Abstract
Objective
Obesity variably disrupts human health, but molecular-based patients' health-risk stratification is limited. Adipose tissue (AT) stresses may link obesity with metabolic dysfunction, but how they signal in humans remains poorly-characterized. We hypothesized that a transcriptional AT stress-signaling cascade involving E2F1 and ASK1 (MAP3K5) molecularly defines high-risk obese subtype.
Methods
ASK1 expression in human AT biopsies was determined by real-time PCR analysis, and chromatin immunoprecipitation (ChIP) adopted to AT explants was used to evaluate the binding of E2F1 to the ASK1 promoter. Dual luciferase assay was used to measure ASK1 promoter activity in HEK293 cells. Effects of E2F1 knockout/knockdown in adipocytes was assessed utilizing mouse-embryonal-fibroblasts (MEF)-derived adipocyte-like cells from WT and E2F1−/− mice and by siRNA, respectively. ASK1 depletion in adipocytes was studied in MEF-derived adipocyte-like cells from WT and adipose tissue-specific ASK1 knockout mice (ASK1-ATKO).
Results
Human visceral-AT ASK1 mRNA (N = 436) was associated with parameters of obesity-related cardio-metabolic morbidity. Adjustment for E2F1 expression attenuated the association of ASK1 with fasting glucose, insulin resistance, circulating IL-6, and lipids (triglycerides, HDL-cholesterol), even after adjusting for BMI. Chromatin-immunoprecipitation in human-AT explants revealed BMI-associated increased occupancy of the ASK1 promoter by E2F1 (r2 = 0.847, p < 0.01). In adipocytes, siRNA-mediated E2F1-knockdown, and MEF-derived adipocytes of E2F1-knockout mice, demonstrated decreased ASK1 expression and signaling to JNK. Mutation/truncation of an E2F1 binding site in hASK1 promoter decreased E2F1-induced ASK1 promoter activity, whereas E2F1-mediated sensitization of ASK1 promoter to further activation by TNFα was inhibited by JNK-inhibitor. Finally, MEF-derived adipocytes from adipocyte-specific ASK1-knockout mice exhibited lower leptin and higher adiponectin expression and secretion, and resistance to the effects of TNFα.
Conclusions
AT E2F1 –ASK1 molecularly defines a metabolically-detrimental obese sub-phenotype. Functionally, it may negatively affect AT endocrine function, linking AT stress to whole-body metabolic dysfunction.
Keywords: Obesity, Transcriptional regulation, Sub-phenotypes, Adipose tissue, Adipocytes, Stress response
1. Introduction
Obese sub-phenotypes associated with increased cardio-metabolic risk are characterized by metabolically and endocrinologically dysfunctional AT that is thought to contribute to the development of obesity-associated morbidities [1], [2], [3]. As an underlying mechanism for dysfunctional AT, various AT stresses have been recognized in obesity, including inflammation, endoplasmic reticulum stress, hypoxia, and oxidative stress [4], [5], [6], [7], [8]. Much effort has been invested in understanding which types of besity-induced AT stress is most significant, and the emerging picture is of an inter-connected “network of stresses” [9]. Yet, the molecular pathways activated by these stresses, linking them with the functional alterations they are claimed to induce, are incompletely mapped. The interest in this extends beyond understanding the basic pathogenic mechanisms; such AT stress-response network(s), particularly when identified in human-AT, may constitute a molecular signature that could be used for improved patient stratification and identification of targets of future pharmacological therapies.
Several signaling pathways were reported to be activated in AT in obesity, mostly in mouse models [10], [11], [12], [13], [14], [15], [16], [17]. In human-AT in obesity, one such stress-responsive pathway activated particularly in the intra-abdominal visceral fat is a MAP kinase signaling cascade comprised of MAP3K ASK1 (MAP3K5), MAP2Ks MKK4 and 3/6, MAPKs p38MAPK, and JNK [18], [19]. We demonstrated that ASK1 expression (both mRNA and protein levels) was up-regulated in obesity and, more importantly, that ASK1 mRNA levels in omental fat constituted, by multivariate analysis, a statistical predictor of whole-body insulin resistance independent of age, sex, BMI, and additional confounders [18]. In cellular systems, ASK1 has been recognized to respond to the same stresses that are implicated in AT in obesity, in particular to inflammation, oxidative and ER stresses [20], [21], [22]. Functionally related to obesity, genetic variants of ASK1 that resulted in decreased ASK1 expression in muscle were associated with insulin resistance in Pima Indians [23]. In mice, whole-body knockout of ASK1 predisposed to obesity, potentially by interfering with brown fat function [24]. These two studies assign a potential role for ASK1 in whole-body metabolic (dys)regulation. Yet, the impact of ASK1 over-expression in humans, particularly in white adipose tissue, and the molecular underpinnings for its elevated expression, remain unknown.
As a MAP kinase, ASK1 was mainly shown to be regulated by phosphorylation and/or oxidation of its inhibitory partner thioredoxin [22], [25], [26]. Yet, transcriptional regulation of ASK1 was also reported. The transcription factor E2F1, mainly known as a cell cycle regulator, binds directly to the ASK1 promoter and up-regulates its expression in cancer cells [27], [28], [29]. Recently, we reported significant gene regulatory functions for E2F1 in adipocytes, cells that are post-mitotic [30]. E2F1 was up-regulated in omental AT in obesity in the adipocyte cell fraction, increasing not only the expression of several autophagy genes but also the autophagy process itself [30]. This finding is consistent with other studies implicating non-cell-cycle related, including metabolic, functions of E2F1 (and of other classical cell-cycle regulators) [31], [32], [33], [34], [35]. In the present study, we test the hypothesis that ASK1 expression and its downstream signals are regulated by E2F1 in human-AT in obesity. Both clinically and mechanistically, we systematically investigated the possibility that the E2F1-ASK1 network molecularly defines an obese sub-phenotype characterized by AT stress and metabolic dysfunction.
2. Material and methods
2.1. Study population
Participants were from two cohorts, one in Beer-Sheva, Israel (N = 16) and one in Leipzig, Germany (N = 436), both of which have been described in a previous publication [30]. AT biopsies were analyzed exactly as described in previous publications [18], [19], [36], [37], [38]. All procedures were approved in advance by the ethics committee and were conducted in accordance with Declaration of Helsinki guidelines. In brief, participants were recruited before undergoing abdominal surgery (primarily bariatric surgery and elective cholecystectomy) after providing written informed consent. Paired subcutaneous (Sc) and Om AT biopsies were obtained during surgery and immediately delivered to the laboratory where they were processed for mRNA expression or chromatin immunoprecipitation (ChIP) studies.
2.2. Materials
Tissue culture media were from Biological Industries (Beit-Haemek, Israel). Indomethacin (I7378), dexamethasone (D4902), 3-isobutylmethylxanthine (IBMX) (I7018), and rosiglitazone (R2408) were from Sigma–Aldrich. Recombinant murine/human TNF (410-MT, 210-TA respectively) and murine IL-1β (401-ML) were purchased from R&D Systems Inc.
2.3. Cell culture
Human embryonic kidney (HEK) 293T cells (ATCC, Manassas, VA) were grown in DMEM containing 4.5 mM glucose, 10%FBS, 2 mM l-glutamine, and 100 U/ml penicillin-streptomycin. Medium was changed every other day. For Western blot analysis experiments, HEK293 were treated with TNF (10 ng/ml) with or without SP600125 for 24 h. Mouse embryonal fibroblasts (MEF) from E2F1−/− and WT mice (kindly provided by Prof. Gustavo Leone, Department of molecular virology and genetics, College of Medicine and Public Health, Ohio State University, Ohio, USA) were cultured in DMEM containing 4.5 mM glucose, 10%FBS, 2 mM l-glutamine, and 100 U/ml penicillin-streptomycin. Differentiation into adipocytes was performed as previously described [30], [39]. For Western blot analyses and leptin/adiponectin concentration measurements, adipocyte-like MEFs were treated with TNFα (10 ng/ml) + IL-1β (10 ng/ml) for 24 h. Epididymal pre-adipocytes were grown and differentiated into mature adipocytes as previously described [40]. For quantitative Real-Time PCR experiments, epididymal adipocytes were treated for 4 or 24 h with one of the following treatments: TNFα (10 ng/ml)/Fas ligand (2 ng/ml)/Glucose oxidase (50 mU/ml) and Tunicamycin (5 μg/ml). For ASK1 depletion in adipocyte-like cells, we utilized MEF from adipocyte-specific ASK1-KO mice and control littermates. Targeted embryonic stem cell clones with exon 14 (whose deletion will lead to a frame shift) of ASK1 flanked by loxP sites and a FRT-flanked selection cassette were bought from the European Conditional Mouse Mutagenesis Program (EUCOMM) to generate floxed ASK1 mice. To obtain adipocyte-specific ASK1 depletion, homozygous ASK1 floxed mice were bred to mice that express the Cre enzyme driven by the adipocyte-specific adiponectin promoter (Adipoq-Cre mice) [41].
