Abstract
The hypoxia-regulated tumor-suppressor von Hippel-Lindau (VHL) is an E3 ligase that recognizes its substrates as part of an oxygen-dependent prolyl hydroxylase (PHD) reaction, with hypoxia-inducible factor α (HIFα) being its most notable substrate. Here we report that VHL has an equally important function distinct from its hypoxia-regulated activity. We find that Aurora kinase A (AURKA) is a novel, hypoxia-independent target for VHL ubiquitination. In contrast to its hypoxia-regulated activity, VHL mono-, rather than poly-ubiquitinates AURKA, in a PHD-independent reaction targeting AURKA for degradation in quiescent cells, where degradation of AURKA is required to maintain the primary cilium. Tumor-associated variants of VHL differentiate between these two functions, as a pathogenic VHL mutant that retains intrinsic ability to ubiquitinate HIFα is unable to ubiquitinate AURKA. Together, these data identify VHL as an E3 ligase with important cellular functions under both normoxic and hypoxic conditions.
Introduction
von Hippel-Lindau (VHL) disease is an autosomal dominant inherited cancer syndrome with a high incidence of clear cell renal cell carcinoma (RCC).1 Mutations in the VHL gene are associated with familial disease, and >90% of sporadic cases exhibit biallelic inactivation of this tumor-suppressor gene.2, 3 The protein encoded by the VHL gene serves as the substrate receptor of the Cullin 2-RING-ligase complex. pVHL binds the adaptor proteins Elongin B and Elongin C (Elo B/C) to form a complex with the N-terminus of the scaffold protein Cullin 2, which additionally supports binding of ring finger protein 2 (Rbx2) at its C-terminus to form the VHL-ElonginC-ElonginB complex.4, 5 VHL substrates are recognized when hydroxylated by prolyl hydroxylases (PHDs) at specific proline residues within their oxygen-dependent degradation domains.6, 7, 8, 9, 10
The PHD-family of PHDs, like other dioxygenases, requires oxygen to hydroxylate their targets.11 The best-known VHL substrates are the hypoxia-inducible factors 1 and 2 α (HIF1α and HIF2α), which are hydroxylated at proline residues by PHD proteins under normoxic conditions. PHD-mediated proline hydroxylation target HIF1α and HIF2α for VHL-mediated ubiquitination and proteasome-mediated degradation,7 inhibiting transcription of HIFα targets that increase glucose uptake and angiogenesis.12 Under hypoxic conditions, PHD-mediated hydroxylation and VHL-mediated ubiquitination are inhibited, resulting in HIFα stabilization. Other oxygen-dependent VHL substrates have been identified including epidermal growth factor receptor,13 atypical protein kinase C,14 Sprouty2,6 β-adrenergic receptor II,15 myb-binding protein 160,16 and RNA polymerase subunits Rpb19, 10 and Rpb7.17 More recently, the differential and often compromised capability of pathogenic VHL mutants to stabilize microtubules has highlighted an additional function thought to contribute to VHL’s function as a tumor suppressor.18, 19 However, the exact mechanism(s) by which VHL regulates microtubules is incompletely understood.
We report here that VHL has an equally important function in cells that is hypoxia independent. We find VHL directly ubiquitinates Aurora kinase A (AURKA) independent of oxygen-dependent PHD activity to regulate formation of the primary cilium in quiescent cells. Furthermore, in contrast to other known targets, VHL multi-monoubiquitinates AURKA to target this kinase for proteasome-mediated degradation. These data establish a new tumor-suppressor activity for the VHL E3 ligase distinct from polyubiquitination of its oxygen-dependent targets such as HIFα, and points to AURKA as a potential target for therapy in VHL-deficient tumors.
Results
VHL modulates AURKA protein levels
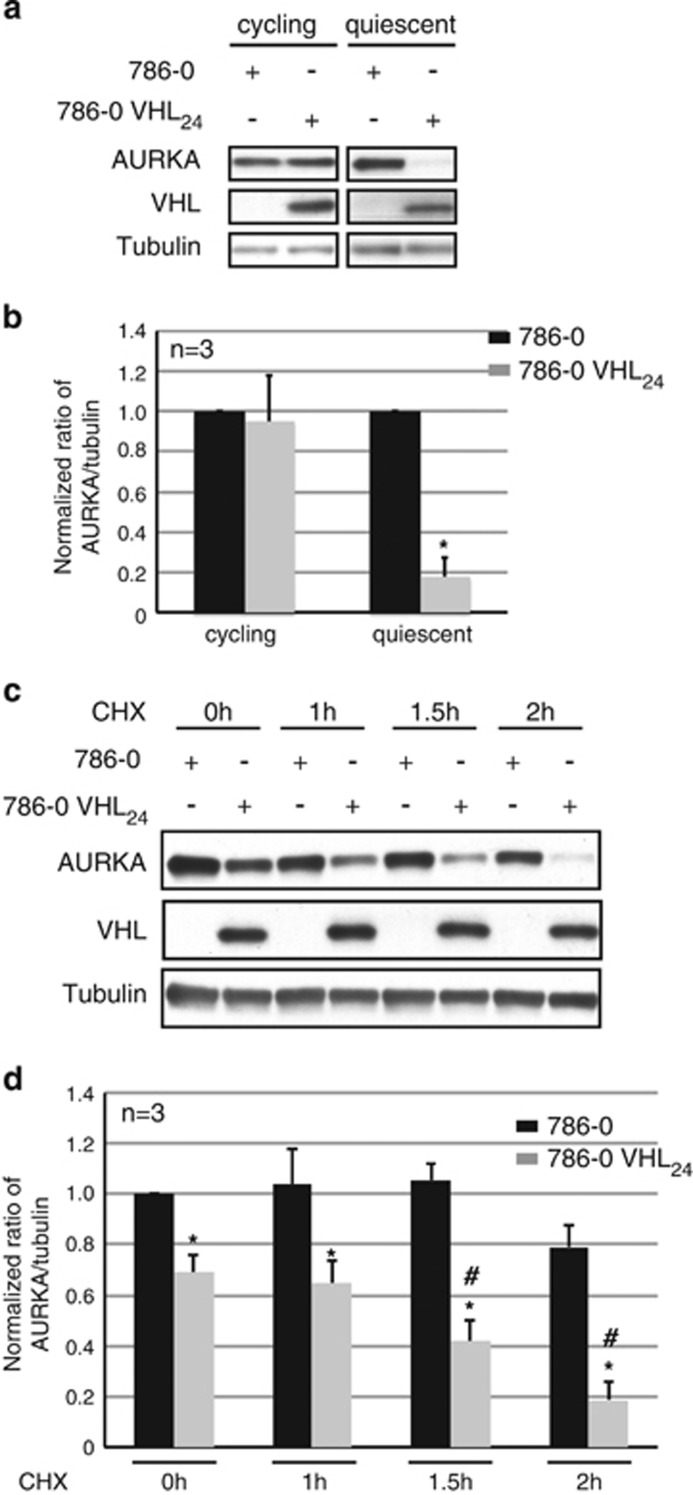
In VHL-null RCC 786-0 cells, re-expression of VHL significantly decreased AURKA in quiescent cells, whereas in cycling 786-0 cells re-expressing VHL AURKA levels remained unchanged (Figures 1a and b). A similar decrease in non-mitotic AURKA was observed in quiescent cultures of A-498 (VHL-deficient RCC cells) re-expressing the two known isoforms of VHL; VHL24, and VHL19 (Supplementary Figures 1a and 1b). In VHL-proficient human telomerase reverse transcriptase (hTERT) immortalized retinal pigmented epithelial (RPE-1) cells, overexpression of exogenous VHL resulted in a dose-responsive decrease in AURKA protein levels in quiescent cells (Supplementary Figure 1c).
Figure 1.
VHL modulates AURKA protein levels. (a) Lysates from 786-0 to isogenic 786-0 cells overexpressing VHL (VHL24) cultured at sub-confluent (cycling) or confluent and serum starved conditions (48 h) probed as indicated. (b) Densitometric quantitation of the average ratio of AURKA to tubulin expression from 786-0 (black bars) to 786-0 VHL24 (gray bars) cells. *P<0.01. (c) Lysates harvested from 786-0 to 786-0 VHL24 cells cultured to confluence and serum starved for 24 h before treatment with CHX for the indicated time points probed as shown. (d) Densitometric quantitation showing an average ratio of AURKA to tubulin in 786-0 (black bars) and 786-0 VHL24 (gray bars) cells treated with CHX. *P<0.01. # Denotes significant differences exclusively in the 786-0 VHL24 cell line at the each of the indicated CHX treatment time points compared with the 0- h time point (P<0.05). Error bars denote s.e.m.
Demonstrating VHL-regulated AURKA at the protein level, treatment with the protein synthesis inhibitor cycloheximide (CHX) following serum starvation for 24 h led to a marked decrease in AURKA in VHL-proficient cells over a 2- h time course (Figure 1c). Quantitation of AURKA levels in VHL-deficient 786-0 cells and isogenic VHL-proficient 786-0 cells treated with CHX revealed a significant decrease in AURKA in 786-0 expressing VHL with time following CHX treatment, whereas AURKA abundance was unchanged by CHX treatment in VHL-deficient cells (Figure 1d).
