S1 Alignment of cDNA sequences of AmFLC, AauFLC and PEP1 used for designing primers for qPCR.
Fig. S2 Maximum Parsimony (MP) phylogenetic reconstruction calculated in MEGA5 (Tamura et al. 2011) based on the orthologues of ADH, CHS and At2g13360.
Fig. S3 Vernalization response of (A) A. montbretiana and (B) A. auriculata measured in leaf number.
Fig. S4 Biological replicates of expression data shown in Fig. 2.
Fig. S5 Genotypes of introgression lines (A. montbretiana in A. alpina acc. Pajares background) used in the expression analysis of AmFLC and PEP1 for confirming the results obtained from the two cuttings of the original F1 plant (A. montbretiana x A. alpina acc. Pajares).
Fig. S6 (A) Dot plot of the AmFLC genomic region (last exon upstream gene until the end of the COOLAIR promoter) vs. AmFLC cDNA; 7 exons can be detected, no exon has been duplicated (B, C, D) Comparison of the PEP1, AmFLC and FLC sequences by GATA plots.
Fig. S7 Statistical test on the SNP distribution in the alignment of AmFLC and PEP1.
Fig. S8 Alignment of the proximal promoter region including UTR of FLC, AmFLC and PEP1; the proximal promoter is defined as the region homologous to what was described as FLC minimal promoter (Sheldon et al. 2002).
Fig. S9 Alignment of COLDAIR and A. montbretiana and A. alpina COLDAIR ‐like 1 and COLDAIR ‐like 2; yellow positions indicate ancestral positions maintained in COLDAIR ‐like 1 while pink positions indicate conservation in COLDAIR ‐like 2; more ancestral positions are maintained in COLDAIR ‐like 1; COLDAIR‐likes are defined as regions showing homology to the Arabidopsis COLDAIR sequence (Heo & Sung 2011).
Fig. S10 Alignment of VRE and A. montbretiana and A. alpina VRE ‐like 1 and VRE ‐like 2; VRE‐like sequences are defined as sequences showing homology to the Vernalization Response Element of FLC in Arabidopsis thaliana (Sung et al. 2006).
Fig. S11 Alignment of the A. montbretiana and A. alpina region homologous to the nucleation region in A. thaliana defined by the ChIP primers from (Yang et al. 2014).
Fig. S12 Arabis montbretiana life cycle monitored under greenhouse conditions.
Fig. S13 Alignment of PEP1 exon 1A and 1B and AmFLC exon 1 along with adjacent intron sequences; the 416 bp used to differentiated copy 1A and copy 1B are indicated by $ while the SNPs differentiating the duplicated segment of intron 1 in PEP1 are marked by *
Fig. S14 Kmer abundance distribution.
Fig. S15 Ends of concatenated contigs which were found to show homology to FLC and which were shown to be part of the same molecule (AnFLC B) based on PCR and subsequent Sanger sequencing; All contigs showed overlapping ends.
Fig. S16 Statistical test on the SNP distribution in the alignment of (A) AmFLC and PEP1 and (B‐C) FLC and AlFLC1. The test measures whether the observed SNP number in a window is different from a random distribution (A and B more SNPs than expected, C fewer SNPs than expected); the left panel shows the number of SNPs per window, the right panel shows the –log10 of the associated p‐values; the dashed line is the threshold above which statistical significance was reached; values were corrected for FDR.
Fig. S17 Statistical test on the SNP distribution in the alignment of (A) AnFLC‐A and PEP1 and (B) AauFLC and AmFLC.
Fig. S18 Statistical test on the SNP distribution in the alignment of (A) AnFLC‐A and PEP1 and (B) AauFLC and AmFLC.
Fig. S19 Sequence alignment of six Brassicaceae species in the segment showing more SNPs than expected by chance in the nucleation region from Fig. 5A.
Fig. S20 Sequence alignment of six Brassicaceae species in the segment showing more SNPs than expected by chance upstream/in the region homologous to the VRE from Fig. 5A.
Fig. S21 Sequence alignment of six Brassicaceae species in the segment showing fewer SNPs than expected in a random distribution in the comparison of AlFLC1 and FLC showing homology to (A) the end of the nucleation region and (B) COLDAIR from Fig. 5A; the conserved GGATTTGT‐motif is indicated a red square.
Fig. S22 Sequence alignment of six Brassicaceae species in the segment showing fewer SNPs than expected by chance within the region showing homology to the nucleation region in the annual couple (AauFLC‐AmFLC) from Fig. 5B.
Fig. S23 Sequence alignment of six Brassicaceae species in the segment showing fewer SNPs than expected by chance within the region showing homology to the nucleation region in the perennial couple (PEP1‐AnFLC‐A) from Fig. 5B
Data S1 Ct values and evaluation of qPCR data
Data Availability Statement
Whole‐genome shotgun projects: deposited at GenBank under the Accession nos LNCG00000000 (A. nordmanniana) and LNCH00000000 (A. montbretiana).
DNA sequences: KC814706;JQ919880;GU181934;GU181999;JQ919835;JQ919813;JQ919857;KC814698;JQ919879;HM046191;HM046243;HM046216;KC814699;JQ919881;GU182084;GU181952;GU182017;KC814700;JQ919882;KC814707;JQ919884;HQ646785;HQ646724;FJ187937;FJ188234;FJ188083;KC814701;JQ919885;HM046193;HM046245;HM046218;KC814708;JQ919886;KF547353;KF547826;KF547624;KC814702;JQ919887;JQ919839;JQ919817;JQ919861;KC814709;JQ919888;KC814703;JQ919889;GU181938;GU182003;GU182089;GU181957;GU182022;KC814713;JQ919899;JQ919840;JQ919818;JQ919862;KC814704;JQ919890;HQ646642;HQ646763;HQ646702;KC814705;JQ919891;KF547181;GU202868;GU202800;KC814710;JQ919893;JQ919844;JQ919822;JQ919866;KC814711;JQ919894;JQ919845;JQ919823;JQ919867;KC814712;JQ919896;KU525030;KU525030;KU525030;KU525030;KU525030;KU525030;KU321504;KU321505;KU321506;KU321507;KU321508;KU321509;KU321510;KU321511;KU321512;KU321513;KU321514;KU321515;KU321516;KU321517;KU321518;KU321519;KU321520;KU321353;KU321354;KU321355;KU321356;KU321357;KU321358;KU321359;KU321360;KU321361.


