Abstract
We present an overview of contact-assisted predictions in the eleventh round of Critical Assessment of Protein Structure Prediction (CASP11), which included four categories: predicted contacts (Tp), correct contacts (Tc), simulated sparse NMR contacts (Ts), and cross-linking contacts (Tx). Comparison of assisted to unassisted model quality highlighted a relatively poor overall performance in CASP11 using predicted Tp and crosslinked Tx contact information. However, average model quality significantly improved in the correct Tc and simulated NMR Ts categories for most targets, where maximum improvement of unassisted models reached an impressive 70 GDT_TS. Comparison of the performance in the correct Tc category to CASP10 suggested the improvement in CASP11 model quality originated from an increased number of provided contacts per target. Group rankings based on a combination of scores used in the CASP11 free modeling (FM) assessment for each category highlight four top-performing groups, with three from the Lee lab and one from the Baker lab. We used the overall performance of these groups in each category to develop hypotheses for their relative outperformance in the correct Tc and simulated NMR Ts categories, which stemmed from the fraction of correct contacts provided (correct Tc category) and a reduced fraction of correct contacts offset by an increased coverage of the correct contacts (simulated NMR Ts category).
Keywords: protein structure prediction, CASP11, contact-assisted
INTRODUCTION
The CASP11 contact-assisted structure modeling categories intend to learn how knowledge of long-range contacts improved the quality of tertiary structure prediction models provided by so-called hybrid prediction methods1–3. For a selection of more challenging tertiary structure prediction targets (T0), contact-assisted data were distributed to the CASP community subsequent to the release of the target structure and collection of the initial predictions, but prior to the public release of the experimental coordinates. Four types of contact-assisted data (abbreviated T*) were provided: predicted three-dimensional contacts gathered from the contact prediction category of CASP11 (Tp, subscript ‘p’ for predicted), selected subsets of correct contacts from the contact prediction category (Tc, ‘c’ for correct), simulated sparse NMR contacts (Ts, ‘s’ for simulated), and contacts obtained from cross-linking mass spectroscopy studies (Tx, ‘x’ for crosslinked). These categories expanded on the promising results observed in the CASP10 contact-assisted assessment3, which evaluated only correct contacts (Tc).
An overview of the experimental setup for the CASP 11 contact assisted categories is illustrated in Figure 1. The Prediction Center chose sets of pairwise contacts for the predicted Tp and correct Tc contact-assisted categories from long-range contacts collected in CASP's Residue-Residue Contact Prediction (RR) category. The lists of submitted contacts in the RR category (both true and false positive) were filtered to retain only long-range contacts (separation along the sequence >23 residues), sorted according to the submitted probability, and truncated to the first L/5 contacts if necessary (L- target length in residues). For each predicted Tp target, the processed lists were released for ten CASP11 RR groups that were among the best performers in the previous CASP4. For the correct Tc category contacts, the lists of predicted contacts in the RR category were pre-filtered for correctness by measuring the contact distances in the native structure. Correct contacts were defined as distance between Cβ from each residue of the given pair being less than 8 Å. The correct Tc pairs were then subjected to the procedure used in the predicted Tp category, usually limiting to L/5 contacts, with the number being sometimes smaller (if not enough long-range contacts existed) or larger (to include all contacts with the same probability as that of the bottom, L/5-th contact).
Figure 1. Contact Assisted Category Dataset Schematic.

The leftmost outer box represents all CASP11 T0 targets, with a relatively smaller subset of the difficult targets selected for various contact-assisted categories (circles). The data for predicted Tp contact targets (blue) were selected from the predicted contacts provided for the CASP11 RR category and were filtered for close contacts for data provided in the correct Tc category (red). The data for the simulated NMR Ts targets (green) were provided by the Montelione lab, and the data for the crosslinked Tx targets (orange) were generated experimentally by the Rappsilber lab.
The simulated NMR Ts and crosslinked Tx contact data were generated by the Montelione and Rappsilber labs, respectively. CASP organizers provided coordinates of crystal structures of the selected simulated NMR Ts targets to the Montelione group (Rutgers). These coordinates were used to mimic the data available in the initial stage of an NMR study. First, NOESY cross peaks were assigned to targets using a simulation procedure [G. Montelione, this issue], and then ambiguous distance restraints from these peaks were generated using the Automated Structure Determination Platform ASDP5. CASP organizers arranged for shipment of biological material from CASP target providers to the Rappsilber lab (Technical University of Berlin). The target proteins were cross-linked, and distance restraints were obtained using mass spectrometry [J.Rappsilber, this issue].
A total of 27 targets were selected by the Prediction Center for contact-assisted predictions in CASP11 (Table 1). The targets were divided into the following categories: 24 in the predicted Tp set, 19 in the simulated NMR Ts set, 24 in the correct Tc set, and 4 in the crosslinked Tx set. The targets were designated according to the category abbreviation (Tp, Ts, Tc, or Tx) followed by the 3-digit T0 target number (i.e. 761 from T0761-D0). One target (Tp826) was omitted from evaluation because the simulated NMR Ts contacts were released prior to the predicted Tp contacts.
Table 1.
Summary of contact-assisted targets
| Target ID | Range | Dom | Class | Category | ECOD Architecture |
|---|---|---|---|---|---|
| T0761-D0 | 49–285 | 2 | FM | Tp, Ts, Tc | α+β two layers; duplication |
| T0763-D1 | 31–160 | 1 | FM | Tp, Ts, Tc | α+β two layers |
| T0767-D0 | 39–312 | 2 | TBM;FM | Tp, Ts, Tc, Tx | α+β two layers; a+b two layers |
| T0771-D0 | 26–203 | 1 | FM | Tp | α+β two layers |
| T0777-D1 | 18–362 | 1 | FM | Tp, Ts, Tc | α complex topology |
| T0781-D0 | 34–415 | 2 | FM;TBM-H | Tx | α+β two layers; duplication |
| T0785-D1 | 3–114 | 1 | FM | Tp, Ts, Tc | β-sandwiches |
| T0794-D0 | 1–462 | 2 | TBM;FM | Tp, Ts, Tc | α+β four layers; beta sandwiches |
| T0800-D1 | 36–247 | 1 | TBM-H | Tp, Ts, Tc | β duplicates or obligate multimers |
| T0802-D0 | 5–122 | 1 | FM | Tp, Ts, Tc | β-sandwiches |
| T0804-D0 | 9–202 | 2 | FM | Tp, Ts, Tc | β duplicates or obligate multimers; beta sandwiches |
| T0806-D1 | 1–256 | 1 | FM | Tp, Ts, Tc | α/β three-layered sandwiches |
| T0808-D0 | 19–418 | 2 | TBM;FM | Tx | β sandwiches; duplication |
| T0810-D1 | 24–136 | 1 | FM | Tp, Ts, Tc | α superhelices |
| T0812-D1 | 5–187 | 1 | TBM-H | Ts, Tc, Tx | β-sandwiches |
| T0814-D0 | 23–419 | 3 | FM;TBM-H | Tp, Ts, Tc | β-sandwiches; triplication |
| T0818-D1 | 30–163 | 1 | TBM | Tp, Ts, Tc | α+β two layers |
| T0824-D1 | 2–109 | 1 | FM | Tp, Ts, Tc | few SS elements |
| T0826-D1 | 11–211 | 1 | FM | Tp, Ts, Tc | α bundles |
| T0827-D2 | 212–369 | 1 | FM | Tp, Ts, Tc | α complex topology |
| T0831-D2 | 109–352 | 1 | FM | Tp, Tc | α bundles |
| T0832-D1 | 10–218 | 1 | FM | Tp, Ts, Tc | α+β complex topology |
| T0834-D0 | 2–215 | 2 | FM | Tp, Tc | α+β three layers; alpha bundles |
| T0835-D1 | 21–424 | 1 | TBM | Tp, Ts, Tc | α superhelices |
| T0836-D1 | 1–204 | 1 | FM | Tp, Tc | α bundles |
| T0848-D2 | 172–354 | 1 | TBM-H | Tp, Tc | α+β two layers |
| T0853-D12 | 5–152 | 2 | TBM | Tp, Tc | α+β two layers; duplication |
The CASP11 contact-assisted targets included 17 that were evaluated in the tertiary structure prediction category as single domains, with 12 categorized as FM, two categorized as TBM, and three categorized as TBM-Hard6. The remaining ten targets are multidomain, with four exhibiting duplications of the same domain and one exhibiting a triplication. The multidomain targets were categorized as all TBM (1 target), all FM (3 targets), a combination of TBM and FM (4 targets), and a combination of TBM-Hard and FM (2 targets).
A number of groups participated in the contact-assisted categories in CASP11, including 6 servers and 23 human groups (Table 2). Only 10 groups contributed models for nearly all targets in all of the contact-assisted categories. Five additional groups contributed models for nearly all targets in three of the four categories while one group contributed in two of the four categories. Three groups concentrated on the crosslinked Tx category with the smallest number of targets.
Table 2.
Summary of group participation in contact-assisted categories
| Group Num | Group Name | Type | T0 (27) |
Tp (23) |
Ts (19) |
Tc (24) |
Tx (4) |
|---|---|---|---|---|---|---|---|
| 32 | Legato | human | 27 | 21 | 19 | 21 | 4 |
| 38 | nns | server | 27 | 23 | 19 | 24 | 4 |
| 40 | GoScience | human | 1 | 1 | 8 | 9 | 0 |
| 41 | MULTICOM-NOVEL | server | 27 | 23 | 19 | 24 | 4 |
| 42 | TASSER | human | 27 | 0 | 0 | 0 | 2 |
| 44 | LEER | human | 27 | 3 | 19 | 24 | 0 |
| 64 | BAKER | human | 27 | 10 | 19 | 23 | 4 |
| 65 | Jones-UCL | human | 26 | 21 | 19 | 23 | 4 |
| 80 | MeilerLab | human | 26 | 23 | 19 | 24 | 3 |
| 155 | Cornell-Gdansk | human | 27 | 0 | 0 | 0 | 1 |
| 157 | FLOUDAS_A1 | human | 27 | 0 | 0 | 0 | 4 |
| 162 | McGuffin | human | 27 | 23 | 19 | 24 | 4 |
| 169 | LEE | human | 27 | 23 | 19 | 24 | 4 |
| 186 | Void_Crushers | human | 1 | 1 | 8 | 9 | 0 |
| 219 | Sternberg | human | 0 | 17 | 19 | 0 | 3 |
| 276 | FLOUDAS_A4 | human | 27 | 1 | 18 | 22 | 4 |
| 287 | RBO-human | human | 0 | 0 | 11 | 0 | 4 |
| 300 | PhyreX | server | 27 | 4 | 0 | 0 | 0 |
| 310 | MUFOLD-R | human | 25 | 0 | 15 | 0 | 0 |
| 329 | NMR-I-TASSER | human | 0 | 0 | 4 | 0 | 0 |
| 342 | Anthropic_Dreams | human | 1 | 0 | 8 | 9 | 4 |
| 345 | FUSION | server | 27 | 22 | 19 | 24 | 4 |
| 357 | STAP | human | 27 | 21 | 19 | 21 | 4 |
| 361 | Contenders | human | 1 | 1 | 6 | 7 | 1 |
| 420 | MULTICOM-CLUSTER | server | 27 | 23 | 19 | 24 | 0 |
| 428 | Laufer | human | 1 | 0 | 10 | 8 | 0 |
| 476 | Foldit | human | 1 | 1 | 8 | 9 | 0 |
| 479 | RBO_Aleph | server | 27 | 21 | 9 | 21 | 4 |
| 490 | Wiskers | human | 1 | 0 | 2 | 2 | 0 |
MATERIALS AND METHODS
Improvements over unassisted T0 models
We evaluated the community-wide improvement in performance quality by comparing the contact-assisted models (all T*: Tp, Ts, Tc, and Tx) to the unassisted (T0) models using the GDT_TS score7 that has been used in CASP assessments for over a decade8–13. We considered the differences in both individual performance and absolute performance on a target-wide basis similar to the evaluation of the CASP10 contact-assisted category3. For comparing overall performance improvements on each of the assisted targets, the best unassisted T0 GDT_TS from the group (individual performance) or the best overall unassisted GDT_TS among all groups (absolute performance) was subtracted from the group’s T*model GDT_TS. To include individual performance scores for those groups that did not provide T0 models, the average T0 GDT_TS for all groups participating in the contact-assisted category for that target substituted for the missing T0s. To be consistent with the previous CASP10 assisted evaluation, we estimated the significance of community-wide performance improvement for each target using one-tailed t-tests that compared all assisted T* model GDT_TS scores to all T0 model GDT_TS scores (not only best T0’s). We used one-tailed paired t-tests to evaluate the significance of each group’s performance improvements (absolute and individual) over their unassisted T0 targets. The t-tests compared all of the group’s assisted T* model GDT_TS scores to either the group’s best T0 model scores (substituting missing T0 scores with the average GDT_TS for the corresponding target) or the overall maximum T0 model GDT_TS scores among all participating groups, respectively.
