Abstract
Objective
A significant portion of the heritable risk for complex metabolic disorders cannot be attributed to classic Mendelian genetic factors. At least some of this missing heritability is thought to be due to the epigenetic influence of parental and grandparental metabolic state on offspring health. Previous work suggests that this transgenerational phenomenon is evolutionarily conserved in Drosophila. These studies, however, have all depended on dietary paradigms to alter parental metabolic state, which can have inconsistent heritable effects on the metabolism of offspring.
Methods
Here we use AKHR null alleles to induce obesity in the parental generation and then score both metabolic parameters and genome-wide transcriptional responses in AKHR heterozygote F1 progeny and genetically wild-type F2 progeny.
Results
Unexpectedly, we observe elevated glycogen levels and changes in gene expression in AKHR heterozygotes due to haploinsufficiency at this locus. We also show that genetic manipulation of parental metabolism using AKHR mutations results in significant physiological changes in F2 wild-type offspring of the grandpaternal/maternal lineage.
Conclusions
Our results demonstrate that genetic manipulation of parental metabolism in Drosophila can have an effect on the health of F2 progeny, providing a non-dietary paradigm to better understand the mechanisms behind the transgenerational inheritance of metabolic state.
Keywords: Physiology, Transgenerational inheritance, Obesity, Drosophila metabolism
Abbreviations: AKH, Adipokinetic hormone; AKHR, Adipokinetic Hormone Receptor; GO, Gene Ontology; bmm, brummer
Highlights
-
•
Haploinsufficiency for AKHR affects metabolite levels and gene expression.
-
•
Genetic alteration of parental metabolism leads to heritable effects on offspring
-
•
Parental obesity selectively affects the grandpaternal/maternal lineage.
1. Introduction
Over the past several decades, the incidence of complex metabolic disease has increased at an alarming rate in developed countries. This increase is accompanied by significant social and economic costs, totaling $245 billion for diabetes in the United States in 2012 [1]. Despite this, the known genetic risk factors for diabetes account for only ∼10% of its heritability [2], [3]. Some of this missing heritability can be explained by gene-environment and gene–gene interactions, but the majority of the heritable risk is still unidentified [4], [5]. Over the last decade, however, it has become clear that at least some of this risk can be attributed to the epigenetic inheritance of the parental and even grandparental metabolic state.
Studies in famine-exposed human populations have suggested that parental and developmental caloric restriction can lead to metabolic dysfunction in adult progeny. The best controlled of these studies followed individuals conceived during a period from October 1944 through May 1945 in German-occupied Holland, when civilians were subjected to severe rationing. Compared to sibling controls born before October 1944 or conceived after May 1945, individuals exposed to famine during the first trimester were at a higher risk for diabetes, obesity, cardiovascular disease, and certain types of cancer [6], [7], [8]. Similar results were found in famine-exposed populations from China or Leningrad, especially when exposed individuals had access to a rich, Western-style diet later in life [9], [10]. Furthermore, paternal and multigenerational exposure to caloric restriction can impact the metabolism of adult children and grandchildren, suggesting that these phenotypes are due to more than gestational and developmental effects [11], [12].
This response appears to be conserved through evolution. Dietary conditioning of rodents in the parental generation can influence the metabolic state of their progeny. Both maternal and paternal low protein or high fat diets alter circulating and liver lipid levels, DNA methylation, and gene expression patterns through at least one and often two generations [13], [14], [15], [16], [17].
These molecular changes are accompanied by changes in the risk of diabetes, obesity, and cardiovascular disease [15], [18], [19], [20]. Recent evidence has linked these molecular and physiological phenotypes to changes in the expression of small RNAs in the germline, suggesting that metabolic changes in offspring might be due to the effects of these small RNAs on gene expression [13], [14], [21].
Although promising, the time and financial cost of doing these multigenerational experiments in mammalian models have inspired a move to Drosophila as an ideal system in which to study the effects of parental metabolic state on offspring metabolism [22], [23]. Treatment of either maternal or paternal flies with a high sugar diet leads to changes in glycemic levels and a propensity to develop obesity in progeny generations [24], [25]. Similarly, altering parental dietary protein impacts triglyceride and glycogen levels as well as longevity and fecundity in progeny, and a parental high fat diet can result in changes in body weight and major metabolites in the F1 and F2 generations [26], [27], [28]. These types of dietary manipulations in parents, however, are subject to environmental, sex-specific, and genotypic variation, which can affect the penetrance of progeny phenotypes [24], [28].
An alternative approach to induce metabolic changes in the parental generation is through genetic mutation, which has been used successfully for transgenerational studies in rodent models [29], [30], [31]. In this paradigm, a mutant is crossed to a wild-type individual to generate F1 heterozygous offspring, which are subsequently crossed to wild-type animals to generate genetically wild-type F2 offspring. In one example of this, the presence of one allele at the Obrq2A quantitative trait locus (QTL) in the parental generation is associated with low body weight and insulin sensitivity. Interestingly, the presence of this allele in grandparents leads to low body weight in wild-type F2 offspring, especially when passed through a grandmaternal/paternal pattern of inheritance [29]. This and other findings demonstrate that genetic perturbations in metabolic state of parents can effectively impact the physiology of progeny in rodents.
In this study, we employ a genetic approach to examine the effects of obesity in the parental generation of Drosophila using Adipokinetic Hormone Receptor (AKHR) loss-of-function mutations. Adipokinetic hormone (AKH) is released from neuroendocrine cells of the corpora cardiaca in response to fasting and acts remotely on the fat body to promote the release of stored lipids and carbohydrates for energy production [32]. In this manner, AKH functions analogously to the fasting hormone glucagon in mammals. Consistent with the catabolic activities of its ligand, loss of AKHR results in reduced fat body lipid mobilization and elevated triglyceride stores [33], [34]. We show that grandpaternal obesity resulting from a loss of AKHR is associated with low triglyceride levels in wild-type F2 offspring of heterozygous F1 mothers. In addition, we observed an unexpected effect of AKHR mutant heterozygosity in the F1 generation that results in elevated levels of stored glycogen and effects on gene expression. Our results indicate that genetic manipulation of parental metabolism in Drosophila provides an effective approach to induce transgenerational changes in metabolic state through the F2 generation. This paradigm should allow more reproducible studies of the effects of parental metabolic state on offspring health under controlled environmental and genetic conditions, facilitating the discovery of the molecular mechanisms that underlie this mode of inheritance.
