Summary
The ability of excess Mg2+ to compensate the absence of cell wall related genes in Bacillus subtilis has been known for a long time, but the mechanism has remained obscure. Here, we show that the rigidity of wild‐type cells remains unaffected with excess Mg2+, but the proportion of amidated meso‐diaminopimelic (mDAP) acid in their peptidoglycan (PG) is significantly reduced. We identify the amidotransferase AsnB as responsible for mDAP amidation and show that the gene encoding it is essential without added Mg2+. Growth without excess Mg2+ causes ΔasnB mutant cells to deform and ultimately lyse. In cell regions with deformations, PG insertion is orderly and indistinguishable from the wild‐type. However, PG degradation is unevenly distributed along the sidewalls. Furthermore, ΔasnB mutant cells exhibit increased sensitivity to antibiotics targeting the cell wall. These results suggest that absence of amidated mDAP causes a lethal deregulation of PG hydrolysis that can be inhibited by increased levels of Mg2+. Consistently, we find that Mg2+ inhibits autolysis of wild‐type cells. We suggest that Mg2+ helps to maintain the balance between PG synthesis and hydrolysis in cell wall mutants where this balance is perturbed in favor of increased degradation.
Introduction
Bacterial cell walls are exoskeletal macromolecular structures that protect cells and give them shape and mechanical integrity. Their physiology is characterized by a delicate balance between rigidity, which confers mechanical stability and plasticity, which permits growth and division. The physical basis of the rigidity of bacterial cell walls is a network of polymers whose dominant component is the peptidoglycan (PG) (Turner et al., 2014). The PG meshwork (the sacculus) is composed of linear strands of glycan chains crosslinked via peptide stems. Glycan chains are universally composed of repeating units of β‐(1,4) linked N‐acetyl glycosamine and N‐acetyl muramic acid (Turner et al., 2014). The stem peptides vary between species and are used as a taxonomic criterion for classifying bacteria (Schleifer and Kandler, 1972). They are synthesized as pentapeptide chains containing unconventional D‐amino acid residues attached to the lactyl moiety of muramic acid, and possess a di‐basic amino acid in the third position. Meso‐diaminopimelic acid (mDAP) is characteristic of Gram‐positive bacilli and most Gram‐negative species, L‐Lys of the majority of Gram‐positive organisms and L‐Orn of Spirochetes (Vollmer et al., 2008a). In the rod‐shaped Gram‐positive model organism Bacillus subtilis the pentapeptide consists of L‐Ala‐D‐iso‐Glu‐mDAP‐D‐Ala‐D‐Ala (Atrih et al., 1999). Gram‐positive cell walls further contain carbohydrate‐based anionic polymers, primarily teichoic acids, which account for roughly half of the mass of the cell wall. Their physiological functions remain elusive but they play a major role in divalent cation binding (particularly Mg2+) (Brown et al., 2013; Neuhaus and Baddiley, 2003; Heckels et al., 1977), they serve as scaffolds for a wide range of molecules and they regulate cell wall‐associated enzymes (Yamamoto et al., 2008; Brown et al., 2013).
Plasticity of PG depends upon systems of enzymes with opposing activities. On the one hand, synthetic enzymes make and interlink new glycan chains, and, on the other hand, hydrolytic enzymes cut existing bonds to permit growth and remodeling (Turner et al., 2014). High‐molecular‐weight penicillin binding proteins (PBPs) are membrane‐embedded PG‐synthesizing enzymes that possess transglycosylase and/or transpeptidase activities. In B. subtilis, PG degradative enzymes, termed PG hydrolases, are numerous and highly redundant (Smith et al., 2000; Vollmer et al., 2008b). They possess a range of hydrolytic activities including recognizing and cutting the bonds between the sugar moieties in the glycan chain (glucosaminidases and muramidases/lytic transglycosylases), between N‐acetyl muramic acid and the peptide stem (amidases) and between specific amino acids in the peptide stems or in the peptide bridges (endopeptidases) (Smith et al., 2000). Uncontrolled activity of PG hydrolases could compromise cellular integrity and lead to lysis. Indeed, some PG hydrolases are autolysins that can lyse growing cells when their proton motive force (pmf) is dissipated by depolarization of the membrane by chemical uncouplers, such as azide or CCCP, or by starvation (Jolliffe et al., 1981; Lamsa et al., 2012). In B. subtilis, the autolysins are the amidase LytC, the glucosaminidase LytD, and the DL‐endopeptidases LytE and LytF (Smith et al., 2000). Cells of the mutant strain where genes encoding these four autolysins are absent do not undergo lysis upon dissipation of the pmf, but they still grow and divide, indicating that other PG hydrolases can confer plasticity to the cell wall for these purposes (Lamsa et al., 2012). The rest of the putative PG hydrolase complement of B subtilis includes more than 30 enzymes (Smith et al., 2000) that are not autolysins and are often enmeshed in complex regulatory networks (Meisner et al., 2013). Overall, and despite the important role of PG hydrolases in cell wall metabolism, their regulation remains largely unknown.
In rod‐shaped bacteria, the coordination of PG synthesis and degradation results in controlled cell wall enlargement and division, and produces cells with narrowly defined cell dimensions for a given growth condition. This coordination is effected by two complementary PG‐synthesizing systems that are associated with cytoskeletal elements, and control sidewall elongation and septation respectively. The system involved in cell division is governed by the bacterial tubulin FtsZ and the associated septation‐specific PBPs. The system involved in sidewall elongation is controlled by the Rod system, a system of morphogenetic proteins including the bacterial actin homologue MreB as well as MreC, MreD, RodA, RodZ and elongation‐specific PBPs (Eun et al., 2015). One of the unexplained aspects of the Rod system in B. subtilis is that the lethality and/or morphological defects of the absence of some of its components can be overcome by adding Mg2+ to the growth medium (Formstone and Errington, 2005). mreB and its paralog mbl are essential in standard laboratory conditions. However, when growth media are supplemented with 5–25 mM Mg2+, mreB and mbl mutants grow and divide normally and assume a normal rod‐shaped morphology. When Mg2+ is depleted, the morphological phenotype becomes manifest and they grow as deformed and ballooning cells before eventually lysing (Formstone and Errington, 2005; Chastanet and Carballido‐Lopez, 2012). Mg2+ likewise suppresses the viability and/or morphological defects of several other cell wall related mutants (e.g. mreC, mreD, ponA, ugtP, pgcA or gtaB)(Murray et al., 1998; Chastanet and Carballido‐Lopez, 2012), but the mechanism underlying this rescuing role is currently unknown. Two main hypotheses have been put forth to explain it. It has been suggested that Mg2+ may have a structural role in enhancing the rigidity of the cell wall or, alternatively (or simultaneously), a regulatory role by affecting the activity of cell‐wall embedded enzymes (Formstone and Errington, 2005). It has been known for several decades that Mg2+ binds to cell walls of Gram‐positive bacteria (Beveridge and Murray, 1980; Heckels et al., 1977). Recent solid state NMR studies estimate the dissociation constant at approximately 0.6 mM (Kern et al., 2010).
One of the ways bacteria can regulate the metabolism of their cell walls is by chemical modification of the constituent subunits. For example, a common modification is the deacetylation of N‐acetyl glucosamine. This modification abolishes the binding of LysM domain containing proteins, such as some autolysins and PG hydrolases, and may thereby modulate their activity (Mesnage et al., 2014). A distinct and widespread modification is the O‐acetylation of N‐acetyl muramic acid, which is associated to resistance to lysozyme and endogenous autolysis, as well as to resistance to penicillin and macrophage killing (Bernard et al., 2011a; Davis and Weiser, 2011). Another common modification, only present in Gram‐positive organisms, is the amidation of carboxyl groups of amino acids in the stem peptides or peptide cross‐bridges. Commonly amidated are the α‐carboxyl of the D‐Glu in the second position of the stem peptide, and the ε‐carboxyl group of the mDAP in the third position. In Staphylococcus aureus and Streptococcus pneumoniae where the di‐basic amino acid is L‐Lys instead of mDAP, D‐Glu is amidated to D‐iso‐glutamine. The two enzymes responsible for D‐Glu amidation (the MurT/GatD complex) have been identified (Munch et al., 2012; Figueiredo et al., 2012), and PG transpeptidases have been shown to work more efficiently when D‐Glu is amidated (Munch et al., 2012; Figueiredo et al., 2012). In B. subtilis, mDAP was found to be amidated in most muropeptides of vegetative cell PG, while D‐Glu was not amidated (Atrih et al., 1999; Warth and Strominger, 1971). Amidation of mDAP has also been found in PG of other bacteria (Bernard et al., 2011b; Schleifer and Kandler, 1972; Levefaudes et al., 2015; Mahapatra et al., 2005). AsnB1 and LtsA were recently identified as the amidotransferases responsible for mDAP amidation in Lactobacillus plantarum (Bernard et al., 2011b) and Corynebacterium glutamicum (Levefaudes et al., 2015) respectively. These proteins belong to a large family of glutamine amidotransferases (Raushel et al., 1999), which catalyze the transfer of ammonia from glutamine to various substrates. A canonical member of this family is the asparagine synthase of Escherichia coli, AsnB (Huang et al., 2001). PG recognition by the mammalian host innate immune system has been shown to be affected by amidation of mDAP (Asong et al., 2009; Girardin et al., 2003) but the physiological role of mDAP amidation in the homeostasis of the bacterial cell wall remains unknown. PG synthesizing enzymes seem to work as efficiently on amidated as in non‐amidated mDAP in both L. plantarum and C. glutamicum (Bernard et al., 2011b; Levefaudes et al., 2015), suggesting that mDAP amidation may not be necessary for efficient cell wall synthesis (Levefaudes et al., 2015; Hirasawa et al., 2000). However, in L. plantarum asnB1 seems to be essential and the mutant strains are affected in growth and morphology (Bernard et al., 2011b).
