Abstract
Deubiquitinating enzymes (DUBs) remove ubiquitin and ubiquitin-like modifications from proteins and they have been known to contribute to processes relevant in microbial infection, such as immune responses pathways. Numerous viral and bacterial DUBs have been identified, and activities of several host DUBs are known to be modulated during the infection process, either by a pathogen or by a host. Recently there have been attempts to take advantage of this feature and design therapeutic inhibitors of DUBs that can be used to limit the spread of infection. This review is focused on exploring the potential of DUBs in the treatment of infectious diseases.
Keywords: deubiquitinating enzymes, drug discovery, ubiquitin, infectious diseases, post-translational modifications
1. Introduction
The pathogenic organisms corrupt critical host biochemical pathways in order to enhance their survival within the host. On the other hand, the same molecular functions are often important in the immune response. The lack of antimicrobials for some pathogenic diseases as well as the multidrug-resistance of other pathogens mean that further efforts to design appropriate drugs are necessary, and here detailed understanding of host-pathogen interactions is required [1]. Many of the critical molecular mechanisms are regulated by protein ubiquitination, which is a post-translational modification that is essential for the fate and function of the cellular proteins [2, 3]. Ubiquitin and ubiquitin-like proteins regulate key biological processes, such as protein turnover, receptor internalization, protein localization, enzymatic activity and cell signaling. The removal of this post-translational modification is equally important to its attachment, and is catalyzed by deubiquitinating enzymes (DUBs). In humans, there are approximately ninety-five predicted DUBs [4], and this diversity indicates that these enzymes have precise and non-redundant functions as well as specificities towards different substrates and different ubiquitin chains. There are two main classes of DUBs, cysteine proteases and metalloproteases, The cysteine proteases are further classified into the ubiquitin-specific processing proteases (USP/UBP), the ovarian tumor domain-containing proteases (OTU), the Machado-Josephin domain (MJD) DUBs, and the ubiquitin C-terminal hydrolases (UCH). The pathogens evolved to interfere with this tightly controlled system, especially since many DUBs play important functions in immune responses [5].
We have previously described some of the functions of DUBs in the infectious diseases [6], concentrating on the evolutionary origins of the pathogen-encoded DUBs. Several recent reviews also describe the potential therapeutic use of ubiquitin proteasome pathways and, specifically, deubiquitinating enzymes in anti-cancer drug design [7–10]. In this review we will describe recent research with regards to the function of DUBs in infectious diseases, and we will then discuss possibilities to use this knowledge in the antimicrobial drug design.
2. The host immune responses regulated by DUBs
Innate and adaptive immune responses are both important functions that eliminate pathogens, and where the protein deubiquitination has been shown to be critical [11].
CYLD is a DUB controlling inflammatory responses via regulating such signalling cascades as nuclear factor kappa B (NF-κB) pathway [12–14]. Specifically, it has been shown to down-regulate NF-κB activation through deubiquitination of TRAF2, 6, and 7, therefore preventing the formation of an antiviral response [14–17]. E3 ligase Itch and deubiquitinase CYLD form a complex that cleaves lysine (Lys) 63-linked ubiquitin chains and catalyze Lys48-linked ubiquitination on the kinase Tak1, which is a common substrate for these two proteins, thereby contributing to decreased inflammatory signalling [18]. Furthermore, a conditional gene targeting approach was used to create CyldΔ9 mice, which has enhanced cell resistance to LPS-induced endotoxic shock, while the CYLD-deficient macrophages failed to accumulate to the inflammatory site. These results suggest potential benefits of targeting CYLD with inhibitors in control of certain forms of pathogenic inflammation [19].
The second example of a DUB involved in regulation of inflammation and immunity is A20, which is an NF-κB-responsive gene. Apart from being a DUB, A20 also contains an E3 ubiquitin ligase domain, therefore constituting a ubiquitin-editing protein, which is involved in the negative feedback regulation of the NF-κB signaling [20, 21]. As a ubiquitin-editing protein, A20 deubiquitinates Lys63-linked chains on RIP1 and at the same time via its E3 ubiquitin ligase activity it promotes Lys48-linked RIP1 polyubiquitination, contributing to its degradation, possibly with the aid of Itch (similarly as described in the paragraph above), RNF11, and TAXBP-1, which are all parts of the A20 ubiquitin-editing protein complex [22]. However, A20 also inhibits the E3 ligase activities of TRAF2, TRAF6, and cIAP1 by preventing interactions with the E2 ubiquitin conjugating enzymes Ubc13 and UbcH5c, with which proteins A20 has been shown to interact [23].
Deubiquitinating enzyme A (DUBA) also controls inflammation [24]. It selectively cleaves Lys63-linked ubiquitin chains from TRAF3, and it is upregulated in the absence of IL-1R1 signaling. In cells where expression of DUBA is knocked-down, the TLR9-dependent type interferon (IFN) response was increased, therefore DUBA is a negative regulator of innate immune responses [24, 25].
Another DUB, USP4 has been shown to be a negative regulator of Toll-like/interleukin-1 receptor (TLR/IL-1R) signaling, and inhibit IL-1β and LPS-induced IκBα phosphorylation. USP4 removes Lys63-conjugated polyubiquitin from TRAF6 thus preventing NF-κB activation and proinflammatory responses [26].
Apart from deubiquitinating enzymes, proteases specific for ubiquitin-like molecules have also been shown to regulate immunity. Many viral and bacterial infections lead to expression of interferon stimulated gene interferon-induced 17 kDa protein (ISG15), which is a ubiquitin-like protein, and ISG15-specific protease UBP43 (USP18) [27, 28]. In addition, UBP43 was shown to regulate IFN-beta stimulated genes at a genome level, including genes important in antigen presentation and antiviral responses, as well as genes relevant in chemokine and cytokine production. As such, this DUB could be a drug target for the enhancement of immune responses in microbial infections [29].
Furthermore, small ubiquitin-like modifier (SUMO)-specific protease 1 (SENP1) is important in the development of early T and B cells via regulating sumoylation of STAT5, which controls lymphoid development [30]. Moreover, SENP2, another SUMO-specific protease, also plays function in immunity. SENP2 negatively regulates virus-triggered IFN-β induction by deSUMOylating IRF3 and targeting it for Lys48-conjugated ubiquitination and degradation [31]. Also, SENP2 has been shown to deSUMOylate NEMO, which is an essential modulator of NF-κB, and this deSUMOylation process inhibits NF-κB activation [32], [33].
Finally, another example is Cezanne, an enzyme belonging to OTU-containing domain DUBs. It has been found that TNFα can induce Cezanne, and that Cezanne attenuates NF-κB activation. Specifically, Cezanne has been suggested to deubiquitinate RIP1, thereby suppressing TNFR signalling to NF-κB [34].
3. DUBs in viral infections
Ubiquitination alters the function of a number of viral proteins necessary for infection as well as host anti-viral response mechanisms. In this section we will first describe host DUBs known to target viral proteins, followed by a description of viral DUBs that interact with the host (Table 1 and 2).
Table 1.
Host-encoded deubiquitinating enzymes relevant in infection.
| Pathogen | Host DUB | Pathogen protein | Effects | Reference |
|---|---|---|---|---|
| Bacteria | ||||
| Escherichia coli | CYLD | Negative regulator for NF-kappaB, CYLD is a negative regulator for E. coli pneumonia | 86 | |
| Francisella tularensis | USP22 | USP22 expression is required for bacterial replication in the host | 82 | |
| Helicobacter pylori | USP7 | H. pylori infection is associated with decreased levels of USP7 | 81 | |
| Listeria monocytogenes | UCHL1 | UCHL1 promotes bacterial internalization, induction of actin stress fibers | 80 | |
| non-typeable Haemophilus influenzae | CYLD | Negative regulator for Haemophilus influenza-induced inflammation | 84, 85 | |
| Pseudomonas aeruginosa | USP10 | Cif toxin | Cif indirectly inhibits USP10 by stabilization of the USP10 and G3BP1 interaction, which reduces interaction between USP10 and CFT | 88 |
| Salmonella enterica | UCHL1 | UCHL1 promotes bacterial internalization, induction of actin stress fibers | 80 | |
| Streptococcus pneumoniae | CYLD | CYLD inhibits MKK3-p38 kinase-dependent expression of plasminogen activator inhibitor-1 (PAI-1) in lung, thereby potentiatingI-induced lethality | 87 | |
| Yersinia enterocolitica | OTUB1 | YopO | OTUB1 increases bacterial internalization | 77 |
| Yersinia pseudotuberculosis | OTUB1 | YpkA | OTUB1 increases bacterial internalization | 77 |
|
| ||||
| Protozoa | ||||
| Leishmania donovani | A20 | Leishmania regulates A20 to inhibit the TLR2-mediated proinflammatory gene expression | 110 | |
|
| ||||
| Viruses | ||||
| Dengue Virus (DENV) | USP14 | Inhibits DENV replication | 40 | |
| Epstein-Barr virus (EBV) | USP7 | EBNA-1 | Enhances EBNA1-mediated transcriptional activation | 44–46 |
| Hepatitis B virus (HBV) | UBP43 | Impacts HBV growth | 28 | |
| Herpes simplex type-1 (HSV-1) | USP7, USP7β | ICP0 | Stabilizes ICP0 and terminates TLR dependent antiviral signaling | 41, 42 |
| Human papillomavirus (HPV) | USP11 | E7 | Stabilizes E7 protein by preventing proteasomal degradation | 47 |
| Human papillomavirus (HPV) | USP15 | E6 | Stabilizes E6 protein by preventing proteasomal degradation | 48 |
| Influenza A virus | USP11 | Viral RNP complex, specifically NP | Deubiqutinates NP and results in the inhibition of viral replication | 36 |
| Influenza A virus | A20 | Inhibits RIG-I-induced NF-kB and IRF3 activation, and negatively regulates IAV induced gene expression. | 35 | |
| Kaposi’s sarcoma herpesvirus (KSHV) | USP7 | LANA | Regulates of viral latent replication | 43 |
| Murid herpesvirus 68 (MHV68) and Rhesus monkey rhadinovirus (RRV) | USP7 | LANA homologs | 43 | |
| Poliovirus | A20 | Negative regulator of poliovirus growth | 37 | |
| Porcine reproductive and respiratory syndrome (PRRSV) | USP18 | Restricts viral growth | 38 | |
| Sendai virus (SeV) | USP17 | Required for Sendai virus (SeV)-induced expression of IFN | 51 | |
| Sendai virus (SeV) and Vesicular stomatitis virus (VsV) | OTUB1 and OTUB2 | TRAF3 and TRAF6 | OTUB1 and OTUB2 negatively regulate virus-triggered type I IFN induction by deubiquitinating TRAF3 and TRAF6 | 52 |
| T cell leukemia virus type 1 (HTLV-1) | USP20 | Tax | Negative regulator of Tax-induced NF-κB signaling | 50 |
| T cell leukemia virus type 1 (HTLV-1) | STAMBPL1 | Tax | Controls Tax nuclear export, and Tax mediated cytoplasmic activation of NF-kB | 49 |
Table 2.