2. Materials and methods
2.1. Drosophila stocks and maintenance
Drosophila melanogaster were maintained at 25 °C in an incubator with a 12 h light/dark cycle and transferred every two to four days to fresh media containing 1% agar, 8% yeast, 2% yeast extract, 2% peptone, 3% sucrose, 6% dextrose, 0.05% MgSO4·x6H2O, 0.05% CaCl2·x2H2O, 0.1% p-hydroxybenzoic acid methyl ester, and 0.6% propionic acid [35]. This media was prepared in the lab in large batches and stored for use over multiple generations, providing a stable nutritional environment. Adult males at 10 days of age under ad libitum feeding conditions were used for all experiments unless otherwise indicated. A transheteroallelic combination of the AKHRDsRed (Bloomington 140835) and AKHR1 alleles (a generous gift from R. Kühnlein) were used to generate AKHR null mutants for the parental generation [33], [36]. These alleles had been previously outcrossed to the Canton S wild-type line that was used as a control throughout this study in order to provide a uniform genetic background. The first, third, and Y chromosomes of the mutant stocks were completely exchanged for the Canton S chromosomes, and the second chromosome carrying the mutations was allowed to undergo free recombination for at least three generations. AKHR mutants were crossed to the same Canton S strain to generate the F1 and F2 generations. To generate the F1 and F2 offspring, males and female virgins were crossed and maintained on egg caps, which were replaced every 24 h. The first two egg collections were discarded, after which embryos were collected from egg caps and transferred to fresh vials at a density of approximately 50–100 embryos/vial. Embryos were collected on two consecutive days from each cross. Most studies were performed only in males, except as noted, because the metabolism of adult Drosophila females is heavily dedicated to oogenesis and reproduction.
2.2. Metabolite assays
Five adult male flies were collected at the indicated ages and washed in 1× PBS. Each sample was homogenized in 100 μL 1× PBS, after which 10 μL was reserved for a protein assay and the remainder of the lysate was heat-treated for 10 min at 70 °C. Protein, glucose, glycogen, and triglyceride assays were performed as described [37]. All experiments were replicated at least three times and were consistent across all replicates unless otherwise indicated.
2.3. RNA-seq transcriptional profiling
RNA was isolated from 10 to 11 day-old F1 or F2 male progeny using Trizol extraction (Thermo Fisher) and the Qiagen RNeasy Mini Kit. Library generation (Illumina TruSeq RNA Sample Preparation Kit v2 with oligo dT selection) and sequencing (HiSeq 50 Cycle Single Read Sequencing v3) were performed by the High-Throughput Genomics core facility at the University of Utah. The Bioinformatics Core Facility at the University of Utah aligned this dataset to the genome, utilizing the Genome Build DM3 from April 2006. Data from F2 sequencing was further analyzed using independent filtering approaches to account for batch effects. We restricted our analysis to identify transcripts that display an approximate 10% change in expression level by using a cut-off for significance of Log2 ratio ±0.13 and p-value <0.05. Gene Ontology analysis was performed using DAVID [38], [39]. RNA-seq data from this study can be accessed at NCBI GEO (accession numbers GSE97201 and GSE97525).
2.4. Statistical analysis
GraphPad PRISM 6 software was used to plot metabolite data and to perform statistical analysis. Pairwise comparisons in the P0 generation were performed using a standard Student's T-test with Welch's correction for unequal variances. In the F1 and F2 generations, one-way ANOVA was performed to compare the effects of parental or grandparental obesity on metabolite levels, with multiple comparisons tests to determine the statistical significance of differences between the individual groups by Sidak's multiple comparisons test. A two-way ANOVA was used to determine the effect of experimental replicates on variation between the groups in comparison with the effect of parental/grandparental obesity.
3. Results
3.1. Loss of AKHR function leads to parental obesity
We selected two loss-of-function AKHR alleles to induce metabolic dysfunction in the parental generation. One of these, AKHR1, is a deletion mutation generated by imprecise excision of a P-element [33]. The other allele, AKHRDsRed, carries a piggyBac insertion in the second intron of AKHR along with a splice acceptor and stop codon, resulting in early termination of AKHR translation [36]. Based on the location of the piggyBac insertion, this is predicted to be a null allele for the AKHR locus. The transposon also carries a reporter gene encoding an eye-specific fluorescent DsRed protein, facilitating the detection of this allele [36].
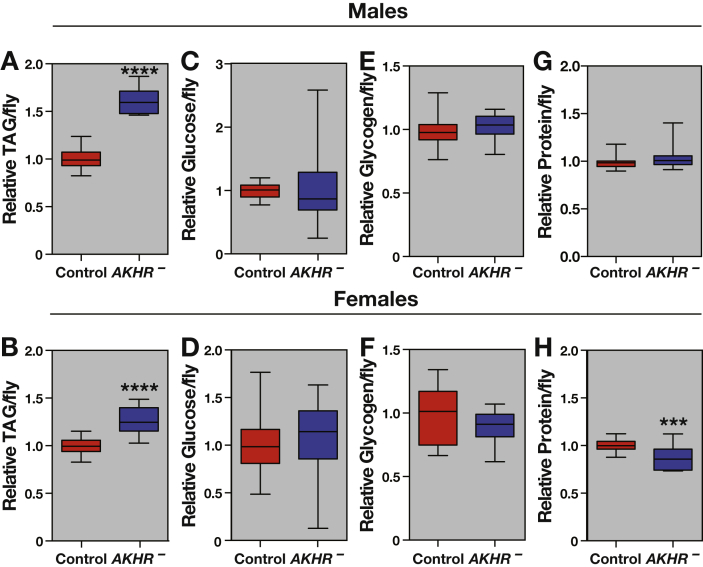
We used a wild-type inbred Canton S line for our studies along with AKHR1 and AKHRDsRed mutants that had been outcrossed into this Canton S genetic background. Metabolite measurements in transheterozygotes carrying the AKHR1 and AKHRDsRed alleles were consistent with the results of earlier genetic studies of AKHR function [33], [34]. Both males and females lacking AKHR have elevated triglycerides (Figure 1A and B), which serve as a marker for obesity in Drosophila, with no apparent changes in either glucose or glycogen levels (Figure 1C–F) [33], [40]. Protein levels are unchanged in males and slightly reduced in females (Figure 1G and H). The disruption in female protein, however, varied among experimental replicates (p < 0.05), while this was not the case for obesity in either males or females. We conclude that AKHR mutants have severe, reproducible defects in triglyceride mobilization.
Figure 1.
AKHR mutants are obese compared to genetically matched controls. Metabolites were measured from AKHRDsRed/AKHR1 mutant males and females (AKHR–) and genetically matched controls (n = 16). Triglycerides in males (A) and females (B) are substantially elevated relative to controls, while glycogen (C, D) and glucose levels (E, F) are unchanged. Protein is unchanged in males (G) but slightly reduced in females (H). Basic comparisons were analyzed using a Student's t-test with Welch's correction for unequal variance, while batch effects were determined using two-way ANOVA. ***p < 0.001, ****p < 0.0001.
