Abstract
Robertsonian translocations resulting in fusions between sex chromosomes and autosomes shape karyotype evolution by creating new sex chromosomes from autosomes. These translocations can also reverse sex chromosomes back into autosomes, which is especially intriguing given the dramatic differences between autosomes and sex chromosomes. To study the genomic events following a Y chromosome reversal, we investigated an autosome‐Y translocation in Drosophila pseudoobscura. The ancestral Y chromosome fused to a small autosome (the dot chromosome) approximately 10–15 Mya. We used single molecule real‐time sequencing reads to assemble the D. pseudoobscura dot chromosome, including this Y‐to‐dot translocation. We find that the intervening sequence between the ancestral Y and the rest of the dot chromosome is only ∼78 Kb and is not repeat‐dense, suggesting that the centromere now falls outside, rather than between, the fused chromosomes. The Y‐to‐dot region is 100 times smaller than the D. melanogaster Y chromosome, owing to changes in repeat landscape. However, we do not find a consistent reduction in intron sizes across the Y‐to‐dot region. Instead, deletions in intergenic regions and possibly a small ancestral Y chromosome size may explain the compact size of the Y‐to‐dot translocation.
Keywords: Dot chromosome, Robertsonian translocation, SMRT sequencing, Y chromosome
Most sex chromosomes evolve from a homologous pair of autosomes, where one chromosome acquires a sex‐determining locus, suppresses recombination with its homolog and eventually degenerates into a differentiated Y (or W in cases of female heterogamety) chromosome (reviewed in Ohno 1967; Charlesworth 1978; Bull 1983; Charlesworth and Charlesworth 2000). While sex chromosomes appear stable in some taxa (e.g., mammals), they can be labile in others (e.g., nonmammalian vertebrates, Schartl 2004, Ezaz et al. 2006; insects, White 1973, Vicoso and Bachtrog 2015). Robertsonian translocations between sex chromosomes and autosomes are important for sex chromosome evolution—the resulting chromosome fusions create new sex chromosomes from autosomes and new autosomes from sex chromosomes (Vicoso and Bachtrog 2013, 2015). Autosome‐sex chromosome translocations are well documented in some mammals (e.g., Indian muntjacs, Wurster and Benirschke 1970; shrews, Ford et al. 1957; gerbils, Wahrman and Zahavi 1955; voles, Fredga 1970; sticklebacks, Ross et al. 2009) and Drosophila species (White 1973; Steinemann 1982; McAllister and Charlesworth 1999; Vicoso and Bachtrog 2015), where autosome‐sex chromosome fusions have created new sex chromosomes (i.e. neo‐sex chromosomes) of independent origins and different ages.
Following the fusion of an autosome to a sex chromosome, the neo‐Y chromosome typically degenerates, losing many of its genes relative to its counterpart, the neo‐X chromosome. This degeneration occurs as a result of suppressed recombination, where a reduced efficacy of natural selection leads to the accumulation of deleterious mutations and repeats (transposable elements and satellite DNAs; reviewed in Charlesworth and Charlesworth 2000; Steinemann and Steinemann 2005a; Bachtrog 2013). The degenerated Y chromosome is a harsh genomic environment for most genes—it is dense in repetitive elements, heterochromatic, and nonrecombining (Bonaccorsi et al. 1988; Bonaccorsi and Lohe 1991). Old differentiated Y chromosomes like D. melanogaster’s are gene poor (e.g., up to 20 genes in 40 Mb; Hoskins et al. 2002). Genes that survive on the Y chromosome tend to retain or evolve male‐related functions; and these genes can have peculiar structures—some Drosophila Y‐linked genes have introns that are megabases long (hereon referred to as mega‐introns) and filled with tandem repeats (Bonaccorsi et al. 1988; Bonaccorsi et al. 1990; Bonaccorsi and Lohe 1991; Kurek et al. 1998, 2000; Reugels et al. 2000; Piergentili 2007).
Sex chromosomes differ from major autosomes in gene regulation and ploidy (reviewed in Steinemann and Steinemann 2005b) but despite these gross differences, sex chromosomes can revert back into autosomes. The small heterochromatic Drosophila autosome (dot chromosome; also known as Muller F) is an old X chromosome that reverted to an autosome in an ancestor of drosophilids (Vicoso and Bachtrog 2013). In many ways, the dot chromosome still has more in common with an X chromosome than other autosomes (Larsson and Meller 2006; Riddle and Elgin 2006; Vicoso and Bachtrog 2013). While researchers are just beginning to understand the process of X chromosome reversal, Y chromosome reversals remain relatively unexplored. D. pseudoobscura offers a unique opportunity to study Y chromosome reversals. Two chromosomal rearrangements occurred 10–15 Mya in an ancestor of D. pseudoobscura: an X‐autosome and an autosome‐Y translocation (Carvalho and Clark 2005; Larracuente et al. 2010). The X‐autosome translocation created a pair of neo‐sex chromosomes: the current Y is not homologous to the ancestral Y chromosome (Carvalho and Clark 2005) and instead may be a neo‐Y derived from the homolog of the autosome that fused to the X chromosome (Larracuente et al. 2010). The ancestral Y chromosome translocated to the dot chromosome (Larracuente et al. 2010). Thus after 60 Myr of paternal transmission, the ancestral Y chromosome reverted to an autosome and is now passed through both sexes. Because there are no known transition stages in any extant obscura group species, we do not know which event—the X‐autosome or autosome‐Y translocation—came first (Larracuente et al. 2010).
Robertsonian translocations are chromosome fusions with clinical and evolutionary significance (reviewed in Misteli 2010; Garagna et al. 2014). Studying how the autosome‐Y translocation occurred could lend insight into the mechanics of Robertsonian translocation formation and the evolutionary events that follow these chromosome fusions. Following the autosome‐Y translocation in D. pseudoobscura, the former Y chromosome (hereon referred to as the Y‐to‐dot region) may have shrunk 100‐fold in size (Carvalho and Clark 2005; Larracuente et al. 2010). Patterns of nucleotide variability in the Y‐to‐dot region are consistent with a history of selective sweeps, suggesting that positive selection may have favored large deletions of intronic sequences after becoming autosomal (Larracuente and Clark 2014). Therefore, D. pseudoobscura offers a window into the evolutionary events following a Y chromosome reversal.
Studying the structural changes that occurred after the Y‐to‐dot translocation has been difficult because this region is poorly assembled (Larracuente et al. 2010). Heterochromatic sequences rich in repeats are often underrepresented in BAC libraries and cloning vectors (Brutlag et al. 1977; Lohe and Brutlag 1986, 1987) used in traditional sequencing methods, making them difficult to assemble (Hoskins et al. 2002). The short reads from next generation sequencing technologies have difficulty spanning and assembling repeats (Treangen and Salzberg 2012). Single molecule real‐time sequencing technologies (SMRT) circumvent some of the problems with traditional and next generation short read sequencing (Eid et al. 2009). SMRT reads from Pacific Biosciences (PacBio) are on average ∼16 Kb long, but reach up to 50 Kb (with current technology). These long read lengths help span repeats and assemble repetitive regions (Schatz et al. 2010; Chaisson et al. 2015).
To infer the origins of the Y‐to‐dot translocation and detail the genomic changes accompanying this Y chromosome reversal, we used PacBio SMRT reads to assemble the entire genic portion of the D. pseudoobscura dot chromosome, including the Y‐to‐dot region. We reveal the breakpoints between the conserved part of the dot chromosome and the Y‐to‐dot region and discover that the region between them spans only ∼78 Kb. Based on the organization of this region, we infer that at least one chromosomal inversion followed a Robertsonian translocation between the ancestral Y and dot chromosomes. We also show that while the Y‐to‐dot is 100‐fold smaller than the D. melanogaster Y chromosome, the distribution of intron sizes does not differ from that of the Y chromosome, outside of the mega‐introns.