2.4. RNA extraction and quantitative real-time PCR
Total RNA from epididymal adipocytes or MEFs adipocyte-like cells was extracted using the RNeasy lipid tissue mini-kit (74804, Qiagen) and quantified using nano-drop. Then, 2 μg of RNA were reverse-transcribed with high capacity cDNA reverse transcriptase kit (4374966, Life Technologies Inc.). Taqman system (4369016, Life Technologies, Inc.) was used for real-time PCR amplification. Extraction of total RNA from paired human-AT biopsies (Leipzig cohort) was done according to a previously described protocol [18]. Relative gene expression was obtained after normalization to endogenous control genes, using formula 2−ΔΔCt and specific primers (Supplementary Table 1).
2.5. E2f1 knockdown by small interfering RNA (siRNA) transfection
E2f1 and non-specific sequence control (NS) siRNAs ON-TARGET plus smart pools were obtained from Dharmacon (Thermo Fisher Scientific Inc., Waltham, MA). Each siRNA was transfected into differentiated epididymal adipocyte cell line at a concentration of 1 nmol/well by electroporation with GenePulser Xcell (BioRad Laboratories Inc., Hercules, CA). Briefly, epididymal adipocytes were trypsinized on the third day (of 6 days) of differentiation process from 150 mm culture plate dish. Cells were rinsed three times with PBS, centrifuged (5 min, 1200 rpm, room temperature), and after last wash supernatant was aspirated. Next, cells were re-suspended in 400 μl of Ingenio™ Electroporation Solution (Mirus Bio LLC, Madison, WI) with 4 nmol of E2f1 or NS siRNA, and electroporation was performed in accordance with the manufacturer's instructions using the optimal program (170 V, 10 ms, two pulses). After electroporation, cells were cultured in complete medium (DMEM 20% FCS). 24 h post-electroporation, cells were incubated with TNF(10 ng/ml) in DMEM 0.5%BSA for additional 24 h. Cells were rinsed 3 times with ice-cold PBS and subjected to either quantitative real-time PCR or Western blot analysis.
2.6. Transient transfection and promoter activity assay
HEK293 cells were seeded in 24 well plate in 1 ml/well of DMEM 10% FCS. 24 h later, medium was changed to a fresh 0.5 ml/well, and cells were transfected using jetPEI reagent (101-01N, Poly-plus transfection Inc.), according to manufacturer's instructions. Briefly, 200 ng/well of the expression plasmid (pCMV-empty/E2F1), 200 ng/well of the firefly luciferase reporter plasmids (ASK1WT/ASK1m1/ASK1m2/ASK1m3), and 2 ng/well of the Renilla luciferase reporter plasmid were used for each transfection. 4 h after transfection, medium was changed to fresh DMEM 10%FBS with or without TNFα (10 ng/ml) and with or without SP600125. Cells were lysed 24 h after incubation with or without TNFα 10 ng/ml and with or without SP600125 treatments by applying 100 μl of Passive Lysis Buffer of the Dual Luciferase Reporter Assay Kit (E-1910, Promega Corporation) into each well of the 24-well plate. 20 μl of cell lysate were used for the luciferase reporter assay with the same kit, according to the manufacturer's protocol. Luminescence intensity was quantified in a GloMax 20/20n Luminometer (Promega Corporation). The experiments were performed at least in triplicate. As a control for transfection efficiency, the firefly luciferase activity values were normalized to the Renilla luciferase activity values.
2.7. Promoter analysis and site-directed mutagenesis
Human ASK1 promoter (1000 bp upstream to the transcription start site-TSS and 500 bp downstream) was analyzed for predicted E2F1 binding sites using MatInspector software (Genomatix Tools). Promoter analysis produced one predicted E2F1 binding site on ASK1 promoter, which was located 384 bp upstream the TSS with a score probability of 0.84 (1-highest probability). Using site-directed mutagenesis kit (Stratagene, Santa Clara, CA), E2F1-predicted binding site on the ASK1-Luc plasmid was mutated at the core sequence (CGCG) generating 3 new mutated ASK1-Luc plasmids, as indicated in Figure 4B. Mutations/deletions were validated by sequencing.
2.8. Leptin and adiponectin concentrations
Leptin and adiponectin were measured in cultured media with or without TNFα (10 ng/ml) + IL-1β (10 ng/ml) treatment for 24 h by ELISA (MOB00, MRP300, respectively, R&D Systems Inc.).
2.9. Chromatin immunoprecipitation (ChIP)
ChIP from fresh human-AT explants was performed following a recently-described protocol [42]. Briefly, 1 g of Sc or Om AT was minced into small pieces using two pairs of sterile scissors. Next, samples were cross-linked with 1% of formaldehyde at 37 °C for 8 min. Cross-linking reaction was quenched using 0.125 M glycine. Samples were washed with ice cold PBS 3 times, and, following the last wash, adipocyte lysis buffer was added. After 15 min of incubation on ice, samples were homogenized using a Dounce homogenizer (loose pestle, 20 strokes, Wheaton). Nuclei were released after 20 additional strokes using a tight pestle. After sonication, the protein-DNA complexes were immunoprecipitated using anti-ICAM (sc-7891X, as negative control), anti-RNA poll 2 (sc-21750X, as positive control) and anti-E2F1 antibodies (sc-193X, all from Santa Cruz Biotechnology). After cross-linking reversal at 65 °C for 4 h, DNA was purified using the phenol-chloroform method. Primers (5′ to 3′) for end-point PCR or quantitative real-time PCR were as follows: ASK1- Fw – GTGCTGGACCGCTTTTACAATGC, Rv – CAGTTCAAGTCGATCGCATGGAC. Input (diluted 1/300) was used as a normalizing control.
2.10. Statistical analysis
Data, expressed as the mean ± SD, and calculations were performed using GraphPad software. Statistically significant differences between two groups were evaluated using paired or unpaired Student's-t-test, as indicated in the figure legends. Correlation between BMI and E2F1 binding to ASK1 promoter was assessed by linear regression. Clinical and biochemical characteristics of the study population were cross-classified across quintiles of visceral (Om) ASK1 mRNA expression in order to control any un-normal variable distribution. p of trend was calculated across quintiles, and ANOVA with Tuckey post-hoc test was used to assess significance between each quintile pairs. We further assessed beta coefficients to Om-ASK1 by performing step-wise multivariate linear regression models using SPSS statistical package.
3. Results
3.1. A putative E2F1-ASK1 pathway in human adipose tissue in obesity
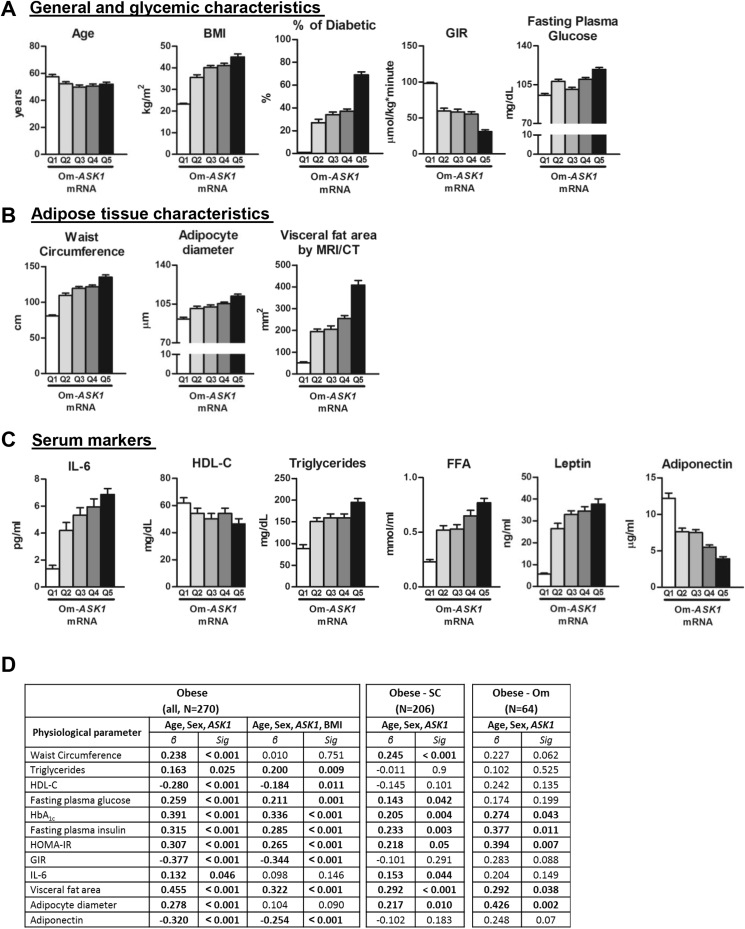
Previously, we reported that omental (Om) AT-ASK1 mRNA levels were independent statistical predictors of whole-body insulin sensitivity by multivariate models that considered multiple potential confounders of this association (age, BMI, serum lipids and FFA, leptin, adiponectin, and IL-6) [18]. Moreover, we demonstrated that although AT-ASK1 expression could be largely attributed in lean persons to the non-adipocyte stromal-vascular cells, the increased expression of this gene that accompanied visceral adiposity was largely contributed by the adipocytes. To assess the relation of Om-ASK1 mRNA levels with additional clinical characteristics of the patients, with emphasis on obesity-related cardio-metabolic parameters, we utilized a larger, previously-described patient dataset [30], stratifying the data into quintiles of Om-ASK1 mRNA (Table S1). Om-ASK1 mRNA levels did not associate with age (Figure 1A). However, highly-significant associations (p of trend<0.05) were observed with BMI, fasting plasma glucose, and insulin resistance (as determined by hyper-insulinemic-euglycemic clamp), supporting the results of our previous work [18] (Figure 1A, Table S1). Furthermore, we found a significant association between Om-ASK1 mRNA levels and adipocyte diameter and correlations with markers of obesity-associated cardio-metabolic risk, such as waist circumference and visceral fat area (Figure 1B). Moreover, high circulating levels of IL-6, triglycerides, free fatty acids (FFA), and leptin and low circulating levels of adiponectin were significantly associated with higher Om-ASK1 mRNA levels (p < 0.001 for all, Figure 1C). Indeed, only 1% of the population in the lower Om-ASK1 mRNA quintile was diagnosed with diabetes, whereas in the top quintile, more than 60% were diabetic patients (Figure 1A). Among the sub-group of obese patients (BMI ≥30 kg/m2, N = 270), Om-ASK1 mRNA associated with fasting glucose, HbA1c, insulin resistance, serum lipids, and with low circulating high-Mw-adiponectin, associations that remained significant even when further adjusting for BMI (in addition to age and sex) (Figure 1D). Furthermore, significant associations could be demonstrated in both obese subtypes categorized by abdominal fat distribution pattern (subcutaneously versus viscerally – obese) (Figure 1D).