Group performance using combined scores, win/loss counting, and head-to-head trials
We calculated Z-score sums (and averages) over all the targets in each category for several different scores. Z-scores were calculated as in previous CASPs10,11 using first and best GDT_TS scores, as well as the combined score used to evaluate CASP 11 tertiary structure predictions (see Kinch et al., Evaluation of CASP11 free modeling targets and CASP ROLL in this issue). Briefly, we calculated Z-scores over each target for first and best GDT_TS, FM-style combined score (GDT_TS, TenS, QCS, ContS, lDDT, and Molprb), and TBM-style combined score (GDT_HA, GDC_ALL, lDDT, SG, and 0.2 × Molprb); and summed (or averaged) the Z-scores for all targets in each contact-assisted category.
The statistical significance of whether each group’s performance differed from that of the other groups was inferred from one-tailed paired t-tests and bootstrap tests10,14,15 on GDT_TS, FM-style and TBM-style scoring schemes. We also carried out a pairwise comparison (head-to-head trials) of the group results, as well as the CASP10-style overall win/loss counts for all-against-all pairwise comparisons3. In head-to-head trails, for each pair of groups, we calculated the fraction of common targets/domains for which one group outperformed the other according to the selected score. In win/loss counts, we performed all-against-all pairwise prediction model comparisons on the selected scores for each target and summed the numbers of win/loss cases for each group. The groups were ranked primarily by the probability that a win/loss record was equal to or better than the observed record that could have been obtained by chance, and secondarily by the fraction of winning comparisons. In GDT_TS comparisons for both head-to-head trials and win/lose count, we extended our comparison to consider models within both 1 and 2 GDT_TS score units as ties to address models with insignificant differences. Due to the registration of multiple groups by a single participant, we studied whether registering multiple groups (as opposed to having a single group) would provide an advantage or disadvantage to the participant’s Z-score and ranking. To address this question, we compared original Z-scores, t-test probabilities, and ranks to those calculated using only one of the multiple groups from the same participant.
Calculating correct contact percentage and correct contact coverage for contact assisted targets
The correct Tc and predicted Tp categories included some duplicated residue pairs that stemmed from overlapping predicted contacts provided by multiple prediction groups. Simulated NMR Ts target contacts included hydrogen atom pairs (as opposed to residue pairs), with some having multiple peak assignments as well as multiple atom counts for some residue pairs. Additionally, contacts in the simulated NMR Ts category and for the cross-linking target Tx781 included residue pairs limited to the same residue (noted as self-contacts). We filtered out duplications and self-contacts, using the numbers for unique and non-self contact pairs. The correct contact percentage (CCP) was calculated as the number of correct residue pairs divided by the number of total residue pairs (times 100 to convert to percentage), with correct contact pairs defined as having Cβ atoms in the target structure no more than 8 Å apart. We also computed the correct contact coverage (CCC) as the correct residue pair count divided by the target length.
Production of dummy structure models using simulated NMR Ts contact restraints
The simulated NMR Ts contacts represent hydrogen pairs from simulated NMR peak assignments, with an indicated distance upper limit (UPL) and its corresponding peak. Due to the ambiguity of the NMR assignments, peaks could be assigned to multiple hydrogen pairs. We produced dummy structure models with the CNS package using different distance restraint sets from the simulated NMR Ts contacts: 1) all contacts, 2) unambiguous contacts, and 3) true contacts. Unambiguous contacts were generated by taking those peaks with only one contact pair. As the UPLs for hydrogen pairs vary, we defined ‘Ts-specific’ contacts as those with distances lower than the corresponding UPLs. Note that the ‘Ts-specific’ contact threshold differs from the contact threshold used in comparison across categories (Cβ atoms within 8 Å).
The simulated annealing protocol of the CNS package16 was used to calculate structures based on provided distance restraints. As these restraints were limited to hydrogen atoms, we assigned the lower limit for distance constraints as 1.5 Å and the upper limit as the UPL given in the contact information. Simulations were performed from both an extended chain (‘anneal.inp’ template option) and an embedded substructure starting model generated for HN, N, CO, Cα, Cβ, and Cγ atoms by distance geometry calculations based on the Nuclear Overhauser Effect (NOE) restraints (‘dg_sa.inp’ template option) and ‘sum’ mode for NOE averaging. Simulations were complete after generating 10 accepted structures or reaching a 48-hour time limit. All simulations using unambiguous contacts, and 7 out of 19 simulations using correct contacts produced 10 accepted structures before reaching the time limit. The simulations generated from 634 to 10815 NMR structure solutions for each target, due to variations in protein length and provided contact numbers. We reported the best GDT_TS score among all the trial structures for each simulated NMR Ts target.
RESULTS AND DISCUSSION
Target-based performance improvements
We used performance improvement measures developed in the previous CASP evaluation3 to assess the CASP community’s ability to use contact information to improve tertiary structure predictions (T0). The first measure, individual performance improvement, represents the difference between the contact assisted (T*) scores and the score of the best unassisted T0 prediction from the same group. If the corresponding unassisted prediction T0 was missing, we used the average GDT_TS score from all unassisted predictions submitted on the target in place of the reference score. The second measure, absolute performance improvement, compares scores of assisted T* models and a gold standard unassisted T0 model (the best among all participating predictors in the specific contact-assisted category). However, the absolute performance unrealistically assumes that each group started with the same best unassisted T0 model. Despite the drawbacks of these measures, the difference distributions for best GDT_TS models on each assisted target (Figure 2) provide insight into the performance improvements of the CASP community as a whole using various types of contact information. Predicted Tp and crosslinked Tx targets exhibited a relatively poor overall performance, with broadly negative absolute improvement values and relatively lower individual improvement values than those calculated for correct Tc and simulated NMR Ts targets, which tended to display positive improvements on most targets.
Figure 2. Absolute and Individual Performance for Assisted Targets.
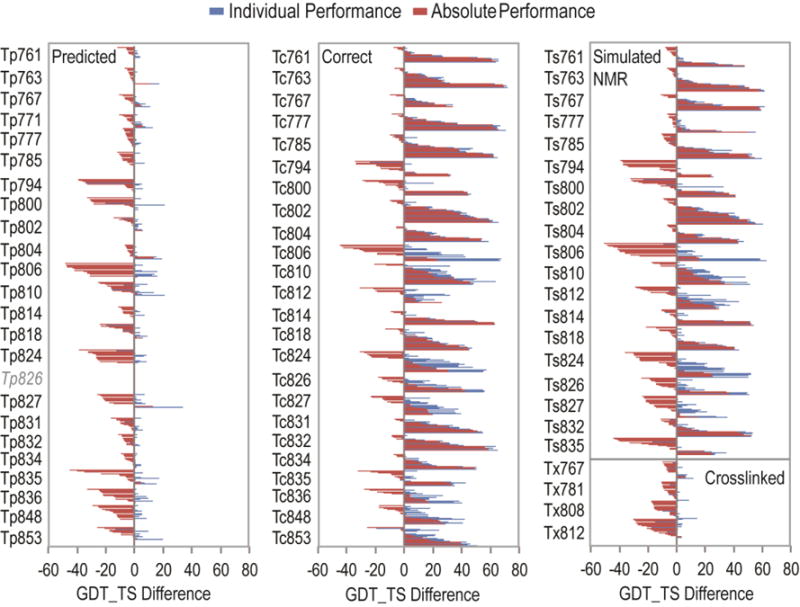
Each group has one bar that corresponds to absolute performance (red), measured by subtracting the gold standard T0 model’s GDT_TS from the best T* model’s GDT_TS for the group; and one bar that corresponds to individual performance (blue), measured by subtracting the best T0 model GDT_TS for each group from their best T* model GDT_TS. The value of the GDT_TS performance difference is indicated below the bar graph, with a grey line drawn through 0. Overall performance on predicted Tp targets (left panel) and crosslinked Tx targets (lower right panel) was worse than on correct Tc targets (middle panel) and simulated NMR Ts targets (upper right panel).
For the predicted Tp and crosslinked Tx categories, the absolute performance is overwhelmingly negative (Figure 2, most red bars in the left panel representing predicted Tp scores and lower part of the right panel representing crosslinked Tx scores are below 0). The average absolute performance difference of best predicted Tp models over all targets was negative (−10.86 GDT_TS), with only 8% of the best models showing positive absolute performance improvement. The individual predicted Tp performance on average differed by −0.19 GDT_TS, and approximately half (51%) of the best predicted Tp models exhibited positive individual performance improvement (Figure 2, blue bars above 0). Similarly, the best crosslinked Tx models had negative averages of −10. 2 GDT_TS (absolute) and −1.9 GDT_TS (individual), beating their unassisted models in 9% (absolute) and 36% (individual) of the cases. The discrepancy between some of the absolute and individual performance improvements suggested that positive individual performance scores might simply reflect poor initial models. Despite this potential caveat, community-wide T-tests as performed in the previous CASP contact-assisted evaluation3 (Table 3) showed marginal, yet significant improvements for 4 of the 23 predicted Tp targets: Tp767-D0, Tp804-D0, Tp806-D1, and Tp834-D0. At the same time, the predictions showed significant deteriorations with respect to their unassisted models on 7 of the 23 predicted Tp targets. Only three predicted Tp targets (Tp763, Tp804 and Tp827) included promising absolute group performance (GDT_TS improvement > 10). Three of four crosslinked Tx targets showed average deterioration in model quality using assisted information, with one (Tx808-D0) being significantly worse.
Table 3.