Materials and Methods
PacBio ASSEMBLIES
We downloaded raw PacBio reads from ftp://ftp.hgsc.bcm.edu/Dpseudoobscura/Towards_finishing_the_D.pseudoobscura_genome/PacBio_Data/FastQ_files/ (data generated in 2014; permission to use these reads was generously granted by Drs. Stephen Schaeffer and Stephen Richards; Table S6). This data set is 10.8 Gb of sequence in total and includes 1.6 million reads with read lengths averaging 6.7 ± 4.2 Kb long (excluding reads < 500 bp). We used the Falcon assembler v0.3.0 (https://github.com/PacificBiosciences/FALCON-integrate) to filter, correct, and assemble reads (FALCON configuration file in Supplemental Doc 1). We polished the assembly using Quiver (SMRT Analysis v2.3.0; Chin et al. 2013). We remapped raw PacBio reads to the polished assembly using bwa v0.7.15 with mem –x pacbio (Li and Durbin 2010). The coverage of unique mapped reads was called by samtools depth (v1.3.1; Li et al. 2009) with –Q 10.
To compare our PacBio assembly with the latest D. pseudoobscura genome assembly (r3.03; English et al. 2012; Flybase), we used MUMMER (v3.23; Kurtz et al. 2004) nucmer with the parameters “–l 200 –D 20–maxmatch –nosimplify”. We also annotated repetitive DNA using RepeatMasker 4.06 with Repbase 20150807 and the parameters “‐species drosophila ‐s ‐pa 10” (Smit et al. 2013–2015).
ANNOTATION AND RNA‐SEQ ANALYSIS
We identified exon‐intron junctions by mapping annotated transcripts (r3.03; Flybase) and transcripts of Y‐linked genes from NCBI (gi|295126506|gb|GU937420.1|, gi|255764727|gb|EU595397.2|, gi|295126512|gb|GU937423.1|) to our assembly using exonerate 2.4.0 (Slater and Birney 2005). To complement these data, we mapped RNA‐seq reads (Chen et al. 2014; Table S6) to our assembly using Tophat v2.0.13 with the parameters “‐p 16 ‐b2‐very‐sensitive” (Trapnell et al. 2009). We annotated transcripts, eliminated small redundant isoforms (gffread parameters “–M –K”) and estimated expression level with cufflinks v2.2.2 (parameters “‐p 16 ‐u ‐b”; Trapnell et al. 2012). We manually modified gene annotations in the Y‐to‐dot region due to their longer introns. We then searched for homologous genes using the predicted transcripts as BLAST queries to all D. melanogaster translations and transposon sequences (r6.07) with NCBIblast (v2.2.30+; Altschul et al. 1990). To survey BAC sequences, the Ppr‐Y primers were obtained from Larracuente and Clark (2014), and the Cadps and Dyrk3 primers are listed in Table S7. Our dot chromosome assembly and GTF file containing annotations are available as supplementary files and the full assembly, GTF file containing annotations and expression data are deposited in the Dryad Digital Repository.
POPULATION ANALYSES AND PHYLOGENETIC ANALYSIS
We downloaded population genomic datasets including 11 D. pseudoobscura individuals from seven different populations and 12 D. miranda individuals from two studies (McGaugh and Noor 2012; Zhou and Bachtrog 2012; Smukowski Heil et al. 2015; NCBI accession numbers in Table S6). After trimming with Trim Galore (v0.4.0; http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/), we mapped all reads to the genome using a pipeline modified from Lack et al. (2015; https://github.com/justin-lack/Drosophila-Genome-Nexus/blob/master/round1_assembly.pl). Briefly, we used bwa mem (v0.7.12; Li and Durbin 2010), picard (v1.114; http://picard.sourceforge.net), and stampy (v1.0.23; Lunter and Goodson 2011). To improve the sensitivity, bam files from the same species were used to call SNPs with GATK UnifiedGenotyper with multisample parameters “‐mbq 10 ‐stand_call_conf 31 ‐stand_emit_conf 31 ‐ploidy 2 –output_mode EMIT_ALL_SITES” and removed heterozygous sites (v3.4‐46; McKenna et al. 2010). We extracted nucleotide information from vcf files using Vcftools (v0.1.14; Danecek et al. 2011) and only used sites with less than four missing genotypes (≥7 individuals in D. pseudoobscura and ≥8 individuals in D. miranda) for the analyses. We calculated Rmin and pairwise nucleotide diversity (π) in 10 Kb windows using compute 0.8.4 (Thornton 2003). We performed statistical analyses and plotted figures in R (v3.1.2) with “agricolae” (v1.2‐3; https://cran.r-project.org/web/packages/agricolae/index.html) and “ggplot2” (v2.1.0; https://cran.r-project.org/web/packages/ggplot2/index.html). We mapped the reads to our assembly and NCBI references (r3.03 of D. pseudoobscura and GCA_000269505.2_DroMir_2.2 of D. miranda). To avoid possible misassemblies outside of the dot‐Y, we report estimates of nucleotide diversity for the dot‐Y from our de novo assembly and the other chromosomes from the NCBI reference assembly. To check the impact of assembly type on our estimates of nucleotide diversity, we also calculated nucleotide diversity using both assembly types and found the same pattern.
We either downloaded PRY, CG30048 and CG33482, and their orthologs in 10 Drosophila species from Flybase (FB2015_04) or extracted them from genome assemblies (this study). We downloaded the Ceratitis capitata (LOC101450228) and Musca domestica sequences (MDOA003316; Scott et al. 2014) from NCBI and Vectorbase, respectively. We used MEGA (v6.06; Tamura et al. 2013) to align amino acid sequences manually, and construct a phylogeny by Maximum likelihood with JTT model and 1000 bootstraps.
KARYOTYPING
We performed brain squashes according to Larracuente and Ferree (2015) with some modifications. All stocks (D. subobsuca:14011–0131.05 and D. bifasaciata:14012–0181.02) were ordered from Drosophila Species Stock center. We dissected 3–5 male third instar larvae and transferred brains to a hypotonic solution of 0.5% sodium citrate for 8–10 min. We fixed the brains in 45% acetic acid for 10–20 min and then squashed, snap froze in liquid nitrogen and dehydrated the slides in absolute ethanol for more than 30 min. Slides were mounted in VectaShield with DAPI (Vector Laboratories), visualized on a Leica DM5500 fluorescence microscope at 100X, imaged with a Hamamatsu Orca R2 CCD camera and analyzed using Leica's LAX software.
Results
ASSEMBLY OF DOT‐Y
We refer to the entire dot chromosome in species that have the translocation as the dot‐Y chromosome. To determine the complete sequence of the dot‐Y chromosome in D. pseudoobscura, we assembled the genome using ∼70X coverage PacBio long reads. Our assembly contains 612 contigs covering 155.9 Mb (N50 = 1.35 Mb, Table 1). All 5 ancestral Y‐linked genes, all 79 conserved dot chromosome genes and the telomeric TART transposons are found on a single 1.92 Mb contig in our assembly (contig 000025F). This single contig appears on 62 different scaffolds in the latest Flybase reference (r3.03; English et al. 2012; Fig. S1. and Table S1). We found all 5 Y‐to‐dot genes in a 320‐Kb centromere‐proximal region of the dot‐Y chromosome (Fig. 1). The most distal gene in our contig is Plex‐A, consistent with a previous study (Villasante et al. 2007). The orientation of the 5 Y‐to‐dot genes is the same as previous reports (Larracuente et al. 2010). Based on this assembly, we define the formerly Y‐linked region from the start of the contig to Ppr‐Y as Y‐to‐dot, and the conserved gene‐dense region from Cadps to Plex‐A as conserved‐dot (Fig. 1). Our assembly of the Y‐to‐dot and conserved‐dot regions is consistent with our expectations with respect to gene content and order. However, the region containing the breakpoint between the two sections of the dot‐Y revealed an interesting structure.
Table 1.
Summary of D. pseudoobscura assemblies
| Assembly size | # contigs | Contig N50 | Scaffold N50 | # dot contigs | |
|---|---|---|---|---|---|
| Pacbio (this article) | 155,857,120 | 612 | 1,350,737 | N/A | 1 |
| Dpse_3.0 (English et al. 2012) | 152,696,384 | 6,823 | 202,541 | 12,541,198 | 62 |
Figure 1.