Figure 1.
Correlation of ASK1 mRNA expression levels in human omental AT with different clinical parameters. (A–C) Clinical parameters of 436 persons were cross-classified across quintiles of Om-ASK1 mRNA expression. p of trend (by linear regression) and ANOVA + post-hoc analysis of inter-quintile significance are presented in Table S1. (D) Multivariate analyses of the association between OM-ASK1 mRNA and various clinical parameters among the entire obese subgroup (N = 270), and in obese patients with predominant subcutaneous obesity (Obese-SC, N = 206, defined as abdominal visceral/subcutaneous area<50% by L3-L4 CT or MRI imaging) or with predominant visceral adiposity (Obese-Om, N = 64). All models are adjusted for age and sex, and for the entire obese subgroup an additional model is presented also adjusted for BMI.
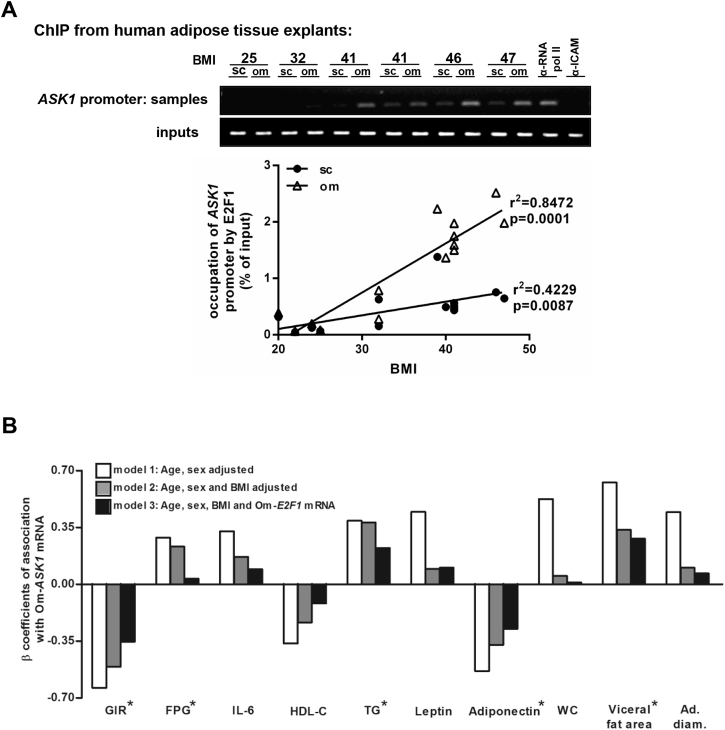
MAP kinases are mainly thought to be regulated by phosphorylation. In cancer cells, transcriptional regulation of ASK1 was also demonstrated by the cell-cycle regulator E2F1 [27], [28], [29]. Recently, we reported that E2F1 expression is increased in Om of intra-abdominally obese humans and may act to activate non-cell-cycle-related genes [30]. Therefore, we assessed whether increased BMI was associated in human-AT with greater occupancy of the ASK1 promoter by E2F1. Using a chromatin immunoprecipitation (ChIP) protocol adopted to human-AT [42], a highly significant correlation between BMI and E2F1 binding to the ASK1 promoter could be observed in both Om and Sc fat, though the effect of BMI was more robust for Om (Om: r = 0.920, p < 0.001; Sc: r = 0.650, p = 0.009, Figure 2A). Next, we adjusted the associations of ASK1 with several obesity-related parameters for BMI and for E2F1 to assess if they may share a common mechanistic pathway. ASK1 remained significantly associated with insulin resistance, high triglycerides, and, importantly, with low adiponectin, even after adjusting the model for BMI and E2F1 (Figure 2B). Yet, the associations of Om-ASK1 with all parameters were attenuated by adjusting for the expression of E2F1 mRNA in Om AT. Collectively, using a large human-AT biobank, we demonstrate that Om-ASK1 expression may share a common path with E2F1 in their association with major clinical parameters indicating AT dysfunction and increased cardio-metabolic risk. Moreover, molecularly, higher BMI associates with increased occupancy of the ASK1 promoter by E2F1.
Figure 2.
Effect of BMI on E2F1 binding to the ASK1 promoter and associations between Om-ASK1 expression levels and different clinical characteristics. (A) Formaldehyde cross-linked chromatin from 16 paired human (Om and Sc) ATs was subjected to ChIP experiments. Immunoprecipitation of E2F1 containing complexes was performed using anti-E2F1 antibody. Anti-POLR2 (polymerase RNA II), was used as positive control and anti-ICAM1 (intercellular adhesion molecule 1) was used as negative control. After isolation of bound DNA, end-point PCR and quantitative real time PCR were performed for a 300 bp region of the endogenous human ASK1 promoter. Quantitative real time PCR results were analyzed using linear regression. Inputs indicate PCR performed on DNA (diluted 1:300) without any immunoprecipitation. (B) Multi variate models to assess associations between Om-ASK1 mRNA levels and parameters shown in Figure 1 as continuous variables. Values are the β coefficient of association, with model 1 adjusted for age and sex, model 2 for age, sex, and BMI, and model 3 adjusted for age, sex, BMI, and Om-E2F1 mRNA expression. * Associations with p values <0.05. GIR; glucose infusion rate during hyper-insulinemic euglycemic clamp, FPG; fasting plasma glucose, HDL-cholesterol; high-density lipoprotein cholesterol, TG; triglycerides, WC; waist circumference, ad. diam.; adipocyte diameter.