In this work, we describe the differential amidation of mDAP as an important chemical modification of the PG of B. subtilis wild‐type cells grown in the presence of high concentrations of Mg2+. We identified AsnB as the enzyme responsible for catalyzing it, and characterized the phenotype of mutant cells. Our results suggest that both Mg2+ and amidation of mDAP are involved in modulating PG hydrolysis.
Results
Excess extracellular Mg2+ causes a decrease in amidation of mDAP in B. subtilis
To gain insight into the effect of magnesium on the cell wall, we examined the muropeptide composition of the PG of wild‐type B. subtilis cells grown in PAB (Penassay broth) in the absence and in the presence of 25 mM MgSO4. The muropeptide profiles (Fig. 1) were similar to those previously reported for the PG of vegetative cells grown in LB medium (Atrih et al., 1999). When no Mg2+ was added, uncleaved dimeric muropeptides containing amidation on both mDAP moieties represented 34% of all muropeptides, while muropeptide dimers with amidation on only one mDAP represented 14% of all muropeptides (the major dimer peaks 12 and 15 are indicated by red arrows in Fig. 1 and highlighted in grey in Supporting Information Table 1). In contrast, cells grown with excess Mg2+ had only 21.6% of dimeric muropeptides with doubly amidated mDAP, while the dimers containing only one amidation represented 23% of all identified muropeptides (Fig. 1 bottom panel, and Supporting Information Table 1). This corresponds to greater than 60% increase of singly amidated dimers in cells grown in the presence of added Mg2+. The effect was identical in different growth media (Fig. 1 for PAB medium, Supporting Information Fig. S1A and B for LB medium), and it was independent of the wild‐type strain used. Data for the strain BSB1 are shown in Fig. 1 and Supporting Information Fig. S1A and B; data for the strain 168ed in Supporting Information Fig. S1C and D.
Figure 1.
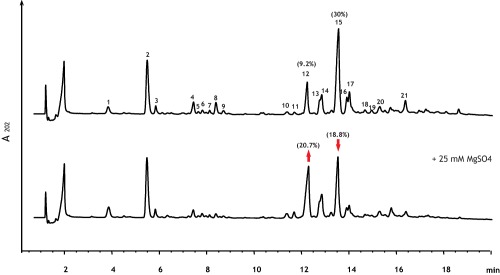
Effect of Mg2+ on muropeptide composition of vegetative PG of wild‐type B. subtilis. RP‐UPLC separation of muropeptides released by mutanolysin digestion of PGs extracted from B. subtilis wild‐type strain BSB1 grown in PAB in the absence (upper chromatogram) and in the presence of 25 mM MgSO4 (lower chromatogram). The major muropeptide dimer peaks with only one (peak 12) or two (peak 15) amidated mDAP moieties are indicated by the red arrow pointing up and down respectively (their percentages of total muropeptide are indicated in parentheses above the peaks). Supporting Information Table 1 lists the masses and the identities of the numbered peaks.
To test whether this effect was produced by a generic increase in the ionic strength in the medium, cells were grown in the presence of 100 mM NaCl. This concentration of NaCl has the same ionic strength as 25 mM MgSO4, since I = , where I is the ionic strength, ci is the molar concentration of ion i and zi is the charge number of that ion. In contrast to cells grown in the presence of high Mg2+, cells grown in medium supplemented with NaCl did not show any changes in the degree of amidation of dimeric muropeptides, nor any other significant change in the muropeptide profile (Supporting Information Fig. S1E). This indicated that Mg2+ specifically affected the level of amidated mDAP in B. subtilis PG.
In addition, we used atomic force microscopy (AFM) to measure the rigidity of the cell wall of living cells in the presence of Mg2+. Excess extracellular Mg2+ had no effect on the rigidity of the cell wall of live hydrated cells (representative cell are shown in Supporting Information Fig. S2B and D). The Young modulus was 40.2 ± 4.9 MPa for cells grown without supplemented Mg2+ and 39.7 ± 4.6 MPa for cells in the presence of 25 mM MgSO4. The topography of cell surfaces likewise remained unchanged (Supporting Information Fig. S2A and C), suggesting that excess Mg2+ does not affect the structure and mechanical properties of the cell wall.
asnB, asnH, asnO are homologues of mDAP amide transferases from L. plantarum and C. glutamicum
We sought to identify the amidotransferase responsible for amidation of mDAP in B. subtilis. BLAST search analysis using the amino acid sequences of AsnB1 from L. plantarum and LtsA from C. glutamicum, revealed three homologues in B. subtilis strain 168: AsnB, AsnH and AsnO (Supporting Information Fig. S3). Their homology with C. glutamicum LtsA has E values below 10e−51 with identities ranging from 23% for AsnH, 31% for AsnO and 56% for AsnB. Similarly, the homologies with L. plantarum AsnB1 are high, with E values below 10e−48 and identities at 28% for AsnH and AsnO and 50% for AsnB. The ansB, asnH and asnO gene products were previously predicted to be asparagine synthetases on the basis of the amino acid sequence similarity to AsnB of E. coli, to which they are all three orthologous to the same extent (∼ 28% similarity) (Yoshida et al., 1999). Each of the three genes complemented an E. coli strain lacking asparagine synthetases, indicating that all encode enzymes capable of synthesizing asparagine (Yoshida et al., 1999). However, while deletion of asnO and/or asnH in B. subtilis had no effect on growth, deletion of asnB was reported to lead to a slow‐growth phenotype even in the presence of high levels of asparagine (Yoshida et al., 1999).
Mg2+‐dependent growth of ΔasnB mutant strain
To investigate whether AsnB, AsnH and/or AsnO are involved in mDAP amidation in B. subtilis, we attempted to inactivate the corresponding genes by backcrossing their previously reported deletions (Yoshida et al., 1999) into our BSB1 wild‐type reference laboratory strain. asnH and asnO null mutants were readily obtained. The colonies of the resulting strains had normal rugose morphology and grew as well as the wild‐type strain. In contrast, the strain carrying a deletion of asnB (asnB::neo) could not be reliably obtained using unsupplemented growth media. Backcrossing the asnB::neo allele gave variable results that were not reproducible between experiments. Sometimes the few neoR transformants produced colonies of different sizes with some being smooth and others rugose, and sometimes no transformants were obtained. This suggested that asnB is essential in B. subtilis, like asnB1 in L. plantarum (Bernard et al., 2011b), and that suppressor mutations arise to alleviate the growth defect associated with the loss of AsnB. To test if asnB plays an essential role in the homeostasis of the cell wall, we tested whether high levels of Mg2+ were able to compensate this defect and permit reliable transformation. In the presence of 25 mM Mg2+, we readily obtained colonies of the same size with homogenously smooth appearance, giving the strain BDA33 (asnB::neo). We were similarly able to construct a triple mutant strain carrying deletions in asnB, asnH, and asnO (BDA329).
The ΔasnB mutant was not able to grow on PAB plates (∼210 μM Mg2+) on which the wild‐type grew normally. Even though AsnB is annotated as an asparagine synthase, addition of asparagine to the medium did not permit the restoration of growth under our experimental conditions (Fig. 2A). However, growth was completely restored when the plates were supplemented with 25 mM Mg2+ (Fig. 2A). Growth of the ΔasnB mutant was also Mg2+‐dependent in liquid media (Fig. 2B and C for PAB medium, and Supporting Information Fig. S4 for LB). In PAB supplemented with 5 mM Mg2+, the mutant grew at significantly slower rate than the wild‐type, and at ≥ 10 mM Mg2+ it grew as well as the wild‐type strain (Fig. 2B and C). Taken together, these results show that asnB is essential in media without additional Mg2+, suggesting that AsnB has a cell wall related function in addition to its previously reported asparagine synthetase activity (Yoshida et al., 1999).
Figure 2.

Conditional lethality and muropeptide analysis of ΔasnB mutant strain. A. Wild‐type B. subtilis BSB1 and ΔasnB (BDA33) strains were streaked on PAB plates unsupplemented (left panel) and supplemented with 100 μg ml−1 of L‐asparagine (central panel) or with 25 mM MgSO4 (right panel). B. and C. Growth curves of the wild‐type strain (B) and the ΔasnB mutant (C) grown at 37°C in PAB medium supplemented of the indicated concentrations of MgSO4. D. RP‐UPLC separation of muropeptides extracted from vegetative PG of the ΔasnB (strain BDA33) grown in PAB supplemented with 25 mM MgSO4. The numbered peaks were identified by MALDI‐TOF and are shown in Supporting Information Table 2.