Significance of target improvement using assisted information
| Target ID | T0 Num | T* Num | MeanT0 GDT_TS | MeanT* GDT_TS | Mean Diff | P-value |
|---|---|---|---|---|---|---|
| A Predicted Contacts Tp | ||||||
| Tp761-D0 | 79 | 53 | 13.69 | 14.08 | 0.39 | 1.77E-01 |
| Tp763-D1 | 99 | 75 | 18.02 | 18.19 | 0.17 | 3.41E-01 |
| Tp767-D0 | 71 | 53 | 12.18 | 13.89 | 1.71 | 4.73E-03 |
| Tp771-D0 | 56 | 59 | 14.87 | 15.58 | 0.71 | 1.33E-01 |
| Tp777-D1 | 71 | 65 | 11.09 | 11.51 | 0.42 | 1.39E-01 |
| Tp785-D1 | 71 | 59 | 21.09 | 19.56 | −1.53 | 5.93E-03 |
| Tp794-D0 | 77 | 65 | 33.71 | 25.51 | −8.2 | 1.11E-03 |
| Tp800-D1 | 60 | 55 | 33.4 | 22.79 | −10.61 | 1.81E-06 |
| Tp802-D0 | 71 | 60 | 20.3 | 21.55 | 1.25 | 5.41E-02 |
| Tp804-D0 | 66 | 65 | 12.35 | 13.61 | 1.26 | 4.83E-02 |
| Tp806-D1 | 65 | 62 | 16.56 | 20.89 | 4.33 | 2.19E-02 |
| Tp810-D1 | 66 | 60 | 23.3 | 23.5 | 0.2 | 4.50E-01 |
| Tp814-D0 | 80 | 58 | 10.25 | 8.94 | −1.31 | 2.13E-02 |
| Tp818-D1 | 67 | 60 | 32.99 | 28.62 | −4.37 | 1.12E-03 |
| Tp824-D1 | 74 | 58 | 26.5 | 24.7 | −1.8 | 3.75E-02 |
| Tp827-D2 | 68 | 60 | 21.26 | 23.5 | 2.24 | 6.61E-02 |
| Tp831-D2 | 65 | 65 | 16.88 | 16.29 | −0.59 | 2.14E-01 |
| Tp832-D1 | 66 | 54 | 16.28 | 15.32 | −0.96 | 2.52E-02 |
| Tp834-D0 | 67 | 64 | 13.26 | 14.33 | 1.07 | 2.46E-02 |
| Tp835-D1 | 67 | 61 | 37.33 | 34.26 | −3.07 | 1.07E-01 |
| Tp836-D1 | 65 | 62 | 22.58 | 23.53 | 0.95 | 2.55E-01 |
| Tp848-D2 | 66 | 61 | 19.23 | 20.96 | 1.73 | 7.22E-02 |
| Tp853-D0 | 66 | 55 | 24.27 | 23.42 | −0.85 | 2.50E-01 |
| B Correct Contacts Tc | ||||||
| Tc761-D0 | 79 | 70 | 13.69 | 39.34 | 25.65 | 1.26E-13 |
| Tc763-D1 | 99 | 86 | 18.02 | 40.57 | 22.55 | 7.04E-14 |
| Tc767-D0 | 71 | 59 | 12.18 | 26.17 | 13.99 | 7.98E-16 |
| Tc777-D1 | 71 | 70 | 11.09 | 38.68 | 27.59 | 2.48E-14 |
| Tc785-D1 | 71 | 101 | 21.09 | 46.68 | 25.59 | 5.70E-14 |
| Tc794-D0 | 77 | 68 | 33.71 | 36.66 | 2.95 | 1.85E-01 |
| Tc800-D1 | 60 | 69 | 33.4 | 42.63 | 9.23 | 8.58E-03 |
| Tc802-D0 | 71 | 100 | 20.3 | 49.51 | 29.21 | 1.90E-18 |
| Tc804-D0 | 66 | 68 | 12.35 | 34.76 | 22.41 | 3.73E-13 |
| Tc806-D1 | 65 | 71 | 16.56 | 43.7 | 27.14 | 8.74E-13 |
| Tc810-D1 | 66 | 94 | 23.3 | 53.69 | 30.39 | 1.47E-22 |
| Tc812-D1 | 81 | 65 | 22.88 | 36.89 | 14.01 | 6.99E-09 |
| Tc814-D0 | 80 | 61 | 10.25 | 35.77 | 25.52 | 2.74E-13 |
| Tc818-D1 | 67 | 96 | 32.99 | 52.5 | 19.51 | 2.53E-12 |
| Tc824-D1 | 74 | 93 | 26.5 | 49.04 | 22.54 | 1.72E-14 |
| Tc826-D1 | 66 | 68 | 18.59 | 41.49 | 22.9 | 2.22E-13 |
| Tc827-D2 | 68 | 85 | 21.26 | 37.82 | 16.56 | 4.02E-15 |
| Tc831-D2 | 65 | 62 | 16.88 | 44.18 | 27.3 | 5.08E-19 |
| Tc832-D1 | 66 | 69 | 16.28 | 45.17 | 28.89 | 7.89E-16 |
| Tc834-D0 | 67 | 66 | 13.26 | 26.22 | 12.96 | 9.67E-11 |
| Tc835-D1 | 67 | 66 | 37.33 | 50.18 | 12.85 | 5.70E-05 |
| Tc836-D1 | 65 | 61 | 22.58 | 39.33 | 16.75 | 1.23E-11 |
| Tc848-D2 | 66 | 81 | 19.23 | 39.59 | 20.36 | 1.36E-18 |
| Tc853-D0 | 66 | 84 | 24.27 | 43.34 | 19.07 | 2.17E-11 |
| C Simulated NMR Contacts Ts | ||||||
| Ts761-D0 | 79 | 86 | 13.69 | 21.78 | 8.09 | 8.01E-07 |
| Ts763-D1 | 99 | 106 | 18.02 | 35.65 | 17.63 | 3.89E-12 |
| Ts767-D0 | 71 | 71 | 12.18 | 34.97 | 22.79 | 4.26E-12 |
| Ts777-D1 | 71 | 80 | 11.09 | 17.88 | 6.79 | 5.60E-05 |
| Ts785-D1 | 71 | 111 | 21.09 | 36.62 | 15.53 | 2.72E-08 |
| Ts794-D0 | 77 | 83 | 33.71 | 26.7 | −7.01 | 1.07E-02 |
| Ts800-D1 | 60 | 85 | 33.4 | 41.61 | 8.21 | 1.34E-02 |
| Ts802-D0 | 71 | 116 | 20.3 | 43.92 | 23.62 | 2.99E-16 |
| Ts804-D0 | 66 | 83 | 12.35 | 28.17 | 15.82 | 1.25E-10 |
| Ts806-D1 | 65 | 80 | 16.56 | 31.13 | 14.57 | 1.02E-05 |
| Ts810-D1 | 66 | 99 | 23.3 | 45.64 | 22.34 | 8.81E-19 |
| Ts812-D1 | 81 | 100 | 22.88 | 38.89 | 16.01 | 3.14E-09 |
| Ts814-D0 | 80 | 73 | 10.25 | 25.98 | 15.73 | 3.64E-08 |
| Ts818-D1 | 67 | 104 | 32.99 | 41.58 | 8.59 | 3.82E-04 |
| Ts824-D1 | 74 | 115 | 26.5 | 40.22 | 13.72 | 2.69E-09 |
| Ts826-D1 | 66 | 80 | 18.59 | 30.72 | 12.13 | 4.43E-07 |
| Ts827-D2 | 68 | 100 | 21.26 | 27.77 | 6.51 | 5.78E-06 |
| Ts832-D1 | 66 | 80 | 16.28 | 32.94 | 16.66 | 1.48E-08 |
| Ts835-D1 | 67 | 77 | 37.33 | 36.9 | −0.43 | 4.51E-01 |
| D Crosslinked Contacts Tx | ||||||
| Tx767-D0 | 71 | 72 | 12.18 | 12.37 | 0.19 | 3.70E-01 |
| Tx781-D0 | 75 | 85 | 9.89 | 9.5 | −0.39 | 2.24E-01 |
| Tx808-D0 | 78 | 84 | 12.81 | 10.02 | −2.79 | 1.98E-03 |
| Tx812-D1 | 81 | 80 | 22.88 | 20.8 | −2.08 | 1.19E-01 |
Positive mean differences and significant P-values are shaded.
In contrast to the poor performance on predicted Tp and crosslinked Tx targets, CASP11 predictors achieved good results modeling correct Tc and simulated NMR Ts targets (Figure 2, center panel for correct Tc and upper left panel for Ts). The average GDT_TS improvement of the best correct Tc models was 12.1 GDT_TS for absolute performance, with the top score improvement approaching 69.2 GDT_TS for Tc763. For individual performance, the average of all best correct Tc models over all targets was 22.9 GDT_TS, with the top score improvement approaching 72.1 GDT_TS for target Tc763. All but one correct Tc target showed significant improvements using community-wide t-tests (Table 3). Similarly, the average performance improvements for best simulated NMR Ts models were both positive (1.5 GDT_TS for absolute and 11.7 for individual), with all but two of the targets (Ts794 and Ts835) showing significant improvements in average model quality by the community-wide t-tests (Table 3).
The correct Tc and simulated NMR Ts score distributions highlight another drawback of comparing assisted scores to initial unassisted T0 scores. Several of the targets exhibited negative absolute performance differences, yet the individual performance differences were generally positive (i.e. Tc/Ts806, Tc/Ts824, and Tc/Ts827). These discrepancies suggested that the gold standard best unassisted T0 models used for calculating absolute performance had unusually high scores. Indeed, one of the manual groups participating in the contact-assisted predictions (Baker, CASP group number 064 – see Table 2 for CASP11 group name-number correspondence) provided outstanding “unassisted” T0 predictions for two of these targets (T0806, see Figure 4, and T0824). We learned that the Baker group had successfully incorporated co-evolution based contact predictions into their T0 tertiary structure predictions17. As such, the top T0 GDT_TS scores did not fairly reflect those of unassisted models, and this incorrect basis for comparison resulted in unusually low community-wide absolute performance scores (and penalized the individual performance scores for group 64 on these two targets).
Figure 4. Examples of Improved Assisted Prediction Models.
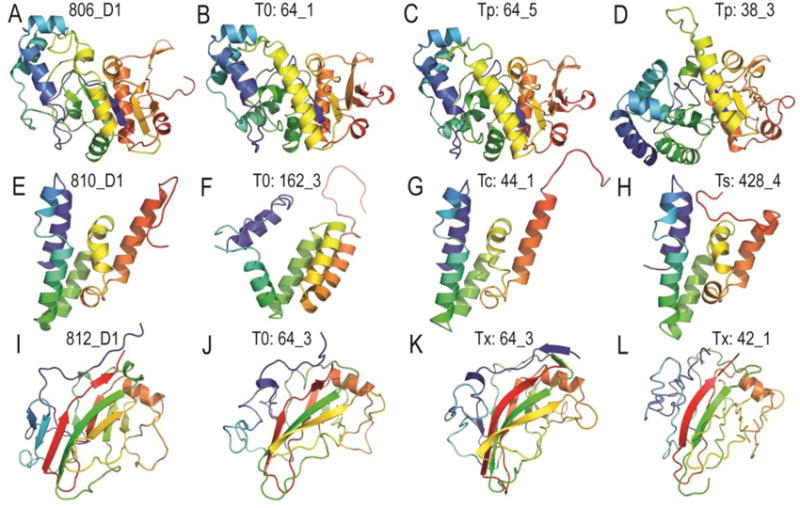
Targets and models are illustrated in cartoon and colored in rainbow from blue (N-terminus) to red (C-terminus). Target 806 D1 (A) is compared to the top unassisted T0 model 64_1 (B) having a GDT_TS of 60.7. The top predicted Tp model 64_5 (C) improves the GDT_TS slightly to 62.5, while the next best predicted Tp group model 38_3 (D) decreases the GDT_TS to 29.5. Target 810 D1 (E) is compared to the top unassisted T0 model 162_3 (F) having a GDT_TS of 40.5. Two identical top correct Tc models 44_1and 169_1 (G) improve the GDT_TS significantly to 86.3, while one top simulated NMR Ts model 428_4 (H) improves the GDT_TS significantly to 79.2. Target 812 D1 (I) is compared to the top unassisted T0 model 64_3 (J) having a GDT_TS of 44.2. The top crosslinked Tx model 64_3 (K) improves the GDT_TS slightly to 47.4, while the next best group model 42_1 (L) decreases the GDT_TS to 40.2.
Group-based performance improvements
We used the same absolute and individual performance improvement measures (with slight alterations) to understand how each group used contact information to improve unassisted T0 models. For the group-based performance improvement evaluation (Figure 3 and Table 4), we considered all assisted models in calculating averages so that the most information possible was included for statistical evaluation, and we compared these models to either the top group unassisted T0 (individual) or the gold standard unassisted T0 (absolute). Most of the groups’ individual and absolute average performance differences were negative for predicted Tp (blue) and crosslinked Tx (orange) targets (Figure 3A). In contrast, the average individual performance differences for both correct Tc and simulated NMR Ts were above 30 GDT_TS for six of the participating groups (three groups from Jooyoung Lee’s lab: Lee, LeeR and NNS server; the Baker group; the Wiskers group; and the Laufer group), with similar trends in the absolute performance (Figure 3B). While the Wiskers group showed one of the most promising GDT_TS difference score trends, they contributed models for only 2 of the 24 correct Tc targets and 2 of the 19 simulated NMR Ts targets (Table 4). In fact, six of the groups contributed models for less than 10 of the 70 total targets in all of the assisted categories (indicated by grey group labels in figure 3) and were ultimately excluded from rankings.
