Abstract
Objective
The loss of skeletal muscle mass and strength are a central feature of traumatic injury and degenerative myopathies. Unfortunately, pharmacological interventions typically fail to stem the long-term decline in quality of life. Reduced Rev-Erb-mediated gene suppression in cultured C2C12 myoblasts has been shown to stimulate myoblast differentiation. Yet the mechanisms that allow Rev-Erb to pleiotropically inhibit muscle differentiation are not well understood. In this study, we sought to elucidate the role of Rev-Erb in the regulation of muscle differentiation and regeneration in vivo.
Methods
Using Rev-Erbα/β shRNAs, pharmacological ligands, and Rev-Erbα null and heterozygous mice, we probed the mechanism of Rev-Erbα/β regulation of muscle differentiation and muscle regeneration.
Results
ChIP seq analysis of Rev-Erb in differentiating myoblasts showed that Rev-Erbα did not transcriptionally regulate muscle differentiation through cognate Rev-Erb/ROR-response elements but through possible interaction with the cell fate regulator NF-Y at CCAAT-motifs. Muscle differentiation is stimulated by Rev-Erb release from CCAAT-motifs at promoter and enhancer elements of a number of myogenesis proteins. Partial loss of Rev-Erb expression in mice heterozygous for Rev-Erbα accelerated muscle repair in vivo whereas Rev-Erb knockout mice showed deficiencies in regenerative repair compared to wild type mice. These phenotypic differences between heterozygous and knockout mice were not apparently dependent on MRF induction in response to injury. Similarly, pharmacological disruption of Rev-Erb suppressive activity in injured muscle accelerated regenerative repair in response to acute injury.
Conclusions
Disrupting Rev-Erb activity in injured muscle accelerates regenerative muscle repair/differentiation through transcriptional de-repression of myogenic programs. Rev-Erb, therefore, may be a potent therapeutic target for a myriad of muscular disorders.
Keywords: Rev-Erb, Nuclear factor-YA, CCAAT-Binding motif, Myogenesis, Muscle regeneration, SR8278
Highlights
-
•
The nuclear receptor Rev-Erb is released from myogenic gene promoter and enhancer regions during myoblast differentiation.
-
•
Rev-Erb regulates myoblast differentiation through interaction with the cell-fate regulator NF-Y at CCAAT-motifs.
-
•
Rev-Erb antagonists accelerate muscle regeneration following acute muscle injury by stimulating myoblast differentiation.
-
•
Human muscle differentiation can be regulated using pharmacological ligands that target Rev-Erb.
1. Introduction
Skeletal muscle comprises about 40–50% of our total body mass and is essential for postural support, locomotion, and breathing. Under normal physiological conditions, skeletal muscle turnover through proteasome and autophagosome-mediated degradation of myofiber proteins proceeds at a low basal rate [1]. Skeletal muscle's high capacity for regeneration is typically sufficient for dynamically maintaining skeletal muscle mass and function in response to minor injuries and normal wear and tear.
In response to muscle injury or physical stress, quiescent muscle stem cells, satellite cells (SCs), form transient progenitor cells that produce myoblasts, which fuse with myofibers in a process known as myogenesis. During this process, SCs also self-renew to replenish the satellite cell pool. In differentiating myoblasts a family of proteins, the myogenic response factors (MRFs), which include myogenic factor 5 (MYF5), myogenic differentiation-1 (MYOD), muscle specific regulatory factor 4 (MRF4), and myogenin (MYOG) are sequentially and transiently expressed to direct myoblast differentiation and fusion with myofibers. MYF5 is considered the master regulator of muscle development, as its expression is sufficient to induce a muscle phenotype in fibroblast cells [2]. The expression of MYF5 induces the expression of MYOD, which drives myoblast proliferation and the subsequent stimulation of MYOG expression, which promotes myoblast fusion into multinucleated myofibrils that comprise skeletal muscle myofibers [1]. Often, with severe injuries (greater than 20% loss of muscle mass), normal muscle regeneration cannot keep pace with the heightened regenerative demands. In this scenario, the loss of skeletal muscle mass can trigger widespread inflammation, fibrosis, and loss of muscle function. Successful regeneration of skeletal muscle after traumatic injury is therefore dependent on an efficient replenishment of muscle fibers through elevated myoblast proliferation and differentiation.
In addition to reduced muscle function, loss of lean mass is a major co-morbidity for severe burns, cancer, HIV infection, congestive heart failure, and chronic obstructive pulmonary disease. In these conditions, skeletal muscle regenerative decline and muscle turnover/catabolism are often untenable through dietary intervention and administration of anabolic steroids or non-steroidal anti-inflammatories (NSAIDs). In addition, the use of anabolic steroids and NSAIDs are accompanied by severe side effects that may further reduce quality of life. Identifying new means of accelerating muscle regeneration has proved a daunting challenge. Therefore, understanding the underlying mechanisms that regulate myofiber regeneration and coordinate regenerative repair could provide future therapeutic options for stymieing the loss of muscle function in the traumatically injured.
The nuclear receptors NR1D1/2 (Rev-Erbα/β) [3], [4] have been shown to be involved in the regulation of myogenic protein expression in cultured myoblasts [5]. Rev-Erbs are nuclear receptors that act as transcriptional repressors that exclusively recruit corepressors (NcoR1 and SMRT) and basally suppress gene expression due to the ready availability of the endogenous ligand heme [6], [7]. A decline in expression of Rev-Erbα has been shown to precede myoblast differentiation and is necessary and sufficient for efficient myotube formation in vitro [5], [8]. Conversely, Rev-Erbα/β expression is involved in the dynamic regulation of mitochondria biogenesis and metabolic function in fully differentiated skeletal muscle. Rev-Erbα knockout mice display dyslipidemia, abnormal structure of skeletal muscle fibers, and reduced mitochondrial oxidative capacity [5], [8], [9], [10]. In addition, mice deficient in the expression of the Rev-Erb target genes brain and muscle ARNT-like-protein 1 (Bmal1) or retinoic acid receptor-like orphan receptor α/β/γ (Rorα/β/γ) exhibit reduced regenerative capacity and diminished myosin heavy chain expression in response to muscle injury [9], [11], [12]. These studies suggest that there is a somewhat dichotomous role of Rev-Erb as both an inhibitor of muscle differentiation and a promoter of mitochondrial function. This apparent paradox suggests that there is a need to explore the regulatory mechanisms underlying Rev-Erb's activity in skeletal muscle in more detail. Crucially, the exact mechanisms of Rev-Erb regulation of muscle differentiation or whether Rev-Erb is integral to muscle regeneration in response to injury in vivo has not been discerned.
Nuclear Factor-Y (NF-Y) is a trimeric transcription factor consisting of NF-YA, B, and C subunits. The NF-Y complex is a well-characterized regulator of myoblast cell fate determination [13], [14], [15]. NF-YA is the only subunit in the NF-Y complex that can bind to CCAAT-motifs directly; therefore the NF-Y complex becomes inactive if NF-YA expression is lost. NF-Y facilitates myoblast proliferation through regulation of cell cycle progression genes cyclin b1, b2 (Ccnb1, Ccnb2), cell division cyclin 25c (Cdc25c), cyclin a2 (Ccna2), and topoisomerase DNAIIα (Top2a) [13], [14], [15]. In differentiated human muscle, NF-YA expression is usually undetectable and loss of the NF-Y complex CCAAT-box binding precedes full muscle differentiation [14]. Gurtner et al. further showed that NF-YA expression is gradually reduced during myoblast differentiation, thereby reducing the binding of the NF-Y complex to CCAAT-motifs in cell cycle transition regulators [14], [15]. Additional studies by Basile and colleagues showed that the long and short isoforms of NF-Y differentially modulate myoblast differentiation [13]. While expression of both isoforms is reduced as myoblasts differentiate, expression of the long isoform, which is predominantly expressed in proliferating myoblasts, is required for the initiation of myotube differentiation. This is primarily due to NF-YA expression being important in proliferating myoblasts as activation of muscle protein specific gene expression (e.g. Myod and Myog), required in the early stages of myoblast differentiation, is reduced when NF-YA expression is inhibited [13]. NF-YA therefore drives both the proliferative burst of myoblasts and also ushers the initiation of myoblast differentiation through the stimulation of expression of MRFs before its activity and expression declines in fully differentiated myotubes.