Complete absence of amidated mDAP in the peptidoglycan of ΔasnB mutant cells
To examine the involvement of AsnB, AsnH and AsnO in amidating mDAP, we studied the chemical composition of the PG of the mutant strains grown in the presence of excess Mg2+. Muropeptide profiles of the wild‐type, ΔasnH, and ΔasnO mutant strains were indistinguishable (Supporting Information Fig. S5). In contrast, the ΔasnB mutant strain (Fig. 2D) displayed a strikingly different muropeptide profile, with a general shift of all major peaks to lower retention times. This is particularly evident for the major monomer (peak b in Fig. 2D) and dimer (peak o in Fig. 2D) peaks. This was consistent with the absence of amidation of the ε‐carboxyl of mDAP since the non‐amidated forms of mDAP‐containing muropeptides are less hydrophobic and thus their retention on the reverse phase (C18) column is reduced. Consistently, mass‐spectrometry revealed masses in accordance with complete absence of amidation on mDAP in the ΔasnB mutant (Supporting Information Table 2).
The calculated crosslinking index of the ΔasnB mutant (39%) was similar to that of the wild‐type (40%) and consistent with previously published estimates (Atrih et al., 1999). This indicated that PG synthetic enzymes involved in transpeptidation are capable of accepting non‐amidated forms of mDAP as substrate as efficiently as the amidated forms.
Altogether, these results show that AsnB is indispensable for amidation of mDAP in B. subtilis and suggest involvement of this chemical modification in cell wall metabolism. The link with cell wall metabolism is further reinforced by the correlation of expression of asnB with cell wall metabolism genes in approximately 200 different growth conditions (Nicolas et al., 2012). All but two of the first twenty genes which are most correlated with asnB are involved in cell wall metabolism, which is not the case for asnH or asnO (http://genome.jouy.inra.fr/cgi-bin/seb/index.py).
Depletion of Mg2+ causes deformation and cell lysis in ΔasnB mutant cells
In the presence of high concentrations of Mg2+ the ultrastructure of the cell envelope of ΔasnB mutant cells was not appreciably different from that of wild‐type cells (Fig. 3A and B). Similarly, their cell surface topography and their rigidity were comparable to those of wild‐type cells when assayed by AFM (Fig. 3C–F). The rigidity of wild‐type was 39.7 ± 4.6 MPa versus 41.6 ± 3 MPa for ΔasnB mutant, suggesting that the absence of mDAP amidation did not affect the structural integrity and mechanics of the cell wall. ΔasnB mutant cells grown in the presence of 25 mM Mg2+ displayed normal rod shape (Fig. 3H), although they were significantly shorter (Fig. 3I and J) and wider (Fig. 3K) than wild‐type cells (mean width was 1.1(±0.05) μm and 0.85(±0.07) μm respectively).
Figure 3.
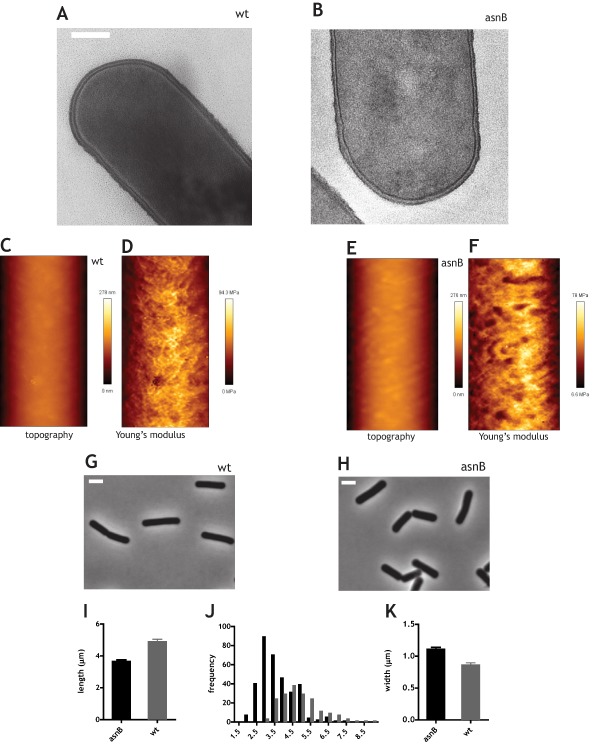
Ultrastructure, mechanics, and morphology of ΔasnB mutant cells grown with excess Mg2+. Cells were grown to mid‐exponential phase in LB medium supplemented with 25 mM Mg2+. A. and B. Transmission electron micrographs of wild‐type (A) and ΔasnB (B) cells. Scale bar is 200 nm. C–F. Atomic force microscopy imaging of the surface topography (C and E) and of rigidity measurement (D and F) of wild‐type (C and D) and ΔasnB mutant cells (E and F). Scale of height is indicated to the right of panels C and E and the scale of Young's modulus is indicated on the right of panels D and E. Scan size was 0.683 by 1.711 µm. G and H. Phase‐contrast micrographs of wild‐type (G) and ΔasnB cells (H). Scale bar is 2 μm. I. Bar graph of average cell length of wild‐type and ΔasnB cells. 95% confidence intervals are shown (> 300 cells were measured). J. Histogram of cell lengths for wild‐type and ΔasnB mutant cells. K. Bar graph of width of wild‐type and ΔasnB mutant cells. 95% confidence intervals are shown (> 300 cells were measured).
To investigate the phenotypes suppressed by the presence of excess Mg2+, we used time‐lapse microscopy. Cells were grown in a microfluidic device in either PAB or LB medium supplemented with 25mM Mg2+ and microscopically followed after removal of Mg2+. When unsupplemented PAB medium was flowed through the microfluidic device, cells lacking AsnB grew for approximately one cell cycle and subsequently started to deform and rapidly lyse (Fig. 4A, Supporting Information Movie 1). Mutant cells grown in batch culture in the presence of limiting concentrations of Mg2+ displayed similar lytic phenotype and morphological defects, with inhomogeneities in their diameter and an undulating appearance of the sidewalls (Fig. 4B). Local thickening of the cell wall was observed in some deformed cells (Fig. 4C). When Mg2+ depletion occurred in LB, where the concentration of Mg2+ is higher than in PAB but still too low to support normal growth of the ΔasnB mutant strain (see above and Supporting Information Fig. S4), growth continued for several cell cycles. The morphological deformations that accompanied growth were more pronounced (Fig. 6B upper row of panels, Supporting Information Movie 3), presumably because cells grew longer before lysing. The most remarkable morphological change was the increase in cell diameter and the appearance of bulges (Fig. 6B and Supporting Information Movie 3). In contrast, wild‐type cells grew and divided normally in the microfluidic device (Fig. 6A and Supporting Information Movie 2).
Figure 4.
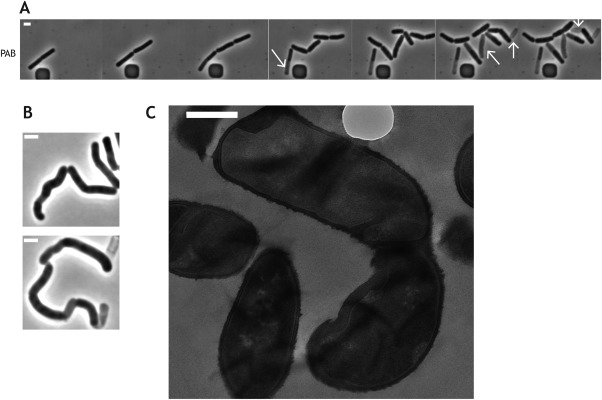
Depletion of excess Mg2+ causes cell deformation and lysis of ΔasnB cells. A. Time‐lapse microscopy capturing the phenotype of ΔasnB cells after removal of Mg2+ in PAB. Cells of strain BDA33 (asnB::neo) were grown in the microfluidics device under continuous flow of PAB medium supplemented with 25 mM MgSO4. The flow was stopped, the microfluidic chamber was flushed with unsupplemented medium and the cells were allowed to resume growth in unsupplemented medium. Cells were imaged every 20 seconds (every 80th image is shown), for full movie see Supporting Information Movie 1. White arrows indicate lysing cells. Scale bar, 2 μm. B. Phase‐contrast micrographs of ΔasnB cells grown in LB medium in the presence of limiting concentrations of Mg2. Top panel, 1 mM MgSO4; bottom panel, 5 mM MgSO4. Scale bar, 2 μm C. Ultrastructure of ΔasnB cells grown in LB without added Mg2+ examined by transmission electron miscroscopy. Scale bar is 500 nm.
Figure 6.