Figure 3. Groupwise performance improvements on Assisted Targets.

The individual performance (A) and absolute performance (B) averages of each indicated group (X coordinate) are plotted for predicted Tp (blue), correct Tc (red), simulated NMR Ts (green), and crosslinked Tx (purple) targets. Groups are ordered from left (highest) to right (lowest) based on the sum of averages over all categories. Groups with less than 5 total predictions (out of 70 possible) are in labeled in grey.
Table 4.
Significance of group performance improvement using assisted information
| Group Num | Individual Performance | Absolute Performance | ||||
|---|---|---|---|---|---|---|
| T* Num | Mean Diff | P-value | T* Num | Mean Diff | P-value | |
| A Predicted Contacts Tp | ||||||
| 32 | 86 | −4.24 | 7.20E-09 | 86 | −15.12 | 3.53E-26 |
| 38 | 115 | 1.21 | 1.88E-02 | 115 | −8.20 | 2.04E-15 |
| 40 | 5 | −1.12 | 7.93E-02 | 5 | −4.38 | 1.22E-03 |
| 41 | 115 | −8.88 | 3.02E-15 | 115 | −19.42 | 3.35E-31 |
| 44 | 15 | −2.28 | 2.57E-03 | 15 | −3.69 | 4.90E-06 |
| 64 | 47 | 1.09 | 2.38E-01 | 47 | −3.48 | 1.71E-04 |
| 65 | 106 | −8.32 | 5.69E-12 | 106 | −14.44 | 2.00E-23 |
| 80 | 115 | −1.96 | 2.22E-04 | 115 | −17.04 | 2.13E-28 |
| 162 | 115 | −2.41 | 6.49E-04 | 115 | −10.89 | 2.70E-20 |
| 169 | 115 | −2.45 | 3.48E-06 | 115 | −8.08 | 4.50E-15 |
| 186 | 5 | −1.19 | 4.34E-02 | 5 | −3.31 | 1.66E-03 |
| 219 | 84 | −10.85 | 1.74E-19 | 84 | −22.54 | 5.93E-28 |
| 276 | 5 | −2.12 | 1.23E-03 | 5 | −6.17 | 1.92E-05 |
| 300 | 17 | −1.78 | 7.33E-03 | 17 | −8.54 | 8.71E-10 |
| 345 | 110 | 3.32 | 4.05E-07 | 110 | −10.10 | 4.95E-17 |
| 357 | 104 | 1.83 | 1.20E-09 | 104 | −18.42 | 2.10E-29 |
| 361 | 5 | −1.38 | 8.48E-03 | 5 | −2.73 | 7.40E-04 |
| 420 | 115 | −5.60 | 9.96E-07 | 115 | −16.72 | 3.00E-26 |
| 476 | 5 | −1.81 | 1.74E-02 | 5 | −3.54 | 1.78E-03 |
| 479 | 105 | −1.83 | 4.86E-03 | 105 | −13.32 | 3.31E-21 |
| B Correct Contacts Tc | ||||||
| 32 | 85 | −2.77 | 8.82E-06 | 85 | −15.24 | 1.04E-21 |
| 38 | 120 | 45.98 | 2.47E-54 | 120 | 36.11 | 4.93E-40 |
| 40 | 45 | 1.50 | 1.45E-01 | 45 | −9.74 | 4.38E-07 |
| 41 | 120 | 13.80 | 4.17E-15 | 120 | 2.47 | 5.58E-02 |
| 44 | 120 | 44.42 | 8.45E-50 | 120 | 38.53 | 4.07E-44 |
| 64 | 115 | 37.56 | 1.35E-47 | 115 | 34.66 | 8.52E-45 |
| 65 | 87 | 3.13 | 4.61E-02 | 87 | −2.12 | 1.17E-01 |
| 80 | 120 | 4.48 | 8.86E-09 | 120 | −11.82 | 7.81E-15 |
| 162 | 120 | 0.14 | 3.95E-01 | 120 | −8.58 | 5.77E-16 |
| 169 | 120 | 45.44 | 1.87E-55 | 120 | 39.43 | 2.85E-48 |
| 186 | 45 | 11.93 | 2.07E-07 | 45 | 0.82 | 3.52E-01 |
| 276 | 106 | 18.00 | 4.16E-27 | 106 | 5.38 | 1.57E-04 |
| 342 | 45 | 18.88 | 3.23E-11 | 45 | 7.68 | 3.69E-03 |
| 345 | 120 | 5.48 | 5.16E-08 | 120 | −9.34 | 3.06E-10 |
| 357 | 98 | 11.20 | 1.03E-20 | 98 | −9.07 | 5.31E-09 |
| 361 | 35 | 7.62 | 1.38E-03 | 35 | −4.89 | 6.11E-02 |
| 420 | 120 | 20.28 | 6.30E-23 | 120 | 7.79 | 3.56E-06 |
| 428 | 40 | 41.58 | 8.22E-28 | 40 | 28.37 | 5.50E-13 |
| 476 | 45 | 16.85 | 3.67E-09 | 45 | 5.78 | 1.94E-02 |
| 479 | 105 | −3.58 | 4.72E-09 | 105 | −15.39 | 2.35E-24 |
| 490 | 2 | 22.46 | 2.41E-01 | 2 | 17.60 | 2.79E-01 |
| C Simulated NMR Contacts Ts | ||||||
| 32 | 84 | −4.95 | 2.37E-08 | 84 | −17.91 | 8.15E-24 |
| 38 | 95 | 37.38 | 3.42E-40 | 95 | 27.69 | 4.25E-28 |
| 40 | 40 | −3.03 | 1.51E-02 | 40 | −14.33 | 2.83E-10 |
| 41 | 95 | 1.20 | 2.67E-01 | 95 | −9.89 | 1.25E-06 |
| 44 | 95 | 35.24 | 1.62E-33 | 95 | 29.5 | 4.54E-28 |
| 64 | 95 | 32.24 | 1.22E-31 | 95 | 28.97 | 4.61E-27 |
| 65 | 91 | −8.65 | 2.76E-07 | 91 | −13.39 | 5.64E-13 |
| 80 | 95 | −2.14 | 5.24E-04 | 95 | −19.33 | 4.41E-24 |
| 162 | 95 | −3.13 | 7.48E-05 | 95 | −12.34 | 6.25E-18 |
| 169 | 95 | 35.29 | 1.24E-33 | 95 | 29.43 | 5.02E-28 |
| 186 | 40 | 3.57 | 1.42E-02 | 40 | −7.59 | 5.72E-06 |
| 219 | 93 | −10.60 | 1.05E-19 | 93 | −21.35 | 2.11E-27 |
| 276 | 90 | 20.57 | 1.85E-22 | 90 | 8.3 | 1.33E-04 |
| 287 | 55 | −2.50 | 1.87E-02 | 55 | −13.57 | 4.05E-09 |
| 310 | 75 | −2.34 | 6.90E-10 | 75 | −10.17 | 6.01E-15 |
| 329 | 4 | 7.21 | 1.52E-01 | 4 | 2.83 | 3.27E-01 |
| 342 | 40 | 8.49 | 5.25E-04 | 40 | −2.76 | 1.68E-01 |
| 345 | 95 | 2.22 | 2.33E-02 | 95 | −12.72 | 2.15E-12 |
| 357 | 95 | 12.53 | 1.61E-08 | 95 | −7.28 | 3.12E-04 |
| 361 | 30 | 1.50 | 2.21E-01 | 30 | −9.38 | 8.76E-04 |
| 420 | 95 | 7.51 | 7.41E-05 | 95 | −4.69 | 5.97E-03 |
| 428 | 50 | 36.16 | 5.61E-22 | 50 | 26.67 | 7.44E-14 |
| 476 | 40 | 8.58 | 2.94E-04 | 40 | −2.53 | 1.71E-01 |
| 479 | 45 | −1.14 | 2.61E-03 | 45 | −17.18 | 3.07E-10 |
| 490 | 2 | 37.85 | 5.21E-02 | 2 | 29 | 1.08E-01 |
| D Crosslinked Contacts Tx | ||||||
| 32 | 15 | −3.55 | 4.57E-03 | 15 | −13.34 | 1.33E-07 |
| 38 | 20 | −3.13 | 3.79E-02 | 20 | −8.35 | 2.29E-03 |
| 41 | 20 | −9.26 | 1.74E-06 | 20 | −17.44 | 5.64E-09 |
| 42 | 10 | −10.26 | 3.13E-03 | 10 | −12.37 | 9.97E-04 |
| 64 | 20 | 1.50 | 2.04E-01 | 20 | −1.97 | 1.06E-02 |
| 65 | 17 | −5.66 | 2.54E-06 | 17 | −9.41 | 5.70E-09 |
| 80 | 15 | 0.69 | 2.33E-02 | 15 | −17.8 | 1.26E-06 |
| 155 | 5 | −2.09 | 1.65E-03 | 5 | −11.39 | 2.20E-06 |
| 157 | 19 | −6.82 | 3.30E-04 | 19 | −13.86 | 1.22E-09 |
| 162 | 20 | −4.60 | 7.72E-03 | 20 | −10.6 | 1.59E-07 |
| 169 | 20 | −6.85 | 4.18E-03 | 20 | −8.4 | 2.15E-03 |
| 219 | 15 | −6.29 | 2.27E-07 | 15 | −12.2 | 1.37E-09 |
| 276 | 20 | −7.45 | 1.94E-06 | 20 | −14.4 | 2.72E-09 |
| 287 | 20 | −4.57 | 2.96E-03 | 20 | −12.83 | 9.52E-06 |
| 345 | 20 | −1.74 | 8.17E-05 | 20 | −16.09 | 5.90E-08 |
| 357 | 20 | 1.00 | 2.59E-03 | 20 | −16.07 | 2.18E-06 |
| 420 | 20 | −5.37 | 1.86E-04 | 20 | −15.79 | 5.75E-08 |
| 428 | 5 | −15.51 | 7.33E-06 | 5 | −30.79 | 4.76E-07 |
| 479 | 20 | −3.08 | 2.45E-05 | 20 | −13.03 | 1.11E-05 |
Positive mean differences and significant P-values are shaded.
According to pairwise Student’s t-tests evaluating the individual and absolute GDT_TS performance improvements for the groups participating in the CASP11 contact assisted categories, only three groups (NNS, Fusion, and Stap) showed significantly positive individual average performances on predicted Tp targets, whereas one additional group (Baker) showed a positive, but insignificant average performance (Table 4A). 15 of the 20 participating groups in the predicted Tp category significantly declined as measured by individual performance differences, and all were significantly worse using absolute performance differences. In the crosslinked Tx category, two groups (Meiler Lab and Stap) showed significant positive individual average performance, one group (Baker) showed positive, but insignificant performance, and the rest showed significantly negative individual average performance (Table 4D).
In the correct Tc and the simulated NMR Ts categories, individual and average performance measures showed significant (by Student’s t-test) improvement over initial models for five groups (Fig 2: Lee, LeeR, NNS, Baker and Laufer). Four additional groups (Floudas, Anthropic Dreams, Multicom-cluster, and Foldit) significantly improved in both individual and average measures for the correct Tc category, and one group (Floudas) showed significant improvements in both measures for the simulated NMR Ts category (Table 4, B and C). The top five performing groups had higher scores on the correct Tc targets than both their individual unassisted T0 scores (average increase of 43.0 GDT_TS) and the gold standard unassisted T0 scores (average increase of 35.4 GDT_TS). They also showed similar average improvements in the simulated NMR Ts category (35.3 GDT_TS for individual and 28.5 GDT_TS for absolute).