Here we investigated the molecular mechanisms of Rev-Erb regulation of myogenesis and regenerative repair in response to acute injury. We observed that shRNA and pharmacological disruption of Rev-Erbα/β activity significantly increased the rate of cultured myoblast differentiation. Surprisingly, in differentiating myoblasts, Rev-Erb almost exclusively regulated MRF (Myf5, Myod, Myog) expression through tethered interaction with NF-Y and not through binding to its cognate Rev-Erb/ROR response elements (ROREs). During differentiation and myotube formation Rev-Erb/NF-Y complexes are released from the regulatory regions of MRFs resulting in the transient induction of MRFs (Myf5, Myod and Myog) myosin heavy chain proteins. In accord with this, pharmacological disruption of Rev-Erb activity using the Rev-Erb antagonist SR8278 accelerated Rev-Erb release from MRF gene promoters and enhancers during myoblast differentiation. We observed that Rev-Erbα knockout mice have elevated basal levels of MRF expression but display a muted MRF induction and reduced regenerative capacity in response to injury. Conversely, mice heterozygous for Rev-Erbα show enhanced regenerative capacity and a more robust induction of myogenic factors in response to injury compared to wild type mice. Notably, Rev-Erb antagonist treatment stimulated MRF expression and enhanced muscle repair and regeneration in vivo. Rev-Erb antagonist treatment similarly enhanced human myotube differentiation through induction of myogenic gene expression suggesting the human REV-ERB may similarly regulate human muscle differentiation and regeneration. These observations collectively suggest that Rev-Erb may be an interesting therapeutic target for the treatment of diverse muscular diseases.
2. Materials and methods
2.1. Mouse models
C57BL/6, B6.Cg-Nr1d1tm1Ven/LazJ (Rev-Erbα+/+, Rev-Erbα+/−, and Rev-Erbα−/−) mice were purchased from Jackson laboratory. All mice were housed in a 12 h–12 h light–dark cycle and received a standard chow diet and allowed food and water ad libitum unless otherwise stated. At the conclusion of the study, mice were sacrificed by CO2 asphyxiation followed by cervical dislocation. Mouse experimental procedures were approved by the Saint Louis University Institutional Animal Care and Use Committee (protocol#2474).
2.2. Immunoblotting
Cells and muscle tissue were lysed by a RIPA with protease inhibitors. The extracted proteins were flash frozen and stored at −80 °C. The extracted proteins were separated by SDS-PAGE and transferred onto PVDF membranes. Immunoblot analyses were performed to standard procedures. A list of antibodies used can be found in Supplemental Table S2.
2.3. Immunoprecipitation (IP)
HEK293 cells were co-transfected with 9 μg of pCMV-Entry (C-terminal Myc and DDK tag), Rev-Erbα (C-terminal Myc and DDK tag), or Nr1d2 (C-terminal Myc and DDK tag) with 1 μg of GFP plasmid. All plasmids were supplied from Origene: pCMV-Entry (PS10001), Rev-Erbα (MR209431), and Rev-Erbβ (MR209045). Six million cells were reversed transfected with Lipofectamine 2000 (Thermofisher Scientfic) and seeded in a 100 mm × 100 mm cell culture dish for 24 h. Transfection efficiency was determined by GFP expression (90%). HEK293s were harvested and lysed by RIPA lysis buffer with protease inhibitor cocktail. Protein concentrations were determined by Peirce BCA Protein Assay Kit (Thermo Scientific). Three micrograms of protein were incubated with ANTI-FLAG M2 Affinity Gel (Sigma) and subjected to immunoprecipitation overnight at 4 °C. Immunoprecipitation was conducted by the manufacturer's protocol (Sigma). Elution of FLAG-fusion proteins was conducted by SDS-PAGE. Rev-Erbα/Rev-Erbβ FLAG protein interaction was determined by incubating blots overnight at 4 °C with select antibodies. List of antibodies utilized are described in Supplemental Table 1. HEK293 lysates were normalized to HSP90 and Beta Actin to verify equal loading. All blots were imaged by ChemiDoc MP (Biorad).
2.4. Cardiotoxin (CTX) induced injury model
CTX injury was induced in 6-week-old C57BL/6 mice by injecting 30 μL of CTX (10 μM) along three points of tibialis anterior (TA) muscle of the left hind limb to produce uniform muscle damage. Sham injections of PBS were injected into the TA muscle of the right hind limb. Mice were sacrificed at day 8 and day 15 after CTX-injury. TA muscles were harvested and frozen in OCT and stored at −80 °C for histology and immunohistochemistry or flash frozen in liquid nitrogen for qPCR analysis. Mice were administered the Rev-Erbα/β antagonist SR8278 (25 mg/kg) by subcutaneous injection. SR8278 was formulated in 10% DMSO: 10% cremophor: 80% PBS vehicle. Control mice were injected with vehicle alone. Both groups of CTX mice were dosed once a day at 12:00 h.
2.5. C2C12 cell culture
Low passage (3–7) C2C12 mouse myoblasts were cultured in 15% FBS in Gibco Dulbecco's Modified Eagle Medium supplemented with GlutaMAX. Cells were incubated at 37 °C in a humidified atmosphere of 5% CO2. Cells were split when 50–60% confluent. Before differentiation, 90% confluent C2C12 cells were exposed to 5 μM of SR8278 or SR9011 for 24 h in 15% FBS. Cells were then induced to differentiate by exchanging the proliferation media with fusion media (2% horse serum in DMEM). Throughout the differentiation process, the cells were exposed to 5 μM of SR8278 or SR9011. Protein from C2C12 cells was isolated by adding 100 μL of RIPA to each well and scraped and frozen at −80 °C for immunoblot assay. RNA extraction was conducted by lysing the cells with 350 μL of Ambion Lysis buffer. Cells were frozen and mRNA extraction and purification conducted in accordance with the manufacturer's protocol.
2.6. Lentiviral infection and induction of shRNAs
SMARTvector Lentiviral Mouse Rev-Erbα (or Rev-Erbβ) mCMV-TurboGFP shRNA by Dharmacon RNAi Technologies was used to infect 1.5 × 104 proliferating C2C12 cells for 48 h. Cells were split and seeded at a density of 1.5 × 104 in 6-well culture plates. Twenty-four hours later, cell selection was conducted by exposing the cells to 4 μg/mL of puromyocin. shRNA mediated knock down of Rev-Erbα or Rev-Erbβ in C2C12 cells was induced by doxycycline (1 μg/mL) exposure for 48 h in select media (fusion or proliferation). Gene expression was utilized to assess targeted knock down when compared to a control shRNA expressing cells.
2.7. Live cell staining and imaging
C2C12 cells under select experimental conditions were allowed to differentiation for 4 days before imaging. Cells were washed with 1×PBS and mitochondrial were stained by being exposed to 50 nM of MitoTracker® Red CMXRos (Molecular Probes™) in FluoroBrite™ DMEM Media (Gibco) for 30 min at 37 °C. Cells were then washed with 1×PBS and incubated with DAPI in FluoroBrite™ media for 20 min at room temperature. Cells were imaged by florescence at 10× magnification. Nuclei counting was conducted in ImageJ. Total differentiation was calculated by counting the number of nuclei within the myotubes divided by the total number of nuclei with in the image (N = 3). The average the level if myoblast fusion was calculated as follows: number of nuclei per myotube/total number of myotubes (N = 3).
2.8. Muscle biopsy and cell culture
Muscle biopsy specimens were taken from the vastus lateralis using the Bergstrom technique [16] and processed the same day for cell culture. Satellite cells (quiescent mononuclear muscle cells) were isolated by trypsin digestion, pre-plated on an uncoated petri dish for an hour to remove fibroblasts, and subsequently transferred to T-25 collagen–coated flasks in Dulbecco's Minimum Essential Medium (DMEM) supplemented with 16% Fetal Bovine Serum (FBS) and human growth factors. Cell lines were grown at 37 °C in a humidified atmosphere of 5% CO2 and expanded in T-150 flasks. Myoblasts at passage 3 were plated in 6-well Corning Cell Bind plates for all assays. Differentiation of myoblasts into myotubes was initiated at approximately 90% confluence, by switching to α-MEM with antibiotics, 2% FBS and fetuin for maximal adhesion and growth. Differentiation media was changed every other day [16], [17].