3.2. Genetic crosses to generate wild-type F2 offspring from AKHR mutant parents
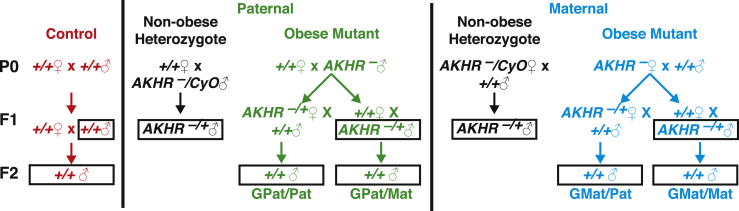
The dominant eye-specific DsRed marker in our AKHRDsRed mutant allowed us to follow this allele and observe the effect of parental obesity on future generations (Figure 2). Transheteroallelic AKHR1/AKHRDsRed mutant parental males and females were crossed to wild-type Canton S females and males to generate the F1 offspring, all of which were heterozygous for one of the two AKHR mutant alleles. F1 progeny were selected that carry the AKHRDsRed allele, and males and females of this genotype were crossed to wild-type females and males to generate wild-type F2 offspring (Figure 2). This crossing scheme allowed us to examine physiological phenotypes in wild-type progeny descended from obese parents when compared to wild-type progeny from parents with no obesity in their genealogical history. As controls, we also set up crosses between male or female AKHR−/+ heterozygotes and wild-type females or males to generate heterozygous AKHR−/+ F1 offspring from non-obese parents. These offspring were derived from heterozygous parents for at least two generations such that there was no expected metabolic dysfunction in either their parents or grandparents. In addition, heterozygotes were selected from crosses in which either the mother or father carried the AKHR mutant allele (Figure 2).
Figure 2.
Crossing scheme to generate F1 and F2 progeny from obese individuals. AKHRDsRed/AKHR1 mutant males or females (AKHR–) were crossed to wild-type females or males to generate heterozygous AKHRDsRed/+ F1 male and female progeny (AKHR−/+). These were subsequently crossed to wild-type females or males to generate genetically wild-type F2 progeny (+/+). As controls for the F1 generation, AKHRDsRed/CyO heterozygote males or females (AKHR−/CyO) were crossed to wild-type females or males to generate heterozygous AKHRDsRed/+ F1 male and female progeny (AKHR−/+). Wild-type control crosses (+/+) were maintained in both the F1 and F2 generations. Descendants from the wild-type control line are colored in red, from the heterozygote control lines in black, from the paternal obese line in green, and from the maternal obese line in cyan. Metabolites were measured in both F1 and F2 males (black boxes).
3.3. AKHR heterozygotes display elevated levels of glycogen
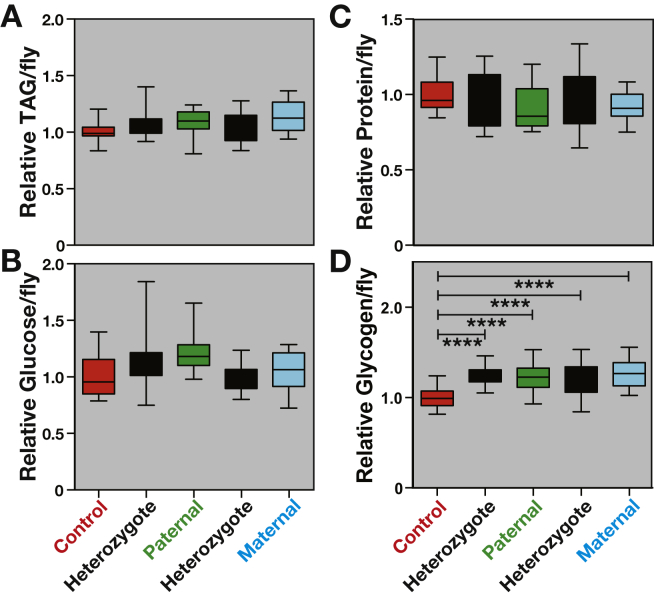
We first examined metabolite levels in the F1 generation of progeny from all five genetic crosses (Figure 2) to determine if any physiological phenotypes are dependent on the parental metabolic state. Interestingly, glycogen levels are significantly increased in all F1 AKHR heterozygotes descended from either obese or non-obese parents (Figure 3D, Supplementary Figure 1). Triglyceride levels are not changed between wild-type and either heterozygous controls or heterozygotes descended from an obese parent (Figure 3A). The same is true for glucose (Figure 3B) and protein (Figure 3C) levels. Our studies thus demonstrate that heterozygosity at the AKHR locus results in a reproducible effect on stored glycogen levels, something that has not been reported in past work [33], [34].
Figure 3.
AKHR heterozygotes have elevated glycogen levels. Metabolites were measured in F1 progeny from parents of the following genotypes: wild-type (red), AKHR−/+ heterozygotes (black), AKHR mutant fathers (green), and AKHR mutant mothers (cyan). Results from F1 progeny (n = 20/each) were pooled across two experimental replicates. F1 progeny from obese maternal or paternal parents have no physiological defects that are independent of genotype. Triglyceride (A), glucose (B), and protein (C) levels are largely unaffected in F1 heterozygous progeny descended from obese parents when compared to heterozygote controls or when compared to wild-type controls. Glycogen levels (D) are elevated in all heterozygous progeny when compared to controls. Comparisons among inheritance lines were performed using one-way ANOVA and Sidak's multiple comparisons test. Batch effects were determined using two-way ANOVA. Brackets are present to indicate the two samples being compared for the p-value presented. Only comparisons resulting in a significant difference that is consistent between the different experimental lines and controls are indicated. ****p < 0.0001.
3.4. Transcriptional profiling of F1 progeny
The lack of an effect on stored metabolites in the F1 generation may be due to the multiple, physiological pathways that maintain energy homeostasis. Because transcription is a more direct read-out of epigenetic state, we hypothesized that changes in mRNA levels might provide a better assessment of phenotypes in F1 offspring. We performed RNA-seq analysis on three biological replicates of F1 progeny from wild-type controls, AKHR−/+ heterozygous controls descended from non-obese parents, AKHR−/+ heterozygotes descended from obese paternal parents, and AKHR−/+ heterozygotes descended from obese maternal parents. Approximately half of the heterozygote controls were descended from paternal parents carrying the AKHRDsRed allele, while the other half were descended from maternal parents carrying the AKHRDsRed allele. RNA was extracted from 60 adult male flies for each replicate at approximately 10–11 days of age. Because we expected that the transcriptional differences caused by parental obesity might be subtle, transcripts were selected that changed more than 9.5% in expression level, with a significance of p < 0.05.