Pathogen-encoded deubiquitinating enzymes.
| Pathogen | Pathogen DUB | Effects | References | |
|---|---|---|---|---|
| Bacteria | ||||
| Chlamydia trachomatis | ChlaDub1 | suppresses NF-kappaB activation and inhibits IkappaBalpha ubiquitination and degradation | 102,103 103 | |
| Burkholderia mallei | TssM | cleaves both Lys48- and Lys63-linked ubiquitin dimers | 105 | |
| Burkholderia pseudomallei | TssM | TssM down-regulates inflammatory responses by inhibiting NF-κB and type I IFN pathway activation; interferes with the ubiquitination of TRAF6 and IkappaBalpha | 104 | |
| Chlamydia trachomatis | ChlaDub2 | unknown | 102 | |
| Escherichia coli | ElaD | present in all intestinal pathogenic E. coli strains, but conspicuously absent from extraintestinal pathogenic strains | 101 | |
| Mycobacterium tuberculosis | Dop | Dop, a depupylase, regulates proteasomal degradation in Mycobacterium | 108,109 | |
| Salmonella enterica | SseL | inhibits selective autophagy of cytosolic aggregates, suppresses IkappaBalpha ubiquitination and modulates innate immune response; required for macrophage killing and virulence | 90–92 | |
| Salmonella enterica | AvrA | mediates bacterial intracellular survival during infection | 93–96 | |
| Yersinia sp. | YopJ | targets TRAF proteins to inhibit TLR-mediated NF-kappaB, MAPK and IRF3 signal transduction | 95, 97–100 | |
|
| ||||
| Fungi and parasites | ||||
| Cryptococcus neoformans (yeast) | Ubp5 | essential for the virulence composite of C. neoformans | 118 | |
| Plasmodium falciparum | PfUCHL3 | essential to parasite survival | 112, 114 | |
| Plasmodium falciparum | PfUCH54 | 115 | ||
| Plasmodium falciparum | UBP-1 | Association between drug resistance in Plasmodium and a locus containing a UBP-1; UBP-1 is homologous to human USP7 | 116 | |
| Plasmodium falciparum | PfSENP1 (PFL1635w) | PfSENP1 is a SUMO-specific protease | 117 | |
| Toxoplasma gondii | TgUCHL3 | 112 | ||
| Trichinella spiralis | TsUCH37 | interacts with the proteasome | 111 | |
|
| ||||
| Viruses | ||||
| Crimean-Congo hemorrhagic fever virus (CCHFV) | L-protein | Targets RIG-1 and inhibits RIG-I-mediated innate immune signaling | 71, 72 | |
| Epstein-Barr virus (EBV) | EBNA3C | Deubiquitinates proto-oncogene Mdm2, affects viral lytic transcription | 56 | |
| Epstein-Barr virus (EBV) | BPLF1 | Disrupts PCNA mediated DNA damage repair, reduces ribonucloetide reductase activity, promotes the selective degradation of cullins by the proteasome, and its deneddylase activity regulates virus production | 57–61 | |
| Equine arteritis virus (EAV) | nsp2 | Targets RIG-1 and inhibits RIG-I-mediated innate immune signaling | 71 | |
| Foot-and-mouth disease virus (FMDV) | Lpro | Acts as an inhibitor of type I IFN signaling | 70 | |
| Hepatitis B virus | HBx | Inhibits type I IFN production | 62 | |
| Hepatitis E virus (HEV) | pORF1 | Deubiquitinates and deISGylates proteins involved in antiviral pathways | 73 | |
| Herpes simplex virus 1 (HSV) | VP1-2 | Autocatalytic DUB property of VP1-2 is required for the stability of this protein | 53, 54 | |
| Human coronavirus (HCoV) | NL63 | Disrupts STING dimerization, and STING-mediated IFN induction | 68 | |
| Human cytomegalovirus (HCMV) | UL48 | Can affect viral growth at low multiplicity of infection | 55 | |
| Kaposi’s sarcoma-associated herpesvirus (KSHV) | Orf64 | Necessary for viral replication, and suppression of RIG-I-mediated host IFN signaling | 64 | |
| Lactate dehydrogenase elevating virus (LDV) | nsp2 | Targets RIG-1 and inhibits RIG-I-mediated innate immune signaling | 71 | |
| Murine gamma herpesvirus 68 (MHV-68) | Orf64 | Necessary for viral replication | 65 | |
| Nairobi sheep disease virus (NSDV) | Lprotein | Delays IFN production and action, inhibits STAT1 and STAT2 phosphorylation, and deISGylates cellular proteins | 76 | |
| Porcine respiratory and reproductive syndrome virus (PRRSV) | nsp2 | Targets RIG-1 and inhibits RIG-I-mediated innate immune signaling, and ISG15 conjugation to cellular proteins | 71 | |
| Porcine respiratory and reproductive syndrome virus (PRRSV) | nsp2 | Inhibits nuclear translocation of IRF-3, antagonizing IFN induction | 74, 75 | |
| Pseudorabies virus (PrV) | pUL36 | Reduces PrV replication and delayed neuroinvasion | ||
| Severe acute respiratory syndrome coronavirus (SARS-CoV) | PLpro | Antagonizes a number of IFN-γ-induced responses including the expression of OAS, IL-6, IL-8, and phosphorylation of STAT-1 and c-Jun | 66–69 | |
| Simian hemorrhagic fever virus (SHFV) | nsp2 | Targets RIG-1 and inhibits RIG-I-mediated innate immune signaling | 71 | |
3.1. Host DUBs relevant in viral infections
Host pattern recognition receptors TLR3 and RIG-I are important sensors of Influenza A virus (IAV) during infection. Of these two, innate antiviral immune response towards IAV depends predominantly on RIG-I mediated signaling (RLR). Ligand binding results in the association of RIG-I with Mitochondrial antiviral-signaling protein (MAVS), which gets inserted in the outer mitochondrial membrane. Downstream signaling involves the activation of NF-κB and IRF3. DUBs negatively regulate RLR signaling by removing Lys63-linked polyubiquitin chains from several signaling molecules. A host DUB A20 has recently been shown to be a negative regulator of IAV-induced antiviral signaling in macrophages. In response to IAV infection, A20 knockout mice survived better and were less morbid compared to the wild type. This effect was marked by increased cytokine and chemokine production as well as enhanced viral clearance [35]. Furthermore, RNAi screening identified a host DUB USP11 that inhibits IAV RNA replication. Knockdown of cellular USP11 increased the virus titer 10-fold, and USP11 was shown to interact with the viral RNP complex, which include NP protein. NP gets monoubiqutinated at Lys184, which was shown to increase its RNA binding affinity. It is NP monoubiquitination that is targeted by USP11’s DUB activity, thus affecting viral RNA replication [36].
Poliovirus inhibits transcription and translation of host cell mRNAs during infection. One plausible mechanism for this inhibition is the cleavage of several cellular transcription factors such as CREB, Oct1 and TBP affecting a number of cellular pathways. While transcription of up to 80% genes is inhibited by poliovirus, certain classes of cellular mRNAs were synthesized, which code for genes relevant to innate immune response. Some of the transcriptionally induced genes include IκBα, A20, CCL2, IL-6, IFIT1, IFIT2, SOD2, JUNB, and ISG15. A host DUB A20 is a known to be a negative regulator of NF-κB and is also itself a target of NF-κB. A20 was shown to have a negative effect on poliovirus growth as its depletion resulted in an increase in the production of infectious virus [37].
Antiviral response against porcine reproductive and respiratory syndrome (PRRSV) involved the host DUB USP18. USP18 is a negative regulator of type-I IFN signaling and recent studies have shown that overexpression of this gene restricts PRRSV growth. The inhibition by USP18 could be the result of cellular re-distribution of the p50 (predominantly cytoplasmic) and p65 (predominantly nuclear) subunits of NF-κB [38] that impact the NF-κB function.
Hepatitis B virus (HBV) infections are confined to humans and chimpanzees. IFN-α/β stimulation during infection up-regulates ISG-15, which is a ubiquitin-like protein. ISG15 can exist in free form or is conjugated to cellular proteins. ISGlyation is similar to ubiquitin conjugation pathway and is important for carrying out antiviral functions in infected cells. An IFN-inducible ubiquitin protease 43 (UBP43) is a known deISGylating protein. Transgenic mice that mimic acute HBV infection have been described and were used to study HBV. Studies with mice deficient in UBP43 and with UBP43-silencing shRNA showed reduced levels of HBV at steady state. This anti-HBV effect of UBP43 was found to be independent of its deISGylation function. Nevertheless, these results demonstrate the therapeutic potential of UBP43 in combating HBV infections, a major health problem worldwide [28].
