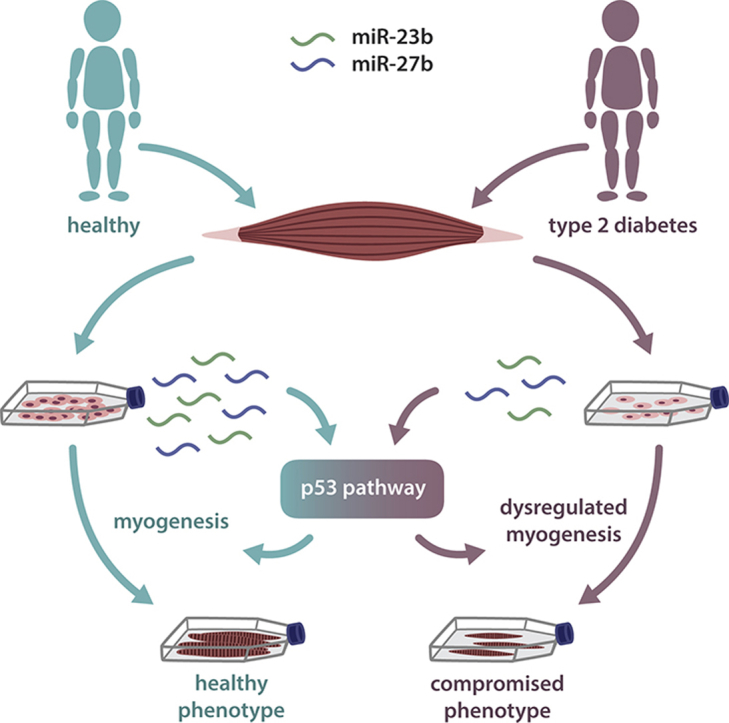
Abstract
Objective
MicroRNAs (miRNAs) are increasingly recognized as fine-tuning regulators of metabolism, and are dysregulated in several disease conditions. With their capacity to rapidly change gene expression, miRNAs are also important regulators of development and cell differentiation. In the current study, we describe an impaired myogenic capacity of muscle stem cells isolated from humans with type 2 diabetes (T2DM) and assess whether this phenotype is regulated by miRNAs.
Methods
We measured global miRNA expression during in vitro differentiation of muscle stem cells derived from T2DM patients and healthy controls.
Results
The mir-23b/27b cluster was downregulated in the cells of the patients, and a pro-myogenic effect of these miRNAs was mediated through the p53 pathway, which was concordantly dysregulated in the muscle cells derived from humans with T2DM.
Conclusions
Our results indicate that we have identified a novel pathway for coordination of myogenesis, the miR-23b/27b-p53 axis that, when dysregulated, potentially contributes to a sustained muscular dysfunction in T2DM.
Keywords: Satellite cell, MicroRNA, Diabetes, Muscle, Human, Myogenesis, muscle stem cells, p53, miR-23b, miR-27b
Graphical abstract
Highlights
-
•
miR-23b and miR-27b are pro-myogenic and are downregulated in T2DM.
-
•
miR-23b and miR-27b regulate myogenesis through the p53 pathway.
-
•
The p53 pathway is concordantly dysregulated in T2DM.
1. Introduction
MicroRNAs (miRNAs) are short, non-coding regulators of protein abundance [1]. Acting at the posttranscriptional level miRNAs quickly alters the biological setting of a cell, making them suitable regulators of tissue development and differentiation [1]. Several recent studies report that miRNAs are induced by physical activity [2], [3] and mediate metabolic control by regulating glucose metabolism and insulin signaling [4], [5], [6], [7] as well as pancreatic beta cell survival [8]. In human skeletal muscle, the global miRNA profile is dysregulated already during the pre-state of type 2 diabetes [9], while specific changes occur following establishment of the disease [9], [10]. It was further reported that non-obese patients with type 2 diabetes (T2DM) had dysregulated miRNAs which associated to genes involved in development and differentiation [9]. We therefore hypothesized that non-obese T2DM patients would have alterations in their skeletal muscle at a stem cell level, resulting in impaired miRNA-mediated muscle cell differentiation. Muscle cells are multinucleated, post-mitotic cells with satellite cells (muscle stem cells) residing between the basal lamina and the sarcolemma of the muscle fiber [11]. Muscle stem cells contribute to muscle hypertrophy [12] and, although debated, could thus have a role in replacing old and damaged muscle cells in a continuous manner [13].
To our knowledge, myogenic differentiation is poorly investigated in T2DM in humans, however, a decline in muscle mass and function in T2DM patients compared with healthy control subjects becomes clearer in aging patients [14], [15], [16]. Previous studies in muscle strength in T2DM in humans have suggested that loss of muscle quality may be due to diabetic neuropathy [17] yet the mechanism behind this loss of muscle quality currently remains unclear. The severe decline in skeletal muscle function during T2DM in terms of deficient glucose metabolism and insulin signaling certainly suggests a possibility for impaired muscle maintenance. We utilized a previously reported model system of isolated muscle stem cells from several donors which had T2DM or were healthy. Insulin resistance, inflammation and dysfunctional growth factor signaling, are among established abnormalities preserved, from an in vivo state of T2DM, in isolated, in vitro differentiated muscle stem cells [18], [19], [20], [21], [22]. Thus, to address our hypothesis, we studied miRNA regulation and differentiation of muscle stem cells derived from humans with T2DM or from healthy controls.
2. Experimental procedures
Additional information can be found in supplemental experimental procedures.
2.1. Human subjects
Human tissue and cell cultures were obtained from a subset of donors included in a previously described study [23]. The clinical characteristics of the human subjects are listed in Table 1. All participants gave written informed consent before inclusion and the study was performed according to the Declaration of Helsinki and approved by The Regional Committee on Biomedical Research Ethics in Denmark (KF 01-141/04).
Table 1.
Clinical characteristics of muscle stem cell donors.
| Healthy (n = 8) | T2DM (n = 8) | |
|---|---|---|
| Age (years) | 58.0 (50–67) | 58.9 (51–65) |
| BMI (kg/m2) | 25.4 ± 1.8 | 26.2 ± 2.8 |
| Fasting glucose (mmol/L) | 5.6 ± 0.6 | 11.3 ± 4.4** |
| OGTT 2-hour glucose (mmol/L) | 5.1 ± 1.8 | 20.2 ± 3.8**** |
| Fasting insulin (pmol/L) | 29.9 ± 16.5 | 42.3 ± 17.0 |
| OGTT 2-hour insulin (pmol/L) | 154. ± 125.9 | 152.6 ± 46.1 |
| HOMA-IR | 1.2 ± 0.7 | 3.9 ± 3.1* |
| VO2 max (L/min) | 3.0 ± 0.6 | 2.3 ± 0.8 |
| VO2 max (mL/kg/min) | 35.7 ± 8.4 | 26.2 ± 8.4* |
Data are means ± SE. BMI = body mass index; OGTT = oral glucose tolerance test. Glucose values are mmol/L, insulin values are pmol/L. Differences between groups was compared using student's unpaired t-test. *P < 0.05, **P < 0.001, ****P < 0.0001.