AURKA is a target for the VHL E3 ligase
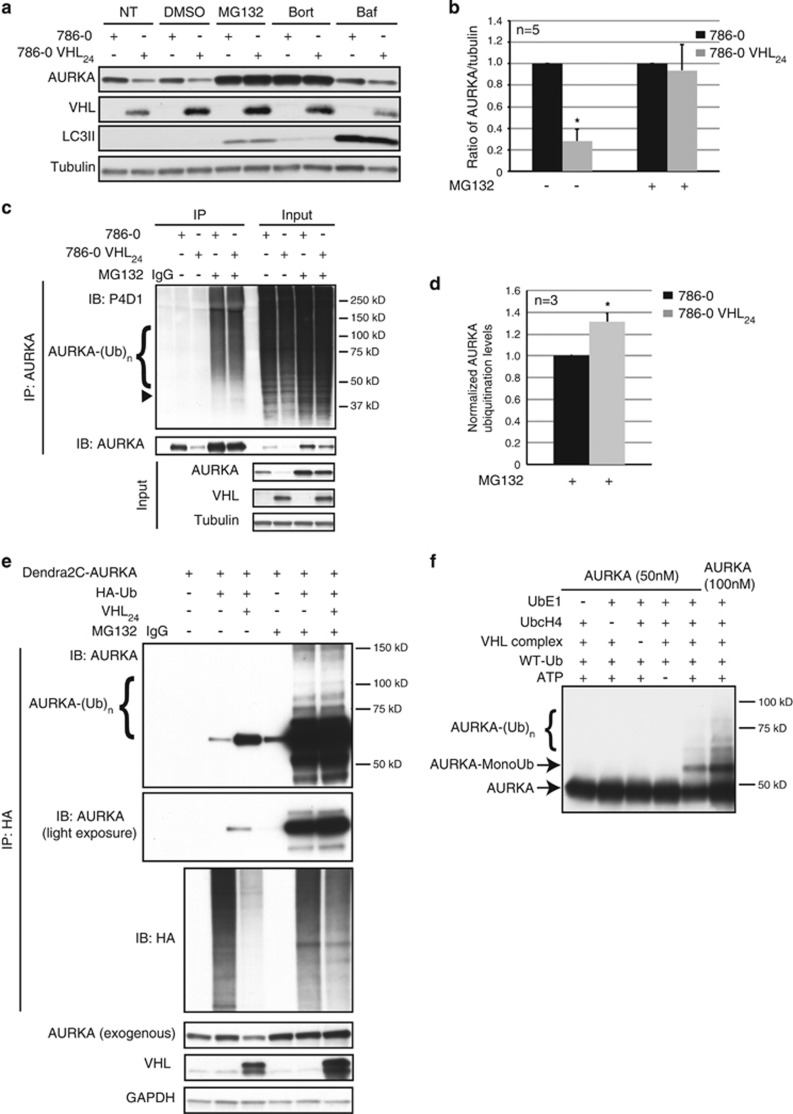
In quiescent 786-0 and VHL-proficient 786-0 cells treated with proteasome inhibitors MG132 or Bortezomib, the decrease in AURKA levels observed upon re-expression of VHL was reversed by proteasome inhibition (Figures 2a and b). The 80% decrease in AURKA protein seen with reintroduction of VHL in 786-0 cells was rescued by MG132, accompanied by stabilization of ubiquitinated AURKA (Figures 2b and c). Endogenous AURKA ubiquitination was evaluated using denaturing immunoprecipitation, which disrupts ternary complexes, but ensures intact retrieval of ubiquitinated AURKA. Accounting for the decrease in AURKA in cells re-expressing VHL, we normalized for the amount of AURKA in 786-0-deficient and -proficient cells, and quantitated pull down efficiencies, which revealed increased ubiquitination of AURKA in cells re-expressing VHL (Figure 2d). Similarly, exogenously expressed AURKA (Dendra2C-AURKA) showed increased ubiquitination when overexpressed with VHL24 in RPE-1 (hTERT RPE-1) cells (Figure 2e). Treatment with Bafilomycin A, an inhibitor of lysosomal degradation, failed to increase AURKA, precluding lysosome-mediated AURKA degradation by VHL (Figure 2a).
Figure 2.
VHL E3 ligase directly ubiquitinates AURKA. (a) 786-0 and isogenic 786-0 cells overexpressing VHL (VHL24) treated with DMSO (vehicle), proteasome inhibitors (MG132 and Bort (Bortezomib)) or lysosome inhibitor (Baf (Bafilomycin A)). Immunoblots probed for the indicated antibodies. (b) Densitometric quantitation from 786-0 (black bars) to 786-0 VHL24 (gray bars) cells showing an averaged ratio of AURKA to tubulin in the absence and presence of MG132. *P<0.00001. (c) In vivo ubiquitination assay using lysates from 786-0 to 786-0 VHL24 cell lines in the absence or presence of MG132. Immunoprecipitated AURKA probed using an anti-ubiquitin (P4D1) antibody, and pull down efficiency measured with an anti-AURKA antibody. Arrowhead indicates the expected molecular weight of monoubiquitinated AURKA. Input shows AURKA, VHL and tubulin. (d) Graphical representation of densitometric quantitation (measured from 50 to 200 kD) showing an average ratio of ubiquitinated AURKA (probed using the P4D1 anti-ubiquitin antibody) to the pull down efficiency (using an anti-AURKA antibody). Black and gray bars represent the ratio in 786-0 and 786-0 VHL24 cells, respectively. *P<0.01. (e) In vivo ubiquitination assay performed using hTERT RPE-1 cells overexpressing Dendra2C-AURKA and HA-Ub with and without overexpressed VHL24 in the absence and presence of MG132. Ubiquitinated AURKA immunoprecipitated under denaturing conditions using an anti-HA antibody and probed with an anti-AURKA antibody. Input lysates immunoblotted for the indicated antibodies. (f) In vitro ubiquitination assay. All error bars denote s.e.m.
These in vivo data were supported by in vitro data demonstrating VHL could directly ubiquitinate AURKA. VHL was recently reported to be phosphorylated by AURKA,20 and we confirmed a VHL–AURKA interaction using co-immunoprecipitation assays (Supplementary Figure 2). In vitro ubiquitination assays using recombinant human AURKA as substrate revealed that an active recombinant VHL-ElonginC-ElonginB complex containing VHL directly ubiquitinated AURKA (Figure 2f). Interestingly, we observed a distinct monoubiquitinated AURKA band, as well as ubiquitinated forms of AURKA with slower mobility (Figure 2f), consistent with either polyubiquitination or multi-monoubiquitination of AURKA by the VHL E3 ligase.
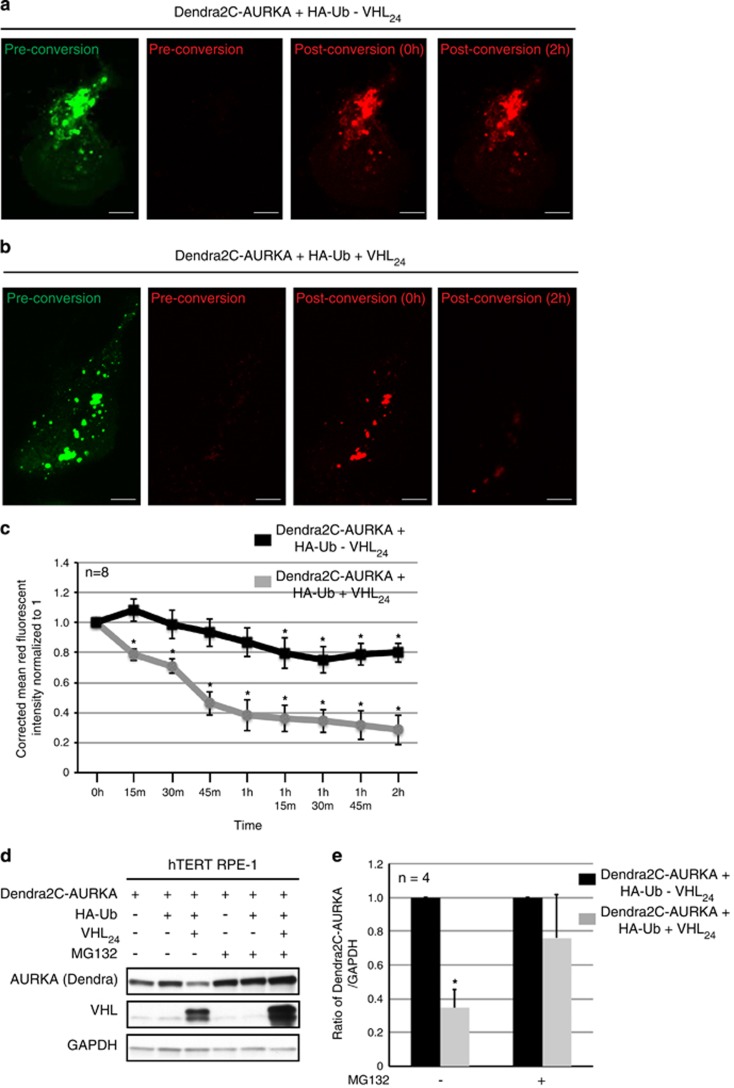
To translate biochemical data to live cells, we used a photoswitchable AURKA (Dendra2C-AURKA) to visualize AURKA protein abundance and half-life. Confluent hTERT RPE-1 cells expressing Dendra2C-AURKA and HA-Ub in the presence or absence of overexpressed VHL were imaged before, immediately after photoconversion, and every 15 min thereafter for 2 h (Figures 3a and b). As shown in Figure 3c, the half-life of Dendra2C-AURKA was reduced markedly from over 2 h (AURKA levels did not reach 50% of the original levels in 2 h) in the absence of exogenous VHL to 45 min in cells overexpressing VHL (Figure 3c) that correlated with a significant increase in Dendra2C-AURKA ubiquitination (Figure 2e), and an overall decrease in Dendra2C-AURKA levels in cells overexpressing VHL that could be rescued with MG132 (Figures 3d and e).
Figure 3.
VHL regulates AURKA protein half-life and abundance. (a, b) Representative images of hTERT RPE-1 cells transfected with Dendra2C-AURKA, and HA-Ub without (a) or with (b) overexpressed VHL (VHL24) before photoconversion (green and red channels), immediately after photoconversion (0 h), and 2 h post conversion. Scale bar, 10 μM (c) Quantitation of the corrected mean red fluorescent intensity of Dendra2C-AURKA measured every 15 min from cells expressing Dendra2C-AURKA and HA-Ub with (gray line), and without (black line) overexpressed VHL24 (averaged from eight independent biological and technical replicates). *P<0.02 (compared with the 0-h time point). (d) Lysates from hTERT RPE-1 cells expressing Dendra2C-AURKA and HA-Ub in the absence or presence of VHL24 probed for the indicated antibodies. (e) Densitometric quantitation showing an average ratio of Dendra2C-AURKA to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) in cells without (black bars) and with (gray bars) overexpressed VHL24. *P<0.01. Error bars denote s.e.m.