2.9. Quantitative PCR
Total RNA was isolated from muscle tissue by using the Trizol reagent (Invitrogen) or a PureLink RNA Mini Kit (Ambion) from C2C12 and human primary cells. Isolated RNA (1 μg) was reverse transcribed into cDNA by using qScript cDNA Synthesis Kit (Quanta Biosciences, Inc.). Quantitative PCR (qPCR) was performed by with SYBR Select Master Mix (Applied Biosystems) with select primers. Gene expression for mouse tissue and cells was normalized to Gapdh. Gene expression from human primary muscle cells was normalized to CYCLOPHILIN.
2.10. ChIP-Seq
ChIP-Seq experiments were performed using C2C12 cells. Cells were fixed and chromatin harvested at 24 h and 120 h after seeding for proliferating and differentiating-Day 5 cells, respectively, at the same time of day when Rev-Erbα/β mRNA expression was lowest, which, as Rev-Erbα auto-inhibits its own gene expression, correlates with a high abundance of nuclear Rev-Erbα/β protein (Supplemental Figure 2A). Soluble chromatin was obtained by fragmentation via partial MNase digestion that yielded chromatin fragments of 1–5 nucleosome in size (∼150 bp/nucleosome), followed by ChIP using a validated in-house made Rev-erbα antibody (Supplemental Figure S2B) and SimpleChIP® Enzymatic Chromatin IP kit (Cell Signaling Technology). A library was constructed using NEBNext® Ultra™ DNA Library Prep Kit for Illumina and NEBNext® Multiplex Oligos for Illumina (New England BioLabs). After ligation with bar-coded adapters, fragments corresponding to an original 150 bp in size were selected using AMPure XP Beads (Beckman Coulter) and PCR-amplified. ChIP-Seq was performed at the Genome Technology Access Center at Washington University School of Medicine in St. Louis. Approximately 50 million reads per sample were generated from single read 1 × 50 sequencing. Demultiplexed FASTQ files were then exported for downstream analysis.
2.11. ChIP-Seq analysis
Raw FASTQ reads were mapped to mouse mm10 genome using STAR (version 2.4.2a) to generate sorted BAM files. Published ChIP-Seq datasets (SRP006743/GSE25308 [18]), and SRP057271/GSE67962 [19] were downloaded from NIH GEO/SRA servers (http://www.ncbi.nlm.nih.gov/sra). FASTQ files were then converted from original SRA files using fastq-dump (version 2.3.5). BAM files were analyzed using HOMER software (version 4.7.2) in combination with R packages (version 3.2.0) to identify binding peaks, quantify binding signals, and produce histogram plots. First, makeTagDirectory program was used to create tag directories from the mapped reads in the BAM file. Next, findPeaks.pl was used for peak calling to identify Rev-erb binding sites across the genome (FDR <= 0.001, fold over input > = 4, Poisson p <= 0.0001). De novo motif discovery and motif enrichment analyses were carried out using findMotifsGenome.pl. Quantitative analyses of motif occurrence and binding signals were performed using annotatePeaks.pl, and the results were imported into R to generate various plots reported in the manuscript. All analyses were performed with default settings. To visualize ChIP-Seq signals, bedgraph files were generated using makeUCSCfile and loaded into IGV (Integrative Genomics Viewer) (https://www.broadinstitute.org/igv/). P-values of gene-list enrichment were calculated using binomial tests with R packages.
2.12. Gene Ontology (GO) analysis
Gene Ontology and gene-list enrichment analysis were performed with ToppGene server (http://toppgene.cchmc.org). ChIP seq data has been deposited in the Gene Expression Omnibus (GEO) database of NIH.
2.13. H&E staining and cross-sectional area (CSA) analysis
Muscle samples (Tibia anterior or gastrocnemius) were fixed overnight in 4% formalin and then embedded into paraffin. Paraffin embedded muscle sections where stained by a standard hematoxylin and eosin (H&E) protocol. Slide images were captured using a Leica MC120 HD attached to a Leica DM750 microscope. Cross-sectional area (CSA) of H&E stained myofibers where quantified by ImageJ. Six images from each experimental animal were utilized to calculate the average CSA per mouse. Images were taken at 10× and 40× for needed calculations.
2.14. Clinical study
Clinical Trials identifier: NCT02226640. Primary human skeletal muscle cells were obtained from 4 lean, healthy, Caucasian female donors. This study (IRB# 238153) was approved by the Florida University Hospital IRB. All participants gave written and verbal consent to all study procedures. Donor characteristics are detailed in the Supplemental Table S1.
2.15. Pharmacokinetics and mass spectrometry
Muscle and plasma samples were collected 4 h after the final dose, flash frozen, and stored at −80 °C until analysis. Muscle samples were then weighed and placed into Eppendorf tubes. Naïve tissue was used to prepare standard curves in muscle tissue matrix. To each sample or standard tube, 3 stainless steel beads (2–3 mm) were added, and the appropriate volume of cold 3:1 acetonitrile:water (containing 100 ng/mL extraction internal standard SR8277) was added to achieve a tissue concentration of 200 mg/mL. Tubes were placed in a bead beater for 1–2 min. Samples and standards were centrifuged at 3200 rpm for 5 min at 4 °C. The supernatant (200 μL) was transferred to a 96-well plate, evaporated to dryness under nitrogen, reconstituted with 100 μL of 0.1% v/v formic acid in 9:1 water:acetonitrile, and vortexed for 5 min. Plasma samples or standards prepared in plasma matrix (100 μL) were added to a 96-well plate. To each well, 400 uL of cold acetonitrile containing 100 ng/mL extraction internal standard SR8277 was added. The plate was vortexed for 5 min at 4 °C. The supernatant (400 μL) was transferred to a second 96-well plate, evaporated to dryness under nitrogen, reconstituted with 100 μL of 0.1% v/v formic acid in 9:1 water:acetonitrile, and vortexed for 5 min. Finally, to each reconstituted muscle or plasma sample, 10 μL of 1000 ng/mL enalapril in acetonitrile was added as an injection internal standard, and the 96-well plate was vortexed, briefly centrifuged, and submitted for LC/MS analysis.
2.16. SR8278 concentration
SR8278 concentrations were determined on a Sciex API-4000 LC/MS system in positive electrospray mode. Analytes were eluted from an Amour C18 reverse phase column (2.1 × 30 mm, 5 μm) using a 0.1% formic acid (aqueous) to 100% acetonitrile gradient mobile phase system at a flow rate of 0.35 mL/min. Peak areas for the mass transition of m/z 362 > 157 for SR8278, m/z 394 > 189 for the extraction internal standard SR8277, and m/z 376 > 91 for the injection internal standard enalapril were integrated using Analyst 1.5.1 software. Peak area ratios of SR8278 area/SR8277 area were plotted against concentration with a 1/x-weighted linear regression. Enalapril was used to monitor proper injection signal throughout the course of LC/MS analysis.
2.17. Statistical analysis
For qPCR, immunoblot and histological quantification of fluorescent signals, statistical significance was determined by subjecting mean values per group to students-t test unless otherwise specified. A value of p ≤ 0.05 is considered statistically significant.