USP14 is a proteasome-associated deubiquitinating enzyme, which can inhibit the degradation of polyubiquitin, and inhibition of USP14 can lead to an enhanced proteasomal activity [39]. Inhibition of USP14 has been found to impair Dengue Virus (DENV) replication. DENV causes fatal Dengue hemorrhagic fever. Nearly 2.5 billion people are at risk of DENV infection. Due to the lack of effective vaccination against this virus, compounds that target DUBs required for DENV replication in host have immense therapeutic potential. One such drug, IU1 targets host DUB USP14 and inhibits DENV replication, although the mechanism of USP14 in this infection is not yet known [40].
Some host DUB interactions with viral proteins promote viral infections. A well-studied host DUB-viral protein interaction is that of USP7 (HAUSP) and ICP0. ICP0 is an immediate early protein of Herpes simplex type-1 (HSV-1), which is RING-finger E3 ubiquitin ligase. ICP0 is a key viral protein that is required for HSV-1 lytic replication as well as for reactivation from latency. The E3 ubiquitin ligase activity of ICP0 targets a number of cellular proteins for proteasomal degradation, including the host DUB USP7, and this in turn could favor or repress transcriptional quiescence. ICP0 can auto catalyze its own ubiquitination. Deubiquitinating activity of USP7 and USP7β, a higher molecular weight isoform stabilize this viral trans-activator during infection [41]. USP7 is also known to regulate TLR signaling, which results in the activation of NF-kB and JNK to combat HSV infections. Upon TLR activation, USP7 is exported from nucleus to the cytoplasm where it deubiquitinates TRAF6 and IKKγ, terminating the TLR-dependent immune response. ICP0 utilizes this regulatory aspect of USP7, mediates the nuclear export of USP7, to abrogate TLR-dependent host antiviral responses [42].
USP7 has been also found to be important in Kaposi’s sarcoma herpesvirus (KSHV) infection. The viral latency-associated nuclear antigen 1 (LANA) is involved in latent replication of the virus, and it has been shown to interact with USP7. This replication occurs via its N-terminal TRAF [tumor necrosis factor (TNF) receptor-associated factor] domain and USP7-LANA interactions could lead to the regulation of viral latent replication. USP7 interaction with LANA homologues in MHV68 and RRV has also been shown, suggesting a widespread function of USP7 in infections with gamma-2 herpesviruses [43].
Moreover, USP7’s function has been also studied in Epstein-Barr virus (EBV) infection. EBV’s protein, Epstein-Barr nuclear antigen-1 (EBNA1), is a regulator of such functions as viral replication, segregation, and transcriptional activation of latent Epstein-Barr virus genomes. USP7 has been shown to interact with EBNA1, and this protein-protein interaction was found important in the regulation of EBNA1 replication activity, although EBNA1 is not stabilized by USP7 [44, 45]. USP7 also contributes to EBNA1-mediated transcriptional activation and stimulates the assembly of EBNA1 on oriP elements in transfected plasmids [46].
Human papillomaviruses (HPVs), specifically high-risk types such as HPV-16 or HPV-18 are linked to human cervical cancer. Two viral proteins E6 and E7, target a number of cellular proteins including p53 and pRb to alter host cell cycle, apoptosis and chromosomal stability, which could contribute to cellular transformation. A yeast two-hybrid based approach identified a cellular DUB USP11 as an interacting partner of HPV-16E7. DUB activity of USP11 stabilizes the E7 protein by preventing its degradation by the proteasome. Stabilization by USP11 can enhance the function of E7 with its target proteins, such as Rb [47]. In another study, a host DUB, USP15 was identified as HPV16 E6 partner via tandem affinity purification method using tagged HPV16 E6. Expression of USP15 resulted in an increase in the expression of E6 protein at least by eight-fold, indicating the host DUB-mediated stabilization of E6. This study did not identify any effects of the E6-USP15 interaction on p53 signaling pathway [48].
Human T-cell leukemia virus type 1 (HTLV-1) is the etiological agent of adult T-cell leukemia [49]. Persistent activation of NF-κB, a key mediator of host immune response, could result in disease. NF-κB activation is the mechanism utilized by Tax, a HTLV-1 oncoprotein for leukomagenesis, which plays an essential role in regulating viral gene expression and activation of host NF-κB signaling pathways that result in the expression of genes involved in antiviral response. Tax translocates between the cytoplasm and nucleus. Its sub-cellular localization that dictates its function is regulated by post-translational modifications including ubiquitination. Ubc13-dependent Lys63-linked polyubiquitination of Tax is necessary for its binding to IKKγ and persistent activation of IKK. Using siRNA library screen, STAMBPL1 (also known as AMSH-LP), a host DUB belonging to a metalloprotease family has been identified as a positive regulator of NF-κB activation mediated by Tax, by both canonical and non-canonical pathways. STAMBPL1 stabilized Tax indirectly by promoting its export from the nucleus to the cytoplasm. In the absence of STAMBPL1, Tax was degraded in the nucleus by the proteasome. Therefore, by controlling Tax trafficking in the cell, this DUB indirectly triggers IKK and NF-κB activation in the cytoplasm [49]. Furthermore, host DUBs that interact with HTLV-1Tax oncoprotein were investigated, and consequently ubiquitin-specific peptidase USP20 was found to deubiquitinate Tax and inhibit Tax-induced NF-κB activation [50].
A cDNA library screen encoding DUBs has identified USP17 as a DUB required for Sendai virus (SeV)-induced expression of IFN. The overexpression of USP17 inhibited SeV-induced activation of IFN-stimulated response element (ISRE), NF-κB, and the IFN-β promoter. Also, in cells where USP17 was knocked down, the polyubiquitination of RIG-I and MDA5 was increased, and the SeV-induced expression of RIG-I and MDA5 was decreased. Therefore it is possible that USP17 acts as a DUB towards these proteins, although indirect effects are also likely. Consequently, USP17 has been found necessary in Sendai virus-triggered type I IFN signaling [51].
Finally, OTUB1 and OTUB2, two DUBs belonging to OTU family have been found to be negative regulators of virus-triggered type I IFN induction. Knockdown of OTUB1 and OTUB2 inhibited vesicular stomatitis virus (VSV) replication. In contrast, overexpression of these proteins led to inhibition of Sendai virus (SeV)-induced activation of IRF3 and NF-κB activation. OTUB1 and OTUB2 interact with and deubiquitinate TRAF3 and TRAF6, which are required for virus-triggered IRF3 and NF-κB activation [52].
3.2. Viral DUBs
Viral encoded DUBs are important for viral replication and many of them are critical in circumventing host antiviral response. The tegument protein in alpha herpesviruses, UL36 has a conserved domain with ubiquitin-specific cysteine protease activity. Comparison of replication of wild type and C26S mutant pseudorabies virus (PrV) in showed that PrV-UL36 (C26S) plaque diameter was reduced by~40% and that activity of this DUB was important for the morphogenesis of the virus, specifically cytoplasmic maturation. Infection studies in mice showed an increase in the mean survival time by two fold with PrV-UL36 (C26S) compared to the wild type or rescue virus. This delay in death correlated with the delay in neuroinvasion of trigeminal ganglia. VP1-2, the product of the UL36 gene in Herpes simplex virus 1 (HSV), is a large multifunctional protein and one of a subset of tegument proteins, which is essential for virus replication and is conserved across the entire herpesvirus family. It contains N-terminal ubiquitin hydrolase domain (DUB) [53]. Autocatalytic property of VP1-2‘s ubiquitin-specific protease was shown to be required for the stability of this protein. However, HSV-1 DUB activity did not appear to have any effect on viral growth [54] (Table 2).
Human cytomegalovirus (HCMV) open reading frame UL48, is a tegument protein with ubiquitin-specific protease activity. Characterization of this DUB showed that it is more specific for Lys63-linked polyubiquitin substrates. UL48 active site mutants resulted in ten-fold fewer progeny at low multiplicity of infection, implicating significance of this DUB in viral replication [55].
More than 90% of adult population worldwide is infected by Epstein-Barr virus (EBV), a gamma herpesvirus, which can immortalize infected primary B cells. EBV encodes Epstein-Barr virus nuclear antigen 3C (EBNA3C), a latency protein with deubiquitinating activity that interacts with a cellular proto-oncogene Mdm2 for the transformation of EBV infected cells [56]. Mdm2 is overexpressed in 5–10% of human tumors, and this overexpression negatively regulates p53-mediated cell cycle arrest and apoptosis. Mdm2 binds to the trans-activation domain of p53 tumor suppressor protein, and it also has an E3 ubiquitin ligase activity towards p53, thus targeting it for degradation. EBNA3C stabilizes Mdm2 by deubiquitination and therefore enhances its E3 ligase activity for ubiquitination and degradation of p53, consequently enhancing EBV-mediated lymphomagenesis [56].
EBV late lytic gene BPLF contains the DUB activity within the first 205 amino acids. It is one of three DUBs identified in EBV by bioinformatics analyses [57]. An EBV interactome map built by using yeast two-hybrid screening identified EBV ribonucleotide reductase enzyme small subunit (RR2) as BPLF1 interaction partner. It was shown that BPLF1 deubiquitinates the large subunit of the ribonucloetide reductase (RR1) reducing RR activity. Thus BPLF1 can indirectly regulate viral replication, as a functional RR is important for virulence. In fact, expression of short hairpin RNA against BPLF1 was found to reduce the genomic copy number of EBV [58]. Recent evidence points to the role of BPLF1 in modulating host DNA damage repair response during translesion synthesis pathway (TLS), in response to stalled replication forks. Monoubiquitination of proliferating cell nuclear antigen (PCNA) enables the recruitment of polymerase η (Polη), for the replicative bypass of lesions. PCNA’s involvement in viral replication has been long known, although the specific mechanisms of PCNA in this process are not well characterized. BPLF1 deubiquitinates PCNA and disrupts the recruitment of TLS polymerase, thereby rendering cells more susceptible to the UV-induced DNA damage [59]. Also, the NEDD8-specific protease activity of BPLF1 has been shown to be important for the inactivation of cullin-RING ubiquitin ligases (CRLs), a class of E3 ligases in eukaryotes targeted by viral proteins for degrading cellular proteins. Assembly of CRL modular complex structure requires the modification of a conserved lysine residue in the cullin with NEDD8, which is a ubiquitin-like protein. Binding of active BPLF1 promotes the proteasomal degradation of deneddylated cullins by interfering with the recruitment of regulators such as CAND1 [60]. BPLF1 inhibition during the productive virus cycle inhibits cellular DNA re-replication and virus replication. Finally, BPLF1 and its homologues in other herpesviruses induce replication-permissive S-phase-like state in the cells, conducive for viral replication [61].