VHL ubiquitination of AURKA is hypoxia independent
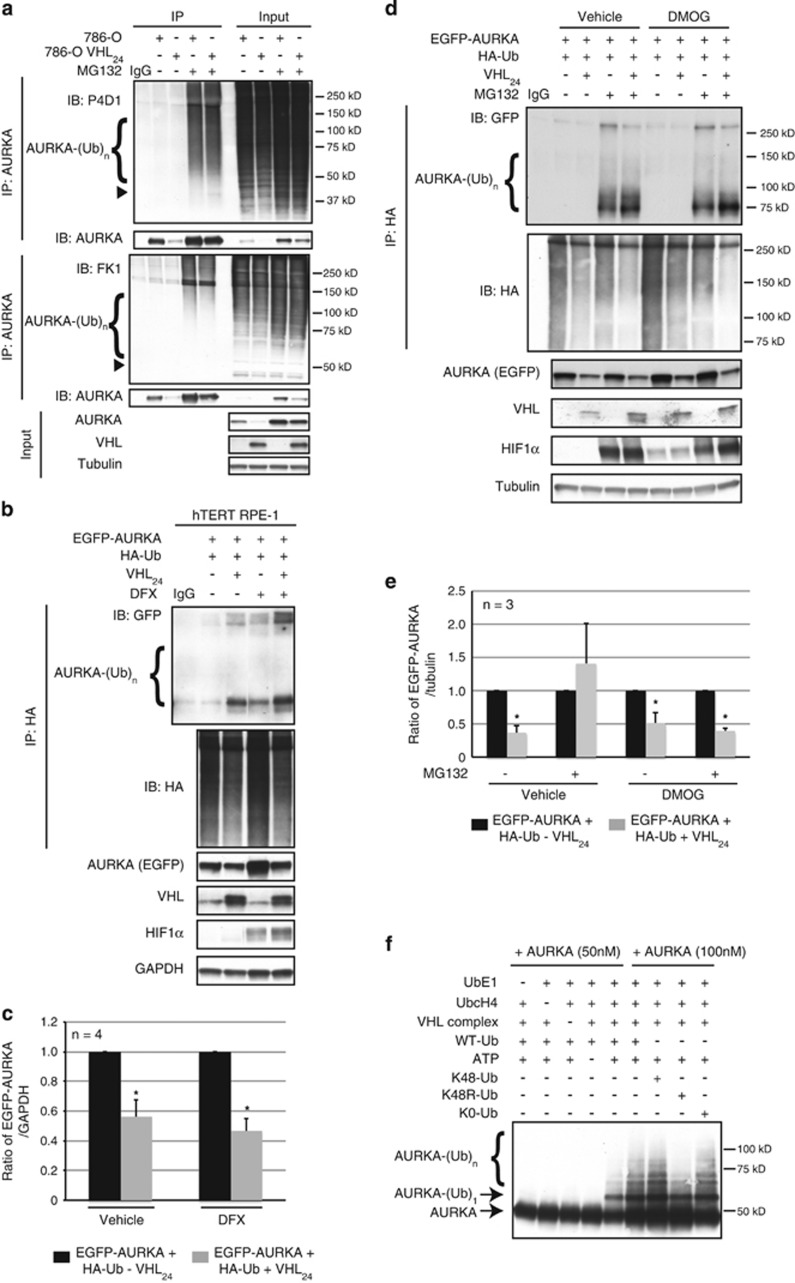
In response to proline hydroxylation by PHDs, VHL is well documented to polyubiquitinate its substrates.21, 22, 23, 24 However, we found that endogenous AURKA immunoprecipitated from quiescent cells was recognized by the P4D1 anti-ubiquitin antibody, which detects both mono- and polyubiquitination, but not the FK1 anti-ubiquitin antibody, which exclusively recognizes only polyubiquitination (Figure 4a). This suggested that VHL was multi-monoubiquitinating AURKA rather than polyubiquitinating this kinase.
Figure 4.
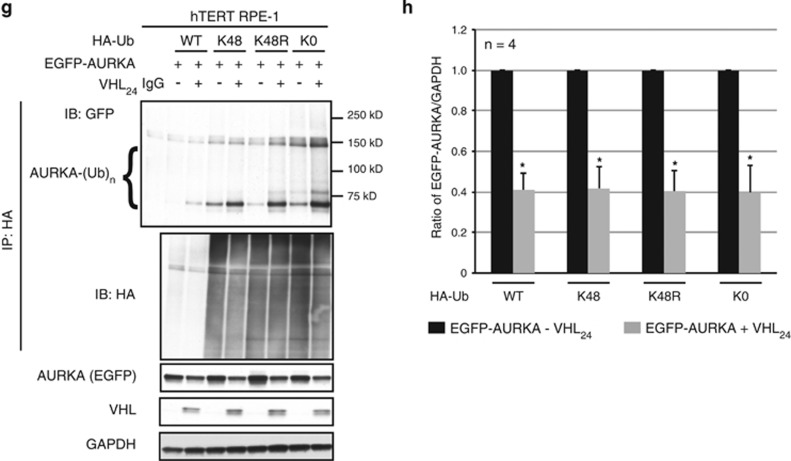
VHL multi-monoubiquitinates AURKA independent of prolyl hydroxylation. (a) In vivo ubiquitination assay showing ubiquitinated AURKA from 786-0 to isogenic 786-0 cells overexpressing VHL (VHL24) probed using anti-P4D1 (recognizes both mono and polyubiquitination) or FK1 (exclusively recognizes polyubiquitination) antibodies. Arrowheads indicate the expected molecular weight of monoubiquitinated AURKA. Input lysates probed with AURKA, VHL and tubulin. (b) In vivo ubiquitination assay showing ubiquitinated AURKA from cells overexpressing EGFP-AURKA and HA-Ub with and without overexpressed VHL24, treated with defroxamine (DFX). Input lysates immunoblotted with the indicated antibodies. (c) Densitometric quantitation showing an average ratio of EGFP-AURKA to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) without (black bars) and with (gray bars) overexpressed VHL24 treated with DFX. *P<0.05. All error bars denote s.e.m. (d) In vivo ubiquitination assay showing ubiquitinated AURKA from cells overexpressing EGFP-AURKA and HA-Ub with and without overexpressed VHL24, treated with dimethyloxaloylglycine (DMOG). Input lysates immunoblotted with the indicated antibodies. (e) Densitometric quantitation showing an average ratio of EGFP-AURKA to tubulin without (black bars) and with (gray bars) overexpressed VHL24 treated with DMOG. *P<0.05. All error bars denote s.e.m. (f) In vitro ubiquitination assay performed with either WT or mutant recombinant ubiquitin. (g) In vivo ubiquitination assay showing ubiquitinated AURKA from cells overexpressing EGFP-AURKA and WT or mutant ubiquitin in the absence and presence of VHL24. (h) Densitometric quantitation showing an average ratio of EGFP-AURKA to GAPDH in cells without (black bars) and with (gray bars) overexpressed VHL24 in the presence of WT or mutant ubiquitin. *P<0.01.
To determine if hydroxylation of AURKA was essential for recognition and ubiquitination by VHL, we performed in vivo denaturing ubiquitination assays to assess AURKA ubiquitination after PHD inhibition. In contrast to the hypoxia-regulated target HIF1α, the hypoxia mimetic deferoxamine (DFX) failed to rescue AURKA levels or ubiquitination (Figures 4b and c), although AURKA levels were found to be generally higher in cells treated with DFX. This increase in AURKA expression could in part be driven by activation of HIFα and regulation of β-catenin-driven transcription of AURKA as previously reported.25, 26 Treatment with the PHD inhibitor dimethyloxaloylglycine similarly failed to block ubiquitination of AURKA or rescue AURKA protein levels (Figures 4d and e).
To establish that AURKA was multi-monoubiquitinated by VHL, we performed in vitro ubiquitination assays using either K48 (lysine most commonly associated with polyubiquitination and proteasome-mediated degradation) or K0 (lysine defective in poly-chain formation allowing the addition of a single (monoubiquitination) or multiple single (multi-monoubiquitination) ubiquitin moieties) ubiquitin mutants. Comparable AURKA ubiquitination occurred with wild-type (WT) ubiquitin, and the K48 (all lysine’s except K48 mutated to arginine), K48R (only lysine 48 mutated to arginine) and K0 (all lysine’s mutated to arginine) ubiquitin mutants (Figure 4f), indicating multi-monoubiquitination of AURKA by VHL. Multi-monoubiquitination was also seen with immunoprecipitation of ubiquitinated AURKA in hTERT RPE-1 cells expressing enhanced green fluorescent protein (EGFP) tagged-AURKA and WT (HA-Ub) or ubiquitin mutants (HA-K48, HA-K48R or HA-K0). No significant changes in AURKA ubiquitination were seen once normalized to pull down efficiencies between the ubiquitin mutants (K48, K48R and K0) and WT ubiquitin (Figure 4g), confirming AURKA multi-monoubiquitination, rather than polyubiquitination. Importantly, multi-monoubiquitination of AURKA (established using K48, K48R and K0 ubiquitin mutants) resulted in the same decrease in AURKA abundance seen with WT ubiquitin (Figures 4g and h). Together, these data show that VHL recognition of AURKA occurs independent of prolyl hydroxylation, and results in multi-monoubiquitination (rather than polyubiqitination) of AURKA.