2.2. Human muscle stem cells
Muscle stem cells were isolated from vastus lateralis muscle biopsies and cultured and differentiated as previously described [18].
2.3. Flow cytometry
Isolated muscle stem cells were propagated in growth medium as described above until 70% confluence. Cells were detached using TrypLETM Express, and subsequently washed twice in wash buffer (phosphate buffered saline (PBS) containing 2% FBS and 0.01% NaN3) and once in staining buffer (PBS containing 2% FBS, 1% human serum, and 0.01% NaN3). Cells were stained with anti-human CD56-APC, CD90-PerCP-Cy5.5, CD31-PE, and CD45-BV421 (all from BD Bioscience) for 20 min and subsequently washed twice in wash buffer. Data was acquired using a FACSFortessa (BD Biosciences). For compensation, single stain was used with one drop of negative control beads and anti-mouse IgG beads (BD Biosciences). The gating strategy is shown in Figure S1. Data analysis was performed using Kaluza software version 1.2 (Beckman Coulter).
2.4. RNA isolation and qPCR
Total RNA was extracted from myocytes using TRIzol according to manufacturer's instructions. Quantitative real-time PCR (qPCR) was performed in triplicate using the ViiA™ 7 Real-Time PCR platform. The sequences of the target primers are listed in Supplementary experimental procedures. Data analysis was performed using the comparative method (ΔΔCT). All endogenous control genes utilized in the study showed significant difference CT values between healthy and T2DM groups at one or more but not all time points in differentiation; to ensure that observed differences in gene expression were not due to differences in endogenous control gene expression we therefore included several endogenous control genes.
2.5. miRNA array analysis
A miRCURY LNA™ microRNA Array (6th gen - hsa, mmu & rno) (Exiqon, Denmark) was utilized for global miRNA detection between human muscle stem cells derived from T2DM subjects and healthy controls during differentiation. Data has been submitted to GEO: GSE86069.
2.6. MiRNA target prediction analysis
Prediction analysis of KEGG pathways targeted by the identified miRNAs was performed using the DIANA miRPath online software [24] employing the microT-CDS algorithm.
2.7. miRNA inhibitor/mimic transfection in human myoblasts
Transient transfections of human myoblasts were performed using pools of either miRNA inhibitor oligonucleotides targeting miR-23b or miR-27b, or oligonucleotides mimicking endogenous miR-23b and miR-27b (Exiqon, Denmark). Transfections were performed by incubating myoblasts at day 0 or day 5 of differentiation with 50 nM miRNA inhibitor and RNAimax Lipofectamine (Invitrogen) for 48 h. Control conditions were incubated with either a scrambled oligonucleotide sequence predesigned to not target any miRNA or Lipofectamine with no oligonucleotide added.
2.8. Immunoflourescence microscopy
In vitro differentiated human muscle stem cells were fixated with 4% Formaldehyde (Sigma) and then permeabilized with 0.5% Triton X-100. Cells were then incubated with mouse monoclonal anti-sarcomeric Alpha Actinin antibody (Abcam ab9465) or incubated with an anti-myosin antibody (DSHB; MF 20 was deposited to the DSHB by Fischman, D.A. (DSHB Hybridoma Product MF 20)), and counterstained with 4′, 6-diamidino-2-phenylindole (DAPI). Alexa Flour 488 goat anti-mouse antibody was used as the secondary antibody (Molecular probes). Nuclear counterstaining was performed with Nucblue Fixed Cell stain ReadyProbes (Molecular probes). Fluorescence microscopy was performed with an EVOS FL (Thermo Fisher).
2.9. Statistical analyses
The quantified miRNA array signals were background corrected and then normalized using the global LOWESS (LOcally WEighted Scatterplot Smoothing) regression algorithm. A two-tailed t-test assuming unequal variance identified microRNAs with a p-value below the Bonferroni cut-off. For visualizing differentially expressed miRNAs during differentiation a Heatmap was performed using the online tool CIM miner. For visualizing the effect of T2DM, the online tool Plotly was used. Statistical analyses were performed using GraphPad Prism 6.0 (GraphPad Software Inc., La Jolla, CA, USA). All data within figures are presented as means ± SEM. Data in tables are presented as means ± SD. For comparisons between two groups, a Student's t-test was used. For multiple comparisons, statistical analysis was performed using two-way ANOVA with Sidak post hoc testing.
3. Results and discussion
3.1. Reduced myogenic capacity in cells derived from humans with T2DM
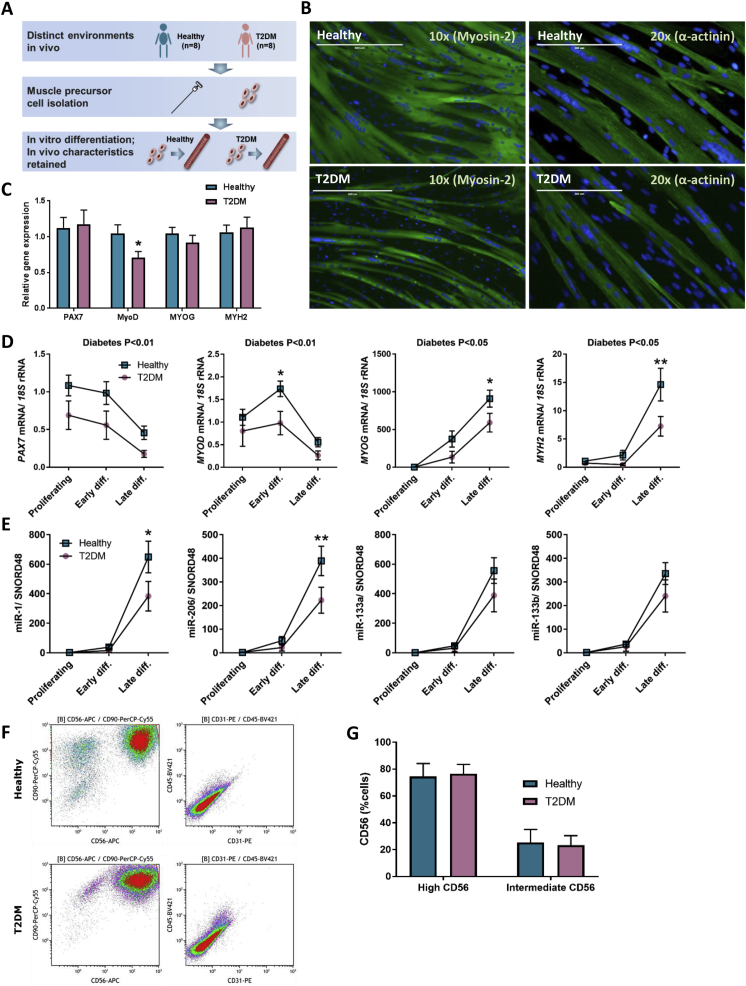
We compared the myogenic capacity of muscle stem cells obtained from eight non-obese humans with T2DM that were age- and BMI-matched with eight healthy, normal glucose tolerant control subjects. A schematic figure of the experimental setup is shown in Figure 1A. Phase contrast pictures of the cells during the different developmental stages were recorded (Figure S2).