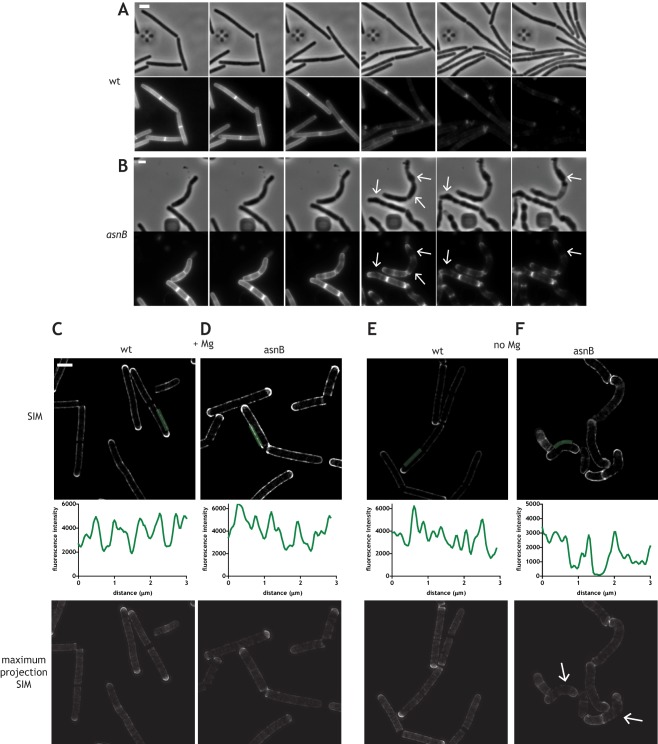
Imaging of depletion of TDL label in growing cells. A. and B. Time‐lapse pulse‐chase experiments in a microfluidic device (CellAssic). Disappearance of the TDL label from the PG of wild‐type (A) and ΔasnB mutant cells (B) during growth in LB without added Mg2+. Images were taken every 5 minutes (every third image is shown in the panels) after a 20 minute labeling pulse done in the presence of added Mg2+. Phase contrast images are shown above of their corresponding fluorescence images. Deformations of cells and the corresponding anisotropic loss of the label in the ΔasnB mutant are indicated by white arrows. See also Supporting Information Movies 2 and 3. Scale bars, 2 μm. C–F. SIM images of the depletion of TDL label in batch cultures of the wild‐type (C and E) and the ΔasnB mutant (D and F) grown in LB medium in the presence (C and D) and in the absence (E and F) of 25 mM MgSO4. SIM images of the medial plane of the cell are shown in the upper panel, quantifications of fluorescence along the green line in SIM images are shown in the middle panel, and maximum projections of the SIM images are shown in lowest panels. White arrows in panel F indicate regions of significant loss of label. Scale bars, 2 μm. Epifluorescence images corresponding to panels C to F are shown in Supporting Information Fig. S7.
Ordered insertion of peptidoglycan in the sidewall of the ΔasnB mutant
The cell deformations in the ΔasnB mutant could be produced by deregulated PG synthetic activity, by uncontrolled PG hydrolysis, or by both. To determine whether cell deformations caused by the absence of AsnB were accompanied by normal PG synthesis, we visualized PG insertion in Mg2+‐depletion experiments using TAMRA red D‐lysine (TDL) (Kuru et al., 2012, 2015) by incubating cells + for ¼ of cell doubling time (5–6 minutes). This non‐toxic fluorescent D‐amino acid derivative is incorporated in the fifth position of the stem peptide in B. subtilis (Kuru et al., 2012). TDL labeling of wild‐type cells produced a regular punctuate fluorescent pattern along the sidewall, revealing the sites of PG insertion (Fig. 5A), as previously described (Kuru et al., 2012, 2015). This punctuate labeling was ordered in either the presence or the absence of Mg2+ (Fig. 5A). Super‐resolution imaging of the TDL‐labeled cells by structured illumination microscopy (SIM) confirmed the regularity of the pattern along the length of the cell (Fig. 5B and C). Peaks of fluorescence, corresponding to the sites of PG insertion, were evenly distributed with an average spacing between approximately 0.5 and 1 μm, consistent with the reported spacing between the tracks followed by MreB‐associated PG synthetic machineries along the sidewalls of B. subtilis (Dominguez‐Escobar et al., 2011; Garner et al., 2011).
Figure 5.
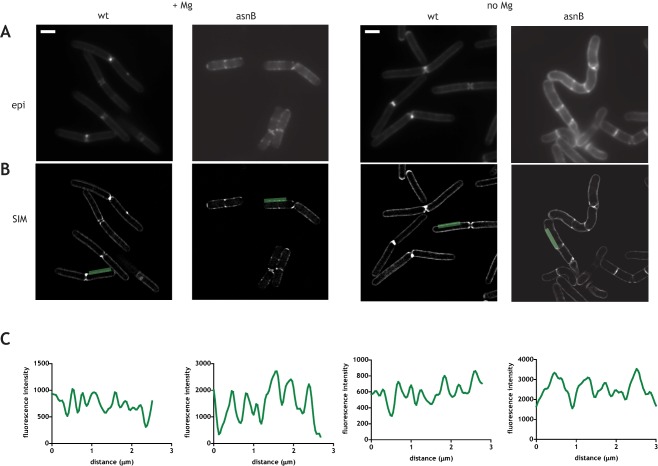
Isotropic insertion of PG along the sidewalls of the wild‐type and ΔasnB mutant cells. Epifluorescence (A) and structured illumination microscopy (B) imaging of insertion of peptidoglycan by TDL labeling in the presence of added Mg2+ (two panels on the left) and in the absence of added Mg2+ (two panels on the right). Structured illumination microscopy (SIM) images of the medial plane of the cell are shown. C. Quantification of fluorescence along the green line shown in SIM images. For SIM images of additional cells see Supporting Information Fig. S6.
Very similar labeling was observed for ΔasnB mutant cells, whether they were grown in the presence or in the absence of Mg2+ (Fig. 5). Even in the zones where ΔasnB cells were becoming deformed, there were no notable differences of the distribution of the fluorescent signal with respect to other regions of the cell where there were no signs of deformations (Fig. 5 and Supporting Information Fig. S6).
In summary, PG synthesis assayed by TDL labeling was ordered in the wild‐type and in the ΔasnB mutant, even under conditions which led to deformed cell morphology in the mutant. This suggested that the organization of PG synthesis is not defective in cells lacking AsnB and thus that the deformations may be caused by another mechanism.
Anisotropic peptidoglycan degradation in the ΔasnB mutant
We then investigated cell wall degradation that accompanies sidewall elongation using TDL pulse‐chase microlufidic experiments. Cells were labeled with TDL for one entire doubling time (∼20 min) in the presence of added Mg2+, washed, and the disappearance of the label was followed in real time allowing the cells to grow in fresh medium without added Mg2+. We reasoned that the appearance of dark zones could indicate zones of PG degradation (either loss/breakup or turnover/maturation), and/or of new PG insertion. Since the pattern of PG synthesis was regular in the ΔasnB mutant (Fig. 5 and Supporting Information Fig. S6), the appearance of irregularities in the loss of fluorescence could therefore be attributed to anisotropic degradation of PG.
Growth of wild‐type cells was accompanied by an isotropic loss of the TDL label (Fig. 6A, see also Supporting Information Movie 3). In contrast, loss of TDL label in the ΔasnB mutant was unevenly distributed (Fig. 6B). Fluorescence loss appeared concentrated in regions where the cells were beginning to deform and increase in diameter (white arrows in Fig. 6B, see also Supporting Information Movie 3). To quantify the loss of label along the sidewall, we performed a similar experiment in batch cultures and imaged the cells using SIM and epifluorescence after one quarter to one half of cell cycle of depletion (∼10 min under these conditions). In the presence of excess Mg2+, the disappearance of the label was isotropic in both wild‐type cells and ΔasnB mutant cells (Fig. 6C and D respectively; see also Supporting Information Fig. S7A and B for the corresponding epifluorescence images). Fluorescence intensity profiles along the sidewalls showed evenly distributed gaps in fluorescence intensity whose average distance was again approximately 0.5–1 μm. (Fig. 6C and D), suggesting that such gaps corresponded to MreB‐associated PG insertion and/or degradation. Wild‐type cells displayed the same pattern of fluorescence loss upon Mg2+ depletion (Fig. 6E and Supporting Information Fig. S7C, see lower panel of Supporting Information Fig. S7C for comparison of patterns obtained from SIM and epifluorescence images). In contrast, fluorescence intensity profiles along the sidewalls of ΔasnB mutant cells in the absence of supplemented Mg2+ showed extensive areas of loss of the TDL label which appeared more pronounced in zones where the cells were becoming deformed from their normal rod‐shape morphology (Fig. 6F and Supporting Information Fig. S7D, see lower panel of Supporting Information Fig. S7D for comparison of patterns obtained from SIM and epifluorescence images). This is particularly obvious in 3D projections of SIM images where the entire cell body is more easily visualized (white arrows in Fig. 6F and Supporting Information Fig. S8).
Because PG insertion was regular in the ΔasnB mutant in the absence of Mg2+ even in regions where the cells were becoming deformed (Fig. 5), these results suggest that the degradation of PG is anisotropic in the ΔasnB mutant. Such degradation could result from either breakup (loss) of PG through PG hydrolase activity or from PG maturation. A key PG maturation enzyme in B. subtilis is the D,D‐carboxypeptidase DacA, which removes the fifth amino acid from stem peptides and contributes to the loss of fluorescent D‐amino acids after labeling of PG (Kuru et al., 2015). Although spatial deregulation of DacA activity could explain the anisotropic loss of TDL in ΔasnB mutant cells, it could not account for the accompanying lysis and deformation phenotype. This phenotype requires the activity of PG hydrolases which would affect the connectivity of the PG meshwork and act on its structural bonds. Therefore, the lysis phenotype of ΔasnB and the associated loss of label in regions of cell deformation is consistent with deregulation of one, or a combination of, PG hydrolases. In conclusion, the absence of amidated mDAP and Mg2+ leads to anisotropic PG hydrolysis and associated morphological deformation, ultimately causing cell lysis.
Mg2+ inhibits cellular autolysis
We found that Mg2+ inhibited the lysis of the ΔasnB mutant, and it was also previously shown to prevent the lysis of ΔmreB mutants (Formstone and Errington, 2005). Furthermore, Mg2+ inhibits cell lysis induced by diverse conditions in unrelated Gram‐negative microorganisms (Rayman and MacLeod, 1975; Leduc et al., 1982; Gschwender and Hofschneider, 1969). Thus, we tested whether Mg2+ may have an inhibitory effect on PG hydrolases in B. subtilis. The autolysins responsible for cell lysis in B. subtilis are LytC, LytD, LytE and LytF (Jolliffe et al., 1981). The rate of cell autolysis upon depolarization of the membrane by addition of uncouplers can be therefore used as a readout of their activity.