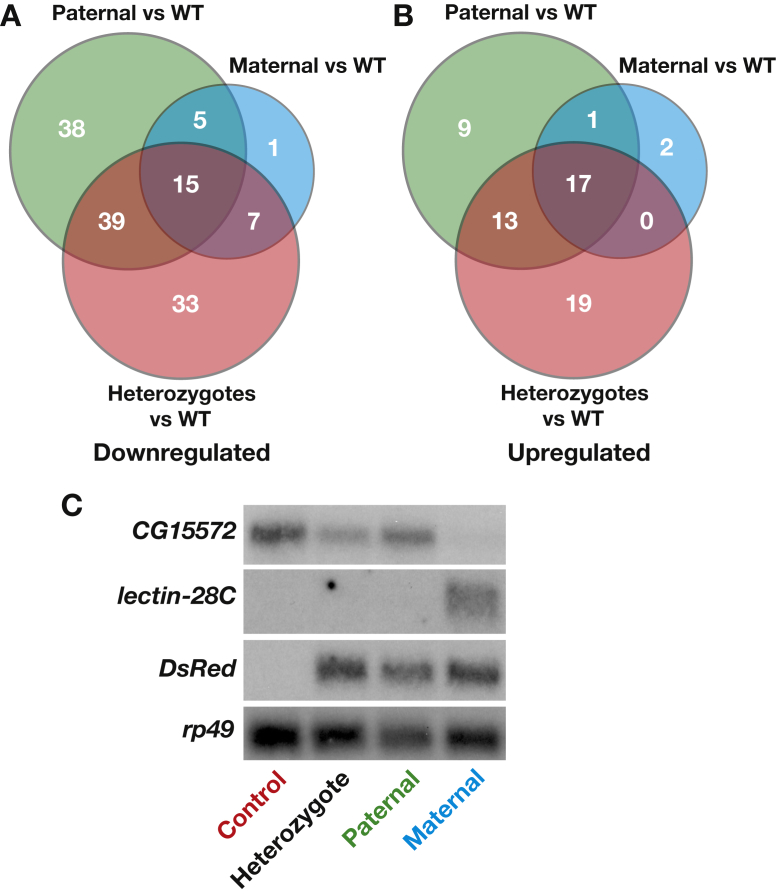
Consistent with the reproducible effects of AKHR heterozygosity on glycogen levels in F1 offspring, we detected a number of genes that are significantly affected by the loss of one copy of this receptor (Supplementary Table 1; Figure 4). There are 143 mis-regulated genes (94 down and 49 up) in heterozygotes descended from non-obese parents compared to wild-type controls. Gene Ontology (GO) analysis of these gene sets revealed that the down-regulated genes affect reproduction, while the up-regulated genes act in carbohydrate metabolic pathways, consistent with the increased stored carbohydrates in AKHR−/+ heterozygotes (Supplementary Tables 2 and 3).
Figure 4.
Transcriptional profiling of F1 offspring. Venn diagrams are presented that depict overlaps in genes that are either down-regulated (A) or up-regulated (B) in AKHR−/+ heterozygotes and F1 flies descended from obese paternal or maternal parents. (C) RNA was isolated from F1 controls, AKHR−/+ heterozygotes, or F1 flies descended from obese paternal or maternal parents and analyzed for CG15572 or lectin-28C expression by northern blot hybridization, as described [48]. Probes to detect DsRed and rp49 mRNA were used as controls. The changes in CG15572 and lectin-28C expression parallel those seen by RNA-seq.
There are 38 genes that are uniquely down-regulated in AKHR−/+ heterozygotes derived from obese fathers compared to wild-type controls (Figure 4A, Supplementary Tables 4 and 5). These include genes involved in sperm function and mating behavior, including seminal fluid proteins. These could indicate differences in the fecundity and germline function in F1 descendants of obese fathers, which may contribute to the lineage-specific F2 phenotypes described below. One gene is uniquely down-regulated in the maternal lineage: CR43494. This presumptive non-coding RNA, however, has no known function [41]. Five genes are expressed at reduced levels in both paternal and maternal obese lineages (Figure 4A, Supplementary Table 4). Interestingly, these genes include three gut-specific predicted serine proteases, suggesting a possible effect on dietary nutrient uptake (Supplementary Table 4).
There are nine genes that are uniquely up-regulated in AKHR−/+ heterozygotes descended from obese fathers compared to wild-type controls (Figure 4B, Supplementary Tables 4 and 5). These genes include Larval serum protein 1α (Lsp1α), which is thought to provide a source of amino acids and energy during metamorphosis, as well as MtnB, which is involved in metal ion homeostasis. Two genes are uniquely up-regulated in the maternal lineage: lectin-28C and CG5966 (Figure 4B, Supplementary Tables 4 and 6). Interestingly, lectin-28C encodes a predicted galactose-binding protein and CG5966 encodes a presumptive triglyceride lipase that is highly expressed in the fat body [42], [43]. The mis-regulation of these genes could be indicative of metabolic changes in the F1 that are buffered by compensating pathways. Finally, only one up-regulated gene, Gpdh, is shared between the paternal and maternal obese lineages (Figure 4B, Supplementary Table 4). This gene encodes glycerol-3-phosphate dehydrogenase, which plays an important role in lipid biosynthesis as well as in coupling NADH production during glycolysis with mitochondrial oxidative phosphorylation through the glycerol-phosphate shuttle. The mis-regulation of this gene could thus contribute to the F2 metabolic defects described below.
Several transcripts were selected for validation by northern blot hybridization based on their statistically significant differences in expression between the heterozygous controls and the maternal or paternal F1 lineages. The testes specific gene CG15572 is up-regulated in the paternal lineage when compared to heterozygous progeny of non-obese parents (∼1.2-fold increased, p = 10−73), while it is down-regulated in the maternal lineage (∼1.4-fold decreased, p = 10−143). When comparing CG15572 levels between heterozygous progeny of non-obese parents with wild-type controls, however, it is still significantly down-regulated (∼1.2-fold decreased, p = 10−67). This pattern of expression is confirmed by northern blot analysis (Figure 4C). Only one other gene, the predicted galactose-binding lectin-28C, is significantly up-regulated in the maternal line compared to heterozygous progeny of non-obese parents (∼1.4-fold increased, p = 10−156). This was confirmed by northern blot analysis (Figure 4C). While the transcriptional differences between the heterozygous progeny of obese and non-obese parents are limited, the few we have identified could be indicative of broader defects that cannot be detected above the more dramatic effects of heterozygosity at the AKHR locus.