Pathogenic VHL mutants discriminate between hypoxia-independent and hypoxia-regulated targets
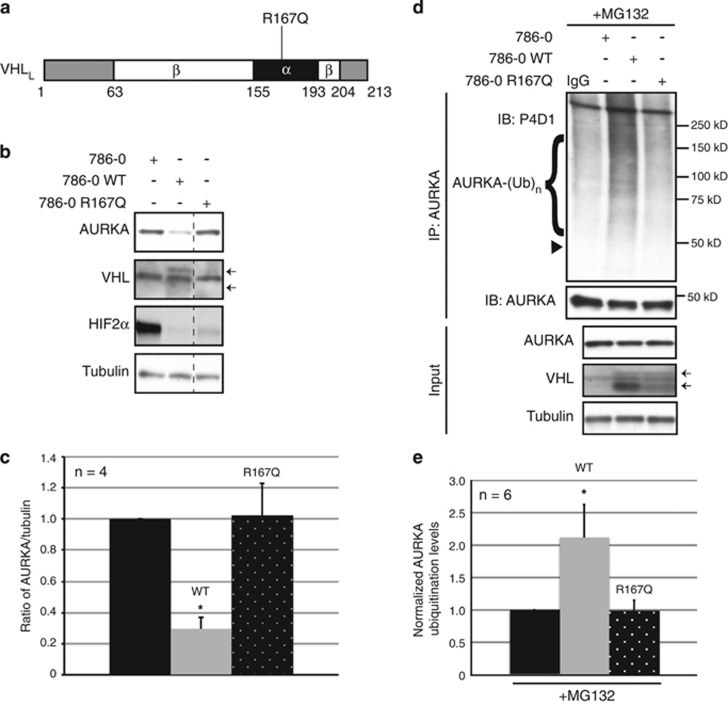
Missense mutations are commonly associated with VHL disease and clear cell RCC.27 The R167Q mutation is the most common pathogenic variant of VHL (Figure 5a). Although VHL-R167Q mutant protein is unstable, if overexpressed or stabilized VHL-R167Q has been shown to retain its intrinsic PHD- and oxygen-dependent ability to ubiquitinate HIFα.28 Generating stable lines of 786-0 cells re-expressing either WT VHL-WT or a VHL-R167Q revealed that the VHL-R167Q mutant when overexpressed reduced HIF2α levels (Figure 5b), corroborating previous studies.28 However, this mutant failed to even marginally reduce AURKA levels compared with VHL-WT expressing cells (Figures 5b and c). In addition, after immunoprecipitating ubiquitinated AURKA, no increase in AURKA ubiquitination was seen in VHL-R167Q rescue cells in contrast to VHL-WT rescue cells, which showed increased AURKA ubiquitination (normalized to AURKA levels) (Figures 5d and e and Supplementary Figures 3a and 3b). Thus, this pathogenic mutant retained the ability to recognize HIFα, but had lost the ability to recognize/ubiquitinate AURKA, suggests ubiquitination of AURKA may participate in VHL’s activity as a tumor suppressor.
Figure 5.
VHL R167Q mutant fails to ubiquitinate and degrade AURKA. (a) Schematic of VHL24 (long isoform of VHL, VHLL) showing the β-domain (amino acid 63–204) and α-domain (amino acid 155–193) and the R167Q mutation. (b) Lysates from 786-0 (parental) to isogenic 786-0 cells stably re-expressing WT (VHL24), or R167Q VHL mutant probed with the indicated antibodies. Arrows indicate the expected bands for VHL. (c) Densitometric quantitation showing an average ratio of AURKA expression to tubulin. Parental-black bar, WT-gray bar, R167Q-black bar with white dots. *P<0.0001. (d) In vivo ubiquitination assay using lysates from 786-0 to isogenic 786-0 VHL24 (WT), and R167Q cell lines treated with MG132. Ubiquitinated AURKA probed using anti-ubiquitin P4D1 antibody. Arrowhead indicates the expected molecular weight of monoubiquitinated AURKA. Input lysates probed with AURKA, VHL and tubulin. Arrows indicate the expected bands for VHL. (e) Graphical representation of densitometric quantitation (measured from 50 to 200 kD) showing an averaged ratio of ubiquitinated AURKA to the pull down efficiency. Black bar (parental 786-0), gray bar (WT), black bar with white dots (R167Q). *P<0.01 All error bars denote s.e.m.
VHL targets AURKA for degradation in quiescent cells
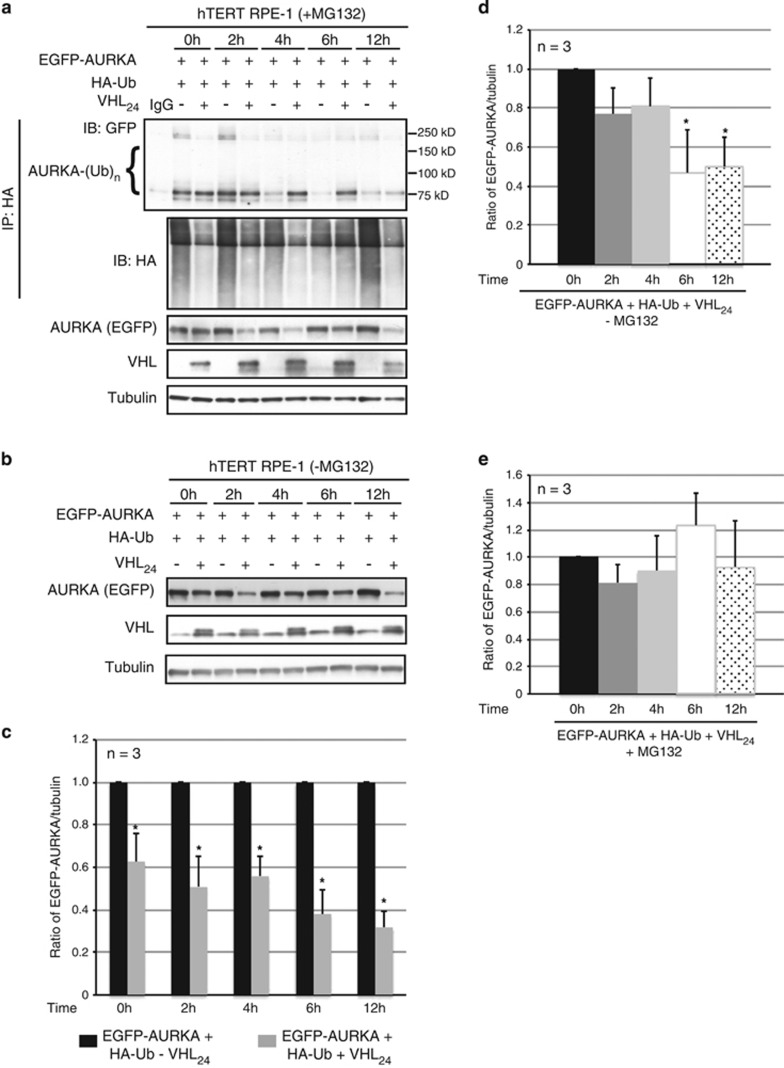
Given that our data showed VHL promotes AURKA degradation in confluent hTERT RPE-1 cells, we investigated the timing and extent of AURKA ubiquitination in cells induced to enter quiescence. In vivo ubiquitination assays were performed to assess AURKA ubiquitination in cells expressing EGFP-AURKA, and HA-Ub in the presence or absence of VHL at specific time points (2, 4, 6 and 12 h) after serum withdrawal. AURKA ubiquitination increased, and total levels decreased, after serum withdrawal in cells expressing exogenous VHL (Figures 6a–c). Importantly, ubiquitination of AURKA (normalized to pool-size of AURKA) increased in cells overexpressing VHL as a function of time (Figure 6a and Supplementary Figure 4) commensurate with decreased AURKA abundance at 6 and 12 h following serum withdrawal (Figure 6d). Blocking proteasome-mediated protein degradation with MG132 blocked this decrease (Figure 6e) demonstrating VHL-mediated ubiquitination of non-mitotic AURKA targets this kinase for degradation upon entering quiescence.
Figure 6.
VHL promotes AURKA degradation as cells enter quiescence. (a) In vivo ubiquitination assay using hTERT RPE-1 cells overexpressing EGFP-AURKA and HA-Ub with and without overexpressed VHL24 in the presence of MG132, harvested following serum withdrawal at the indicated time points. Ubiquitinated AURKA immunoprecipitated using an anti-HA antibody and probed with an anti-AURKA antibody. Input lysates immunoblotted for the indicated antibodies. (b) Lysates from hTERT RPE-1 cells overexpressing EGFP-AURKA and HA-Ub with and without overexpressed VHL24 in the absence of MG132 probed for the indicated antibodies. (c) Densitometric quantitation showing an average ratio of EGFP-AURKA to tubulin in cells without (black bars) and with (gray bars) overexpressed VHL24 at the indicated time points. *P<0.01. (d, e) Densitometric quantitation of EGFP-AURKA to tubulin expression averaged from three independent experiments at each of the indicated time points normalized to the 0-h time point in the absence of MG132 treatment (d) and the presence of MG132 treatment (e). *P<0.01 (compared with the 0-h time point). All error bars denote s.e.m.
VHL ubiquitination of AURKA functions in ciliogenesis
VHL has been shown to have a role in stabilizing microtubules, including the microtubule axoneme of the primary cilium, with loss of this tumor suppressor severely attenuating the ability of cells to make a primary cilium.25, 29, 30 Interestingly, the role of VHL in promoting ciliogenesis has been thought to be hypoxia independent, although the mechanism by which VHL functioned in this hypoxia-independent capacity has not been elucidated. Based on our data that AURKA ubiquitination by VHL was hypoxia independent, we asked if the inability of VHL-deficient cells to target AURKA for degradation was responsible for defective ciliogenesis observed in VHL-deficient cells.