Mg2+ reproducibly inhibited the rate of autolysis of wild‐type cells (a typical autolysis curve for cells in mid‐exponential growth is shown in Fig. 7A). We noticed that the rates of cell autolysis of wild‐type B. subtilis reported in the literature are variable, and this variability was evident in our initial experiments. Thus, we hypothesized that the rate of autolysis is dependent on the phase of growth of the cells. We therefore examined it in various stages of growth, from the exit from the lag phase to entry into stationary phase (see Materials and Methods). To correlate it with the phase of growth, we measured the apparent instantaneous rate of growth of the cells five minutes before addition of the uncoupler and plotted it against the rate of lysis. This approach revealed a linear relationship between the apparent instantaneous growth rate of the cells and the rate of cell autolysis (Fig. 7B), possibly explaining the variability of the reported lysis rates in the literature. Importantly, the presence of 25 mM Mg2+ inhibited the rate of lysis at all stages of growth (Fig. 7B). This suggests that Mg2+ exerts an inhibitory effect on PG hydrolysis catalyzed by autolysins.
Figure 7.
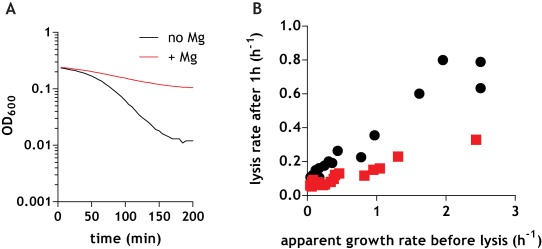
Mg2+ reduces the rate of autolysis. A. Rate of autolysis of wild‐type strain grown to mid‐exponential phase in the absence (black line) or in the presence (red line) of 25 mM MgSO4 added to cultures at the same time as azide. B. Apparent instantaneous growth rate of cells in various stages of exponential growth is plotted against their autolysis rate with (red squares) or without (black circles) 25 mM MgSO4.
Increased sensitivity of the ΔasnB mutant to lysozyme and cell wall active antibiotics
Deregulation of the homeostasis of PG hydrolysis in the ΔasnB mutant, with the resulting imbalance between PG synthesis and degradation, could sensitize ΔasnB cells to perturbations of the cell wall, such as the action of lysozyme and antibiotics that target the cell wall. Furthermore, PG hydrolysis has been implicated in the mechanism of action of many antibiotics that target the cell wall (Rogers et al., 1983; Tomasz et al., 1970; Goodell et al., 1976; Luo and Helmann, 2012).
Presence of lysozyme in the growth medium totally inhibited the growth of ΔasnB mutant strain at concentrations (1 mg ml−1) where the growth of the wild‐type strain was unaffected (Fig. 8A). This increased sensitivity to lysozyme is consistent with the results obtained for the ΔltsA mutant in C. glutamicum (Levefaudes et al., 2015).
Figure 8.
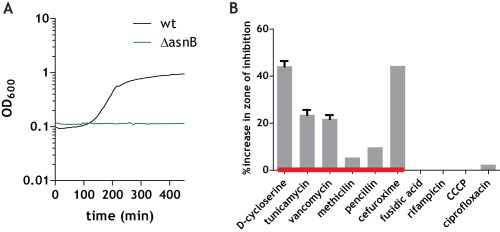
Increased sensitivity of the ΔasnB mutant to lysozyme and antibiotics that target the cell wall. A. Growth curves for wild‐type (black line) and ΔasnB (green line) grown in LB (+25 mM MgSO4) in the presence of 1 mg ml−1 lysozyme. B. Results of disk diffusion assays with the indicated antibiotics. Bars indicate the percent increase in zone of inhibition for the ΔasnB mutant relative to the wild‐type. Error bars are 95% confidence intervals (where not indicated, error bars are smaller than what can be represented graphically by the Prism plotting software).
We then used disk diffusion assays to compare the sensitivity of the ΔasnB mutant strain and the wild‐type to various antibiotics targeting different cellular processes. As expected, the ΔasnB mutant was more sensitive than the wild‐type to antibiotics which target different steps of cell wall synthesis, including D‐cycloserine, vancomycin, tunicamycin, methicillin, penicillin and cefuroxime. It nevertheless had the same sensitivity as the wild‐type strain to the DNA gyrase inhibitor ciprofloxacin, the RNA polymerase inhibitor rifampicin, and the uncoupler CCCP (Fig. 8B).
Discussion
Magnesium holds a very particular place in cell biology of B. subtilis since it can compensate the absence of a number of otherwise essential cell wall related genes. Hypotheses put forward to explain this effect include the idea that Mg2+ may somehow rigidify the cell wall, or that it may affect the activity of cell wall associated enzymes, or both (Formstone and Errington, 2005). We have tested the effect of Mg2+ on the rigidity of the cell wall of wild‐type B. subtilis cells using AFM and have found no effect. However, we identified a significant decrease in the degree of amidation of mDAP in cells grown in the presence of high concentrations (25 mM) of Mg2+, but not in the presence of equivalent ionic strength of NaCl. Amidation of mDAP in B. subtilis has been known for a long time, but its significance has remained obscure. In B. subtilis and other species where it has been reported, mDAP amidation is extensive, but not total, and it seems to slightly differ in different growth conditions (Atrih et al., 1999), suggesting a possible regulatory role in cell wall homeostasis (see also below).
We identified asnB, a gene encoding a member of glutamine amidotransferase family (Massiere and Badet‐Denisot, 1998), as indispensable for this amidation. AsnB is predicted to be a cytoplasmic protein indicating that mDAP amidation occurs on peptidoglycan precursors inside the cell, consistent with previous results with C. glutamicum LtsA (Levefaudes et al., 2015). Surprisingly, the ΔasnB mutant strain was only viable in the presence of high concentrations of Mg2+. Under these conditions, the cells were shorter and wider, but their ultrastructure and mechanics were not different from wild‐type. This width and length phenotype is similar to the phenotype of cells in which the DL‐endopeptidase CwlO is inactivated (Meisner et al., 2013; Salzberg et al., 2013), evoking a possible disruption of normal functioning of the PG hydrolytic system in the ΔasnB mutant. Indeed, when the concentration of Mg2+ was lowered in the growth medium, ΔasnB mutant cells began to deform and lyse, revealing the essential role of mDAP amidation in cell wall homeostasis.
We found that spatial organization of PG insertion is not affected in the ΔasnB mutant. In addition, the degree of crosslinking of peptide stems, which is a proxy for the activity of PG transpeptidases, was unchanged in ΔasnB relative to wild‐type. Transpeptidases are therefore not affected by the absence amidated mDAP in B. subtilis, even though this amino acid participates in crosslinks. This is similar to the results obtained in C. glutamicum where the AsnB homologue LtsA has been shown not to be required for efficient transpeptidation activity (Levefaudes et al., 2015).
In contrast to the normal spatial organization of PG insertion, the zones of PG degradation were anisotropically distributed in the ΔasnB mutant in the absence of excess Mg2+. There were extensive zones of uncontrolled degradation that correlated with the zones of deformation from normal rod‐shaped morphology. Thus, the genesis of the deformed morphology of ΔasnB mutant cells lies in the loss of homeostasis of PG hydrolysis. Uncontrolled regional PG hydrolysis leads to a disequilibrium between synthesis and degradation, causing deformation and, ultimately, lysis. This phenotype suggests a surprising degree of uncoupling between the synthetic and hydrolytic activities in this mutant when Mg2+ concentration is lowered.
It is generally accepted that the synthesis of new PG chains is somehow coupled with hydrolysis of the existing meshwork. According to the prevailing model, growth of the multilayered Gram‐positive sacculi follows a push‐up ‘inside‐to‐outside’ mechanism (Koch and Doyle, 1985), where new PG is inserted at the innermost face of the sacculus, pushing outward pre‐existing PG meshwork, which is then degraded and released into the medium by the action of PG hydrolases. The model is termed ‘make before break’ since the physiological coupling between synthesis and hydrolysis involves the polymerization and crosslinking of glycan chains prior to degradation (Koch et al., 1981). In the absence of amidated mDAP and without excess Mg2+ uncontrolled hydrolysis occurs in the regions of the cell where we are able to detect orderly PG insertion. Our results therefore suggest that mDAP amidation allows physiological control over activity of one or more PG hydrolytic enzymes and their coupling with the PG synthetic machinery.
The masking of the ΔasnB lysis phenotype by Mg2+ indicates that this ion inhibits the lethal activity of certain PG hydrolases that become deregulated in the absence of amidated mDAP. The only PG hydrolases whose kinetics can be measured in whole cells are the autolysins, and we showed that Mg2+ indeed inhibits their activity. B. subtilis has a complement of about 40 putative PG hydrolases (Smith et al., 2000), nearly twenty of which are expressed under vegetative growth conditions. However, it remains to be determined which of these PG hydrolases is inhibited by Mg2+ and which become deregulated in the absence of amidation of mDAP. Nevertheless, the effect of Mg2+ on inhibition of lysis has now been shown both in Gram‐positive (present study) and in phylogenetically distant Gram‐negative organisms (Gschwender and Hofschneider, 1969; Rayman and MacLeod, 1975; Leduc et al., 1982), possibly suggesting the existence of a common inhibitory mechanism. The reported affinity of wild‐type cell wall for Mg2+ is on the order of approximately 0.6 mM (Takahashi et al., 2013; Kern et al., 2010). At milimolar concentrations the vast majority of binding sites on the cell wall are occupied by Mg2+. It is therefore unlikely that changing the concentration of Mg2+ in the medium from 5 to 10 mM significantly affects the degree of Mg2+ bound to cell wall polymers. Nevertheless, this is precisely the concentration range at which the ΔasnB mutant is rescued by Mg2+. This suggests that the inhibitory activity of Mg2+ on PG hydrolases may not depend on its binding to cell wall polymers, but rather may be a result of direct interaction of Mg2+ with the enzymes. Further biochemical studies on purified enzymes will be necessary to elucidate this question.