Figure 1.
Myogenic transcription factors in differentiating muscle stem cells. (A) Schematic overview of the experimental setup. (B) Muscle stem cells from healthy (n = 8) or T2DM donors (n = 8) were isolated and differentiated in vitro. Differentiated myocytes were stained for DAPI (blue) and α-actinin (green) or Myosin-2 (green), representative pictures are shown. (C) qPCR analysis of PAX7, MYOD, MYOG and MYH2 in skeletal muscle biopsies. (D) qPCR analysis of PAX7, MYOD, MYOG and MYH2 in differentiating muscle cells. (E) qPCR analysis of miR-1, miR-206, miR-133a and miR-133b. F) Flow cytometry analysis of healthy and T2D muscle stem cells, stained for CD56 (APC), CD90 (PerCP-Cy5.5), CD31 (PE) and CD45 (BV421). Data are means ± SEM. * indicates P < 0.05; **P < 0.01.
Using immunofluorescence analysis we observed a severely impaired differentiation of the muscle stem cells derived from T2DM donors, as demonstrated by staining for either Myosin-2 or α-actinin (Figure 1B). Myogenesis is regulated by several transcription factors including Paired Box 7 (PAX7), Myoblast determination protein 1 (MYOD) and myogenin (MYOG) [25] as well as muscle-specific miRNAs, (myomiRs), including miR-1-3p (miR-1), miR-133a-3p (miR-133a), miR-133b and mir-206 [26], [27], [28]. To further compare the differentiation capacity between the two groups, we assessed the expression of the above mentioned myogenic regulators, as well as the structural muscle marker, Myosin-2. We observed a reduced expression of all myogenic markers measured in T2DM muscle cells (Figure 1D) as well as of two of the myomiRs: miR-1 and miR-206 (Figure 1E). A coordinated expression of both myogenic transcription factors and myomiRs is required for myogenesis, and in line with previous findings that MyoD regulates myomiR expression [28] we found that a decreased expression of MyoD in the T2DM cells preceded the observed decreased expression of miR-1 and miR-206 (Figure 1D, E). We next assessed whether differences in myogenic markers also were present in skeletal muscle tissue biopsies, and found that MYOD was lower expressed in humans with T2DM compared to healthy control subjects (Figure 1C). This finding supports prior reports [14], [15] on loss of muscle quality in T2DM and demonstrates a previously undescribed loss of myogenic capacity in these patients. To assure that the loss of myogenic gene expression observed in the T2DM cells was not due to a higher degree of infiltration by non-muscle cell types in this group, flow cytometry was applied. CD56, which is specifically displayed on human myoblast but not on adipocytes or fibroblasts [29], [30], was expressed on all cell cultures used in the study at either high or intermediate levels (Figure 1F,G). All cells were also CD90-positive, reflecting their mesenchymal origin [31], whereas cells were negative for the hematopoietic marker CD45 and the endothelial marker CD31 (Figure 1F). Thus, both cell groups were CD56+CD90+CD45−CD31–, with no observed difference between groups, suggesting equal purity of isolations and that any differences in differentiation capacity would not be related to isolation of different cell types.
In conclusion, we observed a reduced myogenic capacity of muscle precursor cells derived from humans with T2DM, despite a comparable myogenic purity of the cell strains between groups, suggesting a functional decline within the actual muscle stem cells.
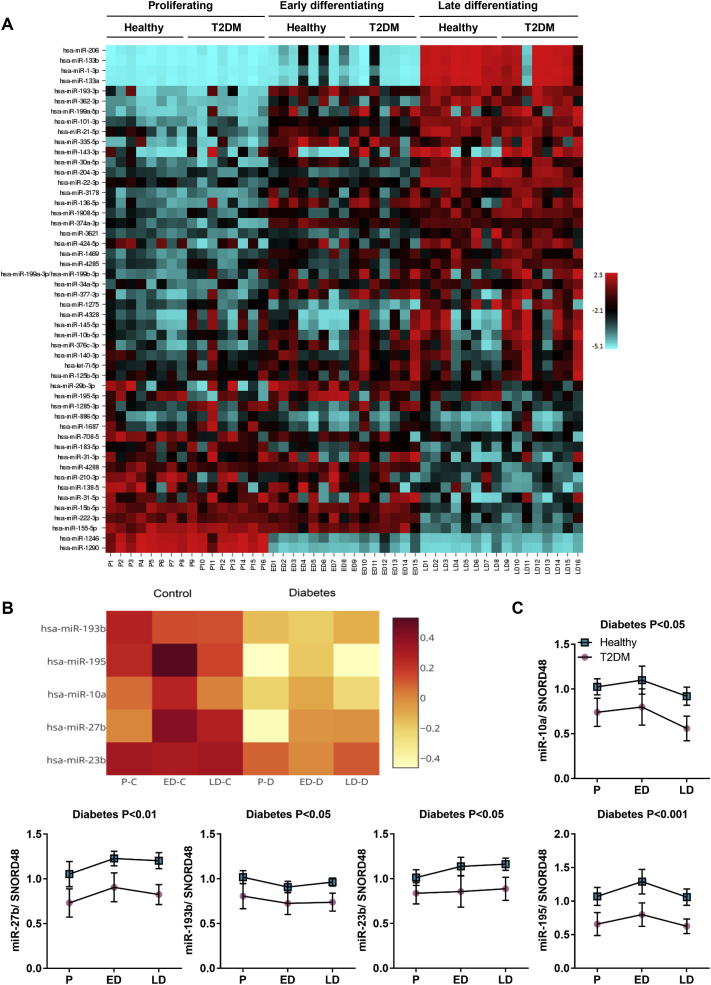
3.2. The miRNA signature of differentiating muscle stem cells from humans with T2DM
We measured the global miRNA expression at three time points during differentiation in T2DM and healthy muscle stem cells, using a miRCURY LNA™ microRNA Array (6th gen - hsa, mmu & rno) (Exiqon, Denmark). Consistent with the findings of a previous study analyzing in vitro differentiated human muscle cells [32], unsupervised clustering analysis of the miRNA expression showed a marked upregulation of myomiRs miR-1, miR-133a, miR-133b and miR-206 in differentiated compared to undifferentiated myoblasts. Interestingly, miR-1246 and miR-1290, two miRNAs previously undescribed in skeletal muscle, were regulated in a distinct temporal expression pattern opposite of the myomiRs; they were highly expressed in proliferating muscle stem cells but were downregulated as the cells went through differentiation, indicating a role of these two miRNAs as novel regulators of muscle stem cell differentiation (Figure S3A). We thus identified a time-dependent miRNA expression signature in human muscle cells, whereas no differences between the T2DM and healthy groups were observed at individual time points (Figure 2A). However, when combining all three time-points within each group, we found five miRNAs that all were downregulated in the T2DM muscle cells (Figure 2B). These miRNAs included: miR-10a-5p (miR-10a), miR-23b-3p (miR-23b), miR-27b-3p (miR-27b), miR-193b-3p (miR-193b) and miR-195-5p (miR-195), ranging from a 0.62- to a 0.84-fold decrease in the T2DM group which was confirmed by qPCR analysis (Figure 2C).