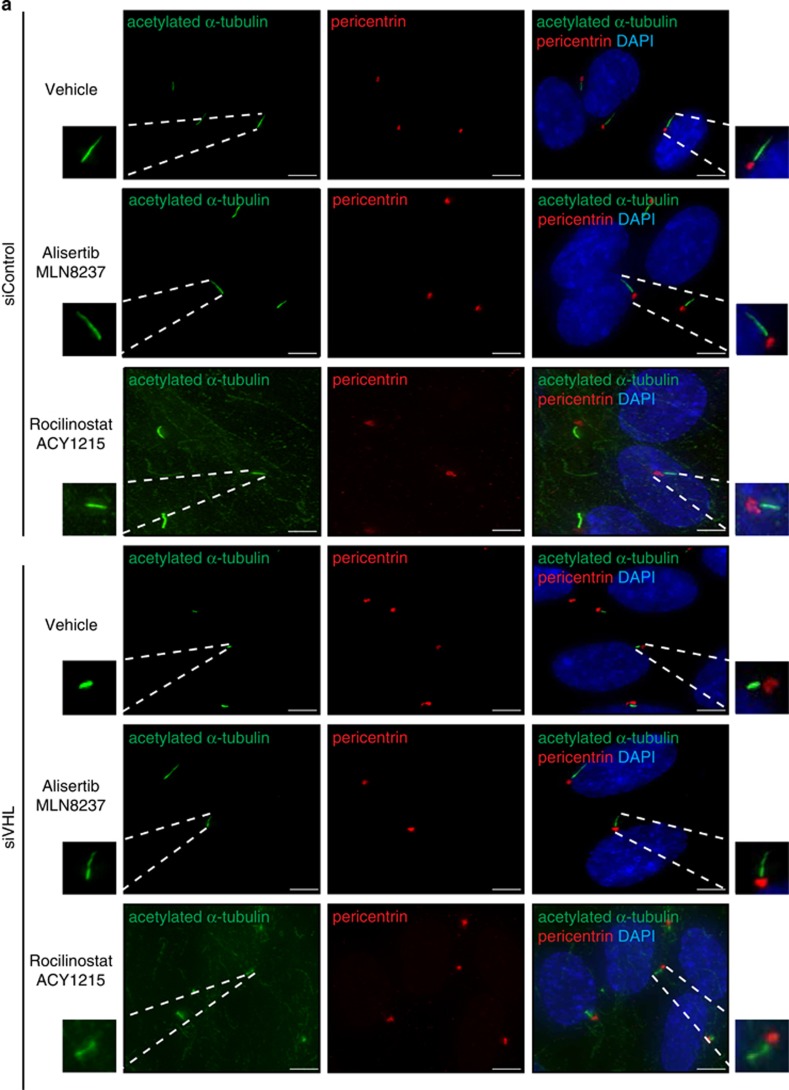
Ciliogenesis was monitored in hTERT RPE-1 cells with an acute loss of VHL using immunofluorescence staining for acetylated α-tubulin (cilia marker) and pericentrin (basal body marker). As demonstrated by us, and others,25, 29, 30 a 60% knockdown of VHL using siRNA (Supplementary Figure 5a) resulted in an increase (50% increase at minimum) in the number of unciliated cells confirming a reduced frequency of ciliation (Figures 7a, b and d). In addition, the cells that did ciliate exhibited significantly shorter (~40% shorter) cilia compared with control cells (Figures 7a, c and e).
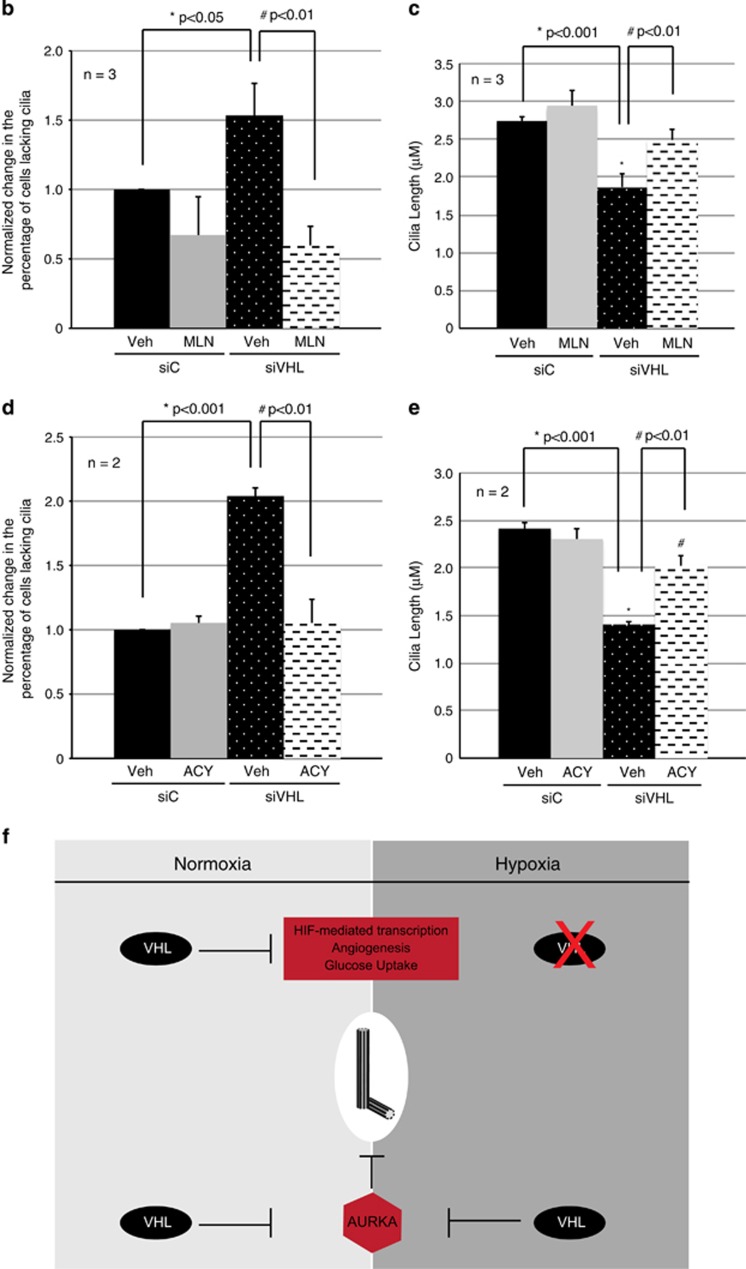
Figure 7.
AURKA and HDAC6 inhibition rescues ciliogenesis in VHL-deficient cells. (a) Representative images from hTERT RPE-1 cells transiently transfected with siControl (siC) or siVHL, treated with vehicle (DMSO), alisertib (MLN8237) or rocilinostat (ACY1215) at the time of serum withdrawal for 48 h. Ciliation monitored by immunofluorescent staining using acetylated α-tubulin (cilia marker) and pericentrin (basal body marker). Nuclei counterstained using DAPI. Highlighted boxes show magnified cilia. Scale bar, 3 μM. (b, d) Percentage of cells that fail to ciliate (unciliated) normalized to 1, from siC or siVHL transfected cells treated with (b) alisertib or (d) rocilinostat. * and # denote statistical significance. (c, e) Cilia length (μM) from siC or siVHL transfected cells treated with (c) alisertib or (e) rocilinostat. At least 100 cells were counted from each replicate. * and # denote statistical significance. All error bars indicate s.e.m. (f) Model depicting a role for VHL’s ubiquitin ligase function in conditions of normoxia targeting HIFα and hypoxia targeting AURKA.
The ciliary axoneme is made up of highly acetylated α-tubulin, and AURKA is known to phosphorylate and activate HDAC6,31 which promotes α-tubulin deacetylation. Therefore, we next turned to explore a possible VHL–AURKA–HDAC6 signaling axis that regulated ciliogenesis. Direct inhibition of AURKA with alisertib (MLN8237) or HDAC6 using rocilinostat (ACY1215) rescued deficient ciliogenesis (Figures 7a, b and d) and increased cilia length (Figures 7a, c and e) in VHL-deficient cells. Direct inhibition of HDAC6 increased levels of acetylated α-tubulin as expected, but did not alter levels of AURKA (Supplementary Figure 5b). AURKA inhibition with alisertib had no effect on total levels of acetylated α-tubulin in the cell (Supplementary Figure 5c), consistent with a more localized AURKA–HDAC6 axis acting at the primary cilium.31 These data provide a mechanism by which VHL can stabilize microtubules of the ciliary axoneme under conditions of both normoxia and hypoxia by inhibiting the AURKA–HDAC6 axis and disassembly of the primary cilium (Figure 7f).
Discussion
In this study, we have identified a new function for the VHL ubiquitin ligase that is hypoxia independent, and distinct from its well-established role in modulating HIFα in normoxia. VHL multi-monoubiquitinates AURKA in quiescent cells and targets it for proteasome-mediated degradation independent of oxygen-dependent PHD activity. VHL-mediated ubiquitination of AURKA provides a mechanism by which VHL protects the stability of axoneme microtubules to regulate the primary cilium. Importantly, the inability of the most common pathogenic variant of VHL, which retains an intrinsic ability to degrade HIFα, to ubiquitinate and degrade AURKA, suggests an equally important role for this hypoxia-independent function of VHL in tumorigenesis.
Hydroxylation of proline residues by PHDs in normoxia is critical for VHL to recognize several of its substrates including HIFα,7, 8 Sprouty2(ref. 6) and RNA polymerase subunit Rpb1,9, 10 although it is not yet clear if this is the case for all VHL substrates. Hydroxylated substrates are recognized, and polyubiquitinated by VHL, which targets them for proteasome-mediated degradation. K48 (lysine 48) linkages are the canonical signal for proteasomal degradation,32 and VHL-mediated K48 linkage was specifically demonstrated for the VHL substrate epidermal growth factor receptor,13 analogous to that demonstrated for HIFα. Polyubiquitination was even reported in the case of nuclear clustrin, a non-proteasomal target of VHL, which was ubiquitinated using a K63 linkage, which dictated the nuclear translocation of clustrin.33 Importantly, our studies demonstrate a unique ability of VHL to multi-monoubiquitinate its substrate to target it for degradation. Historically, polyubiquitination of proteins with an ubiquitin chain made of at least four ubiquitin moieties on a single lysine was believed to be a prerequisite for recognition by the 26S proteasome,34 although more recent evidence has shown that monoubiquitination, in addition to generating structural diversity to control diverse cellular responses, also targets proteins for degradation,35 as exemplified by paired box 3(refs 36, 37) and syndecan 4.38 Data presented here that VHL has the ability to multi-monoubiquitinate its substrate, distinguishes VHL’s oxygen-dependent (polyubiquitination) and oxygen-independent (multi-monoubiquitination) targets.