Two of the top-performing groups in the contact-assisted prediction (Baker and LeeR) also performed well in the FM tertiary structure prediction evaluation of unassisted T0 models18. Since most (21 out of 27) of the contact-assisted targets belong at least in part to the FM category (Table 1), above the average GDT_TS scores of these two groups on unassisted T0 targets could introduce negative bias in difference scores. Thus in theory, evaluation of groups that outperform on T0 targets by their individual GDT_TS difference tests might be unfair. Indeed, the average best T0 GDT_TS score (29.5) on all contact assisted targets for the Baker and LeeR groups was significantly different than the average best T0 GDT_TS score for the remaining groups (22.5) using a two-sample, one-tailed t-test. Given these drawbacks to the performance improvement scores, we chose to rank groups using alternate scores (see Performance evaluation section below).
Examples of top assisted target predictions from top-performing groups
Target Tp806 exhibited the highest overall significant mean difference (4.3 GDT_TS) reflecting performance improvement for the predicted Tp category (Table 3). The FM-categorized T0806 target protein (Figure 4A) adopts an α/β three-layered sandwich architecture in the Evolutionary Classification Of protein Domains (ECOD) database19 that is distantly related by structure (top LGA_S 25.0 to 2q07A) to folds in the X-group “other Rossmann-like structures with the crossover”. The Rossmann-like domain in the target is interrupted by a unique 3-helix insertion that is not present in any structurally related templates. The relatively high GDT_TS score of 60.7 for this target’s top T0 model (64_1, by the Baker group) reflected a correct overall topology for the prediction (Figure 4B) that was significantly closer to the target than the top templates. Despite this impressive top T0 prediction, the mean GDT_TS was much lower (16.56) for T0 models from groups participating in the contact-assisted categories. The best model for this target in the predicted Tp category (also the Baker’s group model 64_5, Figure 4C) slightly improved the GDT_TS score (to 62.5). The next best group prediction (38_3 by the NNS server, Figure 4D) retained the correct topology of the Rossmann fold, but incorrectly oriented the helical insertion with respect to the β-sheet.
Target Tc810-D1 exhibited the highest overall significant mean difference (30.4 GDT_TS) reflecting performance improvement for the correct Tc category, and Ts810-D1 exhibited the third highest mean difference (22.3 GDT_TS) for the simulated NMR Ts category (Table 3). The ECOD database19 classifies the FM-categorized target T0810-D1 as an α-superhelices architecture with a somewhat irregular ARM-repeat fold (Figure 4E). This target domain is fused to a C-terminal domain exhibiting an α/β-barrel architecture fold that is homologous to a TIM barrel in ECOD. This C-terminal domain was categorized as TBM and was excluded from the contact-assisted predictions. The top unassisted prediction model among contact-assisted predictors for this domain (TS162_3, from McGuffin group) displayed a roughly similar topology (GDT_TS 40.5), except the N-terminal helices did not pack against the subdomain formed by the C-terminal helices (Figure 4F). The two top Tc prediction models (44_1 and 169_1 from J. Lee’s lab) were identical and improved over the top T0 model by 45.8 GDT_TS (Figure 4G), while the top simulated NMR Ts prediction model by another group (Laufer, 428_4, Figure 4H) improved over the top T0 model by 38.7 GDT_TS. The top correct Tc and simulated NMR Ts prediction models for T0810-D1 adopted the correct overall topology of the ARM-repeat fold, with the main differences stemming from an extended C-terminal linker sequence with no secondary structure.
The single-domain target T0812-D1 (Figure 4I) was categorized as TBM-hard, and displayed a β-sandwiches ECOD architecture that is homologous to Concanavalin A-like folds. The top T0 prediction model (64_3 from the Baker group, Figure 4J) retained the same overall fold as the target domain, except for the N-terminal residues (5–56) corresponding to the first three β-strands. The overall mean difference for the target T0812-D1 was negative (−2.1 GDT_TS), yet the top performing crosslinked Tx model improved over the T0 model by 3.2 GDT_TS (64_3 from Baker, Figure 4K). The next best group prediction model (42_1 from the Tasser group, Figure 4L) decreased by 4 GDT_TS, as compared to the T0 model. While the top performing crosslinked Tx model only improved by 3.2 GDT_TS, it correctly placed the three N-terminal β-strands and attained the entire fold topology. The next best group model also predicted the correct overall fold topology, but the model exhibited gaps and incorrectly structured β-strands.
Performance evaluation without unassisted models: combining scores for ranks
Due to the potential biases of using unassisted models for the contact-assisted evaluation, we chose to assess group performance using similar score combinations as were used in the FM (see Kinch, this issue) and TBM (see Roland, this issue) evaluations. We generated Z-score sums and averages over all contact-assisted (T*=Tp, Tc, Ts, or Tx) targets for the combined scores on each group’s best or first submitted models. We evaluated all categories using the FM-style combined scores (GDT_TS, ContS, QCS, TenS, lDDT, and MolProb). However, the relative high performance of groups in the correct Tc and simulated NMR Ts categories prompted additional evaluation using TBM-style score combinations to better distinguish models that are closer to their targets (GDT_TS>50).
Group performance was ordered by best FM-style Z-score sum (Table 5, includes also FM-style average, first models and win/loss counts). All groups that could not be distinguished from the top ranked group according to t-test and bootstrap significance (for FM-style Z-score sum) are bolded. The top-performing groups in the contact-assisted categories according to the FM-style and win/loss scoring schemes (Lee, LeeR, NNS, and Baker) were similar to those that outperformed in performance improvement scores (Figure 3). As three of these groups correspond to a single CASP11 participant (Jooyoung Lee - groups 38, 44, and 169), we investigated whether having multiple groups (i.e. submitting as multiple groups) tended to alter the Z-score ranks or significance scores of the participant when compared to having a single group (i.e. submitting as a single group). To check for this case, we omitted two of the three J. Lee’s groups in turn, and recalculated all the relative scores for all the participating groups in these three scenarios. With the exception of the crosslinked Tx category, which had too few targets, the ranks and significance estimates of any single group from the same CASP11 participant did not change, although the absolute values of the Z-scores did (See prodata.swmed.edu/casp11/contact for tables).
When compared to group performance ranks determined by the GDT_TS Z-score sums, the FM-style Z-score sums produced the same ranks for the four top-performing groups in the predicted Tp category (Lee, NNS, McGuffin, and Fusion, in ranked order). However, tests of statistical significance in the predicted Tp category suggested that one of the groups (Baker) that predicted significantly fewer targets (10 out of 23) tied with the two top-performing groups (Lee and NNS). In win/loss counts, the same four groups rank at the top, with the Baker group holding 3rd place.
For the correct Tc category, all scoring methods (GDT_TS, FM-style, and win/loss counts) rank groups LeeR and Lee as first and second, correspondingly. Because the top prediction models in this category were similar to the target (GDT_TS score >50), we also examined TBM-style scoring and significance estimates that were designed to evaluate such similarities. TBM-style scoring ranked the same two groups at the top. These two groups tied in many of the head-to-head trials (10 out of 24 targets), and the performance of the two groups could not be distinguished by significance estimates of TBM-style scoring. The third-place group (Baker) tied with the top-performing group according to significance of FM-style scores, but not TBM-style scores or GDT_TS only scores.
For the simulated NMR Ts category, the same group (Baker) placed as first for all three Z-score-style scoring methods (GDT_TS, FM, and TBM). Two additional groups (LeeR and Lee) tied for top-performance by all statistical measures. The fourth ranked group, NNS server (as well as the Laufer group that predicted less targets), tied with the top groups only using significance from T-tests on TBM-style scoring. Interestingly, win/loss counts with GDT_TS, FM-style, and TBM-style scoring placed the Lee and LeeR groups above the top-ranked Baker group. The cause of this apparent discrepancy in rankings is discussed in the following section (Head-to-Head Comparisons).
For the crosslinked Tx category, the top-performing Baker group was ranked first by GDT_TS and FM-style scoring methods, as well as in win/loss counts. The top group tied with Lee and NNS groups using T-test significance estimates, while it significantly outperformed by FM-style bootstraps. The differences in significance likely originated from the low number of targets in this category (4 targets).
Head-to-Head Comparisons of Top-Performing Groups
To help clarify the performance of the top ranked groups in each category that tied by any of the significance estimates, we plotted their head-to-head GDT_TS scores (Figure 5). For these head-to-head comparisons, we chose the top performing Lee lab group (among Lee, LeeR and NNS) according to FM-style Z-score ranks for each assisted category. For illustrative purposes, we combined the head-to-head results from the Baker and Lee groups for the predicted Tp and crosslinked Tx categories into a single graph (Figure 5A). The predicted Tp targets were limited to only 10 of the 23, since the Baker group did not predict the remaining targets. Most of the predicted Tp targets clustered near the identity line below 40 GDT_TS. However, the Baker group submitted three predicted Tp prediction models above GDT_TS 40 that outperformed (Tp806, Tp818, and Tp827), while the Lee group submitted one (Tp825) that outperformed. This relative outperformance of the Baker group on the reduced target subset likely explains their elevated performance according to significance estimates and their win/loss rank just under the top-performing Lee and NNS server groups (Table 5). Similarly, three out of the four targets in the crosslinked Tx category clustered near the identity line below 25 GDT_TS. The Baker group outperformed on a single crosslinked Tx target (Tx812), while the Lee group outperformed marginally on two of the crosslinked Tx targets. Thus, the outperformance of group Baker on a single target Tx812 established their position at the top of all ranking methods for the crosslinked Tx category (Table 5).
Figure 5. Head-to-Head Plots for Top-Performing Groups.

Top-performing groups according to significance tests were chosen for comparison. FM-style Z-scores were used to select the top group number among multiple submissions from the same prediction team. GDT_TS scores were plotted for A) Baker Group 64 against Lee group 169 for the predicted Tp (blue) and the crosslinked Tx categories (orange), B) Baker group 64 against LeeR group 44 for the correct Tc category, and C) Baker group 64 against LeeR group 44 for the simulated NMR Ts category.
Table 5.