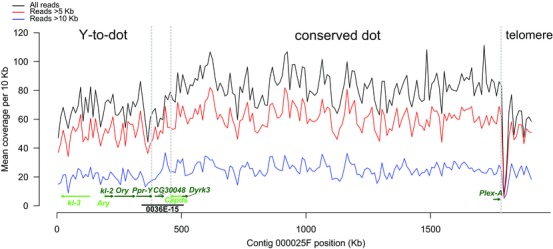
The structure and gene orientation of the dot‐Y chromosome in D. pseudoobscura. The lines correspond to the coverage of all reads, reads >5 Kb, and reads >10 Kb across the entire dot‐Y chromosome. The Y‐to‐dot, conserved‐dot and telomeric regions are delineated by gray dotted lines. All 5 Y‐to‐dot genes, the duplicated gene between the Y‐to‐dot and conserved dot, 3 conserved‐dot genes (the most proximal and distal to the centromere) and their orientations are indicated with green lines. The coordinates of the D. persimilis BAC (36E‐15) are indicated with a black line. [Color figure can be viewed at wileyonlinelibrary.com]
First, the region between the Y‐to‐dot and the conserved‐dot is only ∼78 Kb. This is surprising, given that we initially expected the centromere to fall between these two parts of the chromosome if it originated from a Robertsonian translocation. The region between the Y‐to‐dot and conserved‐dot has two recent large duplications containing a 3193‐bp exon from a testis‐specific gene—an ortholog of CG30048 in D. melanogaster (the three fragments are 5194 bp, 10,765 bp, and 10,548 bp long and are > 95% identical; Fig. S2). Interestingly, CG30048 (TCONS_00043496 and TCONS_00043497 in our annotation) is a parental copy of an ancestral Y‐linked gene—Polycystein‐related Y, or PRY (Fig. S3). In D. pseudoobscura, PRY (GA23077) is on chromosome XL and has testis‐specific expression (Chen et al. 2014). No substitutions differentiate the two duplicates of CG30048. While one copy of CG30048 is highly expressed in testes (FPKM = 58 in testes but < 3 in male carcass or females, Table S2), the other copy lacks the first two exons and is not significantly expressed (all transcriptomes FPKM < 3, Table S2). These recent large duplications likely prevented the assembly of this region using Illumina and Sanger sequencing reads.
To confirm our assembly, we surveyed BACs from D. pseudoobscura and its sister species, D. persimilis. We found 4 BACs (CH226‐45K5, CH226‐45K6, CH226‐6C6, CH226‐55K10) and three fosmids (CH1226‐51D24, CH1226‐62C1, CH1226‐33C10) from D. pseudoobscura that extend into the region between the Y‐to‐dot and conserved dot (Table S3). However, we found that a single BAC from D. persimilis (TSC#14011‐0111.49 0036E‐15) spans the entire region and contains at least part of three dot‐Y genes, including Ppr‐Y, Cadps, and Dyrk3 (Fig. 1). This BAC agrees with the structural arrangement of our assembly—the average insert size of this BAC library is 151 Kb (Song et al. 2011) and according to our assembly, the PCR fragments from three genes span 187 Kb in D. pseudoobscura. It also further supports that the region between the Y‐to‐dot and conserved‐dot is conserved in D. pseudoobscura and its closely related species, D. persimilis.
CHROMOSOMAL REARRANGEMENT
The movement of Y‐linked genes could have been the result of a large segmental duplication to the dot chromosome, a Robertsonian translocation, or another chromosome fusion event. Because the organization of the Y chromosome appears to change rapidly over short periods of evolutionary time, it is difficult to infer the initial event. It is unlikely that a large segmental duplication moved the Y‐linked genes to the dot chromosome because we do not find other copies of these genes in the assembly. Additionally these genes span the centromere and are found on both arms of the D. melanogaster Y chromosome. Though the ancestral Y‐linked genes are on the dot‐Y, the rDNA and their intergenic spacer (IGS) sequences typically found on Drosophila Y chromosomes are absent from this region cytologically (Larracuente et al. 2010) and in our assembly. Instead, the rDNA IGS repeats are on the current Y chromosome of D. pseudoobscura, suggesting that these sequences transferred to the Y chromosome either from the Y‐to‐dot or from the X chromosome (Larracuente et al. 2010).
Directly following a Robertsonian translocation, the position of the centromere should fall between the Y‐to‐dot and conserved‐dot regions. To ask if the region between the Y‐to‐dot and conserved‐dot contains sequences with centromere features, we determined the distribution of repetitive elements (satellite DNAs and transposons) across the dot‐Y chromosome. Typical Drosophila centromeres are large (∼400 Kb) and enriched in satellite DNA and transposable element‐derived sequences (Karpen and Allshire 1997). The Y chromosome centromere in the melanogaster group is derived from telomeric retrotransposons (Abad et al. 2004). However, the 78 Kb region between the Y‐to‐dot and conserved‐dot is not enriched in any repeat class compared to the Y‐to‐dot (Fig. 2; MWU, P > 0.05)—we found no contiguous runs of tandem repeats longer than 5 Kb or TART/HeT‐A arrays in this region (Table S4). Given its small size and relatively low density of repeats, we infer that the centromere is not located in the region between the Y‐to‐dot and conserved‐dot.
Figure 2.
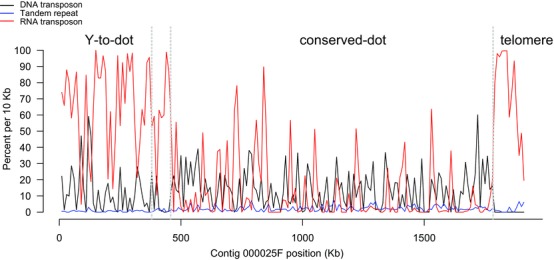
Repeat landscape across D. pseudoobscura dot‐Y chromosome. The density of repeat types as categorized by RepeatMasker— RNA transposons (Class I), including LTR retrotransposons and LINEs, DNA transposons (Class II) including subclass I and subclass II, and simple tandem repeats —are plotted across the dot‐Y chromosome. See materials and methods for details. [Color figure can be viewed at wileyonlinelibrary.com]
REORGANIZATION OF THE Y‐TO‐DOT REGION
The Y‐to‐dot region appears 100‐fold smaller than the D. melanogaster Y chromosome. The 5 Y‐to‐dot genes span a total of ∼320 Kb on the D. pseudoobscura dot chromosome, whereas their orthologs span both chromosome arms on the ∼40 Mb D. melanogaster Y chromosome (Hoskins et al. 2002). This drastic difference in size could be due to deletions in intergenic regions or deletions in both introns and intergenic regions following the translocation. We found a low density of tandem repeats in the Y‐to‐dot region (0.54% of total sequences; Fig. 2), indicating that the deletion of repetitive DNA in intergenic regions contributes to the compact size of the Y‐to‐dot region. However, we surveyed the relative size of the Y and X chromosomes in karyotypes of obscura group species and found that some species have a small Y chromosome (Fig. 3). Therefore, it is possible that the ancestral Y chromosome involved in the translocation was small.
Figure 3.
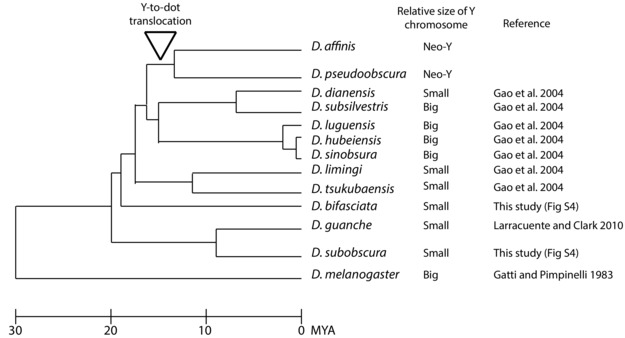
Evolution of Y chromosome size in the obscura group. The phylogeny is modified from Gao et al. (2007). The relative size of the ancestral Y chromosome compared to the ancestral X chromosomal arm (i.e. Muller A) in spreads of mitotic chromosomes (Fig S4; Gao et al. 2004; Larracuente et al. 2010). The ancestral obscura group Y chromosome may have been small.