3.2. Molecular links between E2F1 and ASK1 regulation
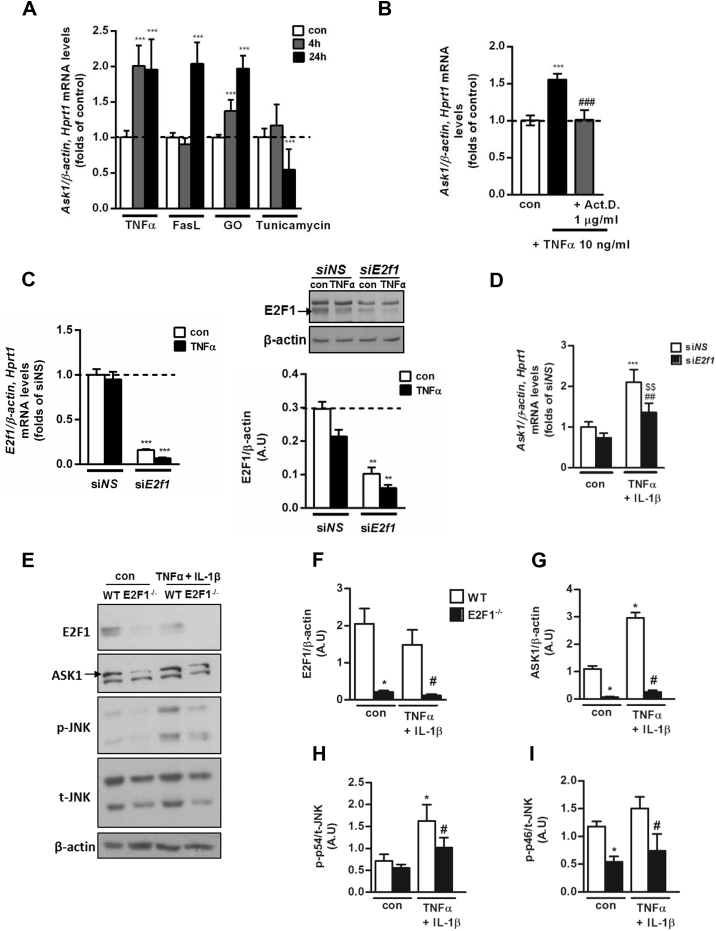
To better define the molecular mechanisms and triggers for increased adipocyte ASK1 expression and to evaluate the contribution of adipocytes, we utilized cellular models. A differentiated murine epididymal adipocyte cell line was utilized to determine potential individual triggers relevant to the AT milieu in obesity, which could increase ASK1 mRNA levels. To this end, inflammatory stimuli such as the prototypical pro-inflammatory cytokine TNFα, and FasL, and an oxidative environment induced by adding the H2O2 generating enzyme glucose oxidase (GO) to the medium, increased Ask1 mRNA by ∼2-fold at 24 h (Figure 3A). At 4 h stimulation the responses differed, likely representing unique signaling components of the different stimuli that resulted in different activation kinetics. Interestingly, tunicamycin, an inducer of ER stress, did not elicit a similar effect. Supporting the notion that the increased steady-state mRNA levels could be attributed to increased transcription (rather than to enhanced mRNA stability), we observed that actinomycin D, a global transcription inhibitor, fully prevented the increase in Ask1 mRNA induced by TNFα (Figure 3B). Next, we set to dissect a putative role for E2f1 in adipocytes by siRNA-mediated knockdown and by utilizing MEFs from E2F1-KO (E2F1−/−) mice that were differentiated into adipocyte-like cells. siRNA targeting E2f1 decreased the mRNA and protein levels of this gene product in mature epididymal adipocytes by ∼90% and 75%, respectively, compared to a non-targeting (NS) siRNA (Figure 3C). Under these conditions, TNFα-induced increase in Ask1 mRNA was significantly inhibited by ∼50% (Figure 3D). We previously demonstrated that despite the role of E2F1 in adipogenesis [35], under in-vitro conditions, MEFs from E2F1−/− mice can differentiate into adipocyte-like cells comparably to MEFs from wild-type (WT) mice, as assessed morphologically and by the similar induction of adipocyte-specific genes (Pparγ, Cebp4α, Fabp4, and Glut4) [30]. Utilizing these cells, we observed that in the absence of E2F1, “basal” ASK1 expression was markedly attenuated (Figure 3E–G). In addition, ASK1 expression was stimulated by 2.72 ± 0.20-fold by TNFα + IL-1β in the WT cells, but only by 1.93 ± 0.08-fold in the E2F1−/− adipocyte-like cells (p = 0.029). Consistently, both basal and TNFα + IL-1β-stimulated phosphorylation of JNK (a MAP kinase downstream of ASK1) was markedly attenuated (Figure 3E,H–I). Thus, in adipocyte cellular models, decreased E2F1 results in lower ASK1 expression and attenuated downstream signaling.
Figure 3.
Increase in transcription of ASK1 gene by mimickers of obesity-associated AT stresses is attenuated by absence of E2F1 in adipocytes. (A) Epididymal adipocytes were treated with several stress inducers (TNFα 10 ng/ml, FasL 1 ng/ml, GO 50 mU/ml, and Tunicamycin 5 μg/ml) for 4 or 24 h and subjected to quantitative real-time PCR analysis. The expression was normalized to β-actin and Hprt1. Results are mean ± SD of n = 5 independent experiments. ∗∗∗p < 0.001 4/24 h of each treatment vs. con. (B) Epididymal adipocytes were incubated with TNFα (10 ng/ml) or TNFα (10 ng/ml) + Act. D. (1 μg/ml) for 4 h and subjected to quantitative real-time PCR analysis. The expression was normalized to β-actin and Hprt1. Results are mean ± SD of n = 3 independent experiments. ∗∗∗p < 0.001 TNFα only versus con. ###p < 0.001 Act. D. (C, D) Epididymal adipocytes were transfected with. E2f1 siRNA (siE2f1) or non-specific siRNA (siNS) using electroporation. 24 h after transfection, adipocytes were incubated with TNFα (10 ng/ml) for an additional 24 h and analyzed using either quantitative real-time PCR or Western blot. Each transcript expression was normalized to β-actin and Hprt1 mRNA levels. β-actin was used as a loading control on the gel. Results are mean ± SD of 3 independent experiments. ∗∗∗p < 0.001 siE2f1con and TNFα vs. siNS con (middle graph), siNS TNFα vs. siNS con (right graph). ∗∗p < 0.01 siE2f1 TNFα vs. siE2F1 con. ##p < 0.01 siE2f1 TNFα vs. siNS TNFα. MEF-derived adipocyte-like cells from WT and E2F1−/− were treated with TNFα 10 ng/ml + IL-1β 10 ng/ml for 24 h and subjected to Western blot analysis. Shown are representative blots (E) and densitometry graphs (F–I) of n = 3 independent experiments. β-actin was used as a loading control. ∗p < 0.05 E2F1−/− con or WT TNFα 10 ng/ml + IL-1β 10 ng/ml vs. WT con. #p < 0.05 E2F1−/− TNFα 10 ng/ml + IL-1β 10 ng/ml vs. E2F1−/− con.
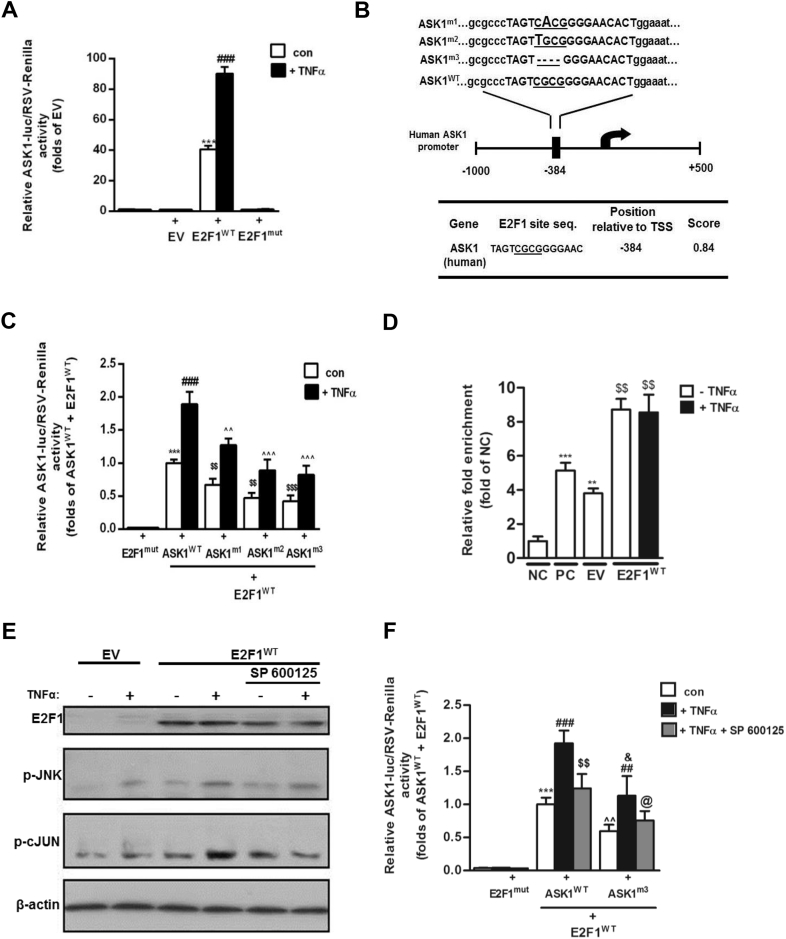
To elucidate how elevated E2F1 might increase ASK1 transcription, a luciferase reporter assay utilizing the human ASK1 promoter region (−1000 to +500 bp) in HEK293 cells was performed. When cells were co-transfected with an empty vector plasmid (EV), promoter activity was low, and it was unresponsive to TNFα stimulation. Upon E2F1 over-expression, two phenomena were observed; basal ASK1 promoter activity was greatly enhanced (by ∼40-fold, Figure 4A), consistent with previous reports [28], [29], and E2F1 over-expression permissively sensitized cells to respond to TNFα stimulus by “super-activating” ASK1 promoter activity >2-fold (Figure 4A). Both the basal and TNFα-stimulated promoter activity required E2F1 binding to DNA, since expression of a mutant E2F1 that lacks DNA binding capacity failed to stimulate luciferase activity (Figure 4A). To further verify the role of direct binding of E2F1 to the ASK1 promoter, 3 mutants in the human ASK1 promoter were generated targeting a single putative E2F1 binding site (see Methods for prediction approach) (Figure 4B). Both site mutants and a 4 bp deletion mutant significantly diminished ASK1 promoter activity (Figure 4C). Yet, residual E2F1-related activity was still evident, and in particular, the 2-fold “super-induction” of ASK1-promoter –mediated luciferase activity by TNFα above basal activity was still evident. Intriguingly, ChIP analysis revealed that E2F1 over-expression indeed increased E2F1 binding to the ASK1 promoter (Figure 4D). Yet, TNFα-induced “super-activation” of ASK1 promoter activity under conditions of E2F1 over-expression (Figure 4A) was not associated with further binding of E2F1 to the ASK1 promoter (Figure 4D). This suggests that although being permissive for ASK1 promoter “super-activation,” this effect of E2F1-overexpression is likely indirect, possibly being mediated by TNFα-induced JNK phosphorylation/activation. To address this possibility, we assessed whether a pharmacological JNK inhibitor, SP600125, could alleviate the TNFα-mediated activation of ASK1 promoter activity under E2F1 overexpression. Indeed, TNFα induced JNK phosphorylation, and the phosphorylation of its substrate, c-JUN (Figure 4E). SP600125 inhibited JNK activity, since it did not affect TNFα-induced p-JNK while preventing the phosphorylation of its substrate c-Jun. Under these conditions, TNFα-induced super-activation of the ASK1 promoter under conditions of E2F1 overexpression was nearly fully inhibited (Figure 4F). Collectively, we confirm that E2F1 over-expression activates ASK1 promoter activity and identify a putative binding site for E2F1 in the human ASK1 promoter. Importantly, we demonstrate that E2F1 plays a permissive, JNK-mediated, sensitization of the ASK1 promoter to TNF-induced “super-activation.”