Group Ranks and Significance
| Grp | Group Name | Num Target | Best FM-Style Scoring
|
First FM-style Scoring
|
Win/Loss Scoring
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sum Z | Sum R | Avg Z | Avg R | Sum Z | Sum R | Avg Z | Avg R | Win F | P-value | Win R | ||||
| A | Tp | |||||||||||||
| 169 | LEE*1,2 | 23 | 16.54 | 1 | 0.72 | 2 | 14.85 | 2 | 0.68 | 6 | 0.86 | 1.3E−32 | 1 | |
| 38 | nns*1,2 | 23 | 15.98 | 2 | 0.69 | 3 | 15.64 | 1 | 0.68 | 5 | 0.86 | 7.8E−32 | 2 | |
| 162 | McGuffin* | 23 | 8.85 | 3 | 0.38 | 7 | −0.93 | 5 | −0.04 | 12 | 0.67 | 2.5E−08 | 4 | |
| 345 | FUSION* | 22 | 3.25 | 4 | 0.24 | 10 | 7.44 | 3 | 0.43 | 7 | 0.59 | 3.1E−03 | 5 | |
| 420 | MULTICOM-CLUSTER | 23 | −0.24 | 5 | −0.01 | 12 | −2.42 | 6 | −0.11 | 13 | 0.48 | 6.7E−01 | 8 | |
| 479 | RBO Aleph | 21 | −2.58 | 6 | 0.07 | 11 | 2.33 | 4 | 0.30 | 9 | 0.56 | 4.3E−02 | 6 | |
| 65 | Jones-UCL | 21 | −5.27 | 7 | −0.06 | 13 | −2.96 | 7 | 0.05 | 10 | 0.52 | 2.5E−01 | 7 | |
| 80 | MeilerLab | 23 | −5.73 | 8 | −0.25 | 14 | −5.41 | 8 | −0.24 | 14 | 0.43 | 9.8E−01 | 9 | |
| 64 | BAKER1,2 | 10 | −13.73 | 9 | 1.23 | 1 | −14.99 | 9 | 1.10 | 1 | 0.96 | 1.4E−26 | 3 | |
| 357 | STAP | 21 | −16.97 | 10 | −0.62 | 15 | −15.40 | 10 | −0.54 | 15 | 0.24 | 1.0E+00 | 10 | |
| 32 | Legato | 21 | −17.24 | 11 | −0.63 | 16 | −17.90 | 11 | −0.66 | 17 | 0.24 | 1.0E+00 | 11 | |
| 41 | MULTICOM-NOVEL | 23 | −21.00 | 12 | −0.91 | 19 | −19.92 | 12 | −0.87 | 19 | 0.16 | 1.0E+00 | 12 | |
| 219 | Sternberg | 17 | −27.11 | 13 | −0.89 | 18 | −25.97 | 13 | −0.82 | 18 | 0.14 | 1.0E+00 | 13 | |
|
| ||||||||||||||
| B | Tc | |||||||||||||
| 44 | LEER*1,2,3,4 | 24 | 29.94 | 1 | 1.25 | 1 | 30.37 | 1 | 1.27 | 1 | 0.95 | 7.8E−79 | 1 | |
| 169 | LEE*3,4 | 24 | 28.47 | 2 | 1.19 | 3 | 28.51 | 2 | 1.19 | 2 | 0.92 | 7.6E−66 | 2 | |
| 64 | BAKER1,2 | 23 | 25.30 | 3 | 1.19 | 2 | 24.68 | 4 | 1.16 | 3 | 0.88 | 4.0E−48 | 3 | |
| 38 | nns | 24 | 24.70 | 4 | 1.03 | 4 | 25.15 | 3 | 1.05 | 4 | 0.83 | 2.8E−38 | 4 | |
| 420 | MULTICOM-CLUSTER | 24 | 3.57 | 5 | 0.15 | 8 | 3.73 | 5 | 0.16 | 8 | 0.62 | 6.9E−06 | 8 | |
| 41 | MULTICOM-NOVEL | 24 | −10.29 | 6 | −0.43 | 13 | −10.35 | 6 | −0.43 | 14 | 0.38 | 1.0E+00 | 12 | |
| 276 | FLOUDAS A4 | 22 | −10.36 | 7 | −0.29 | 11 | −10.67 | 8 | −0.30 | 11 | 0.42 | 1.0E+00 | 10 | |
| 65 | Jones-UCL | 23 | −11.37 | 8 | −0.41 | 12 | −10.36 | 7 | −0.36 | 12 | 0.39 | 1.0E+00 | 11 | |
| 80 | MeilerLab | 24 | −15.92 | 9 | −0.66 | 17 | −15.83 | 9 | −0.66 | 17 | 0.24 | 1.0E+00 | 18 | |
| 345 | FUSION | 24 | −16.69 | 10 | −0.70 | 18 | −16.82 | 10 | −0.70 | 18 | 0.26 | 1.0E+00 | 17 | |
| 162 | McGuffin | 24 | −16.87 | 11 | −0.70 | 19 | −19.21 | 12 | −0.80 | 20 | 0.27 | 1.0E+00 | 16 | |
| 357 | STAP | 21 | −17.74 | 12 | −0.56 | 16 | −17.30 | 11 | −0.54 | 15 | 0.35 | 1.0E+00 | 13 | |
| 479 | RBO_Aleph | 21 | −23.16 | 13 | −0.82 | 20 | −21.05 | 13 | −0.75 | 19 | 0.20 | 1.0E+00 | 19 | |
| 428 | Laufer | 8 | −26.15 | 14 | 0.73 | 5 | −26.30 | 16 | 0.71 | 5 | 0.74 | 1.3E−08 | 5 | |
| 476 | Foldit | 9 | −26.28 | 15 | 0.41 | 6 | −26.06 | 14 | 0.44 | 6 | 0.70 | 1.3E−07 | 6 | |
| 342 | Anthropic Dreams | 9 | −26.66 | 16 | 0.37 | 7 | −26.25 | 15 | 0.42 | 7 | 0.69 | 1.7E−06 | 7 | |
| 186 | Void_Crushers | 9 | −28.97 | 17 | 0.11 | 9 | −30.68 | 17 | −0.08 | 10 | 0.58 | 2.8E−02 | 9 | |
| 32 | Legato | 21 | −31.74 | 18 | −1.23 | 21 | −30.83 | 18 | −1.18 | 21 | 0.05 | 1.0E+00 | 20 | |
| 40 | GoScience | 9 | −34.98 | 19 | −0.55 | 15 | −35.79 | 19 | −0.64 | 16 | 0.33 | 1.0E+00 | 14 | |
| 361 | Contenders | 7 | −37.33 | 20 | −0.48 | 14 | −36.66 | 20 | −0.38 | 13 | 0.33 | 1.0E+00 | 15 | |
|
| ||||||||||||||
| C | Ts | |||||||||||||
| 64 | BAKER*1,2,3,4 | 19 | 25.96 | 1 | 1.37 | 1 | 25.21 | 1 | 1.33 | 1 | 0.90 | 2.9E−55 | 3 | |
| 44 | LEER1,2,3,4 | 19 | 22.24 | 2 | 1.17 | 2 | 22.33 | 3 | 1.18 | 3 | 0.92 | 2.8E−60 | 1 | |
| 169 | LEE1,2,3,4 | 19 | 22.17 | 3 | 1.17 | 3 | 22.52 | 2 | 1.19 | 2 | 0.92 | 2.8E−60 | 2 | |
| 38 | nns3 | 19 | 21.11 | 4 | 1.11 | 4 | 21.69 | 4 | 1.14 | 4 | 0.85 | 1.6E−41 | 4 | |
| 420 | MULTICOM-CLUSTER | 19 | 1.55 | 5 | 0.08 | 10 | 0.82 | 6 | 0.05 | 10 | 0.62 | 1.3E−05 | 7 | |
| 276 | FLOUDAS A4 | 18 | 0.98 | 6 | 0.17 | 9 | 1.41 | 5 | 0.19 | 7 | 0.65 | 1.3E−07 | 6 | |
| 357 | STAP | 19 | 0.33 | 7 | 0.02 | 11 | −0.25 | 7 | −0.01 | 11 | 0.60 | 3.0E−04 | 8 | |
| 428 | Laufer3 | 10 | −7.75 | 8 | 1.02 | 5 | −8.04 | 8 | 1.00 | 5 | 0.87 | 1.2E−26 | 5 | |
| 345 | FUSION | 19 | −10.38 | 9 | −0.55 | 18 | −10.28 | 10 | −0.54 | 20 | 0.32 | 1.0E+00 | 18 | |
| 41 | MULTICOM-NOVEL | 19 | −11.26 | 10 | −0.59 | 20 | −10.18 | 9 | −0.54 | 19 | 0.34 | 1.0E+00 | 17 | |
| 65 | Jones-UCL | 19 | −12.33 | 11 | −0.65 | 21 | −11.60 | 11 | −0.61 | 21 | 0.25 | 1.0E+00 | 19 | |
| 162 | McGuffin | 19 | −12.65 | 12 | −0.67 | 22 | −16.73 | 14 | −0.88 | 23 | 0.25 | 1.0E+00 | 20 | |
| 80 | MeilerLab | 19 | −13.55 | 13 | −0.71 | 23 | −13.24 | 12 | −0.70 | 22 | 0.24 | 1.0E+00 | 21 | |
| 310 | MUFOLD-R | 15 | −13.92 | 14 | −0.39 | 16 | −13.60 | 13 | −0.37 | 17 | 0.39 | 1.0E+00 | 13 | |
| 32 | Legato | 19 | −18.52 | 15 | −0.97 | 24 | −18.44 | 16 | −0.97 | 24 | 0.12 | 1.0E+00 | 22 | |
| 287 | RBO-Human | 11 | −18.80 | 16 | −0.25 | 13 | −18.38 | 15 | −0.22 | 13 | 0.49 | 5.6E−01 | 12 | |
| 219 | Sternberg | 19 | −20.19 | 17 | −1.06 | 25 | −20.23 | 17 | −1.06 | 25 | 0.08 | 1.0E+00 | 23 | |
| 476 | Foldit | 8 | −20.39 | 18 | 0.20 | 7 | −20.81 | 19 | 0.15 | 9 | 0.59 | 1.4E−02 | 10 | |
| 342 | Anthropic_Dreams | 8 | −20.53 | 19 | 0.18 | 8 | −20.73 | 18 | 0.16 | 8 | 0.61 | 3.8E−03 | 9 | |
| 186 | Void_Crushers | 8 | −22.71 | 20 | −0.09 | 12 | −22.77 | 20 | −0.10 | 12 | 0.50 | 5.0E−01 | 11 | |
| 479 | RBO_Aleph | 9 | −24.98 | 21 | −0.55 | 19 | −24.47 | 21 | −0.50 | 18 | 0.35 | 1.0E+00 | 16 | |
| 40 | GoScience | 8 | −25.06 | 22 | −0.38 | 14 | −24.81 | 22 | −0.35 | 16 | 0.36 | 1.0E+00 | 14 | |
| 361 | Contenders | 6 | −28.70 | 23 | −0.45 | 17 | −28.10 | 23 | −0.35 | 15 | 0.35 | 1.0E+00 | 15 | |
|
| ||||||||||||||
| D | Tx | |||||||||||||
| 64 | BAKER*1,2 | 4 | 6.20 | 1 | 1.55 | 1 | 5.85 | 1 | 1.46 | 1 | 0.95 | 3.1E−14 | 1 | |
| 287 | RBO-Human | 4 | 3.31 | 2 | 0.83 | 2 | 3.07 | 2 | 0.77 | 2 | 0.78 | 6.1E−06 | 3 | |
| 169 | LEE1 | 4 | 3.06 | 3 | 0.76 | 3 | 2.91 | 4 | 0.73 | 4 | 0.82 | 3.8E−07 | 2 | |
| 38 | nns1 | 4 | 3.01 | 4 | 0.75 | 4 | 2.95 | 3 | 0.74 | 3 | 0.75 | 6.7E−05 | 4 | |
| 162 | McGuffin | 4 | 1.96 | 5 | 0.49 | 5 | −0.74 | 7 | −0.19 | 9 | 0.65 | 1.4E−02 | 6 | |
| 479 | RBO Aleph | 4 | 0.76 | 6 | 0.19 | 7 | 1.45 | 5 | 0.36 | 5 | 0.53 | 3.5E−01 | 8 | |
| 65 | Jones-UCL | 4 | 0.49 | 7 | 0.12 | 8 | 0.21 | 6 | 0.05 | 7 | 0.58 | 1.2E−01 | 7 | |
| 420 | MULTICOM-CLUSTER | 4 | −1.45 | 8 | −0.36 | 10 | −1.35 | 9 | −0.34 | 11 | 0.42 | 8.8E−01 | 10 | |
| 357 | STAP | 4 | −1.55 | 9 | −0.39 | 11 | −0.96 | 8 | −0.24 | 10 | 0.30 | 1.0E+00 | 14 | |
| 276 | FLOUDAS A4 | 4 | −1.58 | 10 | −0.40 | 12 | −1.46 | 10 | −0.37 | 12 | 0.38 | 9.5E−01 | 11 | |
| 345 | FUSION | 4 | −1.86 | 11 | −0.47 | 13 | −1.77 | 12 | −0.44 | 15 | 0.35 | 9.9E−01 | 13 | |
| 32 | Legato | 4 | −1.91 | 12 | −0.48 | 14 | −1.70 | 11 | −0.43 | 13 | 0.38 | 9.5E−01 | 12 | |
| 157 | FLOUDAS_A1 | 4 | −2.33 | 13 | −0.58 | 16 | −2.97 | 14 | −0.74 | 17 | 0.27 | 1.0E+00 | 15 | |
| 80 | MeilerLab | 3 | −2.55 | 14 | −0.18 | 9 | −1.89 | 13 | 0.04 | 8 | 0.44 | 7.7E−01 | 9 | |
| 42 | TASSER | 2 | −3.49 | 15 | 0.25 | 6 | −3.33 | 15 | 0.33 | 6 | 0.73 | 8.1E−03 | 5 | |
| 219 | Sternberg | 3 | −4.01 | 16 | −0.67 | 17 | −3.93 | 16 | −0.64 | 16 | 0.23 | 1.0E+00 | 16 | |
| 41 | MULTICOM-NOVEL | 4 | −4.70 | 17 | −1.18 | 18 | −3.95 | 17 | −0.99 | 19 | 0.00 | 1.0E+00 | 17 | |
Same ranking by GDT_TS
Significant Best FM score by T-test
Significant Best FM score by Bootstrap
Significant Best TBM score by T-test
Significant Best TBM score by Bootstrap
Positive FM-style scores and win/loss fraction >=0.5 are shaded; top-ranked groups by best model scores (and any groups not significantly different using Bootstraps and T-tests on FM-style and TBM-style scores for Tc and Ts) are bolded.