Hepatitis B virus (HBV) causes acute and chronic inflammatory liver diseases in humans. Despite the availability of a vaccine, there are close to 350 million persistent carriers of HBV. This virus interferes with host interferon production at multiple levels. HBx, a non-structural viral protein with DUB activity is HBV effector molecule. Due to its broad substrate specificity for Lys63-conjugated polyubiquitinated signaling molecules, HBx inhibits type-I IFN production induced by SeV infection. Binding of HBx resulted in the decoupling of retinoic acid-inducible gene I (RIGI) and TNF receptor-associated factor 3 (TRAF3) from the downstream adaptor CARDIF or TBK1 kinase. HBx was shown to interact with RIG I and TRAF3, as well as CARDIF, TRIF, NEMO, TBK1, inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon, and interferon regulatory factor 3 (IRF3), thereby affecting multiple signaling cascades involved in type I IFN induction [62].
The etiological agent of Kaposi’s sarcoma, Kaposi’s sarcoma-associated herpesvirus (KSHV) encodes a lytic protein shown to function as a DUB [63]. Orf64 encoded by KSHV is a viral DUB that targets both Lys48 and Lys63-linked ubiquitin chains. Functional analysis of Orf64 DUB was carried out by inducing lytic replication in latently infected cells. Orf64 knockdown by siRNA resulted in reduced lytic transcription and protein expression. Orf64 appeared to be important for KSHV lytic replication. KSHV persistency is observed in retinoic acid-inducible gene I (RIG-I)-deficient cells. ORF64 was shown to suppress RIG-I-mediated host IFN signaling by deubiquitinating RIG-I which prevents its activation and alter host response to KSHV [64]. Even in murine gamma herpesvirus 68 (MHV-68), a rodent pathogen, the DUB activity of ORF64 was demonstrated to be important for viral replication [65].
Papain-like protease (PLpro) is a viral DUB encoded by severe acute respiratory syndrome coronavirus (SARS-CoV) that causes highly contagious respiratory disease. Antiviral activity of type I interferon α/β involves JAK-STAT signaling pathways-mediated expression of antiviral genes such as PKR, OAS and a ubiquitin-like protein ISG15. Ubiquitination of key signaling molecules including JAK, STAT and ERK1 during cellular antiviral innate immune responses is an important regulatory step. Specific role of SARS-CoV PLpro in type I IFN response has been studied using human promonocytes [66]. SARS-CoV PLpro-expressing cells displayed reduced expression of IFN-α induced antiviral effector molecules, such as PKR and OAS. PLpro also affected AP-1 mediated responses to IFN-γ, as reflected in the reduced expression of IL-15 and IL-8 genes. Downregulation of ERK1 and modulation of its interaction with STAT1 marked type I interferon antagonism mediated by PLpro. Papain like protease encoded by the nsp3 protein of mouse hepatitis virus A59 (MHV-A59), a group II coronavirus phylogenetically related to SARS-CoV, was shown to inhibit IRF3 ubiquitination and prevent its nuclear translocation, thereby reducing type1 IFN induction and resulting in an increase in viral replication [67]. In contrast, studies with human coronavirus NL63 PLpro showed that the DUB activity of this protein was not required for interferon antagonism in the host cells [68], demonstrating the variegated nature of the function of this DUB in different viral infections. Another route that coronavirus PLpro proteins take to circumvent the host type-1 interferon response is via the disruption of STING mediated signaling [69]. Human coronavirus (HCoV) NL63 and SARS PLpro prevented the assembly and/or promoted the dissociation of STING dimers. Ubiquitination of STING dimers is required for its activation, and PLpro DUB activity targets this ubiquitination. This negative regulation of STING activation by PLpro inhibits STING-mediated activation of host antiviral responses, specifically the activation of IRF3 signaling and expression of IFN-β. In addition to STING, PLpro DUB activity targets other important host antiviral signaling molecules, such as RIG-I, TBK-1 and IRF3.
The leader proteinase (Lpro), a papain like protease is important for the pathogenesis of Foot-and-mouth disease (FMD). FMD is a highly contagious viral disease of cloven-hoofed wild and domestic animals, and is caused by FMD virus (FMDV). Lpro is known to inhibit the induction IFN-β and block the host innate immune response. Recent studies [70] showed that the DUB activity of Lpro targets key antiviral molecules involved in IFN signaling, such as RIG-ITBK1TRAF6, and TRAF3, thereby inhibiting the activation of type I IFN signaling in the host cells.
Papain-like protease family (PLP) of viral DUBs belonging to the ovarian tumor (OTU) domain containing subclass, were described for members of arterivirus and nairovirus families, and shown to target innate immune signaling in the host [71]. Arterivirus family has four species Equine arteritis virus (EAV), Porcine respiratory and reproductive syndrome virus (PRRSV), Lactate dehydrogenase-elevating virus (LDV), and Simian hemorrhagic fever virus (SHFV), of which PRRSV outbreaks in pigs cause significant economic losses. Crimean-Congo hemorrhagic fever virus (CCHFV), which causes hemorrhagic fever and results in 30% mortality in humans, belongs to the nairovirus family. In arteriviruses the DUB domain is in the N-terminal region of nonstructural protein 2 (nsp2 and is conserved an all members of this family. The nairovirus DUB domain resides in the N-terminus of the L protein. Since these DUBs belong to the OTU-domain containing DUBs, they were predicted to have a role in immune evasion supporting viral replication, and their function in modulating the pattern recognition receptor-mediated expression of type I interferons and proinflammatory cytokines was carried out. Induction of IFN–β in mouse embryonic fibroblasts by arterivirus RNA (EAV) is dependent on its recognition by pattern recognition receptor MDA5 and requires the adaptor protein MAVS. Overexpression of PLP DUBs from arterivirus and nairovirus families inhibited the ubiquitination of the pattern recognition receptor RIG-1, thus regulating RIG-1 mediated signaling. Unlike eukaryotic OTU-domain containing proteases, OTU DUBs from nairoviruses and arteriviruses also exhibit deISGylating activity. This viral subclass of DUBs can remove both ubiquitin and ISG15 from host proteins. In order to determine the structural basis for ISG15 cross–reactivity, the crystal structures of CCHFV OTU-domain protease domain covalently bound to Ub and to ISG15 was determined and compared to the eukaryotic OTU domain from yeast. This analysis clearly showed structural determinants that enable binding of ubiquitin and ubiquitin-like protein ISG15. These data can be used to design small-molecule-based inhibitors selective for viral, and not host OTUs [72].
Papain-like cysteine protease (PCP) of Hepatitis E virus (HEV) ORF1 protein (pORF1), a methyltransferase, was also shown to have both deubiquitinating and deISGylating activities. Interestingly, for pORF1, deISGylation activity was shown to be more pronounced than its deubiquitinase activity [73].
PRRSV non-structural protein 2 (nsp2) has a cysteine protease domain that strongly inhibits IFN-β production. The specific mechanism of this inhibition includes the regulation of the IFN regulatory factor 3 (IRF3) pathway induced by the Sendai virus (SeV). The protease domain of Nsp2 was found to be necessary for SeV-induced nuclear translocation of IRF3, and inhibition of IRF3 responsive gene expression [74]. A recent study with a panel of deletion mutants of PRRSV nsp2 DUB evaluated the effects on interferon I induced genes, specifically in counteracting the antiviral function of host ISG15 protein [75]. ISG-15 is induced by type I interferon. Overexpression of ISG15 and ISGylating enzymes negatively regulate the PRRSV replication. PRRSV DUB inhibited the expression of ISG15 and ISGylated proteins to counter their effects on viral replication. The DUB inhibited IFN I production itself rather than the inhibition of IFN-induced signaling pathways.
Nairobi sheep disease virus (NSDV), a member of the Nairovirus genus causes acute hemorrhagic gastroenteritis in sheep and goats. The N-terminus of the viral L protein has an ovarian tumor-like (OTU) domain that represents a DUB. NSDV inhibits IFN induction and its signaling pathways, and these activities have been attributed to the OTU-containing viral DUBs. NSDV L protein reduced ubiquitination and ISGylation in cells, while its catalytic activity was found to be necessary for its effects on interferon production [76].
4. DUBs in bacterial infections
4.1. Host-encoded DUBs with roles in bacterial infection
Human encoded DUBs have been shown to modulate infection with several pathogenic bacteria (Table 1).
First, we have shown that host cell uptake by Yersinia enterocolitica and Y. pseudotuberculosis depends on expression and catalytic activity of the human DUB OTUB1. We discovered that a human DUB OTUB1 regulates bacterial uptake into the monocytic and epithelial host cells. The mechanism by which OTUB1 increases bacterial uptake is currently not known, but we found that it depends on the OTUB1 substrate RhoA GTPase, and that this OTUB1 function might be obstructed by Yersinia kinase YpkA [77], which binds OTUB1, actin cytoskeleton, and RhoA [77], [78], [79]. Consequently, OTUB1 is potentially an important player in the cellular defense against Yersinia infection [77].
A human ubiquitin C-terminal hydrolase UCHL1 has been shown to promote Listeria monocytogenes and Salmonella enterica epithelial cell invasion and induce spontaneous formation of actin stress fibers, regulating early events in receptor signaling [80]. In particular, UCHL1 controls early membrane-associated pathways related to the L. monocytogenes entry into the host cell, such as activation of downstream ERK1/2- and AKT-dependent signaling in response to the Hepatocyte Growth Factor (HGF).