Previously, hypoxia-independent VHL activity was thought to be distinct from its function as an ubiquitin ligase, which was believed to be oxygen dependent. Our data demonstrate VHL’s ability to recognize and ubiquitinate AURKA independent of the activity of PHDs, revealing VHL’s E3 ligase activity that can function under hypoxic conditions. Although VHL has an established role in modulating microtubule stability during mitosis, and in the ciliary axoneme,18, 19 the mechanism by which VHL promotes microtubule stability was unclear. Our data establishing AURKA as a target for VHL ubiquitination identifies a VHL–AURKA–HDAC6 axis, and provides a mechanism whereby increased AURKA in VHL-deficient cells destabilizes axonemal microtubules of the primary cilium. Importantly, several studies have reported increased AURKA expression in RCC20, 39 and a corresponding lack of primary cilia.25, 40, 41
It is well established that AURKA is polyubiquitinated by the APC complex and targeted for degradation during mitosis.42, 43, 44 Although the majority of AURKA is thus eliminated well before midbody formation and cytokinesis, a small pool of AURKA persists in interphase cells. The role of this non-mitotic AURKA is only beginning to emerge with the phosphorylation of HDAC6 its first reported activity in quiescent cells.31 However, heretofore it was not known how AURKA levels are regulated outside of mitosis. Our finding that VHL targets AURKA for degradation in quiescent cells now provides a regulatory mechanism for non-mitotic AURKA activity. Conversely, AURKA can phosphorylate VHL at serine 72, a priming phosphorylation for GSK3β, which regulates VHL’s role in microtubule stability.20 It would now be interesting to determine if phosphorylation of VHL by AURKA modulates VHL’s hypoxia-independent E3 ligase activity during ciliogenesis.
Does regulation of AURKA contribute to VHL’s function as a tumor suppressor? Loss of the primary cilium is an early event in cystogenesis of the kidney that characterizes VHL patients, and has led to classification of VHL syndrome as a 'ciliopathy'.45 In addition, formation of a primary cilium is an important component of the cilia-centrosome cycle, and acts as a structural checkpoint for mitosis and cell proliferation.46 We found that the pathogenic R167Q VHL mutant, which retains intrinsic ability to induce HIFα degradation,28 is unable to similarly target AURKA for degradation, separating hypoxia-regulated (HIFα ubiquitination) from hypoxia-independent (AURKA ubiquitination) VHL activity. Other pathogenic VHL mutants have also been shown to have differential and/or compromised ability to stabilize microtubules with the identification of two VHL domains spanning residues 1–53 and 95–123 thought to be important for maintaining primary cilia.47 Although HIF-driven progression of tumors arising from defects in metabolism and increased tumor angiogenesis in patients with loss of VHL is clearly established, virtually nothing is known about the earliest events that result in tumorigenesis and the renal cystogenesis common in VHL patients. Our data establishing VHL’s ability to regulate AURKA and modulate microtubule stability at the cilium could shed light on some of the early events in tumorigenesis.
Our data establishing AURKA as a substrate of VHL presents new opportunities for intervention, not only in RCC, but also in other cancers such as colorectal carcinoma, where genomic data from patients reveals the existence of VHL mutations,48, 49 elevated AURKA 50 and ciliary defects.51 Importantly, our demonstration that primary cilia can be rescued with AURKA and HDAC6 inhibitors provides proof-of-concept that targeting the VHL–AURKA–HDAC6 axis may have therapeutic efficacy. Future studies using in vivo cancer models, as well as ciliopathy models such as polycystic kidney disease (PKD), in which AURKA signaling is dysregulated, now become candidates for therapeutic targeting of the VHL–AURKA–HDAC6 axis.
Materials and methods
Cell lines
Human 786-0, VHL-deficient, RCC cell line was maintained in RPMI-1640 media (Life Technologies, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (Sigma-Aldrich, St Louis, MO, USA). Immortalized retinal epithelial (hTERT RPE-1) cells (a kind gift from Dr Gregory Pazour, University of Massachusetts Medical School, Worcester, MA, USA) were maintained in Dulbecco’s modified Eagle’s medium/F-12 media (Life Technologies, Sigma-Aldrich), supplemented with 10% fetal bovine serum. Stable cells lines re-expressing VHL were generated by clonal selection of 786-0 (VHL-deficient) cells transfected with a WT VHL24 construct (G57T3, previously described in Perera et al.52), and maintained under antibiotic selection using 800 μg/ml G418 (Life Technologies, Sigma-Aldrich). R167Q VHL mutant and VHL24 (WT) expressing stables were generated in 786-0 as described previously.28 A-498 (VHL-deficient) stables re-expressing VHL24 (pCR3-VHL24) or VHL19 (pCR3-VHL19) constructs53 (constructs were a gift from Dr Alan Schoenfeld, Adelphi University, Garden City, NY, USA) were generated by transfection and selection using 800 μg/ml G418 (Life Technologies, Sigma-Aldrich). All human cell lines were short tandem repeats fingerprinted and validated using the Characterized Cell Line Core Facility (U.T. M.D. Anderson Cancer Center). In addition, all cells used in these studies were tested and confirmed negative for mycoplasma.
Constructs, transfections and treatments
EGFP-tagged WT AURKA (EGFP-AURKA) (HsCD00036078) was purchased from DNASU Plasmid Repository (Arizona State University, Tempe, AZ, USA). The pDendra2-C vector was purchased from Clontech (Mountain View, CA, USA). Dendra2C-AURKA was generated by PCR amplification of AURKA complementary DNA from the EGFP-AURKA construct using primers (see below) designed to have BamH1 and EcoR1 sticky ends to enable cloning into the multiple cloning site of the pDendra2-C vector.
Forward primer—5′-TCCGCAGAATTCTATGGACCGATCTAAAGAA-3′
Reverse primer—5′-AGTCGATCGTTTGTCAGAATCCCTAGGATT-3′.
PCR amplification (95 °C—2 min, 35 cycles of 95 °C—1 min, 44 °C—30 s, 72 °C—2 min and, 72 °C—5 min) was performed using Pfu polymerase (Promega, Madison, WI, USA). Following PCR amplification, the PCR product was gel purified using a QIA quick gel extraction kit (Qiagen, Germantown, MD, USA). The purified PCR product was cloned into the BamH1/EcoR1 digested pDendra2-C vector. The Dendra2C-AURKA construct was sequence verified using Genewiz Inc (South Plainfield, NJ, USA). In addition, localization of overexpressed Dendra2C-AURKA was confirmed at the centrosomes. HA-tagged VHL30 construct (HA-VHL30) was a kind gift from Dr Wilhelm Krek, (ETH, Institute of Cell Biology, Zurich, Switzerland). The pCR3-VHL24 construct was a kind gift from Dr Alan Schoenfeld (Adelphi University). pRK5-HA-Ubiquitin-WT (HA-Ub) (Addgene plasmid # 17608; Cambridge, MA, USA), pRK5-HA-Ubiquitin-K48 (HA-K48) (Addgene plasmid # 17605), pRK5-HA-Ubiquitin-K48R (HA-K48R) (Addgene plasmid # 17604), pRK5-HA-Ubiquitin-KO (HA-K0) (Addgene plasmid # 17603) were gifts from Dr Ted Dawson and purchased from Addgene.
For overexpression studies, constructs were transfected into cells using Lipofectamine2000 (Thermo Fisher, Waltham, MA, USA) according to the manufacturer’s protocol. For knockdown studies On-Target plus SMART pool siRNAs (nontargeting, and VHL-specific siRNA) were purchased from Dharmacon (Thermo Fisher Scientific, Waltham, MA, USA), and transfected per the manufacturer’s protocol using DharmaFECT1.
Dimethyloxalylglycine (1 mM), and DFX (250 μM) (Sigma-Aldrich) were solubilized in water; MG132 (10 μM) (Sigma-Aldrich), bortezomib (10 ng/ml) (Selleck Chemicals, Houston, TX, USA) and bafilomycin A (200 nM) (Santa Cruz Biotechnology, Dallas, TX, USA) were solubilized in DMSO. All treatments using dimethyloxaloylglycine, DFX, MG132 and bafilomycin A were performed overnight (16 h). CHX (Sigma-Aldrich) was solubilized in ethanol, and cells treated at a final concentration of 20 ng/ml for the indicated time points. Alisertib (MLN8237, Millenium Pharmaceuticals Inc., Cambridge, MA, USA), and rocilinostat (ACY1215, Selleck Chemicals) were solubilized in DMSO, and cells treated at a final concentration of 2 μM and 100 nM, respectively, at the time of serum withdrawal for 48 h.
Cell lysates and antibodies
Cell lysates for whole-cell extracts were collected in cold 1x cell lysis buffer (20 mM Tris (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton-X-100, 2.5 mM sodium pyrophosphate) containing 1x complete protease inhibitor (Roche, Mannheim, Germany), and 1 mM sodium orthovanadate (Na3VO4). The following primary antibodies were used for immunoblotting: anti-AURKA (1:1000), anti-VHL (1:500), anti-HA (1:1000) from Cell Signaling Technologies, Danvers, MA, USA; anti-AURKA (1:1000) from Thermo Fisher; anti-tubulin (1:20 000) from Thermo Fisher Scientific; anti-ubiquitin (P4D1, 1:1000), anti-glyceraldehyde 3-phosphate dehydrogenase (1:5000), and anti-GFP (1:2000) from Santa Cruz Biotechnology. Monoclonal anti-polyubiquitin (FK1, 1:1000) antibody was purchased from Enzo Life Sciences (Farmingdale, NY, USA). Horseradish peroxidase-conjugated goat anti-mouse and goat anti-rabbit secondary antibodies were purchased from Santa Cruz Biotechnology. Immunoblots were visualized using LumiGLO (KPL), Pierce ECL (Thermo Fisher Scientific), or Amersham ECL Prime (GE Life Sciences, Pittsburgh, PA, USA) substrates.