Compared to intergenic regions, large introns should be under stronger selection due to costs of transcription (Prachumwat et al. 2004). The large introns (> 10 Kb) of some Y‐to‐dot genes appeared to shrink drastically after moving to the dot chromosome (Carvalho and Clark 2005; Larracuente et al. 2010; Larracuente and Clark 2014). A previous study suggests that some large intronic deletions may evolve under positive selection in the D. pseudoobscura Y‐to‐dot region (Larracuente and Clark 2014). Studies of an independent Y‐to‐dot gene movement event in the Drosophila testacea group suggests that in this group, there are more slightly deleterious mutations in the dot‐linked copies of kl‐5 compared to the Y‐linked copies in other species (Dyer et al. 2011). Though effective population sizes of dot chromosomes are four times that of Y chromosomes, their high gene density and low recombination rates suggest that background selection is important in shaping the evolution of dot‐linked genes (Kaiser and Charlesworth 2009; Betancourt et al. 2009; Arguello et al. 2010). However, the inferences of selection on Y‐to‐dot introns in D. pseudoobscura made in previous studies were limited by poor assemblies of the Y chromosome in D. melanogaster (at the time 243 Kb) and Y‐to‐dot in D. pseudoobscura (spread across 10 scaffolds totaling 158 Kb, including estimated gaps; Larracuente et al. 2010). To quantify intron size dynamics on introns of all sizes (instead of just large introns), we used the latest D. melanogaster reference Y chromosome (r6.03; ∼4 Mb) and our complete assembly of the D. pseudoobscura dot‐Y chromosome. We compared intron sizes across the dot‐Y to the Y‐to‐dot genes to their orthologs on the D. melanogaster Y chromosome. As expected, introns across the dot‐Y chromosome are larger than the other autosomes (median intron size dot‐Y = 96 bp, autosomes = 68 bp; MWU, P = 5.802 × 10−14, Fig. 4A), suggesting that the lack of crossing over on the dot contributes to larger introns. Within the D. pseudoobscura dot‐Y chromosome, intron sizes differ among the Y‐to‐dot and conserved‐dot regions (median intron size Y‐to‐dot = 262 bp, conserved‐dot = 93 bp; MWU, P = 0.2712; Fig. 4A). The five largest introns on the dot‐Y are all from the Y‐to‐dot region. However, the D. pseudoobscura Y‐to‐dot genes do not have consistently smaller introns than Y‐linked genes in D. melanogaster (Fig. 4B and Table S5). While we fully assembled all 41 of the introns for the 5 Y‐to‐dot genes in D. pseudoobscura, only 22 of the 41 introns of their Y‐linked orthologs in D. melanogaster (r6.03) are fully assembled. Of the 22 assembled Y‐linked introns in D. melanogaster, 12 are larger in D. pseudoobcura (Fig. 4B and Table S5). It is likely that some of the 19 unassembled Y‐linked introns in D. melanogaster are much larger than other introns across the Y chromosome. The introns of 3 Y‐linked genes—kl‐3, kl‐5, and ORY (i.e. ks‐1; Kennison 1981)—contain megabases of simple satellite sequences in D. melanogaster (Gatti and Pimpinelli 1983; Piergentili and Mencarelli 2008). Two of these mega‐intron‐containing genes, kl‐3 and ORY, are located in the D. pseudoobscura Y‐to‐dot region and are significantly smaller (∼113 and 66 Kb, respectively). In our assembly, the largest intron in these genes is 38 Kb and no introns contain stretches of satellite DNA sequences. Therefore, the lack of consistent intron size reduction across the Y‐to‐dot region suggests instead that shrinking intergenic regions, perhaps due to selection for purging satellite DNA sequences in both intergenic regions and introns, has a greater contribution to the reduced size of the dot‐Y chromosome.
Figure 4.
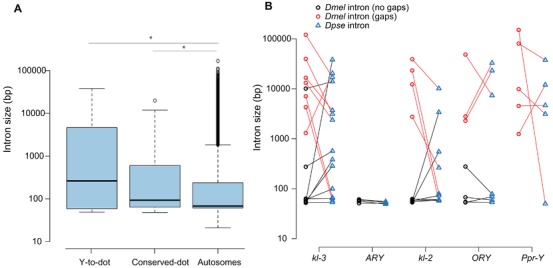
Intron size comparisons. A) Boxplots show intron size differences across the D. pseudoobscura genome. Intron sizes for the dot‐Y chromosome regions are based on our PacBio assembly and annotations; autosomal introns are based on the r3.03 reference genome assembly and annotations (Flybase). Only introns from the first isoforms of each gene were used. For all dot‐Y genes, we only plot those with orthologs in D. melanogaster (Blastx; e < 0.01). Asterisks (*) indicate significantly different means (pairwise MWU; P < 0.05). B) Comparisons of individual orthologous intron pairs between the D. melanogaster Y chromosome (circles) and the D. pseudoobscura Y‐to‐dot translocation (triangles). Introns in the D. melanogaster r6.03 reference Y chromosome with gaps (indicated by Ns; and therefore are minimum intron sizes) are in red and those without gaps are in black. Orthologous introns between the species pair are connected with a line. [Color figure can be viewed at wileyonlinelibrary.com]
Though the intergenic and intronic regions of Y‐linked genes are vastly different between D. pseudoobscura and D. melanogaster, the coding regions are conserved between species (Singh et al. 2014). However, gene content turns over rapidly on Drosophila Y chromosomes (Koerich et al. 2008), so the Y‐to‐dot translocation may have uncharacterized formerly Y‐linked genes. To ask if there are new genes or noncoding RNAs associated with the translocation, we studied gene expression across the dot chromosome. The 5 Y‐to‐dot genes remained testis‐specific after becoming autosomal, as previous studies suggested (Carvalho and Clark 2005). We found some evidence for one new gene in the Y‐to‐dot region—a predicted transcript originating from a duplication of CG9065 (TCONS_00020811) in the intron of kl‐3. This duplication occurred after the Y‐to‐dot translocation because it shares 96% identity with its parental copy. However, this duplicate is not expressed (all RNA‐seq datasets FPKM < 1, Table S2). Other than the original 5 Y‐linked genes, no predicted genes in the translocated region show significant expression (FPKM < 5; Table S2).
POPULATION GENETIC ANALYSES
We estimated levels of nucleotide diversity and population recombination rates across the dot‐Y chromosome for 11 strains of D. pseudoobscura and 12 strains of D. miranda (both species have a dot‐Y). Due to a severe population bottleneck in the demographic history of D. miranda (Bachtrog and Andolfatto 2006; Jensen and Bachtrog 2011), nucleotide diversity is lower than in D. pseudoobscura. In both species, the major autosomes (Muller B and E) harbor more nucleotide diversity than the X (Muller A and D) and dot‐Y chromosomes (median πnondot autosomes = 0.0102 and 0.0014 in D. pseudoobscura and D. miranda, respectively; median πX = 0.0081 and 0.0012 in D. pseudoobscura and D. miranda, respectively; median πdot‐Y = 0.0008 and 0.0002 in D. pseudoobscura and D. miranda, respectively; Kruskal‐Wallis test; multiple comparisons, Bonferroni P < 2.2 x10−16). Within the dot‐Y chromosome, nucleotide diversity is ∼twofold, but not significantly greater on the conserved‐dot compared to the Y‐to‐dot (πconserved‐dot = 0.0009, πY‐to‐dot = 0.0005 in D. pseudoobscura; πconserved‐dot = 0.0002, πY‐to‐dot = 0.0001 in D. miranda; Kruskal‐Wallis test; multiple comparisons, Bonferroni P > 0.05, Fig. 5). Consistent with previous reports (Larracuente and Clark 2014), we detected a low recombination rate on the dot‐Y chromosomes of both D. pseudoobscura (Rmin = 152) and D. miranda (Rmin = 17) along the 1.9 Mb dot chromosome. Recombination rates are even lower within the Y‐to‐dot region (Rmin = 8 from 432 segregating sites in D. pseudoobscura; Rmin = 0 from 102 segregating sites in D. miranda), although low levels of polymorphism may lead us to underestimate recombination. The low recombination rates likely contribute to the reduced nucleotide diversity across these dot chromosomes.