Figure 2.
Global analysis of miRNA expression during muscle stem cell differentiation. Muscle stem cells from healthy (n = 8) or T2DM (n = 8) donors were harvested as proliferating myoblasts or during myocyte differentiation. A miRNA array comprising 2383 capture probes was performed. In early differentiation, T2DM cells are n = 7, as one sample got lost in RNA isolation. A) Heatmap showing temporally regulated miRNAs; red indicates high expression, blue indicates low expression. B) Heatmap of miRNAs that were differentially expressed between healthy and T2DM groups; red indicates high expression, white indicates low expression. C) qPCR analysis of miR-10a, miR-23b, miR-27b, miR-193b and miR-195. Data are means ± SEM.
However, we did not detect any differences between the groups when measuring the miRNAs in the corresponding tissues from which the cells were isolated (Figure S3B), underlining the main weakness with the study; We utilized an in vitro model and thus cannot exclude that alterations in miRNA expression occurred as a secondary event, during in vitro processing. However, muscle tissue is more complex in respect to cell type composition and physiological context than in vitro differentiated cells. Other cell types might contribute to expressional noise that makes it difficult to detect a disease pattern in vivo. With our cell model, this noise is reduced, making it an attractive approach to detect small changes in miRNAs which might have great impact on disease phenotype. Nevertheless, a higher number of study participants might have been sufficient for detecting a difference even in tissue biopsies, as a downregulation of miR-10a and miR-27b in skeletal muscle biopsies of non-obese people with T2DM was previously described [9]. This supports our findings and argues for that the difference observed in miR-27b and miR-23b is a conserved feature from the in vivo T2DM condition. Furthermore, miR-23b, miR-27b and miR-195 were also found to be downregulated in a murine model of cachexia, while miR-27b was actually upregulated in mice with STZ-induced diabetes [33], supporting a role of miR-23b and miR-27b in skeletal muscle integrity and, at the same time, emphasizing the importance of studying these miRNAs in a human model of type 2 diabetes.
3.3. miR-23b and miR-27b are pro-myogenic and target p53 inhibitors
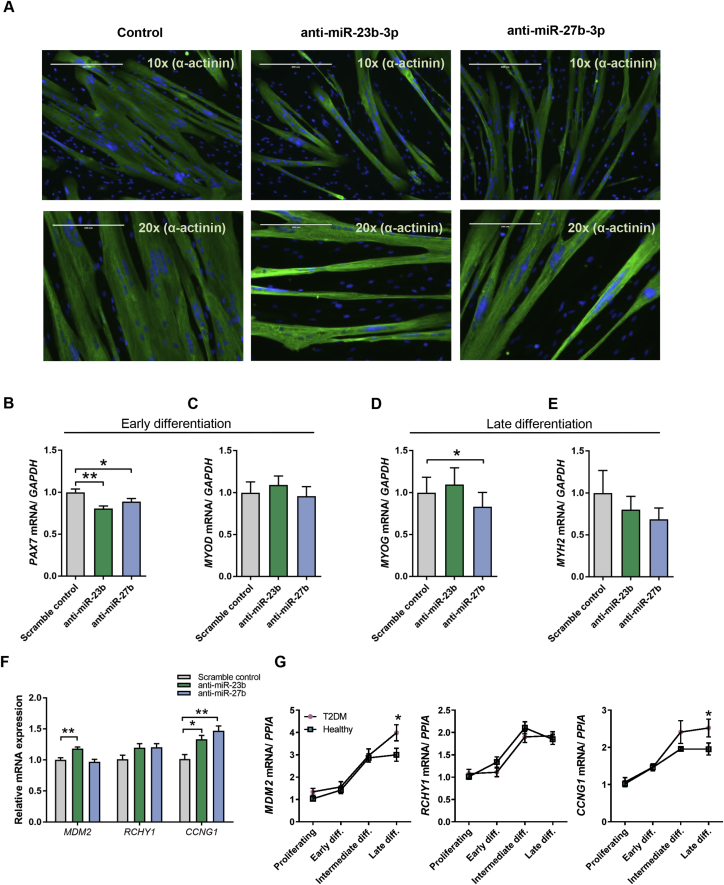
Two of the miRNAs downregulated in the muscle stem cells from humans with T2DM, miR-23b and miR-27b, caught our particular interest. As discussed above, miR-27b was downregulated in muscle tissue of humans with T2DM in a previous study, and miR-23b is transcribed from the same cluster along with miR-24-1 [34]. In fact, miR-24-1 was also downregulated in our data set, but with a p-value of 4.59E-4, this miRNA was just above significance after correcting for multiple testing (data not shown) and was therefore not included in downstream functional studies. Previous studies have suggested a role of miR-23b and miR-27b in the regulation of skeletal muscle tissue quality [35], [36]. In addition, miR-27b has been reported to exhibit pro-myogenic effects in murine cells [37], [38] and miR-23a (which has a high sequence similarity with miR-23b) has been shown to regulate myosin heavy chain isoforms in murine myoblasts [39]. We thus decided to examine the role of miR-23b and miR-27b in the differentiation of muscle stem cells derived from non-obese humans with T2DM and age- and BMI-matched healthy control subjects.