In vitro ubiquitination assay
Human recombinant VHL complex (cat # 23-044), UBE1 (cat # 23-021), UbcH4 (cat # 23-025), AURKA (cat # 14-511) and non-radioactive ATP (cat # 20-306) were purchased from EMD Millipore (Darmstadt, Germany). Human recombinant WT ubiquitin (cat # U-100H), ubiquitin mutant K48 (cat # UM-K480), ubiquitin mutant K48R (cat # UM-K48R) and ubiquitin mutant K0 (cat # UM-NOK) were purchased from Boston Biochem (Cambridge, MA, USA). The in vitro reaction was set up by incubating 10 nM UBE1, 1 μM UbcH4, 1 μg of the VHL complex, 10 μM ATP, and 500 μM ubiquitin (or ubiquitin mutants) with either 50 nM or 100 nM AURKA in a buffer containing 25 mM MOPS (pH 7.5), 0.01% Tween 20 and 5 mM MgCl2. The reaction was initiated by the addition of ubiquitin and following a 30- min incubation at room temperature, the reaction was terminated using a stop solution (25 mM MOPS (pH 7.5) containing 125 mM EDTA, 150 mM NaCl and 0.05% Tween 20) and loading dye.
In vivo ubiquitination assay
To determine ubiquitination of endogenous AURKA in 786-0 and 786-0 VHL24 cell lines, the cells were cultured to confluence and starved for 48 h. To determine ubiquitination of exogenous AURKA, hTERT RPE-1 cells transfected with EGFP-AURKA/Dendra2C-AURKA, HA-Ub and VHL24 were cultured to 100% confluence and treated with 10 μM MG132 16 h before harvesting (without serum starvation). For denaturing immunoprecipitations, cell lysates were collected in cold modified 1X RIPA buffer (10 mM Tris (pH 7.5), 150 mM NaCl, 5 mM EDTA, 1% NP40, 1% sodium deoxycholate, 0.025% SDS) containing 1X complete protease inhibitor (Roche), 1 mM Na3VO4, and 1 μM N-ethylmaleimide (Sigma-Aldrich). The lysates were immunoprecipitated overnight with anti-HA (1:50) and anti-AURKA (1:50) antibodies from Cell Signaling Technologies. The immune complexes were incubated with pre-washed magnetic beads (Pierce, Thermo Fisher) for an hour, and washed three times with modified 1X RIPA buffer before resuspension in loading dye. Densitometric quantitation for ubiquitinated AURKA was conducted by measuring the intensity of the bands (smears) from molecular weight 50–200 kD.
Immunoprecipitation
Cells transfected with EGFP-AURKA and HA-VHL30 were grown to 100% confluence before harvesting in cold 1X cell lysis buffer containing 1X complete protease inhibitor (Roche), and 1 mM Na3VO4. The lysates were immunoprecipitated overnight using an anti-HA (1:50) or anti-GFP (5 μg) antibodies. The immune complexes were incubated with pre-washed magnetic beads (Pierce, Thermo Fisher) for an hour, and washed three times with 1X cell lysis buffer before resuspending the pellet in loading dye and boiling for 5 min at 95 °C. Eluted proteins were immunoblotted with anti-HA (1:1000) and anti-GFP (1:1000) antibodies.
Reverse transcriptase–PCR analyses
Reverse transcriptase–PCR analysis to assess knockdown efficiency of VHL was performed as described previously.25 Briefly, mRNA was isolated from cells transfected with siControl/siVHL, and complementary DNA prepared by reverse transcription. Gene expression was assessed by real-time quantitative PCR using specific TaqMan probes (Thermo Fisher), and a TaqMan Fast Universal master mix on a Viia7 system (Thermo Fisher). mRNA expression was evaluated for VHL, AURKA and glyceraldehyde 3-phosphate dehydrogenase, which was used as an endogenous control. The following set of conditions were used for each real-time reaction: 95 °C for 20 min followed by 40 cycles of 1 s at 95 °C and 20 s at 60 °C. The real-time PCR reactions were all performed in triplicate and were quantified using the −ΔΔ cycle threshold (CT) method. Reverse transcriptase–PCR was performed for each replicate to ensure optimal knockdown of VHL.
Immunofluorescence analyses
Immunofluorescence staining to observe primary cilia was performed as published previously.25 Briefly, hTERT RPE-1 cells were plated on glass coverslips, transfected, starved and treated for 48 h to induce cilia formation. Cells were fixed using 4% paraformaldehyde (15 min), permeabilized with 0.05% Triton-X (10 min), followed by blocking in 3.75% bovine serum albumin solution (1 h). Primary antibodies for acetylated α-tubulin (clone 6-11B-1, 1:5000; Sigma-Aldrich) and pericentrin (1:5000; Abcam, Cambridge, MA, USA) were applied in blocking buffer for 1 h. AlexaFluor 488 and 546 goat anti-mouse or anti-rabbit secondary antibodies (Life Technologies, Sigma-Aldrich) were subsequently applied for another hour. Cells were counterstained using DAPI (1:4000 of 1 mg/ml stock) and visualized using a Deltavision deconvolution microscope (Applied Precision, Pittsburgh, PA, USA) at × 60 magnification. Images were analyzed using the Imaris 3D software (Bitplane, Concord, MA, USA). All experiments were performed in triplicate and image analyses performed on at least 100 individual cells from each replicate.
Photoswitching assay
hTERT RPE-1 cells were co-transfected with the Dendra2C-AURKA construct and HA-Ub in the absence or presence of VHL24 and the cells cultured to confluence. Serum-free media was added to the cells before live cell imaging. Live cell imaging was performed using a Nikon A1 confocal microscope (Melville, NY, USA) with an environmental chamber. Dendra2c-AURKA can be visualized using the 488-laser, which was set to 5% power to prevent and minimize spontaneous conversion of green Dendra2C-AURKA to the red fluorescent state. Cells were scanned (60X) and single cells expressing high Dendra2C-AURKA levels selected for photoconversion. Photoconversion was achieved on a selected region of interest using the 405 laser at 10% power, for 1 s. Images were acquired for green and red channels immediately after conversion, and 15 min thereafter for a minimum of 6 h. As cells moved rapidly out of the selected plane of focus after 2–3 h, all analyses was performed on images obtained no later than 2 h after conversion. Image analysis was performed using Image J (https://imagej.nih.gov) to measure the red fluorescent intensity, which was converted to a corrected mean red fluorescent intensity (background subtraction) of the region of interest.
Statistical analyses
All statistical analyses were performed using the Student’s t-test (one-tailed, assuming equal variance) to determine differences between the average values obtained from quantitation and densitometric analyses of immunoblots, and average values obtained from reverse transcriptase–PCR analyses. The s.e.m. was calculated and P-values of P<0.01 and P<0.05 were considered statistically significant.
Acknowledgments
The authors would like to thank T Berry and S Bai for technical assistance. This work was supported by a Robert A Welch Endowed Chair in Chemistry (BE-0023) to CLW.
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
The authors declare no conflict of interest.