3.5. Grandpaternal/maternal inheritance of metabolic dysfunction in the F2 generation
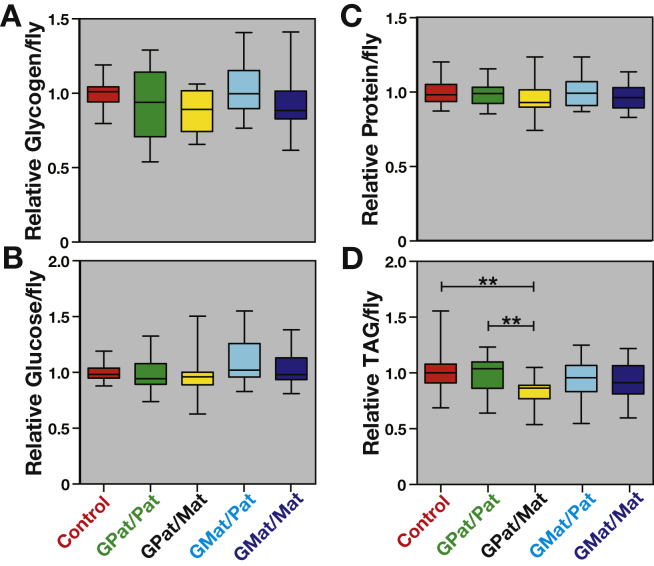
We measured stored metabolites in wild-type F2 offspring in order to determine if parental obesity might lead to reproducible changes in their physiology (Figure 5). Glycogen, glucose, and protein levels are unchanged between controls and all F2 progeny descended from an obese parent (Figure 5A–C). Importantly, however, triglyceride levels are significantly lower in F2 progeny descended from obese grandfathers and heterozygous mothers, or with grandpaternal/maternal inheritance (Figure 5D). This reduction is significant whether the comparison is made between flies of grandpaternal/maternal lineage and controls (p < 0.01) or the grandpaternal/paternal lineage (p < 0.01). Furthermore, there is no significant variation between experimental replicates in triglyceride levels between flies of the grandpaternal/maternal lineage and controls (Supplementary Figure 2).
Figure 5.
F2 progeny descended from obese paternal grandparents and heterozygous mothers are lean. Metabolite levels were measured in wild-type controls (red) and F2 descendants of obese parents of the following lineages: grandpaternal/paternal (GPat/Pat, green), grandpaternal/maternal (GPat/Mat, yellow), grandmaternal/paternal (GMat/Pat, cyan), and grandmaternal/maternal (GMat/Mat, blue). Whole body extracts were prepared at approximately 10 days of adult age and represent combined data from three experimental replicates (n = 25/each). There is no significant difference between controls and any of the F2 descendants of obese parents in glycogen (A), glucose (B), or protein (C) levels. Descendants of the grandpaternal/maternal inheritance line have reduced triglycerides when compared to controls (D). There was no effect of experimental variation across replicates pooled in this dataset when comparing control and grandpaternal/maternal samples. Comparisons between inheritance lines were performed using one-way ANOVA and Sidak's multiple comparisons test. Batch effects were determined using two-way ANOVA. Brackets are present to indicate the two samples being compared for the p-value presented. Only comparisons resulting in a significant difference are indicated. **p < 0.01.
3.6. Transcriptional profiling of F2 progeny
As described above for the F1 offspring, the effects of parental obesity might have a greater impact on the transcriptome of wild-type F2 offspring than it does on stored metabolite levels. We therefore performed RNA-seq analysis on F2 offspring that are genetically identical to the wild-type controls at the AKHR locus. Because we observed changes in triglycerides only in F2 offspring descended through a grandpaternal/maternal lineage, we only analyzed samples with a history of grandpaternal obesity. RNA was again extracted from three biological replicates of 60 adult male flies at approximately 10–11 days of age. Analysis was performed comparing wild-type controls, F2 offspring of the grandpaternal/paternal lineage, and F2 offspring of the grandpaternal/maternal lineage. Because changes might be even subtler in the F2 generation than in the F1 generation, the batches in which samples were collected were taken into account for the analysis. Once again, we expected that the transcriptional changes might be small so we selected transcripts that changed more than 9.5% with a significance of p < 0.05.
We found significantly altered transcript levels at five loci in the grandpaternal/maternal lineage compared with either the wild-type controls or with matched F2 offspring descended from the grandpaternal/paternal lineage (Table 1). This is consistent with the changes in triglyceride levels detected only in F2 progeny descended from a grandpaternal/maternal lineage. The genes ACC (∼1.13-fold increased, p = 0.008), hairy (∼1.13-fold increased, p = 0.009), totA (∼1.17-fold increased, p = 10−6), and totC (∼1.19-fold increased, p = 10−6) are all reproducibly and significantly elevated in F2 descendants of the grandpaternal/maternal lineage when compared to controls, while the gene fst (∼1.2-fold increased, p = 10−6) is significantly elevated in F2 descendants of the grandpaternal/maternal lineage when compared to those of the grandpaternal/paternal lineage. No significant changes were observed when the transcriptomes of the grandpaternal/paternal and control lineages were compared.
Table 1.
Gene altered in expression in grandpaternal/maternal F2 progeny
| Flybase gene ID | Associated gene name | LgR2rt | Fold change | P-value | Function |
|---|---|---|---|---|---|
| Grandpaternal–maternal vs control | |||||
| FBgn0044812 | totC | 0.251 | 1.19 | 1.51E-06 | Stress response |
| FBgn0028396 | totA | 0.232 | 1.17 | 1.95E-06 | Stress response |
| FBgn0033246 | ACC | 0.180 | 1.13 | 8.02E-03 | Fatty acid biosynthesis |
| FBgn0001168 | h | 0.181 | 1.13 | 8.53E-03 | Hairy (Hes family of suppressors) |
| Grandpaternal–maternal vs grandpaternal–paternal | |||||
| FBgn0037724 | fst | 0.262 | 1.20 | 1.68E-06 | Cold/Stress response |
4. Discussion
4.1. Genetic models induce reproducible changes in parental metabolic state
By using AKHR mutants to induce obesity, we were able to generate a reliable and severe metabolic defect in the parental generation. Male triglycerides are significantly increased in AKHR mutants when compared to controls. This degree of obesity is difficult to induce using standard dietary methods. The AKHR model therefore provides a more severe and consistent alteration in the metabolic state of the parental generation under constant nutritional conditions. This consistency likely contributed to the significant changes in metabolite levels seen in wild-type F2 offspring.
4.2. Heterozygosity at the AKHR locus produces unexpected metabolic phenotypes
Previous genetic studies of AKHR have assumed that the existing loss-of-function mutations behave as classic recessive alleles and thus should not display haploinsufficient phenotypes [33], [34]. Here we show that not only is there a reproducible 20–30% increase in glycogen levels in flies lacking one copy of the AKHR locus but also that these changes are accompanied by a transcriptional response. Our RNA-seq study identified 143 genes that change their expression when compared to wild-type levels, all with p-values that are less than 10−14. This demonstrates that the loss of one copy of AKHR leads to significant transcriptional and physiological effects. Moreover, the increased glycogen we report is consistent with the well-known role of AKH in mobilizing glycogen stores in response to nutrient deprivation [34]. Our results suggest that the genetic dose of AKHR is important for maintaining carbohydrate homeostasis in Drosophila under normal feeding conditions.