Moreover, Helicobacter pylori infection is associated with a decrease in the expression/activity of a human DUB, USP7, which coincides with decreased amounts of TRAF6 and p53 [81] partly explaining carcinogenesis during chronic H. pylori infection.
Francisella tularensis, a gram-negative intracellular bacterium that is a causative agent of tularemia (rabbit fever), requires a mammalian DUB USP22 for replication in cytosol of the host cells without affecting phagosomal escape [82]. The direct substrates of USP22 during bacterial infection have not been identified.
Another example is already mentioned CYLD, which is an important inflammatory mediator that deubiquitinates TRAF2/TRAF6, resulting in negative regulation of the NF-κB pathway [12, 13, 17, 83]. This DUB has been studied in several infection models. First, CYLD−/− mice are hypersensitive to infection by Non-typeable Haemophilus influenzae and CYLD expression is induced in cells infected by H. influenzae, down-regulating NF-κB [84, 85]. Similarly, an enhanced innate immune response is observed in CYLD−/− mice affected by Escherichia coli pneumonia [86], but CYLD has an adverse effect in Streptococcus pneumoniae infections, by provoking acute lung injury [87]. The example of CYLD having opposing effects in infection with three different bacteria highlights the importance of understanding mechanisms of action of DUBs before it will be feasible to use any of them as drug targets.
Pseudomonas aeruginosa, a gram-negative bacterium that causes lung infections, produces Cif toxin that reduces cystic fibrosis transmembrane conductance regulator (CFTR)-mediated chloride secretion by human airway epithelial cells. Cif has been found to be a regulator of CFTR deubiquitination in endosomes by indirect inhibition of USP10, which is a human DUB. Cif does this by stabilizing G3BP1-mediated inhibition of USP10, which then reduces USP10-mediated deubiquitination of CFTR and enhances degradation of CFTR in a lysosome [88]. The function of USP10 in endocytosis has been described earlier [89] and it is possible that USP10 might also be targeted by other pathogens that regulate CFTR, and thereby also mucociliary clearance, and as such compromising the immune defenses of the lung.
So far there has been no report of DUBs regulated during Shigella flexneri infection. We therefore profiled DUBs based on their binding to ubiquitin-specific active-site probes during Shigella infections. We infected HeLa cells with Shigella flexneri 2457T (wild-type), S. flexneri BS103 (non-invasive, virulence plasmid-cured) for one hour, or left uninfected. This was followed by preparation of protein extracts and incubation with bromoethyl ubiquitin probe (containing an HA tag), which is reacting with the active site of the DUBs. Samples were subjected to anti-HA immunoprecipitation to pull-down the active DUBs, tryptic digestion and quantitative mass spectrometry analysis via Orbitrap Velos (Figure 1, also see Supportive/Supplementary Material). This experiment revealed that certain DUBs are indeed regulated during early phases of Shigella infection in this epithelial cell model. Particularly, UCHL3 has been found to be significantly downregulated during infection with virulent strain, as compared to control cells, or cells infected with virulence plasmid-cured strain. A close homologue of this DUB, UCHL1, has been already shown to be involved in bacterial entry of Listeria monocytogenes and Salmonella enterica Typhimurium [80], therefore our current finding might be physiologically very interesting and warrants further study.
Figure 1. Proteomic activity profile of deubiquitinating enzymes (DUBs) reveal differential regulation during infection with Shigella virulent strain.
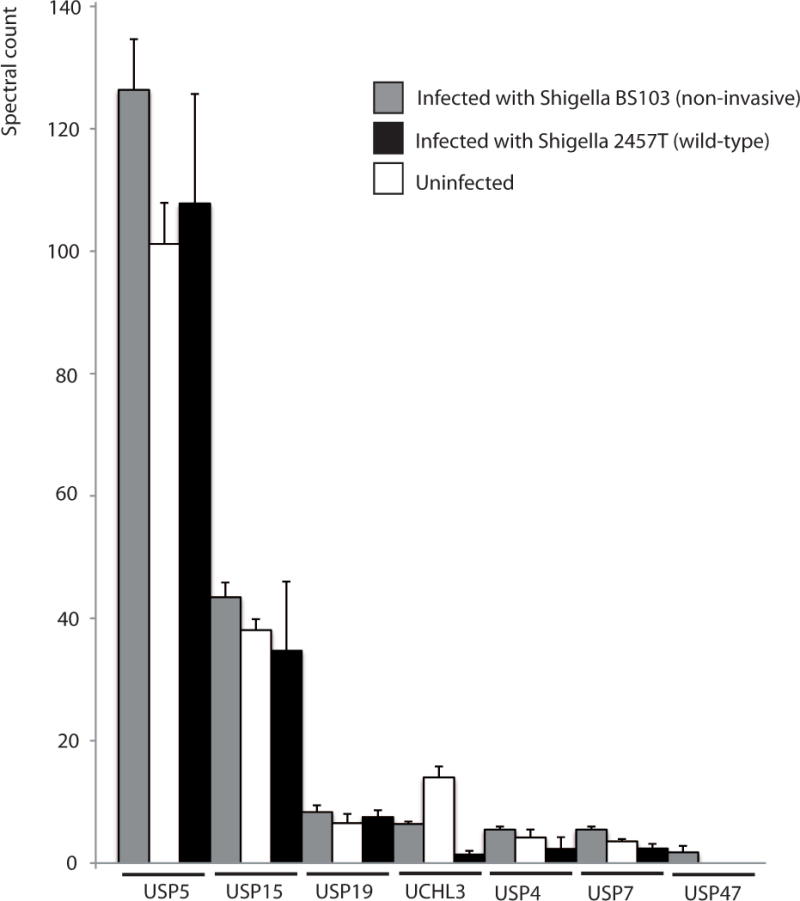
The HeLa cells were infected for one hour with Shigella flexneri virulent 2457T (wild-type) and S. flexneri BS103 (non-invasive, virulence plasmid-cured) strains (or left uninfected for control). The protein extracts from these cells were exposed to reaction with an active-site ubiquitin-specific DUB probe (bromoethyl ubiquitin, HA-tagged) and subjected to anti-HA immunoprecipitation to purify active DUBs bound to a probe, followed by quantitative mass spectrometry analysis using LC-MS/MS LTQ OrbiTrap. The results were analyzed by Proteome Discoverer 1.3 and Scaffold to identify statistically significant changes in DUB expression/activity. UCHL3 has been found to be downregulated in cells infected with wildtype Shigella.
4.2. Bacterial DUBs
Many bacteria also encode for DUBs (Table 2), and these include Salmonella-encoded SseL that hydrolyzes ubiquitinated substrates with preference for Lys63-linked ubiquitin linkages [90]. In terms of its physiological relevance, SseL targets the host cell ubiquitin pathway required for cytotoxicity of macrophages and for bacterial virulence [90]. Secondly, SseL suppresses the IκBα degradation in an unknown fashion, leading to increased inhibition of NF-κB, consequently blocking pro-inflammatory and anti-apoptotic pathways dependent on NF-κB, and allowing the pro-apoptotic effects through TRIF and PKR [91]. Also, SseL has been very recently shown to deubiquitinate as yet unidentified p62-bound proteins and thereby reduce the recruitment of the autophagic machinery [92]. This finding highlights the fact that the function of SseL in Salmonella infection is complex and it requires further characterization, especially in terms of its polyubiquitinated substrates that are part of the p62-complex.
AvrA is another Salmonella-encoded DUB that deubiquitinates IκBα and β-catenin [93], inhibiting the inflammatory responses. Other functions of AvrA are to regulate tight junction protein expression in intestinal epithelial cells. Tight junctions serve as a protective barrier and the role of AvrA might be to act as an anti-inflammatory protein to stabilize tight junctions and enhance bacterial survival in the host [94]. This protein is a homologue of YopJ in Yersinia sp., and both of these enzymes also function as acetyltransferases, and therefore their activity as DUBs has been questioned [95, 96].
In the light of research mentioned above, YopJ from Yersinia sp., with questionable DUB activity, has been nevertheless shown to cleave Lys63-conjugated polyubiquitin chains of TRAF2, TRAF3 and TRAF6 [97], [98], as well as Lys48-polyubiquitinated IκBα, thereby preventing its degradation and translocation of NF-κB [99]. As such, YopJ promotes intracellular survival of Yersinia by inhibiting NF-κB, MAPK and IRF3 signaling pathways [97]. Furthermore, YopJ was proposed to cleave sumoylated proteins [99], and also act as an acetyltransferase [95], [100]. It is possible that YopJ has dual activity and acts as an acetyltransferase and as a DUB, but further studies are necessary for verification.
ElaD is an orthologue of SseL in E. coli with as yet unidentified function. It has been previously misannotated as possible sulfatase/phosphatase. It is likely to be more specific for ubiquitin over ubiquitin-like protein substrates [101].
Chlamydia trachomatis encodes for ChlaDub1 and ChlaDub2, and both of these effectors have deubiquitinating and deneddylating activities [102]. In addition, ChlaDub1 has been shown to inhibit NF-κB activation and block IκBα ubiquitination and thereby its proteasomal degradation [103].
Burkholderia pseudomallei, a causative agent of melioidosis, downregulates inflammatory responses by inhibiting NF-κB and type I IFN pathway activation via its virulence factor TssM. TssM has been shown to be critical for B. pseudomallei infection in mice. TssM interferes with the ubiquitination of signaling proteins, such as TRAF-6 and IκBα, likely by direct deubiquitination of these molecules [104]. B. mallei’s TssM, which is 100% identical to B. pseudomallei’s TssM, has been shown to be an active DUB and cleave both Lys48- and Lys63-linked ubiquitin dimers in vitro [105].