The correct Tc category head-to-head plot highlights a cluster of 23 targets above 48 GDT_TS, with the LeeR group outperforming on most (16 targets). The Baker group appeared to excel at the assisted prediction of target Tc812, while the LeeR group excelled at target Tc794, among a few others. This relative outperformance by the LeeR group on most of the targets resulted in their top ranking by all methods. Their top ranking was also justified by significance tests using the TBM-style scoring scheme, which was chosen by the TBM assessor as distinguishing models that were generally closer to the template (above 50 GDT_TS). The bootstrap and t-test significance estimates using TBM-style scoring suggested the performance of the LeeR group was not distinguishable from the alternate prediction group from the same participants (Lee), yet it was distinguishable from the Baker group (confidence level 0.916).
The simulated NMR Ts category plot comparing LeeR with Baker highlights three outlier targets where Baker outperformed LeeR (Ts761, Ts777, and Ts827), and two targets (Ts794 and Ts826) where LeeR outperformed Baker. Performance scores on the remaining targets clustered closely to the equivalence line, with more favoring the LeeR group, which wins on 10 of 14 remaining targets. Comparison of the Baker group with the Lee group (ranked 2 by GDT_TS Z-score sums) yielded similar results (not shown). Z-score sums tended to emphasize the magnitude of improvements while win/loss counts evaluated the generalization of the methodology on various targets. Therefore, the apparent discrepancy in rankings by the two methods was caused by the Baker group providing more significantly better outlier targets (top Z-score ranking), whereas the LeeR group provided more subtly better winning targets (12 out of 19 targets). Statistical tests, including bootstrap and t-test, suggested that the differences between these two groups were statistically insignificant.
In our above analyses, we treated multi-domain assisted targets as single evaluation units. Besides this treatment, we also calculated scores, rankings, and significance estimates for first model predictions and domain-based predictions (i.e, predictions on multidomain targets were split and evaluated separately). Group performance using first models resembled that of best models with a few exceptions, including (1) LeeR significantly outperformed the other groups on correct Tc targets, and (2) nns tied with the top groups on simulated NMR Ts targets using FM-style scores. The top performing groups performed similarly using best models on a per-domain basis, with a few exceptions. The Baker group tied with the Lee and LeeR groups in the correct Tc category by all significance tests and the NNS server no longer tied with the top performing groups (Baker and Lee) in the simulated NMR Ts category using TBM-style scoring. For first models, Baker TS064 tied with the LeeR group on correct Tc targets by TBM-style scores and Laufer, who predicted less than half (11) targets, tied with the four top groups on simulated NMR Ts targets by TBM- and GDT_TS- style scores. All the evaluation tables are accessible via http://prodata.swmed.edu/casp11/contact.
Performance comparisons to previous contact assisted predictions
The contact-assisted component of CASP11 included several new categories (predicted Tp, simulated NMR Ts, and crosslinked Tx) that had no basis for comparison to the previous assessment. The input data in the only comparable category (designated correct Tc in both CASP10 and CASP11) had some significant differences in both the number and type of provided contacts. The number of provided contacts for CASP10 were restricted to roughly one tenth of the number of residues, and the contacts were only selected if they were present in less than 15% of the unassisted predictions in CASP103. In contrast, in CASP11 the Prediction Center provided a significantly larger number (~10 fold) of correct Tc contacts that were selected among top contact predictors regardless of the contact coverage in the submitted 3D models.
The previous CASP10 contact-assisted correct Tc category showed significant improvements in mean correct Tc GDT_TS scores when compared to mean T0 scores for each target, with the best absolute improvement approaching 40 GDT_TS. The best absolute improvement for CASP11 correct Tc targets was even higher (70 GDT_TS). Even though it is hard to bring the different types of contacts in two different CASPs to the same frame of reference, the data allowed us to notice similar trends in both CASPs, namely improved average performance with increased number of contacts per residue. A scatter plot of CASP11 target-based best absolute GDT_TS improvement against number of unique provided contacts per target residue (ranged from 0.432 to 1.11) highlighted an overall trend of improving performance with enriching contact information (Figure 6A). Although the data showed a relatively low goodness of fit (R2=0.09), extension of the linear fit line (Y=28.20*X+22.18) to the number of contacts released in CASP10 (25.6 GDT_TS difference at 0.12 contacts per residue) suggests a similar trend in CASP10 and CASP11. This extrapolation implied that the apparent CASP11 performance “improvement” stemmed from an increase in the number of given contacts.
Figure 6. Absolute correct Tc performance improves with increasing provided contacts.
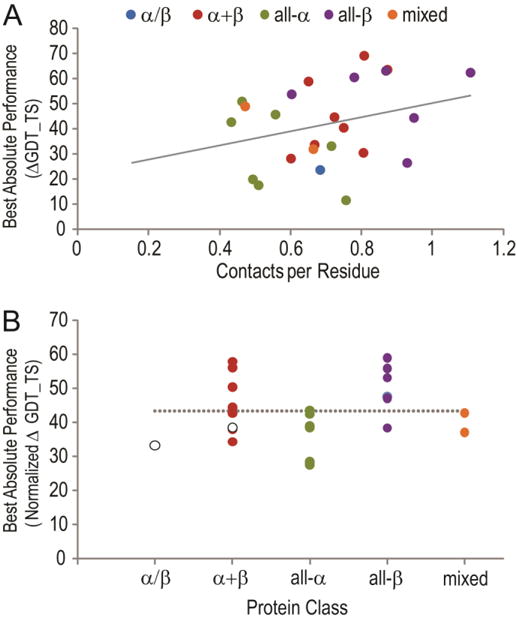
The best absolute performance (top correct Tc GDT_TS – T0max GDT_TS, Y-axis) is plotted against the number of contacts provided per residue in the target (X-axis) in panel (A) and colored according to protein class: α/β (blue), α+β (red), all-α (green), all-β (purple), and mixed (orange). A linear fit to the data has a relatively low goodness of fit R2=0.09. In panel (B) the best absolute performance is normalized by averaging the absolute performance with the expected absolute performance according to the contacts per residue given the linear fit in A. The normalized performance is separated in panel according to protein classes as in panel A. White markers represent data for targets T0806 and T0824 with expected bias in T0. A dashed line indicates the average best absolute performance on all targets.
Two of the correct Tc targets with high outlier T0 predictions (T0806 and T0824, discussed in Target-based performance improvement section above) should have displayed lower than expected best absolute improvements, skewing the trends highlighted in Figure 6A. Indeed, omitting these two targets from linear fit calculations slightly improved the goodness of fit (R2=0.11) and resulted in a somewhat larger slope of the line: Y=30.09*X+22.31, which corresponds to a similar number extended to CASP10 levels (25.9 GDT_TS difference at 0.12 contacts per residue).
Given the relatively high number of correct Tc targets, we examined the performance of predictions on different fold types. We considered the ECOD architecture for each correct Tc target, combining the target architectures into broad categories including α/β, α/β, all-α, all-β, and mixed resulting from the presence of multiple domains. We then plotted the best absolute performance of targets clustered into each category (Figure 6B). Because the targets displayed a trend in performance based on given contacts per residue, we normalized the best absolute performance by averaging it with an estimate of the best absolute performance (Y) based on the given contacts per residue (X) according to the Figure 6A linear fit formula. The results suggest that the provided contacts helped modestly for all-α targets (average normalized performance improvement 35.5 GDT_TS). Only a single target (T0806) populated the α/β category. This target represented an outlier and exhibited a lower than expected absolute difference (33.3 GDT_TS) due to unusually high T0 model quality discussed previously. Indeed, when we used the next-best group T0 target to calculate normalized best absolute performance on the singleton α/β target, the recalculated value (49.9 GDT_TS) exceeded the normalized average best absolute performance value (Figure 6B, dotted line, 43.3 GDT_TS). One possible explanation for the relative contact-assisted outperformance on β-strand-containing targets might involve their more regular interaction in β-sheets dictated by non-local backbone hydrogen bonds. Thus, a single contact provides the correct register for the β-strand with its neighboring β-strands. Alternatively, interactions between α-helices can occur at different angles, requiring more than one contact pair to define their placement.
CONCLUSIONS: PERFORMANCE INSIGHTS AND SUGGESTIONS
Two research labs significantly outperformed the rest using all types of contact-assisted information to enhance prediction model quality: the Lee lab represented by a server NNS, and two manual groups LeeR, and Lee; and the Baker lab with the same-named prediction group. Using contact-assisted information from two different categories, correct Tc and simulated NMR Ts, these top-performing groups provided significantly improved structure predictions. On the other hand, information provided in the predicted Tp and crosslinked Tx categories yielded marginal improvements, despite the success of the Baker group in utilizing contact predictions to significantly improve structure models for several targets (i.e. T0806 and T0824) in the template free modeling category of CASP11 (Baker, personal communication). Unfortunately, the Baker group did not participate in the RR category, from which the assisted Tp category contact data was selected. Thus, the benefit of depth of alignment and improved co-variation methods that led to Baker’s success in residue-residue contact and tertiary structure prediction17,18 could not be evaluated for other groups participating in the predicted Tp category. Moreover, we could not clearly separate the contributions of provided contacts from those embedded in the Baker prediction methodology to their success in the contact-assisted categories. The observation that the Baker group best contact-assisted Tp model (GDT_TS 62.50) was only marginally better than their best unassisted T0 model (GDT_TS 60.65) suggests that the contribution of predicted Tp data from other groups was limited.
Perhaps the most encouraging prediction models came from the simulated NMR Ts category, which aimed to mimic contact information provided by experimental NMR data. The quality of models produced using this information, which albeit only represents a model of real NMR data, approached that of the artificial correct Tc category.
Given the relative outperformance of the Baker and J. Lee’s groups on the contact-assisted categories, we decided to use their average GDT-TS scores for all targets in a given category to represent top performance. We then examined why the predicted Tp and crosslinked Tx categories were much more difficult than the correct Tc and simulated NMR Ts categories (Figure 7A). First, we considered a term that evaluated the quality of provided contacts for each assisted category: the correct contact percentage (CCP). As expected, outperformance in the correct Tc category arose from the high percentage of correct contacts given (100% by definition), with the other three categories having less than 15% of the provided contacts being correct. Interestingly, the CCP average for the simulated NMR Ts category was almost the same as for the predicted Tp category, for which performance was significantly lower. Thus, CCP alone could not account for performance. The given simulated NMR Ts data included far more contacts than in any of the other categories (see paragraph below), so we also calculated the correct contact coverage (CCC) of the target to see if this property could compensate for a lack of correct provided contacts. Indeed, the simulated NMR Ts category displayed a higher CCC average (2.5-fold coverage) than the other three categories (Tp 0.19-fold, Tc 0.7-fold, and Tx 0.16-fold coverage). Thus, the outperformance on the correct Tc targets stemmed from the high percentage of correct contacts, whereas the outperformance in the simulated NMR Ts category stemmed from a reduced percentage of correct contacts that was offset by a much higher coverage of correct contacts. A number of possible explanations for the relatively poor performance in the crosslinked Tx category exist. From our evaluation of contact quality (Figure 7A), the contacts provided by the crosslinked Tx data were only 10.8% correct on average when defined by the 8 Å distance cutoff in the experimental structures. Such poorly defined contacts likely result from the cross-linking agent being too long to represent interacting residues.