Figure 5.
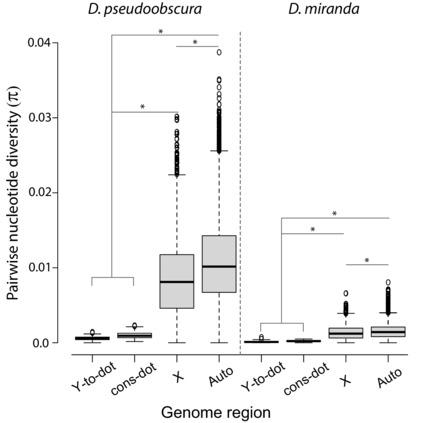
Nucleotide diversity across the genome in D. pseudoobscura and D. miranda. The dot chromosome, including Y‐to‐dot and conserved‐dot regions, have lower pairwise nucleotide diversity than representative autosomes (those chromosome arms not involved in sex‐autosome translocation; i.e. Muller B and E) and the X chromosome (i.e. Muller A and D). Asterisks (*) indicate significantly different means (Kruskal‐Wallis; pair‐wise Bonferroni; P < 0.05)
Discussion
Robertsonian translocations occur through the fusion of two acrocentric or telocentric chromosomes to create one metacentric chromosome. If the Y‐to‐dot event was a Robertsonian translocation, then the fused centromeres of the Y and dot chromosomes would have resided between the two chromosomal arms immediately after the translocation occurred. However, we found that the region between the Y‐to‐dot and conserved‐dot is relatively small, lacks highly repetitive DNA and therefore is unlikely to correspond to the centromere. A pericentric inversion may have moved the centromere to the other side of the translocated region, or the centromere itself may have shifted positions. Without knowing the gene order on the ancestral Y chromosome involved in the translocation, it will be hard to determine definitively—the event is 10–15 Myr old, leaving time for subsequent rearrangements. Alternatively, the dot and Y chromosomes may have fused as a result of an unequal crossover event between the telomeric region of the Y chromosome and the centromeric region of the dot chromosome. While we do not know the identity of the current dot chromosome centromere in D. pseudoobscura, the latter hypothesis would predict that it is derived from the ancestral Y chromosome centromere. Further studies in the obscura group are necessary to reveal the detailed evolutionary history of this event.
Sex‐autosome translocations are often deleterious mutations that do not persist in populations (Ohno 1967). The Y‐to‐dot translocation therefore seems like a relatively rare event in that it was successful and eventually fixed in the ancestral population. For a male‐specific gene, the Y chromosome offers shelter from relaxed or antagonistic selection in females (Fisher 1931; Rice 1987a, b) but at the cost of a reduced efficacy of natural selection (Charlesworth and Charlesworth 2000). Autosomes offer larger effective population sizes and higher recombination rates that should facilitate more efficacious selection (Betancourt and Presgraves 2002; Presgraves 2005). However, these benefits are only realized over long evolutionary timescales, not immediately following a chromosome translocation. What then allowed for the fixation of the Y‐to‐dot translocation? The Y‐to‐dot does not represent the only gene traffic between the dot and Y chromosome in Drosophila species. There are at least three independent cases of individual gene movements or duplications from the Y to dot chromosomes in Drosophila species: kl‐5 (Dyer et al. 2011), JYalpha (Carvalho and Clark 2013), and PRY (Leung et al. 2015). The dot chromosome may offer a suitable environment for a sex‐linked gene. The dot and X chromosomes have similarities: they both have chromosome‐specific gene regulation, are haplosufficient, and feminizing in intersexes (adding an X or dot to a fly with 2X:3A biases development toward females; reviewed in Larsson and Meller 2006; Riddle and Elgin 2006). The dot may also have some similarities to Y chromosomes in chromatin environment—they are both largely heterochromatic (80% of the dot chromosome is heterochromatic) and, at least in some species, they share satellite DNAs (e.g. Lohe et al. 1993). A heterochromatic environment is required for the proper regulation of some autosomal genes (e.g. Eberl et al. 1993), and Y‐linked genes may have similar requirements. The Y chromosome involved in the Y‐to‐dot translocation in D. pseudoobscura was evolutionarily old, with some genes that had likely accumulated large amounts of repetitive DNA in their introns. These introns may have been deleterious if translocated to a typical autosome. This may also explain why it is rare for genes to move from the Y to other chromosomes (Koerich et al. 2008). It is possible that the initial dot‐Y fusion was tolerated because it placed the Y‐to‐dot region in a heterochromatic environment similar enough to the ancestral Y chromosome. While the Y and dot chromosomes may be similar in their heterochromatic regions, the Y‐linked genes are now located relatively close to the conserved‐dot region. In D. melanogaster, the gene‐dense region of the dot is a unique type of chromatin with properties of both heterochromatin and euchromatin (Sun et al. 2000; Leung et al. 2010). Following the translocation, the formerly Y‐linked genes retained testis‐specific expression but their structure changed dramatically: some individual genes are at least 10‐fold smaller than their orthologs on the D. melanogaster Y chromosome due to deletions in very large introns. The shortening of these very large introns following the Y‐to‐dot translocation may have been driven by recurrent selective sweeps (Larracuente and Clark 2014).
Introns across the genome are expected to evolve under natural selection—many contain regulatory sequences and are constrained by splicing requirements and transcription costs (Prachumwat et al. 2004). As such, there is a genome‐wide correlation between intron size and recombination rate in Drosophila (Carvalho and Clark 1999; Comeron and Kreitman 2000). This may be especially true for highly expressed genes (Castillo‐Davis et al. 2002; Urrutia and Hurst 2003). Consistent with this observation, we found that intron sizes are larger on the dot chromosome—where meiotic recombination via crossing over is at most very rare—compared to other autosomes. The mega‐introns of Drosophila Y‐linked genes may be consequences of the reduced efficacy of purifying selection (Carvalho and Clark 1999; Carvalho 2003). Alternatively, these mega‐introns may operate under different selection pressures, as some have hypothesized that they serve a role in spermatogenesis (Bonaccorsi et al. 1990; Pisano et al. 1993; Piergentili et al. 2004). Our analysis lends some general insights into the selection pressures on intron sizes. After becoming autosomal for 10–15 My, outside of lacking the few Y‐linked mega‐introns and very large introns, many introns in the Y‐to‐dot region are not smaller than their Y‐linked orthologs. In some cases, the introns are larger in the Y‐to‐dot region—consistent with the earlier observation that the correlation between recombination rate and intron size does not hold for large introns (Carvalho and Clark 1999). This suggests that in D. pseudoobscura, purifying selection against introns below ∼10 Kb on the Y‐to‐dot is inefficient. D. pseudoobscura presumably has mega‐introns—large lampbrush‐like loops are formed during spermatogenesis (Piergentili 2007)—but we do not find evidence that they come from the homologous introns in the Y‐to‐dot regions. Lampbrush‐like loops originate from different Y‐linked introns in D. melanogaster and D. hydei (Reugels et al. 2000). This suggests that D. pseudoobscura acquired these loops independently, perhaps on the new Y chromosome.