Furthermore, until recently, detection of ubiquitin or ubiquitin-like modifiers in Prokaryota had not been successful. This lead to the conclusion that ubiquitin- or ubiquitin-like-mediated protein degradation is not present in those organisms and equivalent functions are performed via different pathways. However, recent findings suggest that bacteria do encode for a small protein modifier as a marker for degradation, which is a prokaryotic ubiquitin-like protein (Pup; Rv2111c) identified in Mycobacterium tuberculosis [106]. Unlike ubiquitin and its relatives that contain C-terminal glycine, Pup has a di-glycine motif at the penultimate position, but it ends in glutamine that becomes deamidated in the conjugated complex (reviewed in [107]). The enzyme (or one of the enzymes) responsible for removal of this modification is called a deamidase of Pup (Dop). Interestingly, it also functions as an enzyme facilitating pupylation, where it deamidates the carboxy-terminal glutamine of Pup before ligation of Pup to protein substrates. Dop also acts as a depupylase by cleavage of the isopeptide bond [108]. The function of Dop in infection is not characterized, but it does affect proteasomal degradation in Mycobacterium. Since it has no homologous counterparts in Eukaryota [109], it could constitute a good drug target in fighting tuberculosis.
5. DUBs in infections with pathogenic fungi and parasites
5.1. Host DUBs relevant in infection with fungi and parasites
Leishmania donovani is an intracellular pathogen causing visceral leishmaniasis that exploits a host DUB, A20, to inhibit TLR2-mediated signaling in macrophages. A20 was significantly upregulated in infected macrophages. In macrophages infected with Leishmania, an interference with A20 expression led to increased Lys63-linked polyubiquitination of TRAF6, as well as increased protein levels of IL-12 and TNF-α, but decreased levels of IL-10 and TGF-β. All this enables Leishmania to escape the immune responses of the host cell and increase its survival in the host [110] (Table 1).
5.2. Pathogen-encoded DUBs
Trichinella spiralis, a parasitic nematode that is a causative agent of trichnellosis, expresses a DUB, TsUCH37, which is an evolutionarily conserved proteasome-interaction partner and a homologue of a human DUB UCHL5. TsUCH37 associates with putative T. spiralis proteasome components, such as Rpn13 homologue ADRM1. Interestingly, the UCH inhibitor LDN-57444 specifically inhibited recombinant TsUCH37, and it also reduced the viability of cultured parasites if the larvae were directly treated with this drug [111], (Table 2).
A parasitic protozoa Toxoplasma gondii, which causes toxoplasmosis, encode TgUCHL3, an orthologue of human UCHL3. TgUCHL3 most likely has specificity towards ubiquitin and an ubiquitin-like molecule Nedd8 [112]. Three other DUBs have been also identified in this parasite on the basis of their interaction with the ubiquitin-specific active-site probe, but they have not been characterized thus far [112].
A bioinformatics study of Plasmodium falciparum identified approximately 29 DUBs, although the activities of these putative enzymes need to be confirmed [113]. Similarly as T. gondii, P. falciparum also encodes an orthologue of UCHL3, PfUCHL3 with specificities towards ubiquitin and Nedd8 [112]. PfUCHL3 is thought to be essential for parasite survival. Importantly, this DUB could be a possible target for selective drugs due to significant structural differences in ubiquitin binding between PfUCHL3 and its human homologue, as well as unique characteristic of PfUCHL3 to cleave Nedd8, which is absent from human UCHL3 [114]. PfUCH54 is another DUB in P. falciparum. It is an orthologue to human UCH37. PfUCH54 has specificity towards Nedd8 and ubiquitin, although this has only been tested by using the active-site probes [115]. Also, a strong genetic association has been found between drug resistance in Plasmodium and a locus containing a UBP-1, which encodes a DUB homologous to USP7 [116]. Plasmodium also encodes for PfSENP1 (PFL1635w), a SUMO-specific protease, which is likely to control Plasmodium replication in host blood [117].
Cryptococcus neoformans is a parasitic fungus causing meningoencephalitis and it encodes for several putative deubiquitinating enzymes, including Ubp5. A deletion of this gene caused attenuated virulence and elevated sensitivity to external stressors. Ubp5 might be a DUB regulating stress responses in C. neoformans, but other putative deubiquitinase mutants, such as Doa4 and Ubp13 could possibly share some of these functions, since deletion of these genes had some similar effects on a phenotype. Specific inhibitors for Ubp5 and for other Cryptococcus-encoded DUBs could provide benefits in the treatment of this fungal disease [118].
6. The perspectives for the use of DUBs in antimicrobial drug design
The proteasome inhibitor bortezomib (Velcade®) is the only drug targeting ubiquitin pathway system that has been used in clinic to treat malignancies [119–123]. Small molecule inhibitors of DUBs have also been studied for the development of anti-cancer strategies, including inhibitors of UCHL1, UCHL3, or USP7 [8]. On the other hand, the use of drugs targeting DUBs in treatment of infectious diseases has not been well explored. There are, however, several instances where antimicrobial properties of DUB inhibitors have been shown to be promising (Figure 2).
Figure 2.
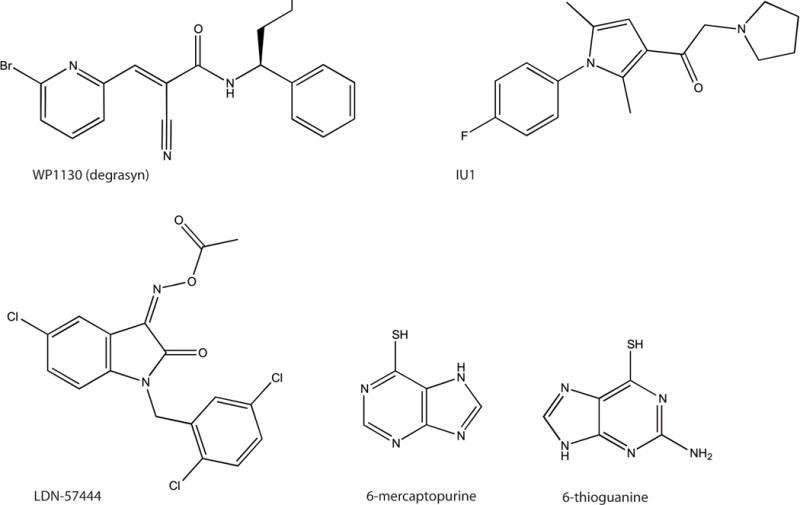
The structures of some of the small-molecule inhibitors that have been used to target specific DUBs (Structures were prepared in ChemDraw Ultra 12.0.3).
A small molecule inhibitor WP1130, also known as degrasyn [(2E)-3-(6-Bromo-2-pyridinyl)-2-cyano-N-[(1S)-1-phenylbutyl]-2-propenamide], has been shown to block activity of several DUBs, and it has been further evaluated in norovirus infection. This drug reduced infection with several RNA viruses, including encephalomyocarditis virus, Sindbis virus, La Crosse virus, and Norwalk virus. The target of WP1130 has been identified to be USP14, and siRNA-mediated knockdown of USP14 inhibited norovirus infection in mouse cells [124]. This study demonstrated that the pharmacological inhibition of DUBs could be used in a wide spectrum of viral diseases.
Moreover, WP1130 has been also tested in Listeria monocytogenes infection, where it increased killing of this intracellular bacterium. In this experimental model the most significant DUB target of WP1130 has not been identified, but it induced killing of bacteria captured by phagosomes, likely by influencing iNOS localization to the phagosome [125]. Interestingly, WP1130 has been previously used in the treatment of malignancies, so it has been rather well characterized as a drug [123, 126, 127].
VEA-260 [3-[[4-[[[(2S,3S)-3-[bis(naphthalen-1-ylmethyl)carbamoyl]oxirane-2-carbonyl]amino] carbamoyl]phenyl]methylcarbamoyl]benzoic acid], a small molecule inhibitor of SUMO-specific protease PfSENP1 (PFL1635w) from Plasmodium falciparum has been described. VEA-260 contains an active aza-epoxide group and it inhibits recombinant PfSENP1 activity and P. falciparum replication in infected blood [117]. This data suggest that PfSENP1 might be a drug target that could be explored for malaria cure.
Treatment of Trichinella spiralis with the UCH inhibitor LDN-57444 [3-(O-acetyloxime), 5-chloro-1-[(2,5-dichlorophenyl)methyl]-1H-Indole-2,3-dione], contributed towards reduced viability of cultured T. spiralis L1 larvae. The direct target of LDN-57444 in T. spiralis is not yet confirmed, but it has been shown to inhibit T. spiralis DUB, TsUCH37, which is one of the substrates of this inhibitor. LDN-57444 has been previously shown to act towards human UCHL1 and UCHL3 [128], and it is expected that T. spiralis encodes for homologues of these DUBs.
IU1 [1-[1-(4-Fluorophenyl)-2,5-dimethyl-1H-pyrrol-3-yl]-2-(1-pyrrolidinyl)ethanon] is a small-molecule inhibitor of the proteasome-associated DUB USP14 and it has been shown to inhibit replication of flaviviruses, especially dengue virus (DENV), which could be possibly related to IU1’s enhancement of the proteasomal activity [40], [39]. The effect of IU1 on replication of other viruses has been also tested, including La Crosse virus (LACV) and Chikungunya virus (CHIKV). IU1 did not have any measurable effect on CHIKV replication, and the replication of LACV was only reduced by three-fold in comparison to twenty-fold reduction of DENV replication using the same concentration of the drug [40].
A high-throughput screen of 50,080 compounds indicated a novel non-covalent inhibitor of papain-like protease PLpro from coronavirus, compound 7724772, which is a racemic mix of 2-methyl-N-[1-(2-naphthyl)ethyl]benzamide. GRL0617 [5-Amino-2-methyl-N-[(R)-1-(1-naphthyl)ethyl]benzamide], a synthetically optimized derivative of 7724772, has been shown to inhibit SARS-CoV viral replication in cells with no cytotoxic effects [129]. GRL0617S has been shown to have no effect on IFN-β and on IFN-stimulated response element (ISRE) reporter activity but it did reverse the inhibition of NF-κB activity. Importantly, GRL0617 did not inhibit such human DUBs like USP7 (HAUSP), USP18, UCH-L1, or UCH-L3 [68].