The presence of defects due to haploinsufficiency may also impact our ability to reliably detect metabolic and transcriptional changes in the F1 progeny of obese parents. The changes we would expect to see from inherited metabolic phenotypes are subtle and therefore likely to be weaker than genotypic effects caused by haploinsufficiency. This is a general concern in any genetic transgenerational paradigm and bears consideration when developing such a system. In addition, heterozygosity in the F1 generation could potentially complicate our interpretation of the F2 phenotypes. It is possible that the elevated glycogen associated with AKHR heterozygosity in the F1 generation could contribute to the phenotypes we report in F2 offspring. Future studies will be necessary to clarify the effects of altered metabolism in F1 AKHR−/+ animals on the F2 generation. These efforts could also include the use of alternative approaches to genetically alter metabolism in the parental generation using mutations that have no demonstrable haploinsufficient phenotype.
4.3. Inheritance of metabolic phenotypes in a lineage specific pattern
Earlier studies that used a dietary paradigm to induce parental obesity found that this led to significantly elevated glycogen and triglyceride levels in F1 offspring [24], [25]. Although we did not observe any metabolic defects in the F1 generation using our genetic paradigm, this could be due to the effects of AKHR heterozygosity, as described above, or a fundamental difference in the nature of parental obesity induced by either a dietary or genetic approach. However, by carrying our study out to the F2 generation, we made the interesting observation that a specific inheritance pattern is required to induce a physiological effect in the genotypically wild-type F2 offspring. Only F2 progeny descended from obese grandfathers through heterozygous mothers have reduced triglycerides when compared to controls. This grandpaternal/maternal pattern of inheritance is reminiscent of genetic models in rodents and humans, in which phenotypes often are seen only in one sex in the offspring or through a specific lineage [11], [12], [29]. It is unclear why the phenotypes we have observed are only inherited in an alternating parental gender pattern. Future studies including both male and female progeny could clarify these inheritance patterns. Furthermore, identifying the mechanism by which metabolic effects induce the transmission of physiological effects to the next generation might help explain this pattern of inheritance.
As a first step toward addressing the mechanisms that underlie transgenerational inheritance, we performed RNA-seq transcriptional profiling on both F1 and F2 offspring. Most notably, we observed only a handful of genes that displayed significant changes in gene expression in F2 descendants of the grandpaternal/maternal lineage. Remarkably, these genes include two of the Tot genes, totA and totC, out of a family of only seven such genes. Although not directly involved in metabolism, these genes, along with fst, contribute to stress responses and may indicate a compensatory response to the physiological state of F2 offspring [44], [45]. Interestingly, we also identified a significant change in the expression of ACC, which encodes a conserved acetyl-CoA carboxylase that acts as the rate-limiting step in fatty acid synthesis [46]. Although the low triglyceride levels observed are the opposite of what would be expected from elevated ACC activity, it is possible that this transcriptional change is a compensatory response to the reduction in stored lipid levels. The hairy (h) locus, on the other hand, encodes a well-characterized transcriptional repressor that is required for embryonic development and has recently been associated with the proper expression of key genes in the glycolytic pathway and tricarboxylic acid cycle [47]. Taken together, these changes in gene expression focus attention on stress responses and specific metabolic pathways, providing directions for future studies that could address how these transcriptional changes might contribute to the metabolic defects observed in F2 offspring.
4.4. Altered F2 physiology is sensitive to environmental effects
It is important to note that the reduced triglycerides observed in F2 offspring from obese grandfathers and heterozygous mothers is subject to the same kind of experimental variation as observed in dietary conditioning paradigms. The metabolic assays for the F2 presented here were derived from three independent biological replicates conducted over a period of two months. However, when a fourth replicate was performed several months later, triglyceride levels were unchanged in the F2 lineages. We were also unable to detect transcriptional changes seen in our RNA-seq by northern blot hybridization studies using RNA samples isolated from animals several months later. This variability is not unexpected because the effects reported here in F2 offspring are relatively small and the environmental conditions likely changed during the intervening period of time. Additionally, we and others have noted that transgenerational metabolic studies using dietary paradigms often result in variable offspring phenotypes [24], [28]. The success of this study likely reflects the effectiveness of the obese phenotype of AKHR parents in the absence of the subtle variations inherent in the use of dietary paradigms to alter parental metabolism. Many other genes that play central roles in metabolism have mutant alleles that carry a dominant marker. For example, a dsRed-marked presumptive null allele for bmm, which encodes the rate-limiting enzyme in triglyceride mobilization, is available from the Bloomington stock center [40]. Future experiments using this or other mutant alleles hold the promise of providing robust changes in parental metabolism that can facilitate more detailed functional analysis of the mechanisms underlying the transgenerational inheritance of metabolic state.
Acknowledgements
We thank M. Sieber for his contributions to the initial stages of this project, R. Kühnlein for the AKHR1 mutant allele, the University of Utah High-Throughput Sequencing and Bioinformatics core facilities, the Bloomington Stock Center for stocks, and FlyBase for informatic support. This research was supported by NIH R01 DK095346 and NIH Metabolism Training Grant support for R.A.S.P. (T32 DK091317).
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molmet.2017.03.012.
Contributor Information
Rebecca A.S. Palu, Email: rsomer@genetics.utah.edu.
Carl S. Thummel, Email: carl.thummel@genetics.utah.edu.
Conflicts of interest
There are no known conflicts of interest associated with the research outlined in this publication.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
AKHR–/+ heterozygotes compared to +/+ wild-type.
GO categories of AKHR–/+ heterozygote down-regulated genes.
GO categories of AKHR–/+ heterozygote up-regulated genes.
Genes in each overlap category from the Venn diagram shown in Figure 4.
Genes that change expression in paternal F1 lineage.
Genes that change expression in maternal F1 lineage.