While the dramatic size reduction of Y‐to‐dot genes was driven mostly by deletions in very large introns (>10 Kb) rather than an overall intron shortening, this intron reduction cannot explain the much smaller size of the Y‐to‐dot region compared to Drosophila Y chromosomes. Deletions of repetitive DNA in Y‐to‐dot intergenic regions must contribute more than introns to the reduced size of the Y‐to‐dot region. However, our view of Drosophila Y chromosomes is melanogaster‐centric. The ancestral obscura Y chromosome could have been structurally different from the D. melanogaster Y chromosome. It is possible that the compact size of the Y‐to‐dot region is not only due to intergenic deletions of a large ancestral Y chromosome, but instead that the ancestral obscura group Y chromosome that fused to the dot chromosome was itself small. Y chromosome size varies among Drosophila species and even within species (White 1973). The karyotypes of other Drosophila species are consistent with the hypothesis that the ancestral obscura group Y chromosome was small (Fig. 3 and Fig. S4). However, without more genomic information from these Y chromosomes, it will be difficult to infer their evolutionary dynamics. Therefore, a combination of selection favoring deletions in large introns and intergenic regions and perhaps even a small ancestral Y chromosome may explain the structural differences between the Y‐to‐dot region and the D. melanogaster Y chromosome.
Y chromosome reversal events like the Y‐to‐dot in D. pseudoobscura may be more common than we currently appreciate. Heterochromatin is generally dense in repeats and rapidly evolving (Charlesworth et al. 1986). If Y chromosome movements tend to be toward heterochromatic regions of other chromosomes, signatures of these movements may be rapidly erased and difficult to detect, leading us to underestimate the frequency of Y‐autosome translocations. Because Y chromosomes typically contain genes essential for male fertility, Y chromosome reversal events may have a role in the turnover of sex chromosomes and sex determining systems in many clades.
Associate Editor: M. Wilson Sayres
Handling Editor: M. Noor
Supporting information
Figure S1. Dotplot comparing the dot‐linked PacBio contig (000025F) against 62 dot‐linked contigs from D. pseudoobscura r3.
Figure S2. Self dotplot of the duplicated CG30048 region on the dot‐linked contig 000025F.
Figure S3. Phylogeny of PRY, CG30048 and CG33482 in Drosophila species
Figure S4. Obscura group karyotypes.
Supporting Tables
Supporting Information
Supporting Information
Supporting Information
AUTHOR CONTRIBUTIONS
A.M.L. and C.‐H.C. designed the experiment. C.‐H.C. assembled the genome and analyzed the data. A.M.L. and C.‐H.C. wrote the article.
ACKNOWLEDGMENTS
We thank Drs. Stephen Richards and Stephen Schaeffer for sharing the D. pseudoobscura PacBio reads—these data were generated under the support of a National Institutes of Health National Institute of General Medical Sciences grant R01 GM098478 to S.W. Schaeffer. This work was supported by National Institutes of Health National Institute of General Medical Sciences grant R35 GM119515‐01 to A.M.L. We also thank the University of Rochester Center for Integrated Research Computing for access to computing cluster resources and two anonymous reviewers and the editor from Axios Review for helpful comments on the manuscript.
DATA ARCHIVING
The doi for our data is 10.5061/dryad.rp949.
LITERATURE CITED
- Abad, J. P. , de Pablos B., Agudo M., Molina I., Giovinazzo G., Martin‐Gallardo A., and Villasante A.. 2004. Genomic and cytological analysis of the Y chromosome of Drosophila melanogaster: telomere‐derived sequences at internal regions. Chromosoma 113:295–304. [DOI] [PubMed] [Google Scholar]
- Arguello, J. R. , Zhang Y., Kado T., Fan C., Zhao R., Innan H., Wang W., and Long M.. 2010. Recombination yet inefficient selection along the Drosophila melanogaster subgroup's fourth chromosome. Mol. Biol. Evol. 27:848–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S. F. , Gish W., Miller W., Myers E. W., and Lipman D. J.. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410. [DOI] [PubMed] [Google Scholar]
- Bachtrog, D. 2013. Y‐chromosome evolution: emerging insights into processes of Y‐chromosome degeneration. Nat. Rev. Genet. 14:113–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog, D. , and Andolfatto P.. 2006. Selection, recombination and demographic history in Drosophila miranda . Genetics 174:2045–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancourt, A. J. , and Presgraves D. C.. 2002. Linkage limits the power of natural selection in Drosophila. Proc. Natl. Acad. Sci. USA 99:13616–13620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancourt, A. J. , Welch J. J., and Charlesworth B.. 2009. Reduced effectiveness of selection caused by a lack of recombination. Curr. Biol. 19:655–660. [DOI] [PubMed] [Google Scholar]
- Bonaccorsi, S. , Gatti M., Pisano C., and Lohe A.. 1990. Transcription of a satellite DNA on two Y chromosome loops of Drosophila melanogaster . Chromosoma 99:260–266. [DOI] [PubMed] [Google Scholar]
- Bonaccorsi, S. , and Lohe A.. 1991. Fine mapping of satellite DNA sequences along the Y chromosome of Drosophila melanogaster: relationships between satellite sequences and fertility factors. Genetics 129:177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaccorsi, S. , Pisano C., Puoti F., and Gatti M.. 1988. Y chromosome loops in Drosophila melanogaster . Genetics 120:1015–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutlag, D. , Carlson M., Fry K., and Hsieh T. S.. 1977. DNA‐sequence organization in Drosophila heterochromatin . Cold Spring Harb. Sym. 42:1137–1146. [DOI] [PubMed] [Google Scholar]
- Bull, J. J. 1983. Evolution of sex determining mechanisms. The Benjamin/Cummings Publishing Company, INC, Menlo Park, CA. [Google Scholar]
- Carvalho, A. B. 2003. The advantages of recombination. Nat. Genet. 34:128–129. [DOI] [PubMed] [Google Scholar]
- Carvalho, A. B. , and Clark A. G.. 1999. Intron size and natural selection. Nature 401:344. [DOI] [PubMed] [Google Scholar]
- Carvalho, A. B. , and Clark A. G. 2005. Y chromosome of D. pseudoobscura is not homologous to the ancestral Drosophila Y. Science 307:108–110. [DOI] [PubMed] [Google Scholar]
- Carvalho, A. B. , and Clark A. G. 2013. Efficient identification of Y chromosome sequences in the human and Drosophila genomes. Genome Res. 23:1894–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo‐Davis, C. I. , Mekhedov S. L., Hartl D. L., Koonin E. V., and Kondrashov F. A.. 2002. Selection for short introns in highly expressed genes. Nat. Genet. 31:415–418. [DOI] [PubMed] [Google Scholar]
- Chaisson, M. J. , Huddleston J., Dennis M. Y., Sudmant P. H., Malig M., Hormozdiari F., Antonacci F., Surti U., Sandstrom R., Boitano M., et al. 2015. Resolving the complexity of the human genome using single‐molecule sequencing. Nature. 517:608–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth, B. 1978. Model for evolution of Y chromosomes and dosage compensation. Proc. Natl. Acad. Sci. USA 75:5618–5622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth, B. , Langley C. H., and Stephan W.. 1986. The evolution of restricted recombination and the accumulation of repeated DNA sequences. Genetics 112:947–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth, B. and Charlesworth D.. 2000. The degeneration of Y chromosomes. Philos. Trans. R Soc. Lond. B Biol. Sci. 355:1563–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Z. X. , Sturgill D., Qu J., Jiang H., Park S., Boley N., Suzuki A. M., Fletcher A. R., Plachetzki D. C., FitzGerald P. C., et al. 2014. Comparative validation of the D. melanogaster modENCODE transcriptome annotation. Genome Res. 24:1209–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin, C. S. , Alexander D. H., Marks P., Klammer A. A., Drake J., Heiner C., Clum A., Copeland A., Huddleston J., Eichler E. E., et al. 2013. Nonhybrid, finished microbial genome assemblies from long‐read SMRT sequencing data. Nat. Methods 10:563–569. [DOI] [PubMed] [Google Scholar]