First, we studied differentiation of muscle stem cells derived from healthy subjects following knockdown of either miR-23b or miR-27b. Confluent myoblasts were transfected with miRNA inhibitors. Immunofluorescence analysis of differentiated myocytes stained for Sarcomeric alpha actinin (α-actinin) or Myosin-2, demonstrated an impaired differentiation of the cells depleted for miR-23b or miR-27b (Figure 3A). In accordance with the imaging analysis, myogenic markers were downregulated upon the reduction of miR-23b and miR-27b (Figure 3B–E). Specifically, knockdown of either miR-23b or miR-27b caused a reduction in PAX7 expression during early differentiation (Figure 3B), and MYOG expression was decreased after knockdown of miR-27b during late differentiation (Figure 3D), suggesting a reduced myogenic determination of cells with reduced expression of miR-23b and 27b.
Figure 3.
miR-23b and miR-27b regulate human myogenesis and target p53 inhibitors. (A–E) Muscle stem cells from healthy donors (n = 6) were transfected with 50 nM miRNA inhibitors against miR-23b or miR-27b or a scrambled control for 48 h. A) Transfected and differentiated myocytes were fixed and stained for α-actinin (green) or Myosin-2 (green) and nuclei (Nucblue, blue). For myogenic gene expression analysis, cells were harvested early in differentiation as aligned myoblasts (B–C) or as differentiated myocytes (D–E). Predicted targets of miR-23b and miR-27b were measured 48 h post transfection during early differentiation (F). Muscle stem cells from healthy (n = 8) or T2DM (n = 8) donors were harvested as proliferating myoblasts or during indicated time points of differentiation. mRNA expression of MDM2, RCHY1 and CCNG1 were measured by using qPCR and data was normalized to PPIA. mRNA expression levels are shown as fold changes from healthy proliferating cells (G). Data are means ± SEM. * indicates P < 0.05; **P < 0.01.
To determine potential targets of miR-23b and miR-27b, we used DIANA miRPath [24], combining a nucleotide base pairing prediction algorithm (microT-CDS) with the KEGG pathway database. This analysis assigned p53 signaling as the most likely pathway to be co-targeted by miR-23b and miR-27b (Table S1). Several genes known to negatively regulate p53 expression were specifically predicted to be targets of miR-23b and miR-27b, including the RING finger and CHY zinc finger domain-containing protein 1 (RCHY1) [40], [41], as well as Cyclin G1 (CCNG1) [42]. We selected these genes for downstream analysis due to their established role in cell cycle regulation as this would be important in any differentiation system. In further support of selecting CCNG1, it was recently demonstrated in cancer cells, that both miR-23b [43] and miR-27b [44], [45] directly target CCNG1 mRNA, providing an anticancer effect by diminishing the negative control of p53. In this respect, we decided to also investigate the ubiquitin-protein ligase Mouse double minute 2 (MDM2) as a potential target for miR-23b and miR-27b, even though this gene was not included in the results from the DIANA miRPath analysis. MDM2 is a major inhibitor of p53 and p21 and thus have an important role in the growth-regulating function of this pathway. Importantly, miR-23b was predicted to target MDM2 mRNA when assessed separately using the same algorithm as applied in the miRPath analysis (microT CDS).
We measured MDM2, RCHY1 and CCNG1 in muscle cells derived from healthy subjects and found an upregulation of MDM2 following transfection with a miR-23b inhibitor and an upregulation of CCNG1 following transfection with either a miR-23b inhibitor or a miR-27b inhibitor, while RCHY1 remained unchanged (Figure 3F). These findings were concordant with the expression of these genes in muscle cells derived from T2DM subjects compared to healthy, as both MDM2 and CCNG1 were higher expressed in cells from subjects with T2DM than in cells from healthy, while no difference was observed in RCHY1 expression (Figure 3G). These data suggest that miR-23b and miR-27b are promyogenic and are targeting MDM2 and CCNGI in human muscle stem cells.
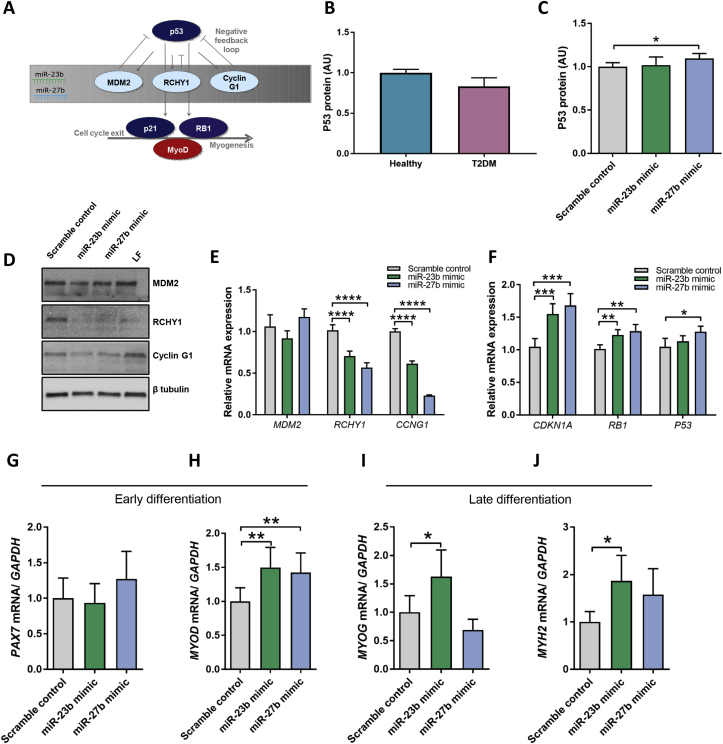
3.4. A bi-layered regulation of p53 by miR-23b/27b and its downstream targets
We next examined the effect of alterations of miR-23b and miR-27b on p53, the master regulator of growth and cell fate. P53 has an established role in myogenesis by inducing the expression of both CDKN1A (p21) and RB1 (retinoblastoma associated protein, Rb) [41], [46]. Induction of these proteins are required to exit the cell cycle in order to continue the myogenic programme [47], [48]. p21 and Rb interact with the myogenic transcription factor, MYOD, to coordinate the early stages of muscle differentiation [49], [50].
We hypothesized that miR-23b and miR-27b would be pro-myogenic by suppressing the negative regulators of p53, which in turn mediate cell cycle exit by p53 induction of p21 and Rb and their interaction with MyoD (Figure 4A). We could not measure any difference in protein expression of p53 in T2DM muscle cells compared to healthy during early differentiation (Figure 4B). However, when transfecting T2DM muscle cells with miR-27b mimics, a modest increase in p53 protein levels was observed (Figure 4C). Considering that an increase in p53 should be associated with a decrease in MDM2, RCHY1 and Cyclin G1 and an increase in p21, Rb and MyoD, we decided to investigate whether transfection with miR-23b and miR-27b mimics could rescue the T2DM muscle cells during early differentiation of muscle.