Taken together, our data lead to a model in which B. subtilis modulates the degree of amidation of mDAP to maintain an appropriate balance between hydrolysis and degradation with changing environmental and physiological conditions. The changes in mDAP amidation induced in the wild‐type cells by the presence of high concentrations of Mg2+ and the lysis phenotype of ΔasnB mutant are consistent this model. Namely, since Mg2+ acts as an inhibitor for certain PG hydrolases, excess Mg2+ in the medium affects the balance between hydrolysis and synthesis and causes it to shift toward decreased hydrolysis. The cell would in turn try to compensate to restore the balance by reducing the extent of amidation of mDAP. Since the expression of asnB is unaffected by the presence of added Mg2+ (our unpublished results), the physiological adaptation occurs through post‐transcriptional means. The decrease in amidation would then permit the cell to increase the activity of hydrolytic enzymes to allow normal growth.
Finally, the altered homeostasis of PG hydrolysis in the ΔasnB mutant likely causes the observed increased sensitivity of the mutant cells to cell wall active antibiotics. Interestingly, mutants of the amidotransferase ltsA responsible for mDAP amidation in C. glutamicum also display higher susceptibility to β‐lactam antibiotics (Levefaudes et al., 2015). This suggests that amidation of mDAP could be used as a target for adjuvant antibiotic therapies aimed to increase the activity of existing antibiotics.
Experimental procedures
Genetics, strain construction and growth conditions
The strains used in this study are described in Supporting Information Table 3. Chromosomal DNA was prepared using the Promega Genomic DNA prep kit according to manufacturer's instructions. Standard genetics techniques were used for transformation (Marston and Errington, 1999). Selection of transformants was done on plates with or without 25 mM MgSO4 as indicated in the text. The strains were verified by PCR. The antibiotic concentrations used were as follows: chloramphenicol 5 μg/mL, erythromycin 1 μg/mL, kanamycin 10 μg/mL and spectinomycin 100 μg/mL. Growth curves were done in 96‐well plates ih Synergy microplate reader (Biotek) with continuous agitation at 37°C. For all experiments, overnight cultures were diluted at least 10 000 in fresh medium and allowed to grow under conditions indicated in the text to early exponential phase before microscopic observation.
Peptidoglycan purification
B. subtilis wild‐type and mutant strains were grown in 1 L cultures (in growth media indicated in the text) to OD600 = 0.5. Cultures were chilled in ice‐ethanol bath for 20 min before harvesting by centrifugation at 5000g for 15 min at 4°C. Cell pellets were resuspended in 40 ml of ice cold water and slowly added to 40 ml of boiling 8% SDS. Cells were boiled for 30 minutes and allowed to cool before centrifugation and repeated (∼15 times) washes with hot water to remove the SDS. The pellets were subsequently resuspended and incubated with 2 mg ml−1 of Pronase (Roche) in 40 mM Tris‐HCl (pH 7) at 60°C for 90 min. After centrifugation, the pellets were washed with water and incubated with trypsin (Sigma) at 200 μg ml−1 in 20 mM Tris‐HCl (pH 7.5) over night with agitation. Samples were boiled for 5 min and washed with water before treatment with DNAase (10 μg ml−1) and RNAase (50 μg ml−1) in 20 mM Tris‐HCl pH 7, 20 mM MgCl2, 10 mM CaCl2 for 2 hours at 37°C with agitation. SDS was added to the samples (1% final concentration) and the samples were incubated in boiling water for 15 min. Samples were washed 3 times with water and resuspended in 10 ml of 8 M LiCl, incubated for 15 min at 37°C and washed with water. The pellets were resuspended in 10 ml of 100 mM EDTA pH 7, incubated at 37°C for 15 min and washed with water. The pellets were resuspended in 10 ml acetone, sonicated for 5 min in ultrasonic bath, and washed with water three times. The samples were then resuspended in small volume of water and transferred to pre‐weighed 2.2 ml tubes whose caps were punctured with a hot needle. The samples were lyophilized overnight and the purified cell wall weighed. The cell walls were resuspended to a final volume of 20 mg ml−1 and stored at −20°C.
To remove the teichoic acids and associated polymers, 10 mg of purified cell walls were incubated with 2 ml of concentrated hydroflyoric acid (46% HF) for 48 hours at 4°C. After this incubation, 15 ml of 100 mM Tris‐HCl pH 7 was added to the sample and the samples were centrifuged and washed repeatedly with 100 mM Tris‐HCl pH7 until the sample pH was neutral. The pellets were then washed three times with water and the pure PG was lyophilized overnight and then resuspended in water to 10 mg ml−1.
Digestion of peptidoglycan with mutanolysin
To digest the purified peptidoglycan, 50 μl of pure peptidoglycan (10 mg ml−1) was incubated in 50 μl of 25 mM NaHPO4 pH 5.5 and 2 μl of mutanolysin (10 mg ml−1) under agitation at 37°C overnight. After this incubation, 100 μl of water was added and the sample was boiled for 5 min to inactivate the enzyme. The sample was centrifuged and the muropeptides found in the supernatant were reduced with NaBH4 (200 μl of muropeptides solution, 200 μl 0.5 M borate buffer pH 9, 25 μl fresh NaBH4 at 50 mg ml−1) for 20 min at room temperature in open eppendorf tubes with occasional agitation to remove the bubbles of H2. The reduction reaction was ended by adjusting the sample pH to approximately 4 with 85% ortho‐phosphoric acid.
UHPLC and mass spectrometry
Muropeptides resulting from mutanolysin digestion were separated by reverse phase ultrahigh‐performance liquid chromatography using a Zorbax RRHD Eclispse Plus column (C18 2.1 × 100 mm 1.8‐μm) at 50°C on Agilent UHPLC129 system. The buffers were at 10 mM ammonium phosphate, pH 5.6 (sodium azide at 180 μg L−1 was added to buffer A to avoid baseline drift at 202 nm). The methanol gradient (from 0% in buffer A to 20% in buffer B) was developed at a flow rate of 0.5 ml min−1. Muropeptides were analyzed directly by matrix‐assisted laser desorption ionization‐time of flight (MALDI‐TOF) mass spectrometry, as previously described (Courtin et al., 2006). The peaks identified by MALDI‐TOF were integrated in the Agilent software to estimate the amount of each muropeptide. The reported crosslinking index (%) was calculated as previously described (Glauner, 1988). The %dimers/(%monomers + 2× %dimers).
Labeling with TDL and microscopy
For TDL labeling, 100 µl of culture in early exponential phase was stained with 1 mM TDL (TAMRA Red D‐lysine) at 37°C for 5 minutes (PG insertion labeling experiments) or 20 min (PG degradation pulse‐chase experiments). Cells were rinsed twice in culture medium and 5 µl of washed cells deposited on 1% agarose pads poured in frames glued on microscope slides (Gene Frame from Thermo Scientific). The imaging was done on Zeiss Elyra PS1 using ZEN software. Cells were imaged in 3D‐SIM mode with a 63× objective, 0.12 µm z step (20 steps), 50 ms exposure time and 20% laser power (561 nm). Image reconstruction was performed in ZEN with automatic parameter setting.
Samples for phase contrast and epifluorescence microscopic observations were prepared as above and imaged by an inverted Nikon microscope (Ti‐E) using a 100× phase lens, or on Zeiss Elyra PS1 as above. For measurement of cell width and length, cells were incubated in the presence of FM4‐64 (1 μg ml−1). Cell width and length measurements were done in a custom program in MATLAB.
Microfluidics
Bacterial growth and pulse chase experiments using TDL dye were realized in the B04A Microfluidic Bacteria Plate (CellASIC ONIX, Millipore) using an Eclipse Ti‐E inverted microscope (Nikon). Experiments were carried out at 37°C. Pressure setting of 5 psi was used to inject the medium into the growth chamber. For growth experiment, phase images were taken every 20 seconds. For TDL pulse‐chase experiments, cells were grown in LB and stained for 20 min in LB + 1 mM TDL in the microfluidic device. Cells were then rinsed in fresh LB and images of phase and fluorescence were taken every 5 min.
Measurement of rate of autolysis
An overnight liquid culture of B. subtilis 168 grown in Difco Antibiotic Medium 3 (PAB) was diluted to an OD600 nm of 1 and subsequently diluted by ≥104 fold in PAB and incubated with continuous shaking at 37°C with OD600 nm measurements being taken every five minutes. Cultures were set up in duplicate and grown to various stages of growth, including early, mid and late exponential phase at which point MgSO4 was added to half of the cultures to a final concentration of 25 mM. Immediately after this addition autolysis was induced by adding sodium azide to all cultures at a final concentration of 75 mM. The OD600 nm was monitored to follow the rate of lysis. To determine the apparent growth rate µ just before adding the uncoupler, we selected the last 20 min (5 time points) of the log‐normalized growth curves, and applied a standard least‐squares linear fit. Similarly, we applied the same regression to the first hour (12 time points) of the log‐normalized lysis curves to obtain the lysis rates reported in Fig. 7.