- Comeron, J. M. , and Kreitman M.. 2000. The correlation between intron length and recombination in drosophila. Dynamic equilibrium between mutational and selective forces. Genetics 156:1175–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek, P. , Auton A., Abecasis G., Albers C. A., Banks E., DePristo M. A., Handsaker R. E., Lunter G., Marth G. T., Sherry S. T., et al. 2011. The variant call format and VCFtools. Bioinformatics 27:2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer, K. A. , White B. E., Bray M. J., Pique D. G., and Betancourt A. J.. 2011. Molecular evolution of a Y chromosome to autosome gene duplication in Drosophila . Mol. Biol. Evol. 28:1293–1306. [DOI] [PubMed] [Google Scholar]
- Eberl, D. F. , Duyf B. J., and Hilliker A. J.. 1993. The role of heterochromatin in the expression of a heterochromatic gene, the rolled locus of Drosophila melanogaster . Genetics 134:277–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eid, J. , Fehr A., Gray J., Luong K., Lyle J., Otto G., Peluso P., Rank D., Baybayan P., Bettman B., et al. 2009. Real‐time DNA sequencing from single polymerase molecules. Science 323:133–138. [DOI] [PubMed] [Google Scholar]
- English, A. C. , Richards S., Han Y., Wang M., Vee V., Qu J., Qin X., Muzny D. M., Reid J. G., Worley K. C., and Gibbs R. A.. 2012. Mind the gap: upgrading genomes with Pacific Biosciences RS long‐read sequencing technology. PLoS One 7:e47768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezaz, T. , Stiglec R., Veyrunes F., and Marshall Graves J. A.. 2006. Relationships between vertebrate ZW and XY sex chromosome systems. Curr. Biol. 16:R736–R743. [DOI] [PubMed] [Google Scholar]
- Fisher, R. A. 1931. The evolution of dominance. Biol. Rev. 6:345–368. [Google Scholar]
- Ford, C. E. , Hamerton J. L., and Sharman G. B.. 1957. Chromosome polymorphism in the common shrew. Nature 180:392–393. [DOI] [PubMed] [Google Scholar]
- Fredga, K. 1970. Unusual sex chromosome inheritance in mammals. Philos. Trans. R Soc. B Biol. Sci. 259:15–36. [DOI] [PubMed] [Google Scholar]
- Gao, J.‐J. , Watabe H.‐A., Zhang Y.‐P., and Aotsuka T.. 2004. Karyotype differentiation in newly discovered members of the Drosophila obscura species group from Yunnan, China. Zool. Res. 25:236–241. [Google Scholar]
- Gao, J. J. , Watabe H. A., Aotsuka T., Pang J. F., and Zhang Y. P.. 2007. Molecular phylogeny of the Drosophila obscura species group, with emphasis on the Old World species. BMC Evol. Biol. 7:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garagna, S. , Page J., Fernandez‐Donoso R., Zuccotti M., and Searle J. B.. 2014. The Robertsonian phenomenon in the house mouse: mutation, meiosis and speciation. Chromosoma 123:529–544. [DOI] [PubMed] [Google Scholar]
- Gatti, M. , and Pimpinelli S.. 1983. Cytological and genetic analysis of the Y chromosome of Drosophila melanogaster. I. Organization of the fertility factors. Chromosoma 88:349–373. [Google Scholar]
- Hoskins, R. A. , Smith C. D., Carlson J. W., Carvalho A. B., Halpern A., Kaminker J. S., Kennedy C., Mungall C. J., Sullivan B. A., Sutton G. G., et al. 2002. Heterochromatic sequences in a Drosophila whole‐genome shotgun assembly. Genome Biol. 3:RESEARCH0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, J. D. , and Bachtrog D.. 2011. Characterizing the influence of effective population size on the rate of adaptation: Gillespie's Darwin domain. Genome Biol. Evol. 3:687–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser, V. B. , and Charlesworth B.. 2009. The effects of deleterious mutations on evolution in non‐recombining genomes. Trends Genet. 25:9–12. [DOI] [PubMed] [Google Scholar]
- Karpen, G. H. , and Allshire R. C.. 1997. The case for epigenetic effects on centromere identity and function. Trends Genet. 13:489–496. [DOI] [PubMed] [Google Scholar]
- Kennison, J. A. 1981. The genetic and cytological organization of the Y chromosome of Drosophila melanogaster . Genetics 98:529–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koerich, L. B. , Wang X., Clark A. G., and Carvalho A. B.. 2008. Low conservation of gene content in the Drosophila Y chromosome. Nature 456:949–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurek, R. , Reugels A. M., Glatzer K. H., and Bunemann H.. 1998. The Y chromosomal fertility factor threads in Drosophila hydei harbors a functional gene encoding an axonemal dynein beta heavy chain protein. Genetics 149:1363–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurek, R. , Reugels A. M., Lammermann U., and Bunemann H.. 2000. Molecular aspects of intron evolution in dynein encoding mega‐genes on the heterochromatic Y chromosome of Drosophila sp. Genetica 109:113–123. [DOI] [PubMed] [Google Scholar]
- Kurtz, S. , Phillippy A., Delcher A. L., Smoot M., Shumway M., Antonescu C., and Salzberg S. L.. 2004. Versatile and open software for comparing large genomes. Genome Biol. 5:R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lack, J. B. , Cardeno C. M., Crepeau M. W., Taylor W., Corbett‐Detig R. B., Stevens K. A., Langley C. H., and Pool J. E.. 2015. The Drosophila genome nexus: a population genomic resource of 623 Drosophila melanogaster genomes, including 197 from a single ancestral range population. Genetics 199:1229–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larracuente, A. M. , and Clark A. G.. 2014. Recent selection on the Y‐to‐dot translocation in Drosophila pseudoobscura . Mol. Biol. Evol. 31:846–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larracuente, A. M. , and Ferree P. M.. 2015. Simple method for fluorescence DNA in situ hybridization to squashed chromosomes. JoVE 95:e52288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larracuente, A. M. , Noor M. A., and Clark A. G.. 2010. Translocation of Y‐linked genes to the dot chromosome in Drosophila pseudoobscura . Mol. Biol. Evol. 27:1612–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson, J. , and Meller V. H.. 2006. Dosage compensation, the origin and the afterlife of sex chromosomes. Chromosome Res. 14:417–431. [DOI] [PubMed] [Google Scholar]
- Leung, W. , Shaffer C. D., Cordonnier T., Wong J., Itano M. S., Slawson Tempel E. E., Kellmann E., Desruisseau D. M., Cain C., Carrasquillo R., et al. 2010. Evolution of a distinct genomic domain in Drosophila: comparative analysis of the dot chromosome in Drosophila melanogaster and Drosophila virilis . Genetics 185:1519–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung, W. , Shaffer C. D., Reed L. K., Smith S. T., Barshop W., Dirkes W., Dothager M., Lee P., Wong J., Xiong D., et al. 2015. Drosophila muller f elements maintain a distinct set of genomic properties over 40 million years of evolution. G3 5:719–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. , and Durbin R.. 2010. Fast and accurate long‐read alignment with Burrows‐Wheeler transform. Bioinformatics 26:589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohe, A. R. , and Brutlag D. L.. 1986. Multiplicity of satellite DNA sequences in Drosophila melanogaster . Proc. Natl. Acad. Sci. USA 83:696–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohe, A. R. , and Brutlag D. L. 1987. Adjacent satellite DNA segments in Drosophila structure of junctions. J. Mol. Biol. 194:171–179. [DOI] [PubMed] [Google Scholar]
- Lohe, A. R. , Hilliker A. J., and Roberts P. A.. 1993. Mapping simple repeated DNA sequences in heterochromatin of Drosophila melanogaster . Genetics 134:1149–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunter, G. , and Goodson M.. 2011. Stampy: a statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res. 21:936–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister, B. F. , and Charlesworth B.. 1999. Reduced sequence variability on the Neo‐Y chromosome of Drosophila americana americana . Genetics 153:221–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGaugh, S. E. , and Noor M. A.. 2012. Genomic impacts of chromosomal inversions in parapatric Drosophila species. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367:422–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna, A. , Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., et al. 2010. The genome analysis toolkit: a MapReduce framework for analyzing next‐generation DNA sequencing data. Genome Res. 20:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli, T. 2010. Higher‐order genome organization in human disease. Cold Spring Harb. Perspect Biol. 2:a000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno, S. 1967. Sex chromosomes and sex‐linked genes. Springer‐Verlag, Berlin, Heidelberg, New York. [Google Scholar]