Supplementary Material
References
- Latif F, Tory K, Gnarra J, Yao M, Duh FM, Orcutt ML et al. Identification of the von Hippel-Lindau disease tumor suppressor gene. Science (New York, NY) 1993; 260: 1317–1320. [DOI] [PubMed] [Google Scholar]
- Hamano K, Esumi M, Igarashi H, Chino K, Mochida J, Ishida et al. Biallelic inactivation of the von Hippel-Lindau tumor suppressor gene in sporadic renal cell carcinoma. J Urol 2002; 167: 713–717. [DOI] [PubMed] [Google Scholar]
- Randall JM, Millard F, Kurzrock R. Molecular aberrations, targeted therapy, and renal cell carcinoma: current state-of-the-art. Cancer Metastasis Rev 2014; 33: 1109–1124. [DOI] [PubMed] [Google Scholar]
- Kamura T, Koepp DM, Conrad MN, Skowyra D, Moreland RJ, Iliopoulos O et al. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science (New York, NY) 1999; 284: 657–661. [DOI] [PubMed] [Google Scholar]
- Stebbins CE, Kaelin WG Jr., Pavletich NP. Structure of the VHL-ElonginC-ElonginB complex: implications for VHL tumor suppressor function. Science (New York, NY) 1999; 284: 455–461. [DOI] [PubMed] [Google Scholar]
- Anderson K, Nordquist KA, Gao X, Hicks KC, Zhai B, Gygi SP et al. Regulation of cellular levels of Sprouty2 protein by prolyl hydroxylase domain and von Hippel-Lindau proteins. J Biol Chem 2011; 286: 42027–42036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M et al. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science (New York, NY) 2001; 292: 464–468. [DOI] [PubMed] [Google Scholar]
- Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science (New York, NY) 2001; 292: 468–472. [DOI] [PubMed] [Google Scholar]
- Kuznetsova AV, Meller J, Schnell PO, Nash JA, Ignacak ML, Sanchez Y et al. von Hippel-Lindau protein binds hyperphosphorylated large subunit of RNA polymerase II through a proline hydroxylation motif and targets it for ubiquitination. Proc Natl Acad Sci USA 2003; 100: 2706–2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikhaylova O, Ignacak ML, Barankiewicz TJ, Harbaugh SV, Yi Y, Maxwell PH et al. The von Hippel-Lindau tumor suppressor protein and Egl-9-Type proline hydroxylases regulate the large subunit of RNA polymerase II in response to oxidative stress. Mol Cell Biol 2008; 28: 2701–2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein AC, Gleadle JM, McNeill LA, Hewitson KS, O'Rourke J, Mole DR et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 2001; 107: 43–54. [DOI] [PubMed] [Google Scholar]
- Schodel J, Grampp S, Maher ER, Moch H, Ratcliffe PJ, Russo P et al. Hypoxia, hypoxia-inducible transcription factors, and renal cancer. Eur Urol 2016; 69: 646–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Yang H. The von Hippel-Lindau tumor suppressor protein promotes c-Cbl-independent poly-ubiquitylation and degradation of the activated EGFR. PLoS One 2011; 6: e23936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda H, Saitoh K, Hirai S, Iwai K, Takaki Y, Baba M et al. The von Hippel-Lindau tumor suppressor protein mediates ubiquitination of activated atypical protein kinase C. J Biol Chem 2001; 276: 43611–43617. [DOI] [PubMed] [Google Scholar]
- Xie L, Xiao K, Whalen EJ, Forrester MT, Freeman RS, Fong G et al. Oxygen-regulated beta(2)-adrenergic receptor hydroxylation by EGLN3 and ubiquitylation by pVHL. Sci Signal 2009; 2: ra33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y, Qiao M, Song M, Weintraub ST, Shiio Y. Quantitative proteomics identifies the Myb-binding protein p160 as a novel target of the von Hippel-Lindau tumor suppressor. PLoS One 2011; 6: e16975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na X, Duan HO, Messing EM, Schoen SR, Ryan CK, di Sant'Agnese PA et al. Identification of the RNA polymerase II subunit hsRPB7 as a novel target of the von Hippel-Lindau protein. EMBO J 2003; 22: 4249–4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hergovich A, Lisztwan J, Barry R, Ballschmieter P, Krek W. Regulation of microtubule stability by the von Hippel-Lindau tumour suppressor protein pVHL. Nat Cell Biol 2003; 5: 64–70. [DOI] [PubMed] [Google Scholar]
- Thoma CR, Toso A, Gutbrodt KL, Reggi SP, Frew IJ, Schraml P et al. VHL loss causes spindle misorientation and chromosome instability. Nat Cell Biol 2009; 11: 994–1001. [DOI] [PubMed] [Google Scholar]
- Martin B, Chesnel F, Delcros JG, Jouan F, Couturier A, Dugay F et al. Identification of pVHL as a novel substrate for Aurora-A in clear cell renal cell carcinoma (ccRCC). PLoS One 2013; 8: e67071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockman ME, Masson N, Mole DR, Jaakkola P, Chang GW, Clifford SC et al. Hypoxia inducible factor-alpha binding and ubiquitylation by the von Hippel-Lindau tumor suppressor protein. J Biol Chem 2000; 275: 25733–25741. [DOI] [PubMed] [Google Scholar]
- Kamura T, Sato S, Iwai K, Czyzyk-Krzeska M, Conaway RC, Conaway JW. Activation of HIF1alpha ubiquitination by a reconstituted von Hippel-Lindau (VHL) tumor suppressor complex. Proc Natl Acad Sci USA 2000; 97: 10430–10435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohh M, Park CW, Ivan M, Hoffman MA, Kim TY, Huang LE et al. Ubiquitination of hypoxia-inducible factor requires direct binding to the beta-domain of the von Hippel-Lindau protein. Nat Cell Biol 2000; 2: 423–427. [DOI] [PubMed] [Google Scholar]
- Tanimoto K, Makino Y, Pereira T, Poellinger L. Mechanism of regulation of the hypoxia-inducible factor-1 alpha by the von Hippel-Lindau tumor suppressor protein. EMBO J 2000; 19: 4298–4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dere R, Perkins AL, Bawa-Khalfe T, Jonasch D, Walker CL. Beta-catenin links von Hippel-Lindau to aurora kinase A and loss of primary cilia in renal cell carcinoma. J Am Soc Nephrol 2015; 26: 553–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romain CV, Paul P, Lee S, Qiao J, Chung DH. Targeting aurora kinase A inhibits hypoxia-mediated neuroblastoma cell tumorigenesis. Anticancer Res 2014; 34: 2269–2274. [PMC free article] [PubMed] [Google Scholar]
- Clifford SC, Cockman ME, Smallwood AC, Mole DR, Woodward ER, Maxwell PH et al. Contrasting effects on HIF-1alpha regulation by disease-causing pVHL mutations correlate with patterns of tumourigenesis in von Hippel-Lindau disease. Hum Mol Genet 2001; 10: 1029–1038. [DOI] [PubMed] [Google Scholar]
- Ding Z, German P, Bai S, Reddy AS, Liu XD, Sun M et al. Genetic and pharmacological strategies to refunctionalize the von Hippel Lindau R167Q mutant protein. Cancer Res 2014; 74: 3127–3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lolkema MP, Mans DA, Ulfman LH, Volpi S, Voest EE, Giles RH. Allele-specific regulation of primary cilia function by the von Hippel-Lindau tumor suppressor. Eur J Hum Genet 2008; 16: 73–78. [DOI] [PubMed] [Google Scholar]
- Thoma CR, Frew IJ, Hoerner CR, Montani M, Moch H, Krek W. pVHL and GSK3beta are components of a primary cilium-maintenance signalling network. Nat Cell Biol 2007; 9: 588–595. [DOI] [PubMed] [Google Scholar]
- Pugacheva EN, Jablonski SA, Hartman TR, Henske EP, Golemis EA. HEF1-dependent Aurora A activation induces disassembly of the primary cilium. Cell 2007; 129: 1351–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabek N, Herman-Bachinsky Y, Buchsbaum S, Lewinson O, Haj-Yahya M, Hejjaoui M et al. The size of the proteasomal substrate determines whether its degradation will be mediated by mono- or polyubiquitylation. Mol Cell 2012; 48: 87–97. [DOI] [PubMed] [Google Scholar]
- Xue J, Lv DD, Jiao S, Zhao W, Li X, Sun H et al. pVHL mediates K63-linked ubiquitination of nCLU. PLoS One 2012; 7: e35848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. EMBO J 2000; 19: 94–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski M, Suryadinata R, Tan AR, Roesley SN, Sarcevic B. Protein monoubiquitination and polyubiquitination generate structural diversity to control distinct biological processes. IUBMB Life 2012; 64: 136–142. [DOI] [PubMed] [Google Scholar]
- Boutet SC, Disatnik MH, Chan LS, Iori K, Rando TA. Regulation of Pax3 by proteasomal degradation of monoubiquitinated protein in skeletal muscle progenitors. Cell 2007; 130: 349–362. [DOI] [PubMed] [Google Scholar]
- Boutet SC, Biressi S, Iori K, Natu V, Rando TA. Taf1 regulates Pax3 protein by monoubiquitination in skeletal muscle progenitors. Mol Cell 2010; 40: 749–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvallo L, Munoz R, Bustos F, Escobedo N, Carrasco H, Olivares G et al. Non-canonical Wnt signaling induces ubiquitination and degradation of Syndecan4. J Biol Chem 2010; 285: 29546–29555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferchichi I, Kourda N, Sassi S, Romdhane KB, Balatgi S, Cremet JY et al. Aurora A overexpression and pVHL reduced expression are correlated with a bad kidney cancer prognosis. Dis Markers 2012; 33: 333–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basten SG, Willekers S, Vermaat JS, Slaats GG, Voest EE, van Diest PJ et al. Reduced cilia frequencies in human renal cell carcinomas versus neighboring parenchymal tissue. Cilia 2013; 2: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehn EW, Walz G, Benzing T. von Hippel-Lindau: a tumor suppressor links microtubules to ciliogenesis and cancer development. Cancer Res 2007; 67: 4537–4540. [DOI] [PubMed] [Google Scholar]
- Castro A, Arlot-Bonnemains Y, Vigneron S, Labbe JC, Prigent C, Lorca T. APC/Fizzy-related targets Aurora-A kinase for proteolysis. EMBO Rep 2002; 3: 457–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coon TA, Glasser JR, Mallampalli RK, Chen BB. Novel E3 ligase component FBXL7 ubiquitinates and degrades Aurora A, causing mitotic arrest. Cell Cycle 2012; 11: 721–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim SK, Gopalan G. Aurora-A kinase interacting protein 1 (AURKAIP1) promotes Aurora-A degradation through an alternative ubiquitin-independent pathway. Biochem J 2007; 403: 119–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger-Nukpezah T, Little JL, Serzhanova V, Golemis EA. Cilia and cilia-associated proteins in cancer. Drug Discov Today Dis Mech 2013; 10: e135–e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa I, Goto H, Kasahara K, Inagaki M. Current topics of functional links between primary cilia and cell cycle. Cilia 2015; 4: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lolkema MP, Mans DA, Snijckers CM, van Noort M, van Beest M, Voest EE et al. The von Hippel-Lindau tumour suppressor interacts with microtubules through kinesin-2. FEBS Lett 2007; 581: 4571–4576. [DOI] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012; 2: 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013; 6: pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasap E, Gerceker E, Boyacioglu SO, Yuceyar H, Yildirm H, Ayhan S et al. The potential role of the NEK6, AURKA, AURKB, and PAK1 genes in adenomatous colorectal polyps and colorectal adenocarcinoma. Tumour Biol 2016; 37: 3071–3080. [DOI] [PubMed] [Google Scholar]
- Basten SG, Giles RH. Functional aspects of primary cilia in signaling, cell cycle and tumorigenesis. Cilia 2013; 2: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera AD, Kleymenova EV, Walker CL. Requirement for the von Hippel-Lindau tumor suppressor gene for functional epidermal growth factor receptor blockade by monoclonal antibody C225 in renal cell carcinoma. Clin Cancer Res 2000; 6: 1518–1523. [PubMed] [Google Scholar]
- Schoenfeld A, Davidowitz EJ, Burk RD. A second major native von Hippel-Lindau gene product, initiated from an internal translation start site, functions as a tumor suppressor. Proc Natl Acad Sci USA 1998; 95: 8817–8822. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.