Figure 4.
ASK1 promoter activity is elevated by E2F1 over-expression and JNK phosphorylation input in HEK293 cells. (A) HEK293 cells were transfected with pA3-ASK1-Luc and an empty pcDNA3 (EV) or pcDNA3-E2F1 (E2F1WT) or pcDNA3-E2F1-mut (E2F1mut). After normalization to endogenous control (RSV-Renilla), an empty pcDNA3 (EV) was determined as basal luciferase activity. ASK1-promoter activity was measured after treatment with TNFα (10 ng/ml) for 24 h using Dual Luciferase Assay. Results are mean ± SD of n = 7 independent experiments. ***p < 0.001 con E2F1WT vs. con EV. ###p < 0.001 TNFα 10 ng/ml E2F1WT vs. con E2F1WT. (B) Wild-type (WT) and mutated constructs of predicted E2F1-binding site on ASK1-promoter area by Met-Inspector software. E2F1 binding to the ASK1 promoter is mediated by physical interactions between E2F1 and a core sequence of the binding site (underlined sequence of four nucleotides). Two point mutations (ASK1m1 and ASK1m2) and one deletion mutation (ASK1m3) of this core sequence were made by using mutagenesis kit as elaborated in the Methods, and their effect on ASK1 promoter activity is presented in (C). After normalization to endogenous control (RSV-Renilla), mutated E2F1 (E2F1mut) was determined as luciferase basal activity. ASK1-promoter activity was measured after treatment with TNFα (10 ng/ml) for 24 h using Dual Luciferase Assay. Results are mean ± SD of n = 5 independent experiments. ***p < 0.001 con ASK1WT vs. E2F1mut . ###p < 0.001 TNFα ASK1WT vs. con ASK1WT. $$p < 0.01 con ASK1mutants vs. con ASK1WT. ˆˆˆ/ˆˆp < 0.001/0.01 respectively, TNFα ASK1mutants vs. TNFα ASK1WT. (D) HEK-293 cells were transfected with an empty pcDNA3 (EV) or pcDNA3-E2F1 (E2F1WT) and treated with TNFα for 24 h. Next, chromatin was cross-linked, fragmented and subjected to ChIP with α-RNA poll (as positive control – PC), α-ICAM (as negative control - NC), and α-E2F1 as the protein of interest, as indicated in the Methods. End-point PCR was done using purified DNA and specific primers for E2F1 binding site on the ASK1 promoter; inputs were diluted 1/300 and used as normalization control. Results are mean ± SD of n = 3 independent experiments. **/***p < 0.005/0.01 respectively, vs. NC. $$p < 0.005 vs. EV. HEK-293 cells were transfected with an empty pcDNA3 (EV) or pcDNA3-E2F1 (E2F1WT) and treated with TNFα (10 ng/ml) and SP 600125 (50 μM). Shown are representative blots of Western blot analysis (E) of cell lysates prepared from HEK-293 cells. β-actin was used as a loading control. (F) HEK-293 cells were transfected with pA3-ASK1WT or ASK1m3 and pcDNA3-E2F1-mut (E2F1mut) or pcDNA3-E2F1 (E2F1WT). After normalization to endogenous control (RSV-Renilla), pcDNA3-E2F1-mut (E2F1mut) was determined as luciferase basal activity. ASK1-promoter activity was measured after treatment with TNFα (10 ng/ml) and SP 600125 (50 μM) for 24 h using Dual Luciferase Assay. Results are mean ± SD of n = 3 independent experiments. ∗∗∗p < 0.001 con ASK1WT (E2F1WT) vs. E2F1mut. ###p < 0.001 TNFα ASK1WT vs. con ASK1WT. $$p < 0.005 TNFα + SP ASK1WT vs. TNFα ASK1WT. ˆˆp < 0.005 con ASK1m3 (E2F1WT) vs. E2F1mut. ##p < 0.01 TNFα ASK1m3 vs. con ASK1m3. &p < 0.05 TNFα ASK1m3 vs. TNFα ASK1WT. @p < 0.05 TNFα + SP ASK1m3 vs. TNFα ASK1m3.
3.3. Functional significance of adipocyte ASK1
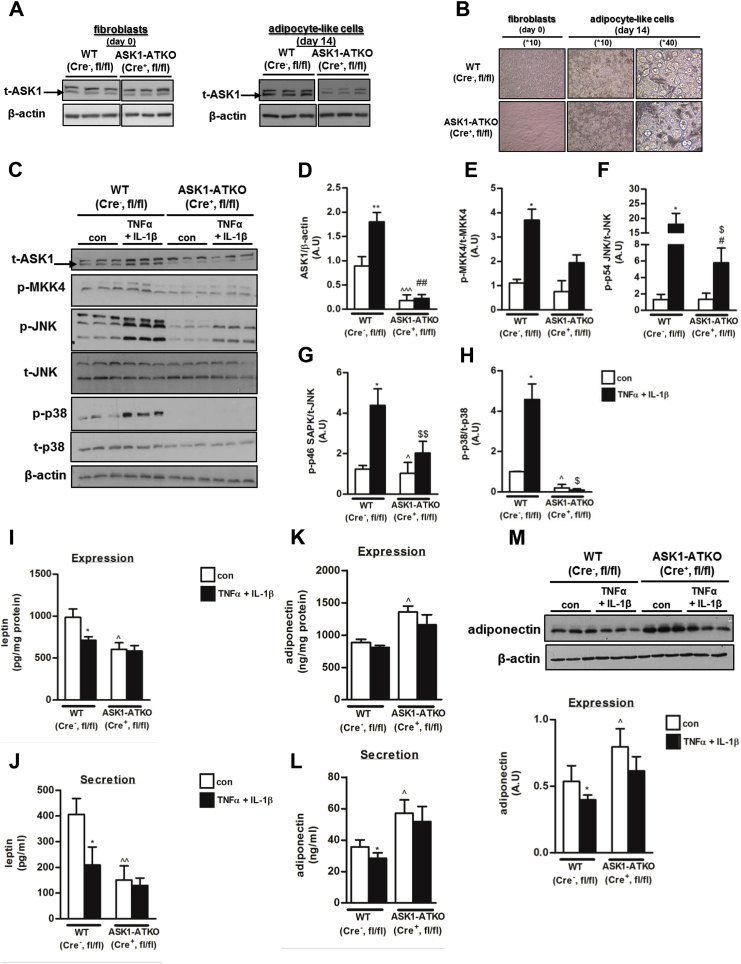
Finally, to unravel the functional role of ASK1 in adipocytes, we utilized MEFs of wild-type (WT, Cre-, ASK1fl/fl) and ASK1-AT-specific KO mice (Cre+, ASK1fl/fl, ASK1-ATKO), which were differentiated into mature adipocyte-like cells. In non-differentiated MEF (before inducing adipogenesis), ASK1 expression was similar between the two genotypes (Figure 5A). After inducing adipogenesis, cells morphologically became adipocyte-like (Figure 5B), and ASK1 expression in the ASK1-ATKO adipocyte-like cells was lower than in the WT under basal conditions (Figure 5C,D). Moreover, induction of ASK1 expression by TNFα + IL-1β-was lower in the ASK1-ATKO adipocyte-like cells (Figure 5C,D). Next, we evaluated the effect of ASK1 absence on several members of the MAP kinase pathway downstream of ASK1. While only mild differences were observed in the expression levels of total JNK and total p-38 between the two types of cells, a significant reduction in basal and TNFα + IL-1β-stimulated phosphorylation of p-MKK4, p-JNK, and p-p38 was evident in the cells with lower ASK1 expression (Figure 5C,D). We next assessed the functional impact of reduced ASK1 expression. No significant differences between the WT and ASK1-ATKO – derived adipocyte like cells were detected in the phosphorylation of both basal and insulin-stimulated Akt (Ser-473, Thr- 308), or total levels of IRS-1 (Supplemental Fig. 1). In contrast, a significant difference in expression and secretion of the key adipokines leptin and adiponectin was observed using both ELISA and Western blot analysis. Whereas ASK1-ATKO adipocyte-like cells secreted lower basal levels of leptin than the WT cells (Figure 5E), the basal adiponectin secretion was significantly higher (Figure 5E). As expected, inflammatory conditions (TNFα + IL-1β) significantly lowered leptin and adiponectin secretion from WT adipocyte-like cells, but less so in the ASK1-ATKO adipocyte-like cells, indicating that ASK1 mediates the negative impact of inflammation on adipokine secretion (Figure 5E–G).
Figure 5.