Sample preparation for AFM
Overnight cultures of the indicated strains were diluted into fresh LB (containing or not 25 mM MgSO4) and grown at 37°C. When cells reached OD600 ∼ 0.4, 1 ml of culture was harvested and allowed to settle for 20 min on a piranha cleaned, cell‐tak modified (Corning), glass bottom Petri dish (WPI, Meyer et al., 2010). Unattached cells were washed away and the attached cells covered with culture medium for further analysis with AFM.
Atomic force microscopy
Imaging and force measurement were realized using a Nanowizard 3 Atomic Force Microscope (JPK Instruments AG, Berlin, Germany) coupled to a Zeiss inverted microscope. Topographic images and elasticity cartography were generated simultaneously using the Quantitative Imaging® (QI) mode with DNP cantilevers (0.06–0.24 N m−1, Bruker) in culture medium. The ramp size and speed were set at 400 µm and 25 µm s−1 respectively and the maximum load was 6 µN, maps of 128 by 128 force curves were realized. Calibration of the probe was carried out by measuring the deflection sensitivity on glass surface and spring constant was determined using the thermal tune method. Young moduli were calculated by fitting force curves with the Sneddon model using the JPK software.
Electron microscopy
For transmission electron microscopy (TEM), cells were harvested at the desired OD600 by centrifugation, washed two times with PBS and fixed in 2% glutaraldehyde and 0.1 M sodium cacodylate buffer pH 7.2, for 2 h at room temperature. Subsequent steps were performed mainly as described in (Rueff et al., 2014). TEM observations were done at the Microscopy and Imaging Platform MIMA2, INRA of Jouy‐en‐Josas, France.
Disk diffusion assays
Disk diffusion assays were performed as follows. Petri plates containing LB medium were covered with 5 ml of soft agar in which 200 μl of exponentially growing cells (OD600 = 0.2–0.4) of the strain being tested were resuspended. The plates were allowed to dry briefly in a laminar flow hood before 6 mm paper disks (Whatman) containing 25 μl of antibiotic solution were placed on the disks (cefuroxime 8 mg ml−1, ciprofloxacin 100 μg ml−1, D‐cycloserine 16 mg ml−1, CCCP 2.25 mg ml−1, fusidic acid, 12.8 μg ml−1, methicillin 12.5 mg ml−1, penicillin 8 mg ml−1, rifampicin 128 μg ml−1, tunicamycin 375 μg ml−1, vancomycin 0.5 μg ml−1). The plates were incubated at 37°C overnight and the zones of inhibition were measured by ImageJ after scanning the plates. Two disks were used per antibiotic and per plate. The values reported in Fig. 8 are an average of three independent experiments.
Funding information
This work was supported by a starting grant from the European Research Council to R.C.‐L. (ERC‐StG 311231). B. Tesson was supported by Marie Sklodowska Curie Individual Fellowship BACTOSHAPE 660935. P. Flores was supported by a doctoral fellowship from INRA.
Author contributions
A.D. conceived the study; A.D., B.T., S.C., P.C., R.K. performed the experiments; A.D., B.T., P.C., P.F. and M.C.‐C. analyzed the data; S.F., C.M., M.C.‐C. contributed equipment and training, A.D., B.T. and R.C.L. wrote the article.
Supporting information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Acknowledgements
We thank Yves Brun, Michael Van Nieuwenhze, and Erkin Kuru for the gift of TDL; Ken‐Ichi Yoshida for the gifts of strains FU340, FU341 and FU342. We thank Christine Longin from the Microscopy and Imaging Platform MIMA2 (INRA, France) for help with TEM observations. MS data were obtained with the instruments located at the INRA Platform for Proteomics Analysis of Paris South West (PAPPSO) and the Mass Spectrometry Laboratory, Analytical Services Unit, Instituto de Tecnologia Química e Biológica, Universidade Nova de Lisboa. The authors declare no conflict of interest.
Contributor Information
Alex Dajkovic, Email: alexdajkovic@gmail.com.
Rut Carballido‐Lopez, Email: rut.carballido-lopez@inra.fr.
References
- Asong, J. , Wolfert, M.A. , Maiti, K.K. , Miller, D. , and Boons, G.J. (2009) Binding and cellular activation studies reveal that toll‐like receptor 2 can differentially recognize peptidoglycan from gram‐positive and gram‐negative bacteria. J Biol Chem 284: 8643–8653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atrih, A. , Bacher, G. , Allmaier, G. , Williamson, M.P. , and Foster, S.J. (1999) Analysis of peptidoglycan structure from vegetative cells of Bacillus subtilis 168 and role of PBP 5 in peptidoglycan maturation. J Bacteriol 181: 3956–3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard, E. , Rolain, T. , Courtin, P. , Guillot, A. , Langella, P. , Hols, P. , and Chapot‐Chartier, M.P. (2011a) Characterization of O‐acetylation of N‐acetylglucosamine: a novel structural variation of bacterial peptidoglycan. J Biol Chem 286: 23950–23958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard, E. , Rolain, T. , Courtin, P. , Hols, P. , and Chapot‐Chartier, M.P. (2011b) Identification of the amidotransferase AsnB1 as being responsible for meso‐diaminopimelic acid amidation in Lactobacillus plantarum peptidoglycan. J Bacteriol 193: 6323–6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge, T.J. , and Murray, R.G. (1980) Sites of metal deposition in the cell wall of Bacillus subtilis. J Bacteriol 141: 876–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, S. , Santa Maria, J.P., Jr. , and Walker, S. (2013) Wall teichoic acids of gram‐positive bacteria. Annu Rev Microbiol 67: 313–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chastanet, A. , and Carballido‐Lopez, R. (2012) The actin‐like MreB proteins in Bacillus subtilis: a new turn. Front Biosci (Schol Ed) 4: 1582–1606. [DOI] [PubMed] [Google Scholar]
- Courtin, P. , Miranda, G. , Guillot, A. , Wessner, F. , Mezange, C. , Domakova, E. , Kulakauskas, S. , and Chapot‐Chartier, M.P. (2006) Peptidoglycan structure analysis of Lactococcus lactis reveals the presence of an L, D‐carboxypeptidase involved in peptidoglycan maturation. J Bacteriol 188: 5293–5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, K.M. , and Weiser, J.N. (2011) Modifications to the peptidoglycan backbone help bacteria to establish infection. Infect Immun 79: 562–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez‐Escobar, J. , Chastanet, A. , Crevenna, A.H. , Fromion, V. , Wedlich‐Soldner, R. , and Carballido‐Lopez, R. (2011) Processive movement of MreB‐associated cell wall biosynthetic complexes in bacteria. Science 333: 225–228. [DOI] [PubMed] [Google Scholar]
- Eun, Y.J. , Kapoor, M. , Hussain, S. , and Garner, E.C. (2015) Bacterial filament systems: toward understanding their emergent behavior and cellular functions. J Biol Chem 290: 17181–17189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo, T.A. , Sobral, R.G. , Ludovice, A.M. , Almeida, J.M. , Bui, N.K. , Vollmer, W. , de Lencastre, H. , and Tomasz, A. (2012) Identification of genetic determinants and enzymes involved with the amidation of glutamic acid residues in the peptidoglycan of Staphylococcus aureus. PLoS Pathog 8: e1002508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formstone, A. , and Errington, J. (2005) A magnesium‐dependent mreB null mutant: implications for the role of mreB in Bacillus subtilis. Mol Microbiol 55: 1646–1657. [DOI] [PubMed] [Google Scholar]
- Garner, E.C. , Bernard, R. , Wang, W. , Zhuang, X. , Rudner, D.Z. , and Mitchison, T. (2011) Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis. Science 333: 222–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardin, S.E. , Travassos, L.H. , Herve, M. , Blanot, D. , Boneca, I.G. , Philpott, D.J. , Sansonetti, P.J. , and Mengin‐Lecreulx, D. (2003) Peptidoglycan molecular requirements allowing detection by Nod1 and Nod2. J Biol Chem 278: 41702–41708. [DOI] [PubMed] [Google Scholar]
- Glauner, B. (1988) Separation and quantification of muropeptides with high‐performance liquid chromatography. Anal Biochem 172: 451–464. [DOI] [PubMed] [Google Scholar]
- Goodell, E.W. , Lopez, R. , and Tomasz, A. (1976) Suppression of lytic effect of beta lactams on Escherichia coli and other bacteria. Proc Natl Acad Sci U S A 73: 3293–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gschwender, H.H. , and Hofschneider, P.H. (1969) Lysis inhibition of phi‐X174‐, M12‐, and Q‐beta‐infected Escherichia coli bacteria by magnesium ions. Biochim Biophys Acta 190: 454–459. [DOI] [PubMed] [Google Scholar]
- Heckels, J.E. , Lambert, P.A. , and Baddiley, J. (1977) Binding of magnesium ions to cell walls of Bacillus subtilis W23 containing teichoic acid or teichuronic acid. Biochem J 162: 359–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirasawa, T. , Wachi, M. , and Nagai, K. (2000) A mutation in the Corynebacterium glutamicum ltsA gene causes susceptibility to lysozyme, temperature‐sensitive growth, and L‐glutamate production. J Bacteriol 182: 2696–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, X. , Holden, H.M. , and Raushel, F.M. (2001) Channeling of substrates and intermediates in enzyme‐catalyzed reactions. Annu Rev Biochem 70: 149–180. [DOI] [PubMed] [Google Scholar]