References
- 1.CDCP . National diabetes statistics report: estimates of diabetes and its burden in the United States. In: U.S. Department of Health and Human Services CfDCaP, editor. 2014. Atlanta, GA 2014. [Google Scholar]
- 2.Imamura M., Maeda S. Genetics of type 2 diabetes: the GWAS era and future perspectives [Review] Endocrine Journal. 2011;58(9):723–739. doi: 10.1507/endocrj.ej11-0113. PubMed PMID: 21778616. [DOI] [PubMed] [Google Scholar]
- 3.Billings L.K., Florez J.C. The genetics of type 2 diabetes: what have we learned from GWAS? Annals of the New York Academy of Sciences. 2010;1212:59–77. doi: 10.1111/j.1749-6632.2010.05838.x. PubMed PMID: 21091714; PubMed Central PMCID: PMCPMC3057517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cordell H.J. Detecting gene-gene interactions that underlie human diseases. Nature Reviews Genetics. 2009;10(6):392–404. doi: 10.1038/nrg2579. PubMed PMID: 19434077; PubMed Central PMCID: PMCPMC2872761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manolio T.A., Collins F.S., Cox N.J., Goldstein D.B., Hindorff L.A., Hunter D.J. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–753. doi: 10.1038/nature08494. PubMed PMID: 19812666; PubMed Central PMCID: PMCPMC2831613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lumey L.H., Stein A.D., Kahn H.S., Romijn J.A. Lipid profiles in middle-aged men and women after famine exposure during gestation: the Dutch Hunger Winter Families Study. The American Journal of Clinical Nutrition. 2009;89(6):1737–1743. doi: 10.3945/ajcn.2008.27038. PubMed PMID: 19386743; PubMed Central PMCID: PMC2682992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Rooij S.R., Painter R.C., Holleman F., Bossuyt P.M., Roseboom T.J. The metabolic syndrome in adults prenatally exposed to the Dutch famine. The American Journal of Clinical Nutrition. 2007;86(4):1219–1224. doi: 10.1093/ajcn/86.4.1219. PubMed PMID: 17921405. [DOI] [PubMed] [Google Scholar]
- 8.Ravelli A.C., van Der Meulen J.H., Osmond C., Barker D.J., Bleker O.P. Obesity at the age of 50 y in men and women exposed to famine prenatally. The American Journal of Clinical Nutrition. 1999;70(5):811–816. doi: 10.1093/ajcn/70.5.811. PubMed PMID: 10539740. [DOI] [PubMed] [Google Scholar]
- 9.Li Y., He Y., Qi L., Jaddoe V.W., Feskens E.J., Yang X. Exposure to the Chinese famine in early life and the risk of hyperglycemia and type 2 diabetes in adulthood. Diabetes. 2010;59(10):2400–2406. doi: 10.2337/db10-0385. PubMed PMID: 20622161; PubMed Central PMCID: PMC3279550, Epub 2010/07/14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stanner S.A., Bulmer K., Andres C., Lantseva O.E., Borodina V., Poteen V.V. Does malnutrition in utero determine diabetes and coronary heart disease in adulthood? Results from the Leningrad siege study, a cross sectional study. British Medical Journal. 1997;315(7119):1342–1348. doi: 10.1136/bmj.315.7119.1342. Epub 1997/12/24. PubMed PMID: 9402775; PubMed Central PMCID: PMC2127836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bygren L.O., Tinghog P., Carstensen J., Edvinsson S., Kaati G., Pembrey M.E. Change in paternal grandmothers' early food supply influenced cardiovascular mortality of the female grandchildren. BMC Genetics. 2014;15:12. doi: 10.1186/1471-2156-15-12. PubMed PMID: 24552514; PubMed Central PMCID: PMCPMC3929550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pembrey M., Saffery R., Bygren L.O., and Network in Epigenetic Epidemiology Human transgenerational responses to early-life experience: potential impact on development, health and biomedical research. Journal of Medical Genetics. 2014;51(9):563–572. doi: 10.1136/jmedgenet-2014-102577. PubMed PMID: 25062846; PubMed Central PMCID: PMCPMC4157403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharma U., Conine C.C., Shea J.M., Boskovic A., Derr A.G., Bing X.Y. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science. 2016;351(6271):391–396. doi: 10.1126/science.aad6780. PubMed PMID: 26721685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Q., Yan M., Cao Z., Li X., Zhang Y., Shi J. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science. 2016;351(6271):397–400. doi: 10.1126/science.aad7977. PubMed PMID: 26721680. [DOI] [PubMed] [Google Scholar]
- 15.Langley-Evans S.C. Nutritional programming of disease: unravelling the mechanism. Journal of Anatomy. 2009;215(1):36–51. doi: 10.1111/j.1469-7580.2008.00977.x. PubMed PMID: 19175805; PubMed Central PMCID: PMCPMC2714637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Warner M.J., Ozanne S.E. Mechanisms involved in the developmental programming of adulthood disease. Biochemical Journal. 2010;427(3):333–347. doi: 10.1042/BJ20091861. PubMed PMID: 20388123. [DOI] [PubMed] [Google Scholar]
- 17.Rando O.J., Simmons R.A. I'm eating for two: parental dietary effects on offspring metabolism. Cell. 2015;161(1):93–105. doi: 10.1016/j.cell.2015.02.021. PubMed PMID: 25815988; PubMed Central PMCID: PMCPMC4465102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Langley-Evans S.C. Fetal programming of cardiovascular function through exposure to maternal undernutrition. The Proceedings of the Nutrition Society. 2001;60(4):505–513. doi: 10.1079/pns2001111. PubMed PMID: 12069404. [DOI] [PubMed] [Google Scholar]
- 19.Schaefer S., Nadeau J.H. The genetics of epigenetic inheritance: modes, molecules, and mechanisms. The Quarterly Review of Biology. 2015;90(4):381–415. doi: 10.1086/683699. PubMed PMID: 26714351. [DOI] [PubMed] [Google Scholar]
- 20.Jimenez-Chillaron J.C., Isganaitis E., Charalambous M., Gesta S., Pentinat-Pelegrin T., Faucette R.R. Intergenerational transmission of glucose intolerance and obesity by in utero undernutrition in mice. Diabetes. 2009;58(2):460–468. doi: 10.2337/db08-0490. PubMed PMID: 19017762; PubMed Central PMCID: PMC2628621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuan S., Oliver D., Schuster A., Zheng H., Yan W. Breeding scheme and maternal small RNAs affect the efficiency of transgenerational inheritance of a paramutation in mice. Scientific Reports. 2015;5:9266. doi: 10.1038/srep09266. PubMed PMID: 25783852; PubMed Central PMCID: PMCPMC4363887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brookheart R.T., Duncan J.G. Modeling dietary influences on offspring metabolic programming in Drosophila melanogaster. Reproduction. 2016;152(3):R79–R90. doi: 10.1530/REP-15-0595. PubMed PMID: 27450801; PubMed Central PMCID: PMCPMC4964793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Somer R.A., Thummel C.S. Epigenetic inheritance of metabolic state. Current Opinion in Genetics and Development. 2014;27:43–47. doi: 10.1016/j.gde.2014.03.008. PubMed PMID: 24846842; PubMed Central PMCID: PMCPMC4125520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ost A., Lempradl A., Casas E., Weigert M., Tiko T., Deniz M. Paternal diet defines offspring chromatin state and intergenerational obesity. Cell. 2014;159(6):1352–1364. doi: 10.1016/j.cell.2014.11.005. PubMed PMID: 25480298. [DOI] [PubMed] [Google Scholar]