3. Results
3.1. Disrupting Rev-Erb expression accelerates myoblast differentiation
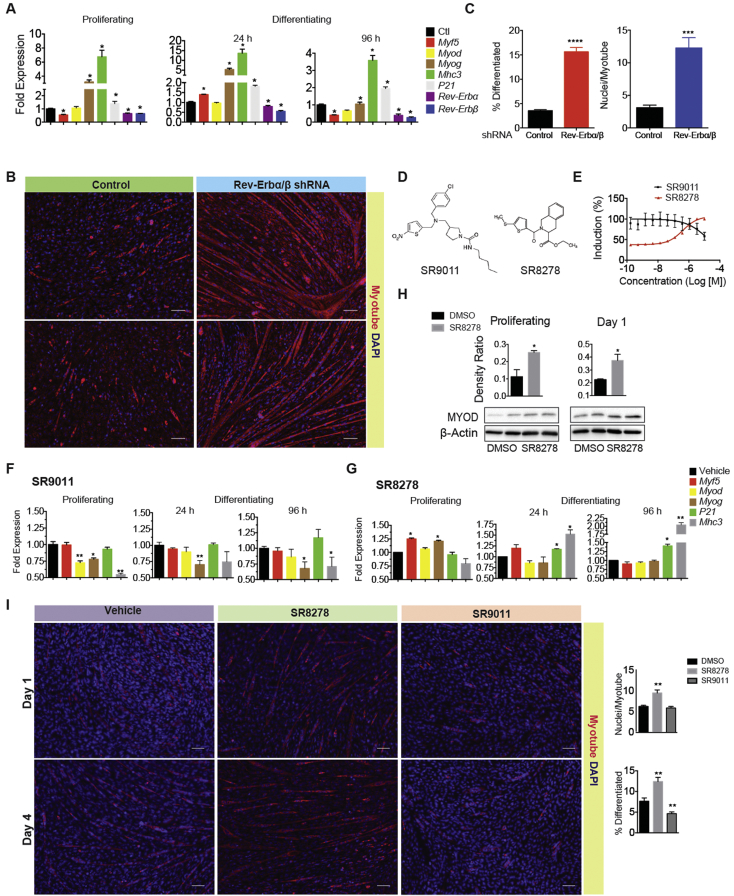
Considering that exogenous Rev-Erbβ expression has been shown to inhibit myoblast differentiation [5], we decided to explore whether reduced Rev-Erbα and β expression significantly enhanced the rate of differentiation of proliferating myoblasts. To investigate this, we stably transfected the mouse myoblast cell line C2C12 with Rev-Erbα/β-specific inducible shRNAs and assessed whether knocking down Rev-Erb expression exclusively in proliferating myoblasts would enhance myogenic gene activation and increase the rate of myotube formation compared to myoblasts expressing normal levels of Rev-Erb. Disrupting Rev-Erbα and/or β expression induced myogenesis genes expression (Myf5, Myod, and MyoG) (Figure 1A, Supplemental Figure S1A and B) and substantially accelerated myotube formation. Myoblasts expressing Rev-Erbα/β shRNAs formed fully differentiated multinucleated myotubes 3 days earlier (Day 4) than cells with intact Rev-Erb expression (Day 7) (Figure 1B and C). These results demonstrate that downregulation of Rev-Erb expression in proliferating myoblasts significantly accelerates myoblast differentiation.
Figure 1.
Disrupting Rev-Erb expression accelerates myoblast differentiation. A) Expression of myogenesis genes in C2C12 myoblasts stably expressing Rev-Erbα/β shRNAs or control (GFP). shRNA expression was induced in 60% confluent cells at 24 h before RNA isolation (proliferating) or at 24 h or 96 h after stimulation of differentiation. B) Differentiation assay showing myotube formation (red) in C2C12 cells expressing Rev-Erb shRNAs after 4 days in differentiating medium. C) Quantification (n = 3) of nuclei/myotube or % differentiated cells as seen in images in (B). *p < 0.05 and ****p < 0.0001 as determined by two-tailed Students t-test. Data represented as mean and ±s.e.m. D) Structure of Rev-Erb agonist SR9011 and SR8278. E) Relative activity of SR8278 and SR9011 in Bmal-promoter driven luciferase reporter assay. F) Myogenesis gene expression in C2C12 cells treated with the Rev-Erb agonist SR9011 and G) antagonist SR8278. H) Immunoblot showing MYOD and Myosin heavy chain-2 (MHC2) expression in C2C12 treated with SR8278. I) Differentiation assay showing relative myotube formation (red) in SR8278 and SR9011 treated C2C12% differentiated cells and nuclear/myotube were quantified using imageJ (see Supplemental Figure S1). Representative images are shown. *p < 0.05 and **p < 0.01 as determined by two-tailed Students t-test. Data represented as mean and ±s.e.m.
3.2. Synthetic Rev-Erb ligands modulate myogenesis
Given that Rev-Erb RNAi-mediated knockdown enhanced myotube formation, we tested whether pharmacological modulation of Rev-Erb transcriptional activity in proliferating myoblasts could similarly accelerate myogenesis. To accomplish this, we treated proliferating and early (2–4 days) differentiating C2C12 cells with the Rev-Erb agonist SR9011 or antagonist SR8278 (Figure 1D and E). The Rev-Erb agonist SR9011 suppressed myogenesis gene expression (Myod, Myog, and Mhc3) in both proliferating and differentiating myoblasts (Figure 1F). Conversely, Rev-Erb antagonism stimulated myogenic gene expression (Figure 1G and H). Interestingly, Myod expression was exclusively enhanced by SR8278 treatment in mitotic and early differentiating myoblasts (Figure 1G and H). In addition, myotube formation was accelerated by Rev-Erb antagonism (SR8278 treatment) and inhibited by Rev-Erb agonism (SR9011 treatment) (Figure 1I). The contrasting effect of Rev-Erb agonists and antagonists suggested that the release of Rev-Erb-mediated transcriptional repression of MRF gene expression might be key to the induction of myoblast differentiation.
3.3. Rev-Erb tethers to NF-Y complexes at CCAAT-boxes in a tissue specific manner
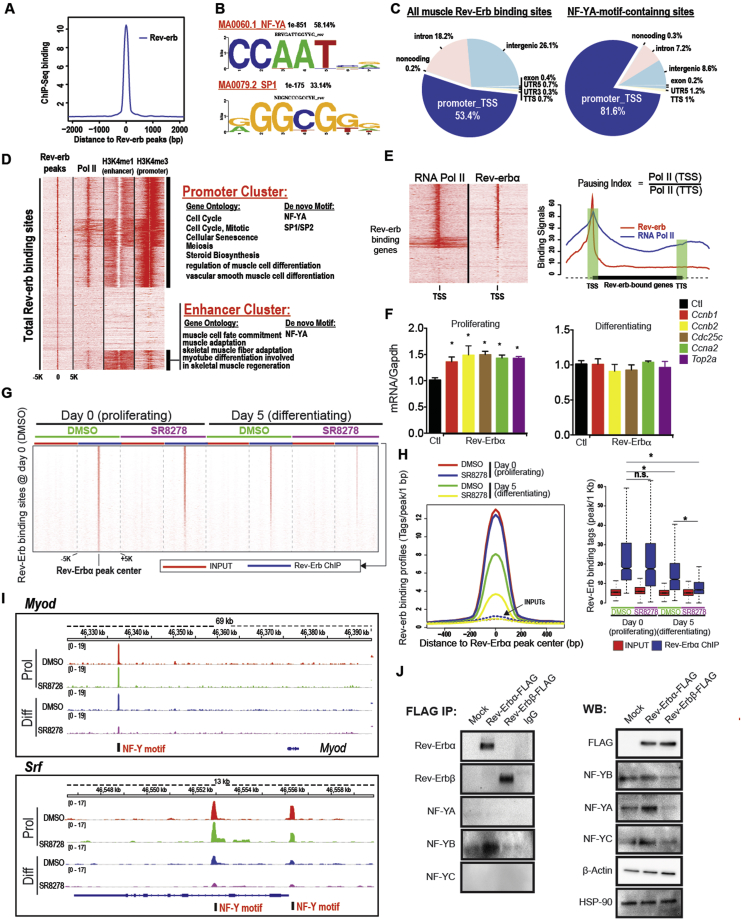
With the evidence that loss of Rev-Erb expression or activity transcriptionally activated the expression of myogenic factors, we performed chromatin immunoprecipitation followed by deep sequencing (ChIP-Seq) in proliferating and differentiating C2C12 myoblasts to identify specific myogenic genes and pathways that Rev-Erb directly modulates during myogenesis. Unexpectedly, Rev-Erb was highly enriched primarily at the cognate promoter binding motifs of NF-Y and SP1/2 but not the classical Rev-Erb response elements (RORE) (Figure 2A–C). Of the total sites occupied by Rev-Erb, only a minor subset (∼1%) contained classical ROREs (Figure 2C, Supplemental Figure S2A–D). Recent reports in which Rev-Erb regulation gene expression was assessed across a number of tissues suggested that the majority of binding sites enriched with Rev-Erb that are common across liver, brain, and white adipose tissue (WAT) are ROREs. However, binding analysis within each tissue revealed that Rev-Erb regulated an unexpectedly large number of genes through RORE-independent interactions with distinct tissues-specific transcription factors [19] (Supplemental Figure S2C), with only a small subset of genes exhibiting Rev-Erb binding at RORE-motifs (Supplemental Figure S2C and D). For example, in the liver, Rev-Erb enrichment at hepatocyte nuclear factor 4a and 6 (HNF4a and HNF6) binding motifs represented the majority of the total binding sites occupied by Rev-Erb in the genome [19] (Supplemental Figure S2C). WAT and brain also showed distinct patterns of Rev-Erb enrichment at binding motifs for tissue specific transcription factors such as CCAAT-enhancer binding protein (CEBP) and nuclear factor 1 (NF1) respectively [19]. Interestingly Rev-Erb enrichment at NF-Y motifs was not unique to muscle as a number of genes (Per1, Bmal) also featured Rev-Erb occupancy at NF-Y motifs at gene promoter and enhancer regions (Supplemental Figure S2E). Conversely, we also observed that Rev-Erb was exclusively enriched at NF-YA motifs for pro-myogenic genes (Srf) exclusively in muscle but not other tissues such as liver (Supplemental Figure S2F). Our Chip-seq results in muscle were analogous to the tissue specific binding characteristics displayed by Rev-Erb in liver, brain, and WAT. Although in comparison to that of brain, WAT, and liver, it is notable that an exceedingly small percentage of Rev-Erb enriched binding sites were ROREs (Figure 2C). Collectively, these results suggest a dominant contributory role of tethered Rev-Erb binding to CCAAT-boxes to Rev-Erb's functions in muscle differentiation.