- Jolliffe, L.K. , Doyle, R.J. , and Streips, U.N. (1981) The energized membrane and cellular autolysis in Bacillus subtilis. Cell 25: 753–763. [DOI] [PubMed] [Google Scholar]
- Kern, T. , Giffard, M. , Hediger, S. , Amoroso, A. , Giustini, C. , Bui, N.K. , Joris, B. , Bougault, C. , Vollmer, W. , and Simorre, J.P. (2010) Dynamics characterization of fully hydrated bacterial cell walls by solid‐state NMR: evidence for cooperative binding of metal ions. J Am Chem Soc 132: 10911–10919. [DOI] [PubMed] [Google Scholar]
- Koch, A.L. , and Doyle, R.J. (1985) Inside‐to‐outside growth and turnover of the wall of gram‐positive rods. J Theor Biol 117: 137–157. [DOI] [PubMed] [Google Scholar]
- Koch, A.L. , Higgins, M.L. , and Doyle, R.J. (1981) Surface tension‐like forces determine bacterial shapes: Streptococcus faecium. J Gen Microbiol 123: 151–161. [DOI] [PubMed] [Google Scholar]
- Kuru, E. , Hughes, H.V. , Brown, P.J. , Hall, E. , Tekkam, S. , Cava, F. , de Pedro, M.A. , Brun, Y.V. , and VanNieuwenhze, M.S. (2012) In Situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent D‐amino acids. Angew Chem Int Ed Engl 51: 12519–12523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuru, E. , Tekkam, S. , Hall, E. , Brun, Y.V. , and Van Nieuwenhze, M.S. (2015) Synthesis of fluorescent D‐amino acids and their use for probing peptidoglycan synthesis and bacterial growth in situ. Nat Protoc 10: 33–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamsa, A. , Liu, W.T. , Dorrestein, P.C. , and Pogliano, K. (2012) The Bacillus subtilis cannibalism toxin SDP collapses the proton motive force and induces autolysis. Mol Microbiol 84: 486–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leduc, M. , Kasra, R. , and van Heijenoort, J. (1982) Induction and control of the autolytic system of Escherichia coli. J Bacteriol 152: 26–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levefaudes, M. , Patin, D. , de Sousa‐D'auria, C. , Chami, M. , Blanot, D. , Herve, M. , Arthur, M. , Houssin, C. , and Mengin‐Lecreulx, D. (2015) Diaminopimelic Acid Amidation in Corynebacteriales: new insights into the role of ltsa in peptidoglycan modification. J Biol Chem 290: 13079–13094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, Y. , and Helmann, J.D. (2012) Analysis of the role of Bacillus subtilis sigma(M) in beta‐lactam resistance reveals an essential role for c‐di‐AMP in peptidoglycan homeostasis. Mol Microbiol 83: 623–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahapatra, S. , Yagi, T. , Belisle, J.T. , Espinosa, B.J. , Hill, P.J. , McNeil, M.R. , Brennan, P.J. , and Crick, D.C. (2005) Mycobacterial lipid II is composed of a complex mixture of modified muramyl and peptide moieties linked to decaprenyl phosphate. J Bacteriol 187: 2747–2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston, A.L. , and Errington, J. (1999) Selection of the midcell division site in Bacillus subtilis through MinD‐dependent polar localization and activation of MinC. Mol Microbiol 33(1): 84–96. [DOI] [PubMed] [Google Scholar]
- Massiere, F. , and Badet‐Denisot, M.A. (1998) The mechanism of glutamine‐dependent amidotransferases. Cell Mol Life Sci 54: 205–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisner, J. , Montero Llopis, P. , Sham, L.T. , Garner, E. , Bernhardt, T.G. , and Rudner, D.Z. (2013) FtsEX is required for CwlO peptidoglycan hydrolase activity during cell wall elongation in Bacillus subtilis. Mol Microbiol 89: 1069–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesnage, S. , Dellarole, M. , Baxter, N.J. , Rouget, J.B. , Dimitrov, J.D. , Wang, N. , Fujimoto, Y. , Hounslow, A.M. , Lacroix‐Desmazes, S. , Fukase, K. , Foster, S.J. , and Williamson, M.P. (2014) Molecular basis for bacterial peptidoglycan recognition by LysM domains. Nat Commun 5: 4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, P. , Gutierrez, J. , Pogliano, K. , and Dworkin, J . (2010) Cell wall synthesis is necessary for membrane dynamics during sporulation of Bacillus subtilis . Mol Microbiol 76(4): 956–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munch, D. , Roemer, T. , Lee, S. , Engeser, H.M. , Sahl, H.G. , and Schneider, T. (2012) Identification and in vitro analysis of the GatD/MurT enzyme‐complex catalyzing lipid II amidation in Staphylococcus aureus. PLoS Pathog 8: e1002509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray, T. , Popham, D.L. , and Setlow, P. (1998) Bacillus subtilis cells lacking penicillin‐binding protein 1 require increased levels of divalent cations for growth. J Bacteriol 180: 4555–4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhaus, F.C. , and Baddiley, J. (2003) A continuum of anionic charge: structures and functions of D‐alanyl‐teichoic acids in gram‐positive bacteria. Microbiol Mol Biol Rev 67: 686–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas, P. , Mader, U. , Dervyn, E. , Rochat, T. , Leduc, A. , Pigeonneau, N. , et al. (2012) Condition‐dependent transcriptome reveals high‐level regulatory architecture in Bacillus subtilis. Science 335: 1103–1106. [DOI] [PubMed] [Google Scholar]
- Raushel, F.M. , Thoden, J.B. , and Holden, H.M. (1999) The amidotransferase family of enzymes: molecular machines for the production and delivery of ammonia. Biochemistry 38: 7891–7899. [DOI] [PubMed] [Google Scholar]
- Rayman, M.K. , and MacLeod, R.A. (1975) Interaction of Mg‐2+ with peptidoglycan and its relation to the prevention of lysis of a marine pseudomonad. J Bacteriol 122: 650–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, H.J. , Thurman, P.F. , and Burdett, I.D. (1983) The bactericidal action of beta‐lactam antibiotics on an autolysin‐deficient strain of Bacillus subtilis. J Gen Microbiol 129: 465–478. [DOI] [PubMed] [Google Scholar]
- Rueff, A.S. , Chastanet, A. , Dominguez‐Escobar, J. , Yao, Z. , Yates, J. , Prejean, M.V. , et al. (2014) An early cytoplasmic step of peptidoglycan synthesis is associated to MreB in Bacillus subtilis. Mol Microbiol 91: 348–362. [DOI] [PubMed] [Google Scholar]
- Salzberg, L.I. , Powell, L. , Hokamp, K. , Botella, E. , Noone, D. , and Devine, K.M. (2013) The WalRK (YycFG) and sigma(I) RsgI regulators cooperate to control CwlO and LytE expression in exponentially growing and stressed Bacillus subtilis cells. Mol Microbiol 87: 180–195. [DOI] [PubMed] [Google Scholar]
- Schleifer, K.H. , and Kandler, O. (1972) Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev 36: 407–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, T.J. , Blackman, S.A. , and Foster, S.J. (2000) Autolysins of Bacillus subtilis: multiple enzymes with multiple functions. Microbiology 146: 249–262. [DOI] [PubMed] [Google Scholar]
- Takahashi, H. , Ayala, I. , Bardet, M. , De Paepe, G. , Simorre, J.P. , and Hediger, S. (2013) Solid‐state NMR on bacterial cells: selective cell wall signal enhancement and resolution improvement using dynamic nuclear polarization. J Am Chem Soc 135: 5105–5110. [DOI] [PubMed] [Google Scholar]
- Tomasz, A. , Albino, A. , and Zanati, E. (1970) Multiple antibiotic resistance in a bacterium with suppressed autolytic system. Nature 227: 138–140. [DOI] [PubMed] [Google Scholar]
- Turner, R.D. , Vollmer, W. , and Foster, S.J. (2014) Different walls for rods and balls: the diversity of peptidoglycan. Mol Microbiol 91: 862–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer, W. , Blanot, D. , and de Pedro, M.A. (2008a) Peptidoglycan structure and architecture. FEMS Microbiol Rev 32: 149–167. [DOI] [PubMed] [Google Scholar]
- Vollmer, W. , Joris, B. , Charlier, P. , and Foster, S. (2008b) Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol Rev 32: 259–286. [DOI] [PubMed] [Google Scholar]
- Warth, A.D. , and Strominger, J.L. (1971) Structure of the peptidoglycan from vegetative cell walls of Bacillus subtilis. Biochemistry 10: 4349–4358. [DOI] [PubMed] [Google Scholar]
- Yamamoto, H. , Miyake, Y. , Hisaoka, M. , Kurosawa, S. , and Sekiguchi, J. (2008) The major and minor wall teichoic acids prevent the sidewall localization of vegetative DL‐endopeptidase LytF in Bacillus subtilis. Mol Microbiol 70: 297–310. [DOI] [PubMed] [Google Scholar]
- Yoshida, K. , Fujita, Y. , and Ehrlich, S.D. (1999) Three asparagine synthetase genes of Bacillus subtilis. J Bacteriol 181: 6081–6091. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Supporting Information
Supporting Information
Supporting Information