Figure 7. Dissecting Prediction Quality. A).

The performance of the top groups (averaged GDT_TS, left panel) is dictated by two components of the provided contact information: the percentage of correct contacts (those within 8 Å in the target structure) over all given contacts (CCP, middle panel) and the fold coverage of correct contacts over the target structure (CCC, right panel). The bars represent the averages over targets from each contact assisted category: predicted Tp (blue), correct Tc (red), simulated NMR Ts (green), and crosslinked Tx (orange). B) The contacts provided for the simulated NMR Ts category can be subdivided into several classes of given information: all provided contacts that include both single peaks and multiple peaks for certain atom pairs (cyan), unambiguous contacts that correspond to given atom pairs with a single peaks (purple), ‘Ts-specific’ true contacts defined as pairs with atomic distance in the target structure within the given upper distance limit (UPL) (green), and correct contacts defined as pairs within 8 Å in the target structure (medium green). Contacts of each subcategory are shown in logarithmic scales and counted by atom (dark colors, labeled ‘Atom’) or by residue (light colors, labeled ‘Res’). C) The various classes of simulated NMR Ts information lead to different levels of performance measured for “dummy” models generated by us using standard NMR structure determination techniques (see Matherials and Methods for details). The GDT_TS scores of these dummy models produced with all contacts, unambiguous contacts, and ‘Ts-specific’ true contacts are colored (from bottom to top) cyan, purple, and green, respectively, and shown as solid lines to aid visualization. Dummy model performance (colored lines) is compared to prediction model performance (GDT_TS) for all groups (blue circles), with the top simulated NMR Ts prediction models (solid red line) outperforming the top dummy models for all targets.
Additionally, the nature of the crosslinking agents could result in an uneven distribution on the structures. This notion might lead to the relatively low average coverage of the correct contacts noted for the category (Figure 7, crosslinked Tx CCC is 0.16). Thus, the crosslinked Tx category experiment provided a fundamentally different type of contact information, as residues must be accessible to the crosslinking reagent (i.e. relatively exposed) and might be more distant (> 8 Å) than the traditional concept of contacting residues. Perhaps including such restrictions in methodology for using crosslinked Tx contacts would improve the quality of structure models.
To gain further insights into the quality of Ts predictions, we compared Ts models generated by predictors to ‘dummy models’ generated by us using standard NMR structure determination software. To generate dummy models, we used one of most cited NMR packages16, the NMR routines in the Crystallography and NMR System (CNS). The CNS package utilizes the distance restraints in simulated annealing protocol to produce a model most compatible with these restraints. The average number of contacts per target given to predictors in Ts category was 14724 hydrogen pairs, corresponding to 9283 residue pairs (Figure 7B, dark and light cyan bars). This number far exceeds that given in other contact-assisted categories. For instance, the largest number of contacts per target from any of the other three categories is only 673 residue pairs (Tp814). However, the overwhelming majority (about 98.5%) of these contacts is “ambiguous”, and the NMP peak is usually assigned to multiple atom pairs. When all given Ts contacts (ambiguous and unambiguous) are used as input, CNS package generated dummy models with approximately random GDT_TS scores for each Ts target (average GDT_TS = 13.56, Figure 7C cyan line), close to some of the worst predictions. Apparently, the ambiguity of the contacts hindered the reconstruction of the structures by CNS, and most predictors found a more clever way to deal with ambiguities.
We next attempted to reduce the ambiguity provided to the CNS software. As the first step, we used only unambiguous contacts, i.e., those for which distance constraint corresponded to a single given pair of atoms. While this method of contact selection does not require the knowledge of the target structure and could have been used by predictors, it comes at the cost of losing most of Ts contact information, because unambiguous assignments corresponded to an average of 1.2% (by atom) / 1.7% (by residue) of the total Ts contacts (Figure 7B, dark and pale purple bars). With unambiguous contacts being the only input, the CNS package generated dummy models with 29.3 GDT_TS score on average (Figure 7C, purple line). Dummy models from five targets predicted the correct fold and achieved GDT_TS above 40 (maximal GDT_TS = 53.8 for target Ts812). Therefore, although the number of unambiguous contacts was limited, those contacts were mostly correct (98.6% of unambiguous atom pairs are correct) and could be used to generate reasonable seed structures for further refinement. Interestingly, many of the CASP simulated NMR Ts predictions (Figure 7C, blue dots) had GDT_TS scores lower than the dummy structures generated from unambiguous contacts by CNS, suggesting that these groups could have benefitted from including standard NMR structure determination software in their methodologies.
Because assessors are granted access to the target structures, we further attempted to disambiguate ambiguous contacts using the knowledge of the target structure. We selected all the correct constraints in the provided simulated NMR Ts contacts to evaluate the theoretical upper limit of the CNS performance. For the purpose of cross-category comparison in previous section calculating CCP and CCC (Figure 7A), the correct contacts were defined as those with Cβ distance no more than 8 Å. Here, we extracted the cutoff for the ‘Ts-specific’ true contacts from the upper limit (UPL) of the atomic distance for individual atom pairs provided by the simulated NMR data, resulting in an average of 1041 correct atom pairs in 625 correct residue pairs (Figure 7B, dark and pale green bars). This definition was slightly higher than the number of correct contacts computed in the cross-category comparison (586 residue pairs, Figure 7B, medium green bar). The dummy models generated by CNS using those ‘Ts-specific’ true contacts produce GDT_TS scores ranging from 43 to 75, with an overall average of 58 (Figure 7C, green line). Impressively, many predictions achieved better performance than the structures built from the true distance constraints selected with the knowledge of the target structure. The best predictions for every target outperform the dummy models obtained by CNS using true contacts. Although the lack of chemical shifts in Ts contacts provided to predictors limits the utilization of the NMR package to its full potential, the structure prediction methods seemed to utilize additional information to push the limit of the NMR methods based purely on the distance constraints. These best prediction methods should be useful for NMR researchers in protein structure determination and may have some advantages over the CNS package.
CASP11 exhibited a number of significant differences in the implementation of the contact-assisted category experiment when compared to the previous CASP10. These differences made evaluation of performance improvement difficult. Performance of the correct Tc categories from both CASPs was roughly dependent on the number contacts given per residue (Figure 6A). Given the artificial nature of the correct Tc category, perhaps future contact-assisted experiments could explore the correlation between given contacts per residue and top structure prediction performance by incrementally providing sets of correct Tc contact pairs over time. At the very least, this category should include more consistently defined contact pairs between CASP experiments to allow methods performance comparisons over time.
References
- 1.Bowers PM, Strauss CE, Baker D. De novo protein structure determination using sparse NMR data. J Biomol NMR. 2000;18(4):311–318. doi: 10.1023/a:1026744431105. [DOI] [PubMed] [Google Scholar]
- 2.Kim DE, Dimaio F, Yu-Ruei Wang R, Song Y, Baker D. One contact for every twelve residues allows robust and accurate topology-level protein structure modeling. Proteins. 2014;82(Suppl 2):208–218. doi: 10.1002/prot.24374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taylor TJ, Bai H, Tai CH, Lee B. Assessment of CASP10 contact-assisted predictions. Proteins. 2014;82(Suppl 2):84–97. doi: 10.1002/prot.24367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Monastyrskyy B, D’Andrea D, Fidelis K, Tramontano A, Kryshtafovych A. Evaluation of residue-residue contact prediction in CASP10. Proteins. 2014;82(Suppl 2):138–153. doi: 10.1002/prot.24340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chance MR, Bresnick AR, Burley SK, Jiang JS, Lima CD, Sali A, Almo SC, Bonanno JB, Buglino JA, Boulton S, Chen H, Eswar N, He G, Huang R, Ilyin V, McMahan L, Pieper U, Ray S, Vidal M, Wang LK. Structural genomics: a pipeline for providing structures for the biologist. Protein Sci. 2002;11(4):723–738. doi: 10.1110/ps.4570102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kinch LN, Li W, Schaeffer RD, Dunbrack RL, Monastyrskyy B, Kryshtafovych A, Grishin IV. CASP 11 Target Classification. Proteins. 2016 doi: 10.1002/prot.24982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zemla A. LGA: A method for finding 3D similarities in protein structures. Nucleic Acids Res. 2003;31(13):3370–3374. doi: 10.1093/nar/gkg571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ben-David M, Noivirt-Brik O, Paz A, Prilusky J, Sussman JL, Levy Y. Assessment of CASP8 structure predictions for template free targets. Proteins. 2009;77(Suppl 9):50–65. doi: 10.1002/prot.22591. [DOI] [PubMed] [Google Scholar]
- 9.Jauch R, Yeo HC, Kolatkar PR, Clarke ND. Assessment of CASP7 structure predictions for template free targets. Proteins. 2007;69(Suppl 8):57–67. doi: 10.1002/prot.21771. [DOI] [PubMed] [Google Scholar]
- 10.Kinch L, Yong Shi S, Cong Q, Cheng H, Liao Y, Grishin NV. CASP9 assessment of free modeling target predictions. Proteins. 2011;79(Suppl 10):59–73. doi: 10.1002/prot.23181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kinch LN, Wrabl JO, Krishna SS, Majumdar I, Sadreyev RI, Qi Y, Pei J, Cheng H, Grishin NV. CASP5 assessment of fold recognition target predictions. Proteins. 2003;53(Suppl 6):395–409. doi: 10.1002/prot.10557. [DOI] [PubMed] [Google Scholar]
- 12.Kopp J, Bordoli L, Battey JN, Kiefer F, Schwede T. Assessment of CASP7 predictions for template-based modeling targets. Proteins. 2007;69(Suppl 8):38–56. doi: 10.1002/prot.21753. [DOI] [PubMed] [Google Scholar]
- 13.Vincent JJ, Tai CH, Sathyanarayana BK, Lee B. Assessment of CASP6 predictions for new and nearly new fold targets. Proteins. 2005;61(Suppl 7):67–83. doi: 10.1002/prot.20722. [DOI] [PubMed] [Google Scholar]
- 14.Cozzetto D, Kryshtafovych A, Fidelis K, Moult J, Rost B, Tramontano A. Evaluation of template-based models in CASP8 with standard measures. Proteins. 2009;77(Suppl 9):18–28. doi: 10.1002/prot.22561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tramontano A, Morea V. Assessment of homology-based predictions in CASP5. Proteins. 2003;53(Suppl 6):352–368. doi: 10.1002/prot.10543. [DOI] [PubMed] [Google Scholar]
- 16.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 17.Monastyrskyy B, D’Andrea D, Fidelis K, Tramontano A, Kryshtafovych A. New encouraging developments in contact prediction: Assessment of the CASP11 results. Proteins. 2015 doi: 10.1002/prot.24943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kinch LN, Li W, Monastyrskyy B, Kryshtafovych A, Grishin IV. Evaluation of free modeling targets in CASP11 and ROLL. Proteins. 2015 doi: 10.1002/prot.24973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng H, Schaeffer RD, Liao Y, Kinch LN, Pei J, Shi S, Kim BH, Grishin NV. ECOD: an evolutionary classification of protein domains. PLoS Comput Biol. 2014;10(12):e1003926. doi: 10.1371/journal.pcbi.1003926. [DOI] [PMC free article] [PubMed] [Google Scholar]