- Piergentili, R. 2007. Evolutionary conservation of lampbrush‐like loops in drosophilids. BMC Cell Biol. 8:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piergentili, R. , Bonaccorsi S., Raffa G. D., Pisano C., Hackstein J. H., and Mencarelli C.. 2004. Autosomal control of the Y‐chromosome kl‐3 loop of Drosophila melanogaster . Chromosoma 113:188–196. [DOI] [PubMed] [Google Scholar]
- Piergentili, R. , and Mencarelli C.. 2008. Drosophila melanogaster kl‐3 and kl‐5 Y‐loops harbor triple‐stranded nucleic acids. J. Cell Sci. 121:1605–1612. [DOI] [PubMed] [Google Scholar]
- Pisano, C. , Bonaccorsi S., and Gatti M.. 1993. The kl‐3 loop of the Y chromosome of Drosophila melanogaster binds a tektin‐like protein. Genetics 133:569–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prachumwat, A. , DeVincentis L., and Palopoli M. F.. 2004. Intron size correlates positively with recombination rate in Caenorhabditis elegans . Genetics 166:1585–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves, D. C. 2005. Recombination enhances protein adaptation in Drosophila melanogaster . Curr. Biol. CB 15:1651–1656. [DOI] [PubMed] [Google Scholar]
- Reugels, A. M. , Kurek R., Lammermann U., and Bunemann H.. 2000. Mega‐introns in the dynein gene DhDhc7(Y) on the heterochromatic Y chromosome give rise to the giant threads loops in primary spermatocytes of Drosophila hydei . Genetics 154:759–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice, W. R. 1987a. The accumulation of sexually antagonistic genes as a selective agent promoting the evolution of reduced recombination between primitive sex chromosomes. Evolution 41:911–914. [DOI] [PubMed] [Google Scholar]
- Rice, W. R. 1987b. Genetic hitchhiking and the evolution of reduced genetic activity of the Y sex chromosome. Genetics 116:161–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle, N. C. , and Elgin S. C.. 2006. The dot chromosome of Drosophila: insights into chromatin states and their change over evolutionary time. Chromosome Res. 14:405–416. [DOI] [PubMed] [Google Scholar]
- Ross, J. A. , Urton J. R., Boland J., Shapiro M. D., and Peichel C. L.. 2009. Turnover of sex chromosomes in the stickleback fishes (gasterosteidae). PLoS Genet. 5:e1000391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schartl, M. 2004. Sex chromosome evolution in non‐mammalian vertebrates. Curr. Opin Genet. Dev. 14:634–641. [DOI] [PubMed] [Google Scholar]
- Schatz, M. C. , Delcher A. L., and Salzberg S. L.. 2010. Assembly of large genomes using second‐generation sequencing. Genome Res. 20:1165–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, J. G. , Warren W. C., Beukeboom L. W., Bopp D., Clark A. G., Giers S. D., Hediger M., Jones A. K., Kasai S., Leichter C. A., et al. 2014. Genome of the house fly, Musca domestica L., a global vector of diseases with adaptations to a septic environment. Genome Biol. 15:466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, N. D. , Koerich L. B., Carvalho A. B., and Clark A. G.. 2014. Positive and purifying selection on the Drosophila Y chromosome. Mol. Biol. Evol. 31:2612–2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater, G. S. , and Birney E.. 2005. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics 6:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit, A. F. A. , Hubley R. and Green P.. 2013–2015. RepeatMasker Open‐4.0. <http://www.repeatmasker.org>.
- Smukowski Heil, C. S. , Ellison C., Dubin M., and Noor M. A.. 2015. Recombining without Hotspots: a comprehensive evolutionary portrait of recombination in two closely related species of Drosophila. Genome Biol. Evol. 7:2829–2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, X. , Goicoechea J. L., Ammiraju J. S., Luo M., He R., Lin J., Lee S. J., Sisneros N., Watts T., Kudrna D. A., et al. 2011. The 19 genomes of Drosophila: a BAC library resource for genus‐wide and genome‐scale comparative evolutionary research. Genetics 187:1023–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinemann, M. 1982. Multiple sex chromosomes in Drosophila miranda: a system to study the degeneration of a chromosome. Chromosoma 86:59–76. [DOI] [PubMed] [Google Scholar]
- Steinemann, S. , and Steinemann M.. 2005a. Retroelements: tools for sex chromosome evolution. Cytogenet. Genome Res. 110:134–143. [DOI] [PubMed] [Google Scholar]
- Steinemann, S. , and Steinemann M. 2005b. Y chromosomes: born to be destroyed. Bioessays 27:1076–1083. [DOI] [PubMed] [Google Scholar]
- Sun, F. L. , Cuaycong M. H., Craig C. A., Wallrath L. L., Locke J., and Elgin S. C.. 2000. The fourth chromosome of Drosophila melanogaster: interspersed euchromatic and heterochromatic domains. Proc. Natl. Acad. Sci. USA 97:5340–5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, K. , Stecher G., Peterson D., Filipski A., and Kumar S.. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30:2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton, K. 2003. Libsequence: a C++ class library for evolutionary genetic analysis. Bioinformatics 19:2325–2327. [DOI] [PubMed] [Google Scholar]
- Trapnell, C. , Pachter L., and Salzberg S. L.. 2009. TopHat: discovering splice junctions with RNA‐Seq. Bioinformatics 25:1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell, C. , Roberts A., Goff L., Pertea G., Kim D., Kelley D. R., Pimentel H., Salzberg S. L., Rinn J. L., and Pachter L.. 2012. Differential gene and transcript expression analysis of RNA‐seq experiments with TopHat and Cufflinks. Nat. Protoc. 7:562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treangen, T. J. , and Salzberg S. L.. 2012. Repetitive DNA and next‐generation sequencing: computational challenges and solutions. Nat. Rev. Genet. 13:36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urrutia, A. O. , and Hurst L. D.. 2003. The signature of selection mediated by expression on human genes. Genome Res. 13:2260–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicoso, B. , and Bachtrog D.. 2013. Reversal of an ancient sex chromosome to an autosome in Drosophila. Nature 499:332–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicoso, B. , and Bachtrog D. 2015. Numerous transitions of sex chromosomes in Diptera. PLoS Biol. 13:e1002078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villasante, A. , Abad J. P., Planello R., Mendez‐Lago M., Celniker S. E., and de Pablos B.. 2007. Drosophila telomeric retrotransposons derived from an ancestral element that was recruited to replace telomerase. Genome Res. 17:1909–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahrman, J. , and Zahavi A.. 1955. Cytological contributions to the phylogeny and classification of the rodent genus gerbillus. Nature 175:600–602. [Google Scholar]
- White, M. J. D. 1973. Animal cytology and evolution. Cambridge Univ. Press, Cambridge. [Google Scholar]
- Wurster, D. H. , and Benirschke K.. 1970. Indian Momtjac, Muntiacus muntiak: a deer with a low diploid chromosome number. Science 168:1364–1366. [DOI] [PubMed] [Google Scholar]
- Zhou, Q. , and Bachtrog D.. 2012. Sex‐specific adaptation drives early sex chromosome evolution in Drosophila. Science 337:341–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Dotplot comparing the dot‐linked PacBio contig (000025F) against 62 dot‐linked contigs from D. pseudoobscura r3.
Figure S2. Self dotplot of the duplicated CG30048 region on the dot‐linked contig 000025F.
Figure S3. Phylogeny of PRY, CG30048 and CG33482 in Drosophila species
Figure S4. Obscura group karyotypes.
Supporting Tables
Supporting Information
Supporting Information
Supporting Information