Figure 2.
Rev-Erb co-regulates myogenic genes through binding at NF-Y and SP1 binding motifs. A) Histogram of Rev-erb ChIP-Seq signals in C2C12 cells at the binding sites of Rev-erb (see Supplemental Figure S2). B) De novo motif discovery analysis showing the two most enriched motifs (NY-F and SP1) at Rev-erb binding sites in C2C12 myoblasts. C) Annotated binding locations of Rev-erb (total binding sites, left) and NF-Y-motif containing sites (right). TSS: transcription start site; TTS: transcription termination site. D) Heatmap showing that Rev-erb binding leads to increased pausing of RNA Pol II at key genes important to myogenesis. Rev-Erb binding signals showing clustering of binding sites at promoter TSS and enhancer regions. Rev-Erb binding sites were clustered based on promoter and enhancer marks – RNA Pol II, mono-methyl H3K4 (H3K4me1, enhancer mark) and tri-methyl-H3K4 (H3K4me3, promoter mark). E) Heatmap and graph showing pausing index of RNA Pol II at Rev-Erb binding sites. Pausing index was measured as the ratio of Pol II occupancies at promoter/TSS and termination/TTS regions, as calculated using meta-genes. F) Expression of NF-Y-target genes (Ccnb1, Ccnb2, Cdc25 cm, Ccna2, and Top2a) in proliferating and differentiating C2C12 (n = 4) myoblasts in response to Rev-Erb shRNA-mediated knockdown relative to control shRNAs. Data were subjected to student's t-test *p < 0.05. G) Heatmap showing Rev-Erb occupancy of NF-YA motifs is reduced during myoblast differentiation (0–5 days) and that Rev-Erb antagonist (SR8278) accelerates Rev-Erb release from regulatory binding sites. H) Histogram and bar graph showing reduced Rev-Erb binding signals in proliferating or differentiating myoblasts treated with Rev-Erb antagonist (SR8278) or vehicle control. I) Rev-Erb binding site enrichment at regulatory regions of key myogenic regulators Myod and Srf in proliferating or differentiating (5 days) myoblasts treated with SR8278 or vehicle. J) Immunoprecipitation of ectopically expressed Rev-Erb in HEK-293 cells showing Rev-Erbα interaction with the B subunit of the trimeric NF-YA/B/C complex. (Right panel) Immunoblot showing expression of NF-Y subunits and Rev-Erbα.
3.4. Rev-Erb interaction with NF-YA causes RNA-Pol II pausing at NF-YA motifs
Our analysis of tethered Rev-Erb transcriptional regulation at NF-YA motifs showed that RNA-polymerase II is paused at NF-Y/Rev-Erb co-occupied promoter CCAAT-boxes (Figure 2D and E). The chromatin regions downstream of Rev-Erb occupied CCAAT-boxes also displayed restrictive confirmations as they lacked histone methylation marks (H3K4me1, H3K4me3) that are characteristic of active transcription (Figure 2D), suggesting that Rev-Erb interaction with NF-Y inhibited NF-Y transcriptional activity. In support of this, we observed that Rev-Erb knockdown specifically enhanced NF-Y target gene expression (Ccnb1, Ccnb2, Cdc25c, Ccna2, and Top2a) in proliferating and not differentiating myoblasts (Figure 2F), suggesting that Rev-Erb repression of NF-Y target genes is dependent on NF-YA interaction with cognate CCAAT-boxes.
We observed that during myoblast differentiation Rev-Erb enrichment decreased at NF-YA regulatory binding sites (Figure 2G and H, Supplemental Figure S2G and H). Importantly, this release of Rev-Erb from NF-YA binding motifs was enhanced by Rev-Erb antagonist treatment (Figure 2G and H and Supplemental Figure 2SG and H). Importantly, Rev-Erb enrichment at NF-Y motifs at Myod and Srf, master regulators of myogenesis was specifically reduced by Rev-Erb antagonist treatment (Figure 2I), suggesting that Rev-Erb antagonists can accelerate de-repression of myogenic gene expression.
3.5. Rev-Erb interacts with the B subunit of the NF-Y complex
We decided to identify the components of the NF-Y complex with which Rev-Erb interacts. Immunoprecipitation of REV-ERB protein revealed that REV-ERBα and not REV-ERBβ selectively interacts with the NF-YB subunit of the NF-Y transcriptional complex (Figure 2J). In addition, enhanced Rev-Erbβ and Rev-Erbα expression differentially modulated the steady-state levels of NF-YA, B, and C protein (Figure 2J). These results suggest that Rev-Erbα interacts with the B-subunit of the NF-Y complex and that Rev-Erbβ expression may influence NF-Y protein subunit stability and expression.
3.6. Reduced Rev-Erb expression enhances muscle repair in response to acute injury in vivo
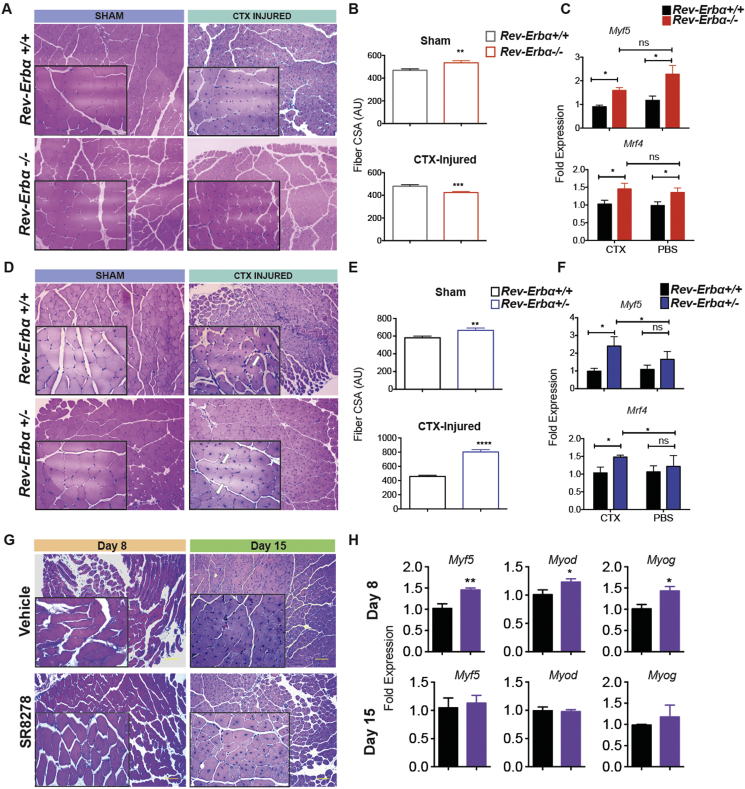
Considering the effect Rev-Erb transcriptional activity had on cultured myoblast differentiation, we went on to test the effect of reduced Rev-Erb expression on skeletal muscle regeneration in vivo. Using a cardio-toxin induced (CTX) acute muscle injury model (Supplemental Figure S2A), we compared regenerative capacity of Rev-Erbα knockout (Rev-Erbα−/−), heterozygous (Rev-Erbα+/−) and wild type mice (Rev-Erbα+/+). Briefly, 8-week old mice were injected in the left and right tibialis anterior (TA) muscle with CTX and vehicle control respectively. In this model, mice injected with CTX develop acute muscle damage that can be assessed for signs of regenerative repair histologically and via myogenic gene profiling. In uninjured muscle, Rev-Erbα−/− mice had larger muscle fiber cross-sectional area (Figure 3A and B). After injury, Rev-Erbα−/− muscle exhibited damaged, poorly differentiated myofibers and a small number of centralized nuclei, a pathological indicator of muscle regeneration, compared to Rev-Erbα+/+ mice. Rev-Erbα−/− mice muscle fiber cross-sectional areas were also significantly decreased by injury in contrast to those observed in wild-type mice (Figure 3A and B).