Another study identified two more PLpro inhibitors, thiopurine analogues, 6-mercaptopurine (6MP) and 6-thioguanine (6TG), which have been previously used in cancer chemotherapies [130]. A structural comparison has shown that both drugs could be potential inhibitors of USP14, based on the docking score and binding energy for 6MP and 6TG [130]. Since USP14 is a cellular DUB also relevant in controlling other viral infections [40], 6MP and 6TG could be tested as potential drugs in several viral illnesses.
As we described above, the function of a host DUB USP7 is hijacked by herpes virus (HSV). In particular, its interaction with ICP0 has a protective function against the host innate responses, since USP7 is directed to deubiquitinate Lys63-conjugated polyubiquitinated TRAF6 and IKKγ [42]. Inhibition of USP7 activity with small molecule drugs could potentially decrease the virulence of HSV. Due to function of USP7 in apoptosis and cancer, a considerable effort has been made towards discovering selective USP7 inhibitors [131–135]. These compounds could then potentially be also used as anti-viral drugs. A few other described here inhibitors have previously found use in anti-cancer therapies, and such a use of old drugs is expected to reduce the cost of drug development.
Importantly, several known DUB inhibitors, including WP1130 (degrasyn), which we described above [124], are not selective towards one specific DUB, but towards a whole range of DUBs. For instance some drugs inhibit a host DUB as well as a viral DUB, exemplified by 6MP and 6TG [130]. This trait could be problematic for treatment of certain pathogenic illnesses, therefore the selectivity of DUB inhibitors must be further tested. Synthetic ubiquitin-specific probes mimicking DUB substrates can be used to facilitate the investigation of specificity of therapeutic inhibitors targeting DUBs, since they measure enzymatic activity of multiple DUBs at once. Labeling of DUBs in cells or in lysate by ubiquitin-specific molecular probes, combined with quantitative mass spectrometry-based proteomics or quantitative immunoblotting represents an approach that can substantially accelerate the drug development process [9, 131] (Figure 1).
Proteasome and many components of ubiquitin pathways are indispensable elements of human cell and inhibition of such might seem problematic. However, as exemplified by a proteasome inhibitor bortezomib (Velcade®), which is used to treat patients with multiple myeloma and mantle cell lymphoma [119–123], the ubiquitin proteasome system constitutes a realistic pathway that can be targeted to treat human disease. The search for antimicrobial drugs directed towards DUBs is rather in its early stages, but the existing data is highly supportive of the potential of such novel antimicrobials.
Supplementary Material
Acknowledgments
The work with WRAIR has been done using the MTA agreement (Award# 013743-001). This work was partially supported by the MAFES Special Research Initiatives (Edelmann) Control of Food-Borne Pathogens (160000-018100-027100-401120), The National Science Foundation (Mississippi EPSCoR-0903787), and Mississippi INBRE funded by grants from the National Center for Research Resources (5P20RR016476-11), and the National Institute of General Medical Sciences (8 P20 GM103476-11) from the National Institutes of Health.
List of abbreviations
- DUB
deubiquitinating enzyme
- IFN
interferon
- Lys
lysine
- NF-κB
nuclear factor kappa B
- OTU
ovarian tumor domain-containing protease
- SUMO
small ubiquitin-like modifier
- MJD
Machado-Josephin domain protease
- UCH
ubiquitin C-terminal hydrolase
- USP/UBP
ubiquitin-specific processing protease
References
- 1.Alksne LE. Virulence as a target for antimicrobial chemotherapy. Expert Opin Investig Drugs. 2002;11:1149–59. doi: 10.1517/13543784.11.8.1149. [DOI] [PubMed] [Google Scholar]
- 2.Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–80. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 3.Welchman RL, Gordon C, Mayer RJ. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat Rev Mol Cell Biol. 2005;6:599–609. doi: 10.1038/nrm1700. [DOI] [PubMed] [Google Scholar]
- 4.Nijman SM, Luna-Vargas MP, Velds A, et al. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–86. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 5.Sun SC. Deubiquitylation and regulation of the immune response. Nat Rev Immunol. 2008;8:501–11. doi: 10.1038/nri2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edelmann MJ, Kessler BM. Ubiquitin and ubiquitin-like specific proteases targeted by infectious pathogens: Emerging patterns and molecular principles. Biochim Biophys Acta. 2008;1782:809–16. doi: 10.1016/j.bbadis.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eldridge AG, O’Brien T. Therapeutic strategies within the ubiquitin proteasome system. Cell Death Differ. 2010;17:4–13. doi: 10.1038/cdd.2009.82. [DOI] [PubMed] [Google Scholar]
- 8.Fraile JM, Quesada V, Rodriguez D, Freije JM, Lopez-Otin C. Deubiquitinases in cancer: new functions and therapeutic options. Oncogene. 2012;31:2373–88. doi: 10.1038/onc.2011.443. [DOI] [PubMed] [Google Scholar]
- 9.Kramer HB, Nicholson B, Kessler BM, Altun M. Detection of ubiquitin-proteasome enzymatic activities in cells: Application of activity-based probes to inhibitor development. Biochim Biophys Acta. 2012 doi: 10.1016/j.bbamcr.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mattern MR, Wu J, Nicholson B. Ubiquitin-based anticancer therapy: Carpet bombing with proteasome inhibitors vs surgical strikes with E1, E2, E3, or DUB inhibitors. Biochim Biophys Acta. 2012 doi: 10.1016/j.bbamcr.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harhaj EW, Dixit VM. Regulation of NF-kappaB by deubiquitinases. Immunol Rev. 2012;246:107–24. doi: 10.1111/j.1600-065X.2012.01100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kovalenko A, Chable-Bessia C, Cantarella G, et al. The tumour suppressor CYLD negatively regulates NF-kappaB signalling by deubiquitination. Nature. 2003;424:801–5. doi: 10.1038/nature01802. [DOI] [PubMed] [Google Scholar]
- 13.Jono H, Lim JH, Chen LF, et al. NF-kappaB is essential for induction of CYLD, the negative regulator of NF-kappaB: evidence for a novel inducible autoregulatory feedback pathway. J Biol Chem. 2004;279:36171–4. doi: 10.1074/jbc.M406638200. [DOI] [PubMed] [Google Scholar]
- 14.Liu S, Lv J, Han L, et al. A pro-inflammatory role of deubiquitinating enzyme cylindromatosis (CYLD) in vascular smooth muscle cells. Biochem Biophys Res Commun. 2012;420:78–83. doi: 10.1016/j.bbrc.2012.02.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoshida H, Jono H, Kai H, Li JD. The tumor suppressor cylindromatosis (CYLD) acts as a negative regulator for toll-like receptor 2 signaling via negative cross-talk with TRAF6 AND TRAF7. J Biol Chem. 2005;280:41111–21. doi: 10.1074/jbc.M509526200. [DOI] [PubMed] [Google Scholar]
- 16.Espinosa L, Cathelin S, D’Altri T, et al. The Notch/Hes1 pathway sustains NF-kappaB activation through CYLD repression in T cell leukemia. Cancer Cell. 2010;18:268–81. doi: 10.1016/j.ccr.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trompouki E, Hatzivassiliou E, Tsichritzis T, et al. CYLD is a deubiquitinating enzyme that negatively regulates NF-kappaB activation by TNFR family members. Nature. 2003;424:793–6. doi: 10.1038/nature01803. [DOI] [PubMed] [Google Scholar]
- 18.Ahmed N, Zeng M, Sinha I, et al. The E3 ligase Itch and deubiquitinase Cyld act together to regulate Tak1 and inflammation. Nat Immunol. 2011;12:1176–83. doi: 10.1038/ni.2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsagaratou A, Kontoyiannis DL, Mosialos G. Truncation of the deubiquitinating domain of CYLD in myelomonocytic cells attenuates inflammatory responses. Plos One. 2011;6:e16397. doi: 10.1371/journal.pone.0016397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wertz IE, O’Rourke KM, Zhou H, et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 2004;430:694–9. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 21.Coornaert B, Carpentier I, Beyaert R. A20: central gatekeeper in inflammation and immunity. J Biol Chem. 2009;284:8217–21. doi: 10.1074/jbc.R800032200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shembade N, Parvatiyar K, Harhaj NS, Harhaj EW. The ubiquitin-editing enzyme A20 requires RNF11 to downregulate NF-kappaB signalling. EMBO J. 2009;28:513–22. doi: 10.1038/emboj.2008.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shembade N, Ma A, Harhaj EW. Inhibition of NF-kappaB signaling by A20 through disruption of ubiquitin enzyme complexes. Science. 2010;327:1135–9. doi: 10.1126/science.1182364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kayagaki N, Phung Q, Chan S, et al. DUBA: a deubiquitinase that regulates type I interferon production. Science. 2007;318:1628–32. doi: 10.1126/science.1145918. [DOI] [PubMed] [Google Scholar]