Figure 4.
miR-23b and miR-27b regulate the p53 signaling pathway in muscle stem cells. Schematic overview of the proposed interactions between the miR-23b/27b cluster, the p53 pathway and myogenesis (A). P53 protein levels were measured by electrochemiluminescent immunoassay (B–C). Muscle stem cells from T2DM donors were transfected with 50 nM miR-23b or miR-27b mimics, a scrambled, non-targeting oligonucleotide control and for immunoblots also a lipofectamine only control (LF). Cells were harvested 48 h post transfection. Protein levels of MDM2, RCHY1, Cyclin G1 and beta tubulin were determined using immunoblotting. A representative blot is shown (D). mRNA expression of MDM2, RCHY1 and CCNG1 (E) and p21 and RB1 and P53 (F) was measured by qPCR and data was normalized to GAPDH mRNA. Myogenic markers including PAX7 (G), MYOD (H), MYOG (I) and MYH2 (J). Data are means ± SEM. * indicates P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Indeed, transfection with miR-23b or miR-27b mimics resulted in robust decrease in at least RCHY1 and CCNG1 at mRNA and protein levels, while MDM2 was not affected (Figure 4D, E, Figure S4). Consistently, mimics of both miR-23b and miR-27b mediated increased mRNA expression of CDKN1A and RB1 as well as p53. Finally, as a secondary indication of rescue, we assessed the myogenic markers that were downregulated in T2DM muscle cells, and observed a substantial upregulation of MYOD during early differentiation following miR-23b or miR-27b mimics transfection. (Figure 4H). MYOG and MYH2 were induced during late differentiation following transfection with miR-23b, while PAX7 remained unaltered (Figure 4G and I, J). Thus, the present data is consistent with the miR-23b/27b cluster regulating p53 during early stages of muscle differentiation, by targeting its negative regulators, and hereby promoting expression of the pro-myogenic cell cycle exit genes p21 and Rb, resulting in progression of the myogenic programme.
In later stages of differentiation, the situation seems to be more complex. Here, p53 was actually upregulated in T2DM muscle cells compared to healthy muscle cells, but failed to upregulate Rb and p21 (Figure S5). In fact, Rb was even downregulated in the T2DM muscle cells compared to healthy control cells, consistent with the impaired myogenesis. Possible explanations to this discrepancy might be the multifaceted function of p53 in cell metabolism and the extensive control mechanisms regulating the abundance, location, activity and degradation of p53 [51], [52]. Importantly, p53 has been described to play dual roles in the regulation of myogenesis. It promotes myogenesis by induced Rb expression [46]. However, under conditions of stress, p53 induces apoptosis or cellular senescence and has been shown to directly inhibit myogenin expression and thus impair late stages of myogenesis [53]. Importantly, we demonstrate in this study that myogenin is downregulated during late differentiation in T2DM muscle cells compared to healthy controls and, additionally is downregulated following transfection with inhibitors of miR-27b.
4. Conclusion
In conclusion, our data constitutes a physiological illustration in human cells of miRNAs, located at the same locus, acting collectively to regulate a whole pathway. We present compelling evidence for a novel miR-23b/27b-p53/Rb pathway, dysregulated in muscle stem cells of non-obese humans with T2DM, resulting in compromised muscle cell differentiation in vitro, and possibly contributing to the impaired muscular phenotype of T2DM in humans.
Author contributions
C.S. designed the study with conceptual advice from M.P., A.V. and B.K.P.; C.S. and S.N. directed the study; T.I.H., S.N., P.K.D., T.J.L., C.S., H.S. and N.H., collected and analyzed data; T.I.H., S.N., and C.S. drafted the manuscript; all authors interpreted data, and edited and approved the final version of the article.
Acknowledgements
This study was supported by a grant from the Novo Nordisk Foundation (NNF12OC1016421) (C.S.), by a PhD scholarship from Faculty of Health Sciences, University of Copenhagen (T.I.H.) and by a research grant from the Danish Diabetes Academy supported by the Novo Nordisk Foundation (T.J.L). The Centre of Inflammation and Metabolism (CIM) is supported by a grant from the Danish National Research Foundation (DNRF55). The Centre for Physical Activity Research (CFAS) is supported by a grant from TrygFonden. CIM is part of the UNIK Project Food, Fitness & Pharma for Health and Disease, supported by the Danish Ministry of Science, Technology, and Innovation. CIM is a member of DD2, the Danish Center for Strategic Research in Type 2 Diabetes (the Danish Council for Strategic Research, grant no. 09-067009 and 09-075724). The Novo Nordisk Foundation Center for Basic Metabolic Research (http://www.metabol.ku.dk) is supported by an unconditional grant from the Novo Nordisk Foundation to University of Copenhagen.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molmet.2017.04.006
Conflict of interest
None declared.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Lee R.C., Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science (New York, N.Y.) 2001;294(5543):862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 2.Nielsen S., Scheele C., Yfanti C., Akerström T., Nielsen A.R., Pedersen B.K. Muscle specific microRNAs are regulated by endurance exercise in human skeletal muscle. The Journal of Physiology. 2010;588(Pt 20):4029–4037. doi: 10.1113/jphysiol.2010.189860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nielsen S., Åkerström T., Rinnov A., Yfanti C., Scheele C., Pedersen B.K. The miRNA plasma signature in response to acute aerobic exercise and endurance training. PLoS ONE. 2014;9(2) doi: 10.1371/journal.pone.0087308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trajkovski M., Hausser J., Soutschek J., Bhat B., Akin A., Zavolan M. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature. 2011;474(7353):649–653. doi: 10.1038/nature10112. [DOI] [PubMed] [Google Scholar]
- 5.Frost R.J.A., Olson E.N. Control of glucose homeostasis and insulin sensitivity by the Let-7 family of microRNAs. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(52):21075–21080. doi: 10.1073/pnas.1118922109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jordan S.D., Krüger M., Willmes D.M., Redemann N., Wunderlich F.T., Brönneke H.S. Obesity-induced overexpression of miRNA-143 inhibits insulin-stimulated AKT activation and impairs glucose metabolism. Nature Cell Biology. 2011;13(4):434–446. doi: 10.1038/ncb2211. [DOI] [PubMed] [Google Scholar]