Figure 3.
Rev-Erb expression modulates muscle repair in response to acute injury in vivo in a dose dependent manner A) Representative H&E stained sections of tibialis anterior (TA) muscles locally injected with cardiotoxin (CTX) from mice Rev-Erbα−/− and Rev-Erbα+/+ mice. *p < 0.05 as determined by One-way ANOVA. B) Quantified cross-sectional areas of H&E stained myofibers from Rev-Erbα−/− and Rev-Erbα+/+ mice with or without CTX injury. C) Q-RTPCR showing the regulation of the myogenic genes Myf5 and Mrf4 in response in muscle from injured and uninjured Rev-Erbα−/− and Rev-Erbα+/+ mice (see Supplemental Figure S3). D) Representative H&E stained sections of tibialis anterior (TA) muscles locally injected with cardiotoxin (CTX) from mice Rev-Erbα+/− and Rev-Erbα+/+ mice. *p < 0.05 as determined by One-way ANOVA. E) Quantified cross-sectional areas of H& E stained myofibers from Rev-Erbα−/− and Rev-Erbα+/+ mice with or without CTX injury. F) Q-RTPCR showing the regulation of the myogenic genes Myf5 and Mrf4 in response in muscle from injured and uninjured Rev-Erbα−/− and Rev-Erbα+/+ mice (see Supplemental Figure S3). G) Representative H&E stained sections of tibialis anterior (TA) muscles locally injected with cardiotoxin (CTX) from mice treated with SR8278 or vehicle control. *p < 0.05 as determined by One-way ANOVA. H) Q-RTPCR showing the regulation of the myogenic genes Myf5 and Mrf4 in CTX-injured muscle in response SR8278 treatment 8 days and 15 days after treatment. Data represented as mean and ±s.e.m.
Interestingly, in Rev-Erbα knockout mice, the basal levels of MRFs (Myf5, Myod, and Myog) were equally elevated in CTX-injured and uninjured muscle. However, the expression of MRFs was not induced specifically in response to injury in Rev-Erbα−/− muscle (Rev-Erbα−/−-CTX vs. Rev-Erbα−/−-PBS groups) (Figure 3C, Supplemental Figure S3B). These results suggested that complete loss of Rev-Erb protein leads to elevation basal levels MRF gene expression. Conversely, wild type mice displayed a relatively slower rate of muscle regeneration compared to that of Rev-Erbα+/− mice. Like Rev-Erbα−/− muscle, Rev-Erbα+/− muscle had larger cross-sectional areas than wild-type mice. However, compared to wild type, heterozygous mice had enhanced regenerative capacity, as they exhibited rapidly restored myofiber structure, more centralized nuclei and significantly increased muscle fiber cross-sectional area after injury (Figure 3D and E). Rev-Erbα+/− mice also displayed a greater induction of Mrf4 and Myf5, but not Myod or Myog, gene expression compared to Rev-Erbα+/+ mice in response to injury (Figure 3F, Supplemental Figure S3B). This suggested that Rev-Erbα+/− have heightened muscle regenerative capacity that may be driven by enhanced Myf4 and Myf5 expression in response to injury. Collectively, these observations suggest that while reduced expression of Rev-Erbα accelerated muscle repair in response to injury, full loss of Rev-Erbα expression has a deleterious effect on muscle regeneration. This implies that Rev-Erbα may regulate coordination of regenerative myogenic gene programs in response to injury in vivo. Therefore, Rev-Erbα protein transcriptional regulation may be important for differentiation in direct response to injury. These factors also suggest that the regenerative deficiencies of Rev-Erbα−/− mice while linked to loss of Rev-Erb regulation is not be due to deficiencies in MRF expression.
3.7. Antagonizing Rev-Erb enhances muscle repair in response to acute injury in vivo
After observing the potent effects of Rev-Erb antagonism on muscle differentiation in vitro and the increased rate of muscle regeneration in Rev-Erbα+/− mice, we then sought to determine if Rev-Erb could be targeted pharmacologically to accelerate muscle repair. In order to ascertain this, we used the Rev-Erb antagonist SR8278 that transiently enriches in muscle and plasma when administered subcutaneously (Supplemental Figure S3C and D). Wild-type mice were injured using CTX as described above and administered SR8278 once daily and assessed for transcriptional and histological indicators of muscle regeneration at 8 and 15 days post injury. Interestingly, SR8278 treated mice showed greater numbers of centralized nuclei and exhibited more fully restored myofiber structures than the vehicle treated groups, suggesting that SR8278 treatment accelerated myofiber regeneration (Figure 3G). In accord with this, SR8278 enhanced expression of the myogenesis factors (Myf5, Myod, and Myog) exclusively in injured muscle (Figure 3H). These results indicate that antagonizing Rev-Erb further activates pro-myogenic programs in response to muscle damage. The differential effects of SR8278 in injured versus intact muscle imply that the drug's activity may be promoting myogenic programs exclusively within the proliferative environment of injured muscle. From these results, it is also apparent therefore, that Rev-Erb can be targeted to modulate the key proliferative and myogenic pathways necessary for efficient muscle regeneration.
3.8. Human muscle differentiation can be regulated by Rev-Erb ligands
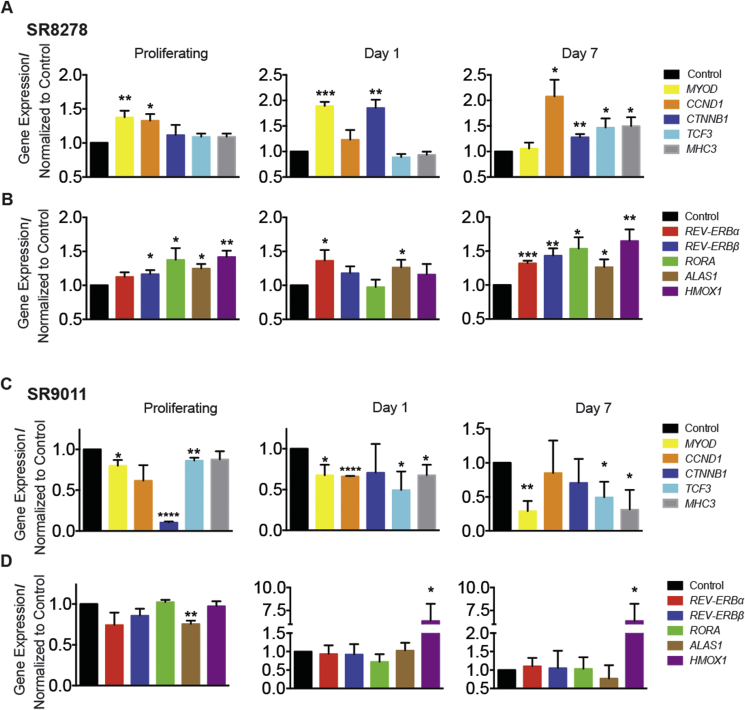
Considering the effects that Rev-Erb antagonists exhibited in mouse myoblasts and differentiated muscle, we decided to investigate if Rev-Erb antagonists had similar pro-myogenic effects on human myoblasts. To accomplish this, we cultured human myoblasts isolated from biopsies from healthy individuals in the presence of SR8278 or SR9011. As observed in mouse myoblasts, SR8278 treatment enhanced the expression of the pro-myogenic master regulator MYOD in proliferating and differentiating human myoblasts (Figure 4A). Myosin heavy chain expression (MHC3) was highly induced in proliferating and early differentiating myoblasts by SR8278 and repressed by SR9011. Interestingly, the induction of Mhc3 by SR8278 in proliferating human myoblasts was analogous to that observed in Rev-Erbα/β shRNA-expressing mouse myoblasts but occurred in proliferating myoblasts, at an earlier stage, than that observed in SR8278 treated mouse myoblasts. This suggested that Rev-Erb antagonists may have distinct potencies in human versus mouse myoblasts and also implied that the role of Rev-Erb in human myoblast differentiation, while similar to that of mice, may have species-specific mechanistic distinctions. The proliferative Wnt-pathway activators β-catenin (CTNNB1) and transcription factor 3 (TCF3) were also increased by SR8278 treatment in proliferating early differentiating and fully differentiated human myoblasts (Figure 4A). In contrast, SR9011 treatment repressed these genes in human myoblasts (Figure 4B). The NF-Y target genes (CCNB1, CCNB2, TOP2A, and TK1) were similarly induced by the Rev-Erb antagonist SR8278 and repressed by the Rev-Erb agonist SR9011 (Supplemental Figure 4). These results show that Rev-Erb may function as master regulators of myoblast differentiation in human muscle (Figure 5). This also suggests that Rev-Erb antagonists may also be able to enhance muscle repair and attenuate muscle degradation in human myopathies (Figure 4).