- 25.Buescher J.L., Musselman L.P., Wilson C.A., Lang T., Keleher M., Baranski T.J. Evidence for transgenerational metabolic programming in Drosophila. Disease Models & Mechanisms. 2013;6(5):1123–1132. doi: 10.1242/dmm.011924. PubMed PMID: 23649823; PubMed Central PMCID: PMC3759332, Epub 2013/05/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xia B., de Belle S. Transgenerational programming of longevity and reproduction by post-eclosion dietary manipulation in Drosophila. Aging. 2016;8(5):1115–1134. doi: 10.18632/aging.100932. PubMed PMID: 27025190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matzkin L.M., Johnson S., Paight C., Markow T.A. Preadult parental diet affects offspring development and metabolism in Drosophila melanogaster. PloS One. 2013;8(3):e59530. doi: 10.1371/journal.pone.0059530. PubMed PMID: 23555695; PubMed Central PMCID: PMC3608729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dew-Budd K., Jarnigan J., Reed L.K. Genetic and sex-specific transgenerational effects of a high fat diet in Drosophila melanogaster. PloS One. 2016;11(8):e0160857. doi: 10.1371/journal.pone.0160857. PubMed PMID: 27518304; PubMed Central PMCID: PMCPMC4982694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yazbek S.N., Spiezio S.H., Nadeau J.H., Buchner D.A. Ancestral paternal genotype controls body weight and food intake for multiple generations. Human Molecular Genetics. 2010;19(21):4134–4144. doi: 10.1093/hmg/ddq332. PubMed PMID: 20696673; PubMed Central PMCID: PMC2951864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Padmanabhan N., Jia D., Geary-Joo C., Wu X., Ferguson-Smith A.C., Fung E. Mutation in folate metabolism causes epigenetic instability and transgenerational effects on development. Cell. 2013;155(1):81–93. doi: 10.1016/j.cell.2013.09.002. PubMed PMID: 24074862.Epub 2013/10/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nelson V.R., Spiezio S.H., Nadeau J.H. Transgenerational genetic effects of the paternal Y chromosome on daughters' phenotypes. Epigenomics. 2010;2(4):513–521. doi: 10.2217/epi.10.26. PubMed PMID: 22121971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim S.K., Rulifson E.J. Conserved mechanisms of glucose sensing and regulation by Drosophila corpora cardiaca cells. Nature. 2004;431(7006):316–320. doi: 10.1038/nature02897. PubMed PMID: 15372035. [DOI] [PubMed] [Google Scholar]
- 33.Gronke S., Muller G., Hirsch J., Fellert S., Andreou A., Haase T. Dual lipolytic control of body fat storage and mobilization in Drosophila. PLoS Biology. 2007;5(6):e137. doi: 10.1371/journal.pbio.0050137. PubMed PMID: 17488184; PubMed Central PMCID: PMC1865564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee G., Park J.H. Hemolymph sugar homeostasis and starvation-induced hyperactivity affected by genetic manipulations of the adipokinetic hormone-encoding gene in Drosophila melanogaster. Genetics. 2004;167(1):311–323. doi: 10.1534/genetics.167.1.311. PubMed PMID: 15166157; PubMed Central PMCID: PMCPMC1470856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Backhaus B., Sulkowski E., Schlote F.W. A semi-synthetic, general-purpose medium for Drosophila melanogaster. Drosophila Information Service. 1984;60:210–212. [Google Scholar]
- 36.Schuldiner O., Berdnik D., Levy J.M., Wu J.S., Luginbuhl D., Gontang A.C. piggyBac-based mosaic screen identifies a postmitotic function for cohesin in regulating developmental axon pruning. Developmental Cell. 2008;14(2):227–238. doi: 10.1016/j.devcel.2007.11.001. PubMed PMID: 18267091; PubMed Central PMCID: PMCPMC2268086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tennessen J.M., Barry W.E., Cox J., Thummel C.S. Methods for studying metabolism in Drosophila. Methods. 2014;68(1):105–115. doi: 10.1016/j.ymeth.2014.02.034. PubMed PMID: 24631891; PubMed Central PMCID: PMCPMC4048761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. PubMed PMID: 19131956. [DOI] [PubMed] [Google Scholar]
- 39.Huang da W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Research. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. PubMed PMID: 19033363; PubMed Central PMCID: PMCPMC2615629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gronke S., Mildner A., Fellert S., Tennagels N., Petry S., Muller G. Brummer lipase is an evolutionary conserved fat storage regulator in Drosophila. Cell Metabolism. 2005;1(5):323–330. doi: 10.1016/j.cmet.2005.04.003. PubMed PMID: 16054079. [DOI] [PubMed] [Google Scholar]
- 41.FlyBase Genome A . 2011. Changes affecting gene model number or type in release 5.41 of the annotated D. melanogaster genome. [Google Scholar]
- 42.Curators F . 2008. Assigning gene Ontology (GO) terms by sequence similarity in FlyBase. [Google Scholar]
- 43.Theopold U., Rissler M., Fabbri M., Schmidt O., Natori S. Insect glycobiology: a lectin multigene family in Drosophila melanogaster. Biochemical and Biophysical Research Communications. 1999;261(3):923–927. doi: 10.1006/bbrc.1999.1121. [DOI] [PubMed] [Google Scholar]
- 44.Ekengren S., Hultmark D. A family of Turandot-related genes in the humoral stress response of Drosophila. Biochemical and Biophysical Research Communications. 2001;284(4):998–1003. doi: 10.1006/bbrc.2001.5067. PubMed PMID: 11409894. [DOI] [PubMed] [Google Scholar]
- 45.Colinet H., Lee S.F., Hoffmann A. Functional characterization of the Frost gene in Drosophila melanogaster: importance for recovery from chill coma. PloS One. 2010;5(6):e10925. doi: 10.1371/journal.pone.0010925. PubMed PMID: 20532197; PubMed Central PMCID: PMCPMC2880008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parvy J.P., Napal L., Rubin T., Poidevin M., Perrin L., Wicker-Thomas C. Drosophila melanogaster Acetyl-CoA-carboxylase sustains a fatty acid-dependent remote signal to waterproof the respiratory system. PLoS Genetics. 2012;8(8):e1002925. doi: 10.1371/journal.pgen.1002925. PubMed PMID: 22956916; PubMed Central PMCID: PMCPMC3431307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou D., Xue J., Lai J.C., Schork N.J., White K.P., Haddad G.G. Mechanisms underlying hypoxia tolerance in Drosophila melanogaster: hairy as a metabolic switch. PLoS Genetics. 2008;4(10):e1000221. doi: 10.1371/journal.pgen.1000221. PubMed PMID: 18927626; PubMed Central PMCID: PMCPMC2556400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karim F.D., Thummel C.S. Ecdysone coordinates the timing and amounts of E74A and E74B transcription in Drosophila. Genes & Development. 1991;5(6):1067–1079. doi: 10.1101/gad.5.6.1067. PubMed PMID: 2044954. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
AKHR–/+ heterozygotes compared to +/+ wild-type.
GO categories of AKHR–/+ heterozygote down-regulated genes.
GO categories of AKHR–/+ heterozygote up-regulated genes.
Genes in each overlap category from the Venn diagram shown in Figure 4.
Genes that change expression in paternal F1 lineage.
Genes that change expression in maternal F1 lineage.