- 7.Kornfeld J.-W., Baitzel C., Könner A.C., Nicholls H.T., Vogt M.C., Herrmanns K. Obesity-induced overexpression of miR-802 impairs glucose metabolism through silencing of Hnf1b. Nature. 2013;494(7435):111–115. doi: 10.1038/nature11793. [DOI] [PubMed] [Google Scholar]
- 8.Belgardt B., Ahmed K., Spranger M., Latreille M., Denzler R., Kondratiuk N. The microRNA-200 family regulates pancreatic beta cell survival in type 2 diabetes. Nature Medicine. 2015;21(6):619–627. doi: 10.1038/nm.3862. [DOI] [PubMed] [Google Scholar]
- 9.Gallagher I.J., Scheele C., Keller P., Nielsen A.R., Remenyi J., Fischer C.P. Integration of microRNA changes in vivo identifies novel molecular features of muscle insulin resistance in type 2 diabetes. Genome Medicine. 2010;2(2):9. doi: 10.1186/gm130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bork-Jensen J., Scheele C., Christophersen D.V., Nilsson E., Friedrichsen M., Fernandez-Twinn D.S. Glucose tolerance is associated with differential expression of microRNAs in skeletal muscle: results from studies of twins with and without type 2 diabetes. Diabetologia. 2015;58(2):363–373. doi: 10.1007/s00125-014-3434-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mauro Satellite cell of skeletal muscle fibers. The Journal of Biophysical and Biochemical Cytology. 1961;9:493–495. doi: 10.1083/jcb.9.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Serrano A.L., Baeza-Raja B., Perdiguero E., Jardí M., Muñoz-Cánoves P. Interleukin-6 Is an Essential Regulator of Satellite Cell-Mediated Skeletal Muscle Hypertrophy. Cell Metabolism. 2008;7(1):33–44. doi: 10.1016/j.cmet.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 13.Pallafacchina G., Blaauw B., Schiaffino S. Role of satellite cells in muscle growth and maintenance of muscle mass. Nutrition. Metabolism and Cardiovascular Diseases. 2013;23(Suppl 1):S12–S18. doi: 10.1016/j.numecd.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 14.Leenders M., Verdijk L.B., van der Hoeven L., Adam J.J., van Kranenburg J., Nilwik R. Patients with type 2 diabetes show a greater decline in muscle mass, muscle strength, and functional capacity with aging. Journal of the American Medical Directors Association. 2013;14(8):585–592. doi: 10.1016/j.jamda.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 15.Park S.W., Goodpaster B.H., Strotmeyer E.S., Kuller L.H., Brodeau R., Kammerer C. Accelerated loss of skeletal muscle strength in older adults with type 2. Diabetes Care. 2007;30(6) doi: 10.2337/dc06-2537. [DOI] [PubMed] [Google Scholar]
- 16.Park S.W., Goodpaster B.H., Strotmeyer E.S., de Rekeneire N., Harris T.B., Schwartz A.V. Decreased muscle strength and quality in older adults with type 2 diabetes: the health, aging, and body composition study. Diabetes. 2006;55(6):1813–1818. doi: 10.2337/db05-1183. [DOI] [PubMed] [Google Scholar]
- 17.Andersen H., Nielsen S., Mogensen C.E., Jakobsen J. Muscle strength in type 2 diabetes. Diabetes. 2004;53(6):1543–1548. doi: 10.2337/diabetes.53.6.1543. [DOI] [PubMed] [Google Scholar]
- 18.Green C.J., Pedersen M., Pedersen B.K., Scheele C. Elevated NF-κB activation is conserved in human myocytes cultured from obese type 2 diabetic patients and attenuated by AMP-activated protein kinase. Diabetes. 2011;60(11):2810–2819. doi: 10.2337/db11-0263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaster M., Petersen I., Hojlund K., Poulsen P., Beck-Nielsen H. The diabetic phenotype is conserved in myotubes established from diabetic subjects: evidence for primary defects in glucose transport and glycogen synthase activity. Diabetes. 2002;51(4):921–927. doi: 10.2337/diabetes.51.4.921. [DOI] [PubMed] [Google Scholar]
- 20.Bouzakri K., Roques M., Gual P., Espinosa S., Guebre-Egziabher F., Riou J.-P. Reduced activation of Phosphatidylinositol-3 kinase and increased serine 636 phosphorylation of insulin receptor Substrate-1 in primary culture of skeletal muscle cells from patients with type 2 diabetes. Diabetes. 2003;52(6):1319–1325. doi: 10.2337/diabetes.52.6.1319. [DOI] [PubMed] [Google Scholar]
- 21.Scheele C., Nielsen S., Kelly M., Broholm C., Nielsen A.R., Taudorf S. Satellite cells derived from obese humans with type 2 diabetes and differentiated into myocytes in vitro exhibit abnormal response to IL-6. PloS One. 2012;7(6):e39657. doi: 10.1371/journal.pone.0039657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Broholm C., Laye M.J., Brandt C., Vadalasetty R., Pilegaard H., Pedersen B.K. LIF is a contraction-induced myokine stimulating human myocyte proliferation. Journal of Applied Physiology. Bethesda, Md.: 1985. 2011;111(1):251–259. doi: 10.1152/japplphysiol.01399.2010. [DOI] [PubMed] [Google Scholar]
- 23.Pedersen M., Pedersen K.K., Bruunsgaard H., Krabbe K.S., Thomsen C., Færch K. Cognitive functions in middle aged individuals are related to metabolic disturbances and aerobic capacity: a cross-sectional study. PloS One. 2012;7(12):e51132. doi: 10.1371/journal.pone.0051132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vlachos I.S., Kostoulas N., Vergoulis T., Georgakilas G., Reczko M., Maragkakis M. DIANA miRPath v.2.0: investigating the combinatorial effect of microRNAs in pathways. Nucleic Acids Research. 2012;40(Web Server issue):W498–W504. doi: 10.1093/nar/gks494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chargé S.B.P., Rudnicki M.a. Cellular and molecular regulation of muscle regeneration. Physiological Reviews. 2004;84(1):209–238. doi: 10.1152/physrev.00019.2003. [DOI] [PubMed] [Google Scholar]
- 26.Chen J.-F., Mandel E.M., Thomson J.M., Wu Q., Callis T.E., Hammond S.M. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nature Genetics. 2006;38(2):228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dey B.K., Gagan J., Dutta A. miR-206 and -486 induce myoblast differentiation by downregulating Pax7. Molecular and Cellular Biology. 2011;31(1):203–214. doi: 10.1128/MCB.01009-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rao P.K., Kumar R.M., Farkhondeh M., Baskerville S., Lodish H.F. Myogenic factors that regulate expression of muscle-specific microRNAs. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(23):8721–8726. doi: 10.1073/pnas.0602831103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Agley C.C., Rowlerson A.M., Velloso C.P., Lazarus N.R., Harridge S.D.R. Human skeletal muscle fibroblasts, but not myogenic cells, readily undergo adipogenic differentiation. Journal of Cell Science. 2013;126(Pt 24):5610–5625. doi: 10.1242/jcs.132563. [DOI] [PubMed] [Google Scholar]