Figure 4.
Rev-Erb regulates developmental gene expression in human myoblast differentiation. A–D) Expression of myogenic regulator MYOD, cell cycle progression regulator CCND1, Wnt target genes; CTNNB1, TCF3, and myosin heavy chain MHC3 and expression of the Rev-Erb regulated genes REV-ERBα/β, RORA, HMOX1 in proliferating, partially differentiated and fully differentiated human myoblasts in response to (A–B) SR8278 or (C–D) SR9011 treatment (n = 4 per group). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.001 were determined by two tailed-student's t-test. Data are expressed as mean ±s.e.m.
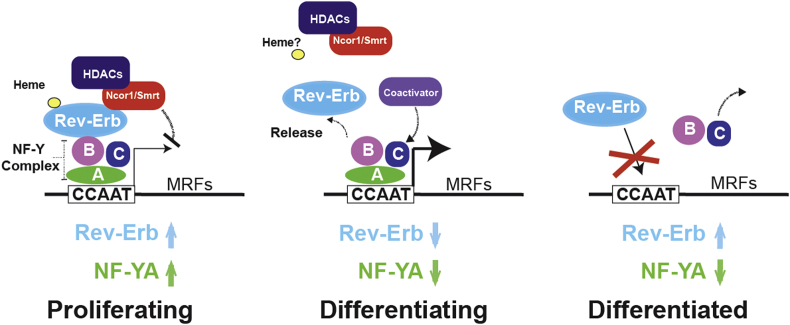
Figure 5.
Illustration of the mechanism of Rev-Erb co-regulation of myogenesis gene expression through interaction with the NF-Y complex at CCAAT motifs in proliferating, differentiating myoblasts, and differentiated myotubes.
4. Discussion
Whether Rev-Erb, a key regulator of metabolism and mitochondrial function, directs muscle regeneration in vivo is unknown. Here we demonstrate that Rev-Erb pleiotropically regulates myogenic gene expression without binding to cognate response elements (ROREs). Instead, we observed that Rev-Erb binds to regulatory regions of myogenic genes, possibly by tethering to a regulator of myoblast differentiation, the NF-Y complex, at CCAAT-boxes. Once myoblast differentiation is initiated, Rev-Erb promoter occupancy is reduced, resulting in de-repression of myogenic genes that drive muscle differentiation. We demonstrate that Rev-Erb ligands can stimulate muscle regeneration upon acute muscle injury in vivo and are able to drive human muscle cell differentiation. Therefore, Rev-Erb ligands may be useful for the clinical treatment of acute muscle injuries and chronic myopathies.
The role of Rev-Erb in skeletal muscle function is paradoxically bifurcated between inhibiting myotube differentiation and stimulating mitochondrial function in differentiated muscle [5], [8], [9], [10], [11], [20]. In adipocytes, Rev-Erb expression is increased during early adipogenesis but suppressed in mature adipocytes, allowing for efficient expression of peroxisome proliferator-activated receptor-γ regulated genes [21], [22]. As observed in adipogenesis, the role of Rev-Erb may be quite distinct in proliferating versus differentiated cells [21], [22]. However, unlike Rev-Erb expression during adipogenesis, in differentiating myoblasts, it declines just prior to differentiation and is restored in differentiated muscle. As Rev-Erb functions exclusively as a transcriptional repressor, it was unclear by what mechanisms it was able to modulate muscle mitochondrial function and differentiation in opposing ways. These studies as well as our recent development of synthetic agonists and antagonists targeting Rev-Erb [23], [24] allowed us to attempt to identify the mechanism of Rev-Erb activity during myogenesis and muscle regeneration.
Our investigation showed that Rev-Erb transcriptional regulation in skeletal muscle may be dependent on the CCAAT binding properties of transcription factors such as of NF-YA and SP1/2, which predominantly facilitates Rev-Erb regulation of myogenesis in myoblasts. Specifically, in differentiated muscle, NF-YA expression and, consequently, the DNA binding activity of the NF-Y complex is lost [14]. Our results suggest that an association between Rev-Erb transcriptional repression and NF-YA inhibition of genes that drive differentiation may exist. However, although we show Rev-Erb is enriched at NF-Y motifs, it is unclear whether Rev-Erb regulation of differentiation is specifically dependent on NF-Y expression and, therefore, more in depth analyses of the mechanisms that dictate Rev-Erb transcriptional activity in muscle are warranted. In line with this, we observed that disrupting Rev-Erb expression and transcriptional activity upregulates myogenic factor expression. Interestingly, while disruption of Rev-Erbα expression alone induced Myod mRNA expression, dual Rev-Erbα/β knockdown only induced Myod protein expression. Similarly, pharmacological disruption of Rev-Erb failed to induce Myod mRNA expression, suggesting that Rev-Erb, despite binding to the Myod enhancer, may not directly regulate Myod expression. We have therefore identified a possible mechanism through which Rev-Erb may differentially regulate gene expression pre and post muscle differentiation.
Here we showed that mice heterozygous for Rev-Erb expression displayed enhanced regenerative capacity, whereas Rev-Erb knockout mice showed deficiencies in muscle repair. Rev-Erb knockout mice were not deficient in MRF expression, suggesting that the phenotypic differences between Rev-Erb heterozygous and knockout mice may be independent of the regulation of MRF expression. Therefore, the underlying, downstream factors that influence the regenerative capacity in Rev-Erb knockout mice are unclear and needs further investigation.
We also show that Rev-Erb is a regulator of muscle differentiation that can be targeted to stimulate muscle regeneration and hence may have utility in treating numerous myopathies including muscular dystrophy, sarcopenia, and cachexia, in addition to acute injury. Rev-Erb is a transcriptional regulator that not only influences circadian behavior but also couples metabolic activity and immune cell function in numerous tissues to diurnal rhythms. The multifaceted roles of Rev-Erb must be factored into any endeavor aimed at design clinically viable treatments for degenerative muscle diseases. For acute injuries potent Rev-Erb drugs with local activity but deficient at accumulating in circulation would be of great utility. One challenge of targeting Rev-Erb activity in genetic/systemic myopathies would be designing drugs that selectively enrich in muscle tissue while avoiding systemic exposure and unwanted side effects in off-target tissues. In particular, clinically viable Rev-Erb antagonists should be intuitively designed to avoid traversing the blood–brain barrier and disrupting important circadian pathways that influence sleep/wake behavior. It is also plausible that Rev-Erb agonists and antagonists could be co-utilized as chronotherapies that restore normal cyclic Rev-Erb-directed metabolic activity once muscle regeneration is complete or lean mass degradation is sufficiently stymied. Toward this end, future studies should focus on understanding the influence of cell specific factors, DNA modifiers, and cofactors on Rev-Erb tissue specific regulation in other metabolic tissues and immune cells.
Author contributions
R.D.W. performed most of the experiments, analyzed data and aided in the design and writing of the manuscript, C.G., M.S., K.J.C., S.D.A, M.J.M, and N.A.S. performed key experiments and analyzed data. J.Z. and S.R.S. analyzed data and aided in the design, writing and editing of the manuscript. C.A.F conceived the study, designed and performed experiments. C.A.F and T.P.B. analyzed data interpreted results and wrote the manuscript.