Effect of ASK1 depletion on phosphorylation of several members of MAP kinase pathway and on leptin/adiponectin expression and secretion in adipocytes. (A) ASK1 expression in MEF-derived fibroblasts (before differentiation into adipocytes) or MEF-derived adipocyte-like cells (after differentiation into adipocytes) from either WT or ASK1-ATKO mice was performed using Western blot analysis. Shown are representative blots from one of 5 independent experiments. Vertical white line indicates splicing of the same membrane for clarity of presentation. (B) Representative images of MEF-derived fibroblasts or MEF-derived adipocyte-like cells were obtained at 10X and 40X by light microscopy. MEF-derived adipocyte-like cells from WT and ASK1-ATKO were treated with TNFα 10 ng/ml + IL-1β 10 ng/ml for 24 h and subjected to Western blot analysis. Shown are representative blots (C) and densitometry graphs (D–H) of n = 6 independent experiments. β-actin was used as a loading control. ∗∗/∗p < 0.01/0.05 WT TNFα 10 ng/ml + IL-1β 10 ng/ml vs. WT con. ˆˆˆ/ˆp < 0.005/0.05 ASK1-ATKO con vs. WT con. ##/#p < 0.01/0.05 ASK1-ATKO TNFα 10 ng/ml + IL-1β 10 ng/ml vs. WT TNFα 10 ng/ml + IL-1β 10 ng/ml $$/$p < 0.01/0.05 ASK1-ATKO TNFα 10 ng/ml + IL-1β 10 ng/ml vs. ASK1-ATKO con. To measure expression and secretion levels of leptin (I, J) or adiponectin (K–M), medium (secretion and expression), and lysates (expression) were analyzed by ELISA (leptin and adiponectin) or by Western blot analysis (adiponectin). The results were normalized to cell protein concentrations. Results are mean ± SD of n = 6 independent experiments. ∗p < 0.05 WT TNFα 10 ng/ml + IL-1β 10 ng/ml vs. WT con. ˆˆ/ˆp < 0.01/0.05 ASK1-ATKO con vs. WT con.
4. Discussion
This study explores the hypothesis that ASK1, via transcriptional upregulation by E2F1, molecularly defines AT that supports a dys-metabolic obese phenotype in humans. We demonstrate associations between increased visceral-AT ASK1 expression and multiple parameters of metabolically-unhealthy obesity, even beyond BMI and patients' sub-phenotyping by fat distribution pattern. Associations of ASK1 with markers of dys-glycemia, dyslipidemia, and systemic inflammation were attenuated by adjusting for E2F1 expression, suggesting that E2F1 and ASK1 share a common mechanistic pathway. At the molecular level, in human-AT, E2F1's occupancy of the ASK1 promoter increases with BMI, more so in visceral than in subcutaneous fat, and two adipocyte cellular models of E2F1 knockdown/knockout demonstrate decreased ASK1 expression and attenuated downstream signaling. Furthermore, an E2F1 binding sequence in the human ASK1 promoter was identified, and elevated E2F1 was shown to contribute to increased ASK1 expression by both a direct transcriptional mechanism and by a JNK-mediated feed-forward loop. These render E2F1 over-expressing cells hyper-sensitive to ASK1 upregulation by inflammatory stimuli. Finally, the functional impact of ASK1 is demonstrated by adipocyte-specific ASK1-KO cells, which exhibit an improved adipokine secretory profile (secreting less leptin and more adiponectin, both basally and in response to an inflammatory stimulus).
Obese sub-phenotypes, particularly based on body-fat distribution patterns (‘apples’ vs ‘pears’), were noticed decades ago [43]. Yet, with its growing prevalence, obesity requires a better characterization of obesity sub-phenotypes for a more stratified/personalized management of the disease. Differences in ATs' adaptation to chronic caloric surplus can serve as an effective means of patient stratification. For example, adipocyte size, indicating the reliance of AT expansion on hypertrophy, has been shown as a potentially clinically-useful tool to stratify obesity-related type 2 diabetes risk [44], [45], [46]. Other histopathological features of a dysfunctional AT include altered cellular composition of the tissue, in particular macrophage infiltration and lipid accumulation (foam cells) [36], [38], [47], [48], and AT fibrosis [49], [50]. Yet, a molecular definition of AT signatures that defines metabolically-unhealthy obese sub-phenotype(s) has not been systematically attempted. In oncology, molecular typing of tumors largely relies on the identification of different tumors' genetic perturbations, increasingly used to guide patients' management [51]. Genetic polymorphisms related to obesity sub-phenotypes and/or responses to therapy are just beginning to be unraveled [52]. Nevertheless, molecular pathways that mediate AT stresses could constitute promising leads for molecular-based obesity sub-phenotyping and personalized treatment.
Along this line, MAP kinases, and in particular the stress-activated MAP kinases [53], are promising candidates. Indeed, different members of this family of kinases, including p38MAP kinase, JNK, and upstream kinases such as the MAP3K's Tpl-2 and ASK1, and even MAP4K's, have been shown to regulate AT/adipocyte biology and to be activated in obesity [14], [15], [16], [17], [18], [54]. MAP kinases are frequently shown to be regulated by rapid phosphorylation-dephosphorylation [55], and, indeed, phosphorylation-based activation of a MAP kinase pathway consisting of ASK1–MKK4,3/6–JNK/p38MAP kinase has been demonstrated in obese humans' visceral-AT [18], [19]. Yet, we previously proposed that a significant level of regulation may be attributed to the expression level of these MAP kinases, reflecting a more chronic adaptation of AT in obesity [18]. Moreover, the association between ASK1 mRNA levels in visceral-AT and insulin resistance, which remained significant even after adjusting for BMI, supported the notion that ASK1 expression levels may define obese sub-phenotypes, i.e. that higher levels of visceral ASK1 may molecularly define obese persons with a more dys-metabolic “type” of obesity [18].
Findings of the present study provide a molecular mechanism for the elevated ASK1 expression in obesity and how it may be functionally linked to whole-body insulin resistance. The role our findings assign to increased E2F1 in upregulating ASK1 parallels the capacity of this transcription factor to regulate the expression of autophagy genes [56], reminiscent of previous observations in cancer cells [27], [28], [29]. The transcriptional activity of E2F1 is regulated at multiple levels, including expression, cellular (nuclear) localization, post-translational modifications, and multiple interacting proteins. Here, increased E2F1 expression parallels with increased E2F1 binding to the ASK1 promoter in human adipose tissue explants. Regardless of potential inter-relations between the ASK1-MAP kinase pathway and autophagy activation, which have yet to be explored, both pathways downstream of E2F1 jointly form a signaling network that molecularly defines dysfunctional AT. The functional outcome of ASK1 activation in AT is linked to decreased adiponectin secretion. Intriguingly, up-regulated autophagy in obese adipose tissue was also shown to result in a dys-metabolic secretory function of adipocytes characterized by low adiponectin release, which induced insulin resistance in liver cells [57].
Our study assigns novel insights on the impact of dysregulated ASK1 expression beyond current literature. In either human muscle [23] or at the whole-body expression level in mice [24], it was decreased expression of ASK1 that was tied to insulin resistance and/or reduced energy expenditure, respectively. Here, we demonstrate upregulated ASK1 expression in visceral AT as a putative link between obesity-related AT stress and metabolic dysfunction.
In summary, we propose a stress signaling network composed of E2F1–ASK1-MAP kinase and E2F1-autophagy, which molecularly defines a metabolically-detrimental obese sub-phenotype in human adipose tissue. Functionally, by negatively affecting AT endocrine function, it may link obesity via adipose tissue stress to whole-body metabolic dysfunction.
Acknowledgments
We would like to thank Prof. Gustavo Leone, Department of Molecular Virology and Genetics, College of Medicine and Public Health, Ohio State University, Ohio, USA, for the E2F1−/− and WT- MEFs. The contribution of Dr. Avi Shtevi for establishing the role of JNK in ASK1 super-activation by TNF is also acknowledged. This study was supported in part by grants from the Deutsche Forschungsge-meinschaft (DFG): SFB 1052/1: “Obesity mechanisms” (project B2 to A.R., project B1 to M.B., and project B4 to N.K.), and the Israel Science Foundation (to A.R., ISF 928-14).
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molmet.2017.05.003.
Conflict of interest
None declared.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
Insulin signaling in MEF-derived adipocyte-like cells of wild-type and ASK1-ATKO mice. MEF-derived adipocyte-like cells were grown as described in the legend for Figure 5. After differentiation, cells were serum-starved for 3h, followed by stimulation with 0.5 nM (sub-maximal) or 100 nM (maximal) insulin, for 7 min. Lysates were assessed for insulin signaling. Shown are representative blots (A) and densitometry analysis (B–D) of n = 4 independent experiments. ∗p < 0.05 vs. WT con. ˆˆp < 0.01 vs. WT con. $$p < 0.01 vs. ASK1-ATKO con.