- 30.Laurens C., Louche K., Sengenes C., Coué M., Langin D., Moro C. 2015. Adipogenic progenitors from obese human skeletal muscle give rise to functional white adipocytes that contribute to insulin resistance. International Journal of Obesity. 2005 doi: 10.1038/ijo.2015.193. [DOI] [PubMed] [Google Scholar]
- 31.Maleki M., Ghanbarvand F., Reza Behvarz M., Ejtemaei M., Ghadirkhomi E. Comparison of mesenchymal stem cell markers in multiple human adult stem cells. International Journal of Stem Cells. 2014;7(2):118–126. doi: 10.15283/ijsc.2014.7.2.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sjögren R.J.O.O., Egan B., Katayama M., Zierath J.R., Krook A. Temporal analysis of reciprocal miRNA-mRNA expression patterns predicts regulatory networks during differentiation in human skeletal muscle cells. Physiological Genomics. 2014;46(0) doi: 10.1152/physiolgenomics.00037.2014. physiolgenomics.00037.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soares R.J., Cagnin S., Chemello F., Silvestrin M., Musaro A., De Pitta C. Involvement of MicroRNAs in the regulation of muscle wasting during catabolic conditions. Journal of Biological Chemistry. 2014;289(32):21909–21925. doi: 10.1074/jbc.M114.561845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu J., Wang F., Yang G.-H., Wang F.-L., Ma Y.-N., Du Z.-W. Human microRNA clusters: genomic organization and expression profile in leukemia cell lines. Biochemical and Biophysical Research Communications. 2006;349(1):59–68. doi: 10.1016/j.bbrc.2006.07.207. [DOI] [PubMed] [Google Scholar]
- 35.Panguluri S.K., Bhatnagar S., Kumar A., McCarthy J.J., Srivastava A.K., Cooper N.G. Genomic profiling of messenger RNAs and microRNAs reveals potential mechanisms of TWEAK-induced skeletal muscle wasting in mice. PloS One. 2010;5(1):e8760. doi: 10.1371/journal.pone.0008760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCarthy J.J., Esser K.A., Peterson C.A., Dupont-Versteegden E.E. Evidence of MyomiR network regulation of beta-myosin heavy chain gene expression during skeletal muscle atrophy. Physiological Genomics. 2009;39(3):219–226. doi: 10.1152/physiolgenomics.00042.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crist C.G., Montarras D., Pallafacchina G., Rocancourt D., Cumano A., Conway S.J. Muscle stem cell behavior is modified by microRNA-27 regulation of Pax3 expression. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(32):13383–13387. doi: 10.1073/pnas.0900210106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McFarlane C., Vajjala A., Arigela H., Lokireddy S., Ge X., Bonala S. Negative auto-regulation of myostatin expression is mediated by Smad3 and MicroRNA-27. PloS One. 2014;9(1):e87687. doi: 10.1371/journal.pone.0087687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang L., Chen X., Zheng Y., Li F., Lu Z., Chen C. MiR-23a inhibits myogenic differentiation through down regulation of fast myosin heavy chain isoforms. Experimental Cell Research. 2012;318(18):2324–2334. doi: 10.1016/j.yexcr.2012.06.018. [DOI] [PubMed] [Google Scholar]
- 40.Honda R., Tanaka H., Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Letters. 1997;420(1):25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 41.Leng R.P., Lin Y., Ma W., Wu H., Lemmers B., Chung S. Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell. 2003;112(6):779–791. doi: 10.1016/s0092-8674(03)00193-4. [DOI] [PubMed] [Google Scholar]
- 42.Ohtsuka T., Jensen M.R., Kim H.G., Kim K.-T., Lee S.W. The negative role of cyclin G in ATM-dependent p53 activation. Oncogene. 2004;23(31):5405–5408. doi: 10.1038/sj.onc.1207693. [DOI] [PubMed] [Google Scholar]
- 43.Yan J., Jiang J., Meng X., Xiu Y., Zong Z. MiR-23b targets cyclin G1 and suppresses ovarian cancer tumorigenesis and progression. Journal of Experimental & Clinical Cancer Research. 2016:1–10. doi: 10.1186/s13046-016-0307-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mu W., Hu C., Zhang H., Qu Z., Cen J., Qiu Z. miR-27b synergizes with anticancer drugs via p53 activation and CYP1B1 suppression. Cell Research. 2015;25(4):477–495. doi: 10.1038/cr.2015.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shang Y., Feng B., Zhou L., Ren G., Zhang Z., Fan X. The miR27b-CCNG1-P53-miR-508-5p axis regulates multidrug resistance of gastric cancer. Oncotarget. 2015 doi: 10.18632/oncotarget.6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Porrello A., Cerone M.A., Coen S., Gurtner A., Fontemaggi G., Cimino L. p53 regulates myogenesis by triggering the differentiation activity of pRb. The Journal of Cell Biology. 2000;151(6):1295–1304. doi: 10.1083/jcb.151.6.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guo K., Wang J., Andrés V., Smith R.C., Walsh K. MyoD-induced expression of p21 inhibits cyclin-dependent kinase activity upon myocyte terminal differentiation. Molecular and Cellular Biology. 1995;15(7):3823–3829. doi: 10.1128/mcb.15.7.3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Halevy O., Novitch B.G., Spicer D.B., Skapek S.X., Rhee J., Hannon G.J. Correlation of terminal cell cycle arrest of skeletal muscle with induction of p21 by MyoD. Science (New York, N.Y.) 1995;267(5200):1018–1021. doi: 10.1126/science.7863327. [DOI] [PubMed] [Google Scholar]
- 49.Huh M.S., Parker M.H., Scimè A., Parks R., Rudnicki M.A. Rb is required for progression through myogenic differentiation but not maintenance of terminal differentiation. The Journal of Cell Biology. 2004;166(6):865–876. doi: 10.1083/jcb.200403004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hawke T.J., Meeson A.P., Jiang N., Graham S., Hutcheson K., DiMaio J.M. p21 is essential for normal myogenic progenitor cell function in regenerating skeletal muscle. American Journal of Physiology. Cell Physiology. 2003;285(5):C1019–C1027. doi: 10.1152/ajpcell.00055.2003. [DOI] [PubMed] [Google Scholar]
- 51.Saldaña-Meyer R., Recillas-Targa F. Transcriptional and epigenetic regulation of the p53 tumor suppressor gene. Epigenetics. 2011;6(9):1068–1077. doi: 10.4161/epi.6.9.16683. [DOI] [PubMed] [Google Scholar]
- 52.Harris S.L., Levine A.J. The p53 pathway: positive and negative feedback loops. Oncogene. 2005;24(17):2899–2908. doi: 10.1038/sj.onc.1208615. [DOI] [PubMed] [Google Scholar]
- 53.Yang Z.J.P., Broz D.K., Noderer W.L., Ferreira J.P., Overton K.W., Spencer S.L. p53 suppresses muscle differentiation at the myogenin step in response to genotoxic stress. Cell Death and Differentiation. 2015;22(4):560–573. doi: 10.1038/cdd.2014.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.