Acknowledgements
We would like to thank Dr. Grant Kolar in the department of Pathology and Saint Louis University for providing assistance with the histological and immuno-histochemical assays conducted herein. This work was supported by a grant from the National Institutes of Health (MH093429, T.P.B.) (R01HL093195, J.Z.).
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molmet.2017.05.001.
Conflict of interest
There are no conflicts of interest to declare.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Braun T., Gautel M. Transcriptional mechanisms regulating skeletal muscle differentiation, growth and homeostasis. Nature Reviews Molecular Cell Biology. 2011;12(6):349–361. doi: 10.1038/nrm3118. published Online First: Epub Date. [DOI] [PubMed] [Google Scholar]
- 2.Braun T., Buschhausen-Denker G., Bober E., Tannich E., Arnold H.H. A novel human muscle factor related to but distinct from MyoD1 induces myogenic conversion in 10T1/2 fibroblasts. EMBO Journal. 1989;8(3):701–709. doi: 10.1002/j.1460-2075.1989.tb03429.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burris T.P., Nawaz Z., Tsai M.J., O'Malley B.W. A nuclear hormone receptor-associated protein that inhibits transactivation by the thyroid hormone and retinoic acid receptors. Proceedings of the National Academy of Sciences of the United States of America. 1995;92(21):9525–9529. doi: 10.1073/pnas.92.21.9525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kojetin D.J., Burris T.P. REV-ERB and ROR nuclear receptors as drug targets. Nature Reviews Drug Discovery. 2014;13(3):197–216. doi: 10.1038/nrd4100. published Online First: Epub Date. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Downes M., Carozzi A.J., Muscat G.E. Constitutive expression of the orphan receptor, Rev-erbA alpha, inhibits muscle differentiation and abrogates the expression of the myoD gene family. Molecular Endocrinology. 1995;9(12):1666–1678. doi: 10.1210/mend.9.12.8614403. published Online First: Epub Date. [DOI] [PubMed] [Google Scholar]
- 6.Raghuram S., Stayrook K.R., Huang P., Rogers P.M., Nosie A.K., McClure D.B. Identification of heme as the ligand for the orphan nuclear receptors REV-ERBalpha and REV-ERBbeta. Nature Structural & Molecular Biology. 2007;14(12):1207–1213. doi: 10.1038/nsmb1344. published Online First: Epub Date. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yin L., Wu N., Curtin J.C., Qatanani M., Szwergold N.R., Reid R.A. Rev-erbalpha, a heme sensor that coordinates metabolic and circadian pathways. Science. 2007;318(5857):1786–1789. doi: 10.1126/science.1150179. published Online First: Epub Date. [DOI] [PubMed] [Google Scholar]
- 8.Burke L., Downes M., Carozzi A., Giguere V., Muscat G.E. Transcriptional repression by the orphan steroid receptor RVR/Rev-erb beta is dependent on the signature motif and helix 5 in the E region: functional evidence for a biological role of RVR in myogenesis. Nucleic Acids Research. 1996;24(18):3481–3489. doi: 10.1093/nar/24.18.3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pircher P., Chomez P., Yu F., Vennstrom B., Larsson L. Aberrant expression of myosin isoforms in skeletal muscles from mice lacking the rev-erbAalpha orphan receptor gene. American Journal of Physiology. Regulatory, Integrative and Comparative Physiology. 2005;288(2):R482–R490. doi: 10.1152/ajpregu.00690.2003. published Online First: Epub Date. [DOI] [PubMed] [Google Scholar]
- 10.Woldt E., Sebti Y., Solt L.A., Duhem C., Lancel S., Eeckhoute J. Rev-erb-alpha modulates skeletal muscle oxidative capacity by regulating mitochondrial biogenesis and autophagy. Nature Medicine. 2013;19(8):1039–1046. doi: 10.1038/nm.3213. published Online First: Epub Date. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chatterjee S., Nam D., Guo B., Kim J.M., Winnier G.E., Lee J. Brain and muscle Arnt-like 1 is a key regulator of myogenesis. Journal of Cell Science. 2013;126(Pt 10):2213–2224. doi: 10.1242/jcs.120519. published Online First: Epub Date. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raichur S., Fitzsimmons R.L., Myers S.A., Pearen M.A., Lau P., Eriksson N. Identification and validation of the pathways and functions regulated by the orphan nuclear receptor, ROR alpha1, in skeletal muscle. Nucleic Acids Research. 2010;38(13):4296–4312. doi: 10.1093/nar/gkq180. published Online First: Epub Date. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Basile V., Baruffaldi F., Dolfini D., Belluti S., Benatti P., Ricci L. NF-YA splice variants have different roles on muscle differentiation. Biochimica et Biophysica Acta. 2016;1859(4):627–638. doi: 10.1016/j.bbagrm.2016.02.011. published Online First: Epub Date. [DOI] [PubMed] [Google Scholar]
- 14.Gurtner A., Manni I., Fuschi P., Mantovani R., Guadagni F., Sacchi A. Requirement for down-regulation of the CCAAT-binding activity of the NF-Y transcription factor during skeletal muscle differentiation. Molecular Biology of the Cell. 2003;14(7):2706–2715. doi: 10.1091/mbc.E02-09-0600. published Online First: Epub Date. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farina A., Manni I., Fontemaggi G., Tiainen M., Cenciarelli C., Bellorini M. Down-regulation of cyclin B1 gene transcription in terminally differentiated skeletal muscle cells is associated with loss of functional CCAAT-binding NF-Y complex. Oncogene. 1999;18(18):2818–2827. doi: 10.1038/sj.onc.1202472. published Online First: Epub Date. [DOI] [PubMed] [Google Scholar]
- 16.Bergstrom J. Percutaneous needle biopsy of skeletal muscle in physiological and clinical research. Scandinavian Journal of Clinical Laboratory Investigation. 1975;35(7):609–616. [PubMed] [Google Scholar]
- 17.Mandillo S., Tucci V., Holter S.M., Meziane H., Banchaabouchi M.A., Kallnik M. Reliability, robustness, and reproducibility in mouse behavioral phenotyping: a cross-laboratory study. Physiological Genomics. 2008;34(3):243–255. doi: 10.1152/physiolgenomics.90207.2008. published Online First: Epub Date. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Asp P., Blum R., Vethantham V., Parisi F., Micsinai M., Cheng J. Genome-wide remodeling of the epigenetic landscape during myogenic differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(22):E149–E158. doi: 10.1073/pnas.1102223108. published Online First: Epub Date. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y., Fang B., Emmett M.J., Damle M., Sun Z., Feng D. GENE REGULATION. Discrete functions of nuclear receptor Rev-erbalpha couple metabolism to the clock. Science. 2015;348(6242):1488–1492. doi: 10.1126/science.aab3021. published Online First: Epub Date. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chatterjee S., Yin H., Nam D., Li Y., Ma K. Brain and muscle Arnt-like 1 promotes skeletal muscle regeneration through satellite cell expansion. Experimental Cell Research. 2015;331(1):200–210. doi: 10.1016/j.yexcr.2014.08.041. published Online First: Epub Date. [DOI] [PubMed] [Google Scholar]
- 21.Wang J., Lazar M.A. Bifunctional role of Rev-erbalpha in adipocyte differentiation. Molecular and Cellular Biology. 2008;28(7):2213–2220. doi: 10.1128/MCB.01608-07. published Online First: Epub Date. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar N., Solt L.A., Wang Y., Rogers P.M., Bhattacharyya G., Kamenecka T.M. Regulation of adipogenesis by natural and synthetic REV-ERB ligands. Endocrinology. 2010;151(7):3015–3025. doi: 10.1210/en.2009-0800. published Online First: Epub Date. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kojetin D., Wang Y., Kamenecka T.M., Burris T.P. Identification of SR8278, a synthetic antagonist of the nuclear heme receptor REV-ERB. ACS Chemical Biology. 2011;6(2):131–134. doi: 10.1021/cb1002575. published Online First: Epub Date. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Solt L.A., Wang Y., Banerjee S., Hughes T., Kojetin D.J., Lundasen T. Regulation of circadian behaviour and metabolism by synthetic REV-ERB agonists. Nature. 2012;485(7396):62–68. doi: 10.1038/nature11030. published Online First: Epub Date. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.