References
- 1.Denis G.V., Obin M.S. ‘Metabolically healthy obesity': origins and implications. Molecular Aspects of Medicine. 2013;34(1):59–70. doi: 10.1016/j.mam.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goossens G.H., Blaak E.E. Adipose tissue dysfunction and impaired metabolic health in human obesity: a matter of oxygen? Frontiers in Endocrinology (Lausanne) 2015;6:55. doi: 10.3389/fendo.2015.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manna P., Jain S.K. Obesity, oxidative stress, adipose tissue dysfunction, and the associated health risks: causes and therapeutic strategies. Metabolic Syndrome and Related Disorders. 2015;13(10):423–444. doi: 10.1089/met.2015.0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bluher M. Adipose tissue inflammation: a cause or consequence of obesity-related insulin resistance? Clinical Science (Lond) 2016;130(18):1603–1614. doi: 10.1042/CS20160005. [DOI] [PubMed] [Google Scholar]
- 5.Greenberg A.S., Obin M.S. Obesity and the role of adipose tissue in inflammation and metabolism. The American Journal of Clinical Nutrition. 2006;83(2):461S–465S. doi: 10.1093/ajcn/83.2.461S. [DOI] [PubMed] [Google Scholar]
- 6.Gregor M.F., Hotamisligil G.S. Thematic review series: adipocyte Biology. Adipocyte stress: the endoplasmic reticulum and metabolic disease. Journal of Lipid Research. 2007;48(9):1905–1914. doi: 10.1194/jlr.R700007-JLR200. [DOI] [PubMed] [Google Scholar]
- 7.Trayhurn P. Hypoxia and adipocyte physiology: implications for adipose tissue dysfunction in obesity. Annual Review of Nutrition. 2014;34:207–236. doi: 10.1146/annurev-nutr-071812-161156. [DOI] [PubMed] [Google Scholar]
- 8.Ruskovska T., Bernlohr D.A. Oxidative stress and protein carbonylation in adipose tissue - implications for insulin resistance and diabetes mellitus. Journal of Proteomics. 2013;92:323–334. doi: 10.1016/j.jprot.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rudich A., Kanety H., Bashan N. Adipose stress-sensing kinases: linking obesity to malfunction. Trends in Endocrinology & Metabolism. 2007;18(8):291–299. doi: 10.1016/j.tem.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 10.Carlsen H., Haugen F., Zadelaar S., Kleemann R., Kooistra T., Drevon C.A. Diet-induced obesity increases NF-kappaB signaling in reporter mice. Genes & Nutrition. 2009;4(3):215–222. doi: 10.1007/s12263-009-0133-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yin M.J., Yamamoto Y., Gaynor R.B. The anti-inflammatory agents aspirin and salicylate inhibit the activity of I(kappa)B kinase-beta. Nature. 1998;396(6706):77–80. doi: 10.1038/23948. [DOI] [PubMed] [Google Scholar]
- 12.Yuan M., Konstantopoulos N., Lee J., Hansen L., Li Z.W., Karin M. Reversal of obesity- and diet-induced insulin resistance with salicylates or targeted disruption of Ikkbeta. Science. 2001;293(5535):1673–1677. doi: 10.1126/science.1061620. [DOI] [PubMed] [Google Scholar]
- 13.Benzler J., Ganjam G.K., Pretz D., Oelkrug R., Koch C.E., Legler K. Central inhibition of IKKbeta/NF-kappaB signaling attenuates high-fat diet-induced obesity and glucose intolerance. Diabetes. 2015;64(6):2015–2027. doi: 10.2337/db14-0093. [DOI] [PubMed] [Google Scholar]
- 14.Ballak D.B., van Essen P., van Diepen J.A., Jansen H., Hijmans A. MAP3K8 (TPL2/COT) affects obesity-induced adipose tissue inflammation without systemic effects in humans and in mice. PLoS One. 2014;9(2):e89615. doi: 10.1371/journal.pone.0089615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perfield J.W., 2nd, Lee Y., Shulman G.I., Samuel V.T., Jurczak M.J., Chang E. Tumor progression locus 2 (TPL2) regulates obesity-associated inflammation and insulin resistance. Diabetes. 2011;60(4):1168–1176. doi: 10.2337/db10-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jager J., Gremeaux T., Gonzalez T., Bonnafous S., Debard C., Laville M. Tpl2 kinase is upregulated in adipose tissue in obesity and may mediate interleukin-1 beta and tumor necrosis factor-{alpha} effects on extracellular signal-regulated kinase activation and lipolysis. Diabetes. 2010;59(1):61–70. doi: 10.2337/db09-0470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirosumi J., Tuncman G., Chang L., Gorgun C.Z., Uysal K.T., Maeda K. A central role for JNK in obesity and insulin resistance. Nature. 2002;420(6913):333–336. doi: 10.1038/nature01137. [DOI] [PubMed] [Google Scholar]
- 18.Bluher M., Bashan N., Shai I., Harman-Boehm I., Tarnovscki T., Avinaoch E. Activated Ask1-MKK4-p38MAPK/JNK stress signaling pathway in human omental fat tissue may link macrophage infiltration to whole-body insulin sensitivity. The Journal of Clinical Endocrinology & Metabolism. 2009;94(7):2507–2515. doi: 10.1210/jc.2009-0002. [DOI] [PubMed] [Google Scholar]
- 19.Bashan N., Dorfman K., Tarnovscki T., Harman-Boehm I., Liberty I.F., Bluher M. Mitogen-activated protein kinases, inhibitory-kappaB kinase, and insulin signaling in human omental versus subcutaneous adipose tissue in obesity. Endocrinology. 2007;148(6):2955–2962. doi: 10.1210/en.2006-1369. [DOI] [PubMed] [Google Scholar]
- 20.Takeda K., Matsuzawa A., Nishitoh H., Tobiume K., Kishida S., Ninomiya-Tsuji J. Involvement of ASK1 in Ca2+-induced p38 MAP kinase activation. EMBO Reports. 2004;5(2):161–166. doi: 10.1038/sj.embor.7400072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishitoh H., Matsuzawa A., Tobiume K., Saegusa K., Takeda K., Inoue K. ASK1 is essential for endoplasmic reticulum stress-induced neuronal cell death triggered by expanded polyglutamine repeats. Genes & Development. 2002;16(11):1345–1355. doi: 10.1101/gad.992302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saitoh M., Nishitoh H., Fujii M., Takeda K., Tobiume K., Sawada Y. Mammalian thioredoxin is a direct inhibitor of apoptosis signal-regulating kinase (ASK) 1. The EMBO Journal. 1998;17(9):2596–2606. doi: 10.1093/emboj/17.9.2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bian L., Hanson R.L., Ossowski V., Wiedrich K., Mason C.C., Traurig M. Variants in ASK1 are associated with skeletal muscle ASK1 expression, in vivo insulin resistance, and type 2 diabetes in Pima Indians. Diabetes. 2010;59(5):1276–1282. doi: 10.2337/db09-1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hattori K., Naguro I., Okabe K., Funatsu T., Furutani S., Takeda K. ASK1 signalling regulates brown and beige adipocyte function. Nature Communications. 2016;7:11158. doi: 10.1038/ncomms11158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu H., Nishitoh H., Ichijo H., Kyriakis J.M. Activation of apoptosis signal-regulating kinase 1 (ASK1) by tumor necrosis factor receptor-associated factor 2 requires prior dissociation of the ASK1 inhibitor thioredoxin. Molecular and Cellular Biology. 2000;20(6):2198–2208. doi: 10.1128/mcb.20.6.2198-2208.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fujino G., Noguchi T., Matsuzawa A., Yamauchi S., Saitoh M., Takeda K. Thioredoxin and TRAF family proteins regulate reactive oxygen species-dependent activation of ASK1 through reciprocal modulation of the N-terminal homophilic interaction of ASK1. Molecular and Cellular Biology. 2007;27(23):8152–8163. doi: 10.1128/MCB.00227-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hershko T., Korotayev K., Polager S., Ginsberg D. E2F1 modulates p38 MAPK phosphorylation via transcriptional regulation of ASK1 and Wip1. Journal of Biological Chemistry. 2006;281(42):31309–31316. doi: 10.1074/jbc.M601758200. [DOI] [PubMed] [Google Scholar]
- 28.Kherrouche Z., Blais A., Ferreira E., De Launoit Y., Monte D. ASK-1 (apoptosis signal-regulating kinase 1) is a direct E2F target gene. Biochemical Journal. 2006;396(3):547–556. doi: 10.1042/BJ20051981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tan J., Zhuang L., Jiang X., Yang K.K., Karuturi K.M., Yu Q. Apoptosis signal-regulating kinase 1 is a direct target of E2F1 and contributes to histone deacetylase inhibitor-induced apoptosis through positive feedback regulation of E2F1 apoptotic activity. Journal of Biological Chemistry. 2006;281(15):10508–10515. doi: 10.1074/jbc.M512719200. [DOI] [PubMed] [Google Scholar]
- 30.Haim Y., Bluher M., Slutsky N., Goldstein N., Kloting N., Harman-Boehm I. Elevated autophagy gene expression in adipose tissue of obese humans: a potential non-cell-cycle-dependent function of E2F1. Autophagy. 2015;11(11):2074–2088. doi: 10.1080/15548627.2015.1094597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blanchet E., Annicotte J.S., Lagarrigue S., Aguilar V., Clape C., Chavey C. E2F transcription factor-1 regulates oxidative metabolism. Nature Cell Biology. 2011;13(9):1146–1152. doi: 10.1038/ncb2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Denechaud P.D., Lopez-Mejia I.C., Giralt A., Lai Q., Blanchet E., Delacuisine B. E2F1 mediates sustained lipogenesis and contributes to hepatic steatosis. The Journal of Clinical Investigation. 2016;126(1):137–150. doi: 10.1172/JCI81542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lagarrigue S., Lopez-Mejia I.C., Denechaud P.D., Escote X., Castillo-Armengol J., Jimenez V. CDK4 is an essential insulin effector in adipocytes. The Journal of Clinical Investigation. 2016;126(1):335–348. doi: 10.1172/JCI81480. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Annicotte J.S., Blanchet E., Chavey C., Iankova I., Costes S., Assou S. The CDK4-pRB-E2F1 pathway controls insulin secretion. Nature Cell Biology. 2009;11(8):1017–1023. doi: 10.1038/ncb1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fajas L., Landsberg R.L., Huss-Garcia Y., Sardet C., Lees J.A., Auwerx J. E2Fs regulate adipocyte differentiation. Developmental Cell. 2002;3(1):39–49. doi: 10.1016/s1534-5807(02)00190-9. [DOI] [PubMed] [Google Scholar]
- 36.Harman-Boehm I., Bluher M., Redel H., Sion-Vardy N., Ovadia S., Avinoach E. Macrophage infiltration into omental versus subcutaneous fat across different populations: effect of regional adiposity and the comorbidities of obesity. The Journal of Clinical Endocrinology & Metabolism. 2007;92(6):2240–2247. doi: 10.1210/jc.2006-1811. [DOI] [PubMed] [Google Scholar]
- 37.Kovsan J., Bluher M., Tarnovscki T., Kloting N., Kirshtein B., Madar L. Altered autophagy in human adipose tissues in obesity. The Journal of Clinical Endocrinology & Metabolism. 2011;96(2):E268–E277. doi: 10.1210/jc.2010-1681. [DOI] [PubMed] [Google Scholar]
- 38.Shapiro H., Pecht T., Shaco-Levy R., Harman-Boehm I., Kirshtein B., Kuperman Y. Adipose tissue foam cells are present in human obesity. The Journal of Clinical Endocrinology & Metabolism. 2013;98(3):1173–1181. doi: 10.1210/jc.2012-2745. [DOI] [PubMed] [Google Scholar]
- 39.Chutkow W.A., Birkenfeld A.L., Brown J.D., Lee H.Y., Frederick D.W., Yoshioka J. Deletion of the alpha-arrestin protein Txnip in mice promotes adiposity and adipogenesis while preserving insulin sensitivity. Diabetes. 2010;59(6):1424–1434. doi: 10.2337/db09-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kovsan J., Osnis A., Maissel A., Mazor L., Tarnovscki T., Hollander L. Depot-specific adipocyte cell lines reveal differential drug-induced responses of white adipocytes–relevance for partial lipodystrophy. American Journal of Physiology-Endocrinology and Metabolism. 2009;296(2):E315–E322. doi: 10.1152/ajpendo.90486.2008. [DOI] [PubMed] [Google Scholar]
- 41.Lee K.Y., Russell S.J., Ussar S., Boucher J., Vernochet C., Mori M.A. Lessons on conditional gene targeting in mouse adipose tissue. Diabetes. 2013;62(3):864–874. doi: 10.2337/db12-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haim Y., Tarnovscki T., Bashari D., Rudich A. A chromatin immunoprecipitation (ChIP) protocol for use in whole human adipose tissue. American Journal of Physiology-Endocrinology and Metabolism. 2013;305(9):E1172–E1177. doi: 10.1152/ajpendo.00598.2012. [DOI] [PubMed] [Google Scholar]
- 43.Thoma M.E., Hediger M.L., Sundaram R., Stanford J.B., Peterson C.M., Croughan M.S. Comparing apples and pears: women's perceptions of their body size and shape. Journal of Women's Health (Larchmt) 2012;21(10):1074–1081. doi: 10.1089/jwh.2012.3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cotillard A., Poitou C., Torcivia A., Bouillot J.L., Dietrich A., Kloting N. Adipocyte size threshold matters: link with risk of type 2 diabetes and improved insulin resistance after gastric bypass. The Journal of Clinical Endocrinology & Metabolism. 2014;99(8):E1466–E1470. doi: 10.1210/jc.2014-1074. [DOI] [PubMed] [Google Scholar]
- 45.Laforest S., Labrecque J., Michaud A., Cianflone K., Tchernof A. Adipocyte size as a determinant of metabolic disease and adipose tissue dysfunction. Critical Reviews in Clinical Laboratory Sciences. 2015;52(6):301–313. doi: 10.3109/10408363.2015.1041582. [DOI] [PubMed] [Google Scholar]
- 46.Petaja E.M., Sevastianova K., Hakkarainen A., Orho-Melander M., Lundbom N., Yki-Jarvinen H. Adipocyte size is associated with NAFLD independent of obesity, fat distribution, and PNPLA3 genotype. Obesity (Silver Spring) 2013;21(6):1174–1179. doi: 10.1002/oby.20114. [DOI] [PubMed] [Google Scholar]
- 47.Cancello R., Henegar C., Viguerie N., Taleb S., Poitou C., Rouault C. Reduction of macrophage infiltration and chemoattractant gene expression changes in white adipose tissue of morbidly obese subjects after surgery-induced weight loss. Diabetes. 2005;54(8):2277–2286. doi: 10.2337/diabetes.54.8.2277. [DOI] [PubMed] [Google Scholar]
- 48.Aron-Wisnewsky J., Tordjman J., Poitou C., Darakhshan F., Hugol D., Basdevant A. Human adipose tissue macrophages: m1 and m2 cell surface markers in subcutaneous and omental depots and after weight loss. The Journal of Clinical Endocrinology & Metabolism. 2009;94(11):4619–4623. doi: 10.1210/jc.2009-0925. [DOI] [PubMed] [Google Scholar]
- 49.Abdennour M., Reggio S., Le Naour G., Liu Y., Poitou C., Aron-Wisnewsky J. Association of adipose tissue and liver fibrosis with tissue stiffness in morbid obesity: links with diabetes and BMI loss after gastric bypass. The Journal of Clinical Endocrinology & Metabolism. 2014;99(3):898–907. doi: 10.1210/jc.2013-3253. [DOI] [PubMed] [Google Scholar]
- 50.Sun K., Tordjman J., Clement K., Scherer P.E. Fibrosis and adipose tissue dysfunction. Cell Metabolism. 2013;18(4):470–477. doi: 10.1016/j.cmet.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.O'Neill A.C., Jagannathan J.P., Ramaiya N.H. Evolving cancer classification in the era of personalized medicine: a primer for radiologists. Korean Journal of Radiology. 2017;18(1):6–17. doi: 10.3348/kjr.2017.18.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Solas M., Milagro F.I., Martinez-Urbistondo D., Ramirez M.J., Martinez J.A. Precision obesity treatments including pharmacogenetic and nutrigenetic approaches. Trends in Pharmacological Sciences. 2016;37(7):575–593. doi: 10.1016/j.tips.2016.04.008. [DOI] [PubMed] [Google Scholar]
- 53.Manieri E., Sabio G. Stress kinases in the modulation of metabolism and energy balance. Journal of Molecular Endocrinology. 2015;55(2):R11–R22. doi: 10.1530/JME-15-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tang X., Guilherme A., Chakladar A., Powelka A.M., Konda S., Virbasius J.V. An RNA interference-based screen identifies MAP4K4/NIK as a negative regulator of PPARgamma, adipogenesis, and insulin-responsive hexose transport. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(7):2087–2092. doi: 10.1073/pnas.0507660103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Plotnikov A., Zehorai E., Procaccia S., Seger R. The MAPK cascades: signaling components, nuclear roles and mechanisms of nuclear translocation. Biochimica et Biophysica Acta. 2011;1813(9):1619–1633. doi: 10.1016/j.bbamcr.2010.12.012. [DOI] [PubMed] [Google Scholar]
- 56.Polager S., Ofir M., Ginsberg D. E2F1 regulates autophagy and the transcription of autophagy genes. Oncogene. 2008;27(35):4860–4864. doi: 10.1038/onc.2008.117. [DOI] [PubMed] [Google Scholar]
- 57.Slutsky N., Vatarescu M., Haim Y., Goldstein N., Kirshtein B., Harman-Boehm I. Decreased adiponectin links elevated adipose tissue autophagy with adipocyte endocrine dysfunction in obesity. International Journal of Obesity (Lond) 2016;40(6):912–920. doi: 10.1038/ijo.2016.5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Insulin signaling in MEF-derived adipocyte-like cells of wild-type and ASK1-ATKO mice. MEF-derived adipocyte-like cells were grown as described in the legend for Figure 5. After differentiation, cells were serum-starved for 3h, followed by stimulation with 0.5 nM (sub-maximal) or 100 nM (maximal) insulin, for 7 min. Lysates were assessed for insulin signaling. Shown are representative blots (A) and densitometry analysis (B–D) of n = 4 independent experiments. ∗p < 0.05 vs. WT con. ˆˆp < 0.01 vs. WT con. $$p < 0.01 vs. ASK1-ATKO con.