- 25.Gonzalez-Navajas JM, Law J, Nguyen KP, et al. Interleukin 1 receptor signaling regulates DUBA expression and facilitates Toll-like receptor 9-driven antiinflammatory cytokine production. J Exp Med. 2010;207:2799–807. doi: 10.1084/jem.20101326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou F, Zhang X, van Dam H, et al. Ubiquitin-specific Protease 4 Mitigates Toll-like/Interleukin-1 Receptor Signaling and Regulates Innate Immune Activation. J Biol Chem. 2012;287:11002–10. doi: 10.1074/jbc.M111.328187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Colonne PM, Sahni A, Sahni SK. Rickettsia conorii infection stimulates the expression of ISG15 and ISG15 protease UBP43 in human microvascular endothelial cells. Biochem Biophys Res Commun. 2011;416:153–8. doi: 10.1016/j.bbrc.2011.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim JH, Luo JK, Zhang DE. The level of hepatitis B virus replication is not affected by protein ISG15 modification but is reduced by inhibition of UBP43 (USP18) expression. J Immunol. 2008;181:6467–72. doi: 10.4049/jimmunol.181.9.6467. [DOI] [PubMed] [Google Scholar]
- 29.Zou W, Kim JH, Handidu A, et al. Microarray analysis reveals that Type I interferon strongly increases the expression of immune-response related genes in Ubp43 (Usp18) deficient macrophages. Biochem Biophys Res Commun. 2007;356:193–9. doi: 10.1016/j.bbrc.2007.02.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van Nguyen T, Angkasekwinai P, Dou H, et al. SUMO-specific protease 1 is critical for early lymphoid development through regulation of STAT5 activation. Mol Cell. 2012;45:210–21. doi: 10.1016/j.molcel.2011.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ran Y, Liu TT, Zhou Q, et al. SENP2 negatively regulates cellular antiviral response by deSUMOylating IRF3 and conditioning it for ubiquitination and degradation. J Mol Cell Biol. 2011;3:283–92. doi: 10.1093/jmcb/mjr020. [DOI] [PubMed] [Google Scholar]
- 32.Lee MH, Mabb AM, Gill GB, Yeh ET, Miyamoto S. NF-kappaB induction of the SUMO protease SENP2: A negative feedback loop to attenuate cell survival response to genotoxic stress. Mol Cell. 2011;43:180–91. doi: 10.1016/j.molcel.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCool KW, Miyamoto S. DNA damage-dependent NF-kappaB activation: NEMO turns nuclear signaling inside out. Immunol Rev. 2012;246:311–26. doi: 10.1111/j.1600-065X.2012.01101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Enesa K, Zakkar M, Chaudhury H, et al. NF-kappaB suppression by the deubiquitinating enzyme Cezanne: a novel negative feedback loop in pro-inflammatory signaling. J Biol Chem. 2008;283:7036–45. doi: 10.1074/jbc.M708690200. [DOI] [PubMed] [Google Scholar]
- 35.Maelfait J, Roose K, Bogaert P, et al. A20 (Tnfaip3) deficiency in myeloid cells protects against influenza A virus infection. PLoS Pathog. 2012;8:e1002570. doi: 10.1371/journal.ppat.1002570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liao TL, Wu CY, Su WC, Jeng KS, Lai MM. Ubiquitination and deubiquitination of NP protein regulates influenza A virus RNA replication. EMBO J. 2010;29:3879–90. doi: 10.1038/emboj.2010.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doukas T, Sarnow P. Escape from transcriptional shutoff during poliovirus infection: NF-kappaB-responsive genes IkappaBa and A20. J Virol. 2011;85:10101–8. doi: 10.1128/JVI.00575-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu D, Lillico SG, Barnett MW, et al. USP18 restricts PRRSV growth through alteration of nuclear translocation of NF-kappaB p65 and p50 in MARC-145 cells. Virus Res. 2012 doi: 10.1016/j.virusres.2012.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee BH, Lee MJ, Park S, et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature. 2010;467:179–84. doi: 10.1038/nature09299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nag DK, Finley D. A small-molecule inhibitor of deubiquitinating enzyme USP14 inhibits Dengue virus replication. Virus Res. 2012;165:103–6. doi: 10.1016/j.virusres.2012.01.009. [DOI] [PubMed] [Google Scholar]
- 41.Antrobus R, Boutell C. Identification of a novel higher molecular weight isoform of USP7/HAUSP that interacts with the Herpes simplex virus type-1 immediate early protein ICP0. Virus Res. 2008;137:64–71. doi: 10.1016/j.virusres.2008.05.017. [DOI] [PubMed] [Google Scholar]
- 42.Daubeuf S, Singh D, Tan Y, et al. HSV ICP0 recruits USP7 to modulate TLR-mediated innate response. Blood. 2009;113:3264–75. doi: 10.1182/blood-2008-07-168203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jager W, Santag S, Weidner-Glunde M, et al. The ubiquitin-specific protease USP7 modulates the replication of Kaposi’s sarcoma-associated herpesvirus latent episomal DNA. J Virol. 2012;86:6745–57. doi: 10.1128/JVI.06840-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Holowaty MN, Sheng Y, Nguyen T, Arrowsmith C, Frappier L. Protein interaction domains of the ubiquitin-specific protease, USP7/HAUSP. Journal of Biological Chemistry. 2003;278:47753–47761. doi: 10.1074/jbc.M307200200. [DOI] [PubMed] [Google Scholar]
- 45.Holowaty MN, Zeghouf M, Wu H, et al. Protein profiling with Epstein-Barr nuclear antigen-1 reveals an interaction with the herpesvirus-associated ubiquitin-specific protease HAUSP/USP7. J Biol Chem. 2003;278:29987–94. doi: 10.1074/jbc.M303977200. [DOI] [PubMed] [Google Scholar]
- 46.Sarkari F, Sanchez-Alcaraz T, Wang S, et al. EBNA1-mediated recruitment of a histone H2B deubiquitylating complex to the Epstein-Barr virus latent origin of DNA replication. PLoS Pathog. 2009;5:e1000624. doi: 10.1371/journal.ppat.1000624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin CH, Chang HS, Yu WC. USP11 stabilizes HPV-16E7 and further modulates the E7 biological activity. J Biol Chem. 2008;283:15681–8. doi: 10.1074/jbc.M708278200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vos RM, Altreuter J, White EA, Howley PM. The ubiquitin-specific peptidase USP15 regulates human papillomavirus type 16 E6 protein stability. J Virol. 2009;83:8885–92. doi: 10.1128/JVI.00605-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lavorgna A, Harhaj EW. An RNA interference screen identifies the Deubiquitinase STAMBPL1 as a critical regulator of human T-cell leukemia virus type 1 tax nuclear export and NF-kappaB activation. J Virol. 2012;86:3357–69. doi: 10.1128/JVI.06456-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yasunaga J, Lin FC, Lu X, Jeang KT. Ubiquitin-specific peptidase 20 targets TRAF6 and human T cell leukemia virus type 1 tax to negatively regulate NF-kappaB signaling. J Virol. 2011;85:6212–9. doi: 10.1128/JVI.00079-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen R, Zhang L, Zhong B, et al. The ubiquitin-specific protease 17 is involved in virus-triggered type I IFN signaling. Cell Res. 2010;20:802–11. doi: 10.1038/cr.2010.41. [DOI] [PubMed] [Google Scholar]
- 52.Li S, Zheng H, Mao AP, et al. Regulation of virus-triggered signaling by OTUB1- and OTUB2-mediated deubiquitination of TRAF3 and TRAF6. J Biol Chem. 2010;285:4291–7. doi: 10.1074/jbc.M109.074971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kattenhorn LM, Korbel GA, Kessler BM, Spooner E, Ploegh HL. A deubiquitinating enzyme encoded by HSV-1 belongs to a family of cysteine proteases that is conserved across the family Herpesviridae. Mol Cell. 2005;19:547–57. doi: 10.1016/j.molcel.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 54.Bolstad M, Abaitua F, Crump CM, O’Hare P. Autocatalytic activity of the ubiquitin-specific protease domain of herpes simplex virus 1 VP1-2. J Virol. 2011;85:8738–51. doi: 10.1128/JVI.00798-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim ET, Oh SE, Lee YO, Gibson W, Ahn JH. Cleavage specificity of the UL48 deubiquitinating protease activity of human cytomegalovirus and the growth of an active-site mutant virus in cultured cells. J Virol. 2009;83:12046–56. doi: 10.1128/JVI.00411-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saha A, Murakami M, Kumar P, et al. Epstein-Barr virus nuclear antigen 3C augments Mdm2-mediated p53 ubiquitination and degradation by deubiquitinating Mdm2. J Virol. 2009;83:4652–69. doi: 10.1128/JVI.02408-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sompallae R, Gastaldello S, Hildebrand S, et al. Epstein-barr virus encodes three bona fide ubiquitin-specific proteases. J Virol. 2008;82:10477–86. doi: 10.1128/JVI.01113-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Whitehurst CB, Ning S, Bentz GL, et al. The Epstein-Barr virus (EBV) deubiquitinating enzyme BPLF1 reduces EBV ribonucleotide reductase activity. J Virol. 2009;83:4345–53. doi: 10.1128/JVI.02195-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Whitehurst CB, Vaziri C, Shackelford J, Pagano JS. Epstein-Barr Virus BPLF1 Deubiquitinates PCNA and Attenuates Polymerase eta Recruitment to DNA Damage Sites. J Virol. 2012;86:8097–106. doi: 10.1128/JVI.00588-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gastaldello S, Callegari S, Coppotelli G, et al. Herpes virus deneddylases interrupt the cullin-RING ligase neddylation cycle by inhibiting the binding of CAND1. J Mol Cell Biol. 2012;4:242–51. doi: 10.1093/jmcb/mjs012. [DOI] [PubMed] [Google Scholar]
- 61.Gastaldello S, Hildebrand S, Faridani O, et al. A deneddylase encoded by Epstein-Barr virus promotes viral DNA replication by regulating the activity of cullin-RING ligases. Nat Cell Biol. 2010;12:351–61. doi: 10.1038/ncb2035. [DOI] [PubMed] [Google Scholar]
- 62.Jiang J, Tang H. Mechanism of inhibiting type I interferon induction by hepatitis B virus X protein. Protein Cell. 2010;1:1106–17. doi: 10.1007/s13238-010-0141-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gonzalez CM, Wang L, Damania B. Kaposi’s sarcoma-associated herpesvirus encodes a viral deubiquitinase. J Virol. 2009;83:10224–33. doi: 10.1128/JVI.00589-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Inn KS, Lee SH, Rathbun JY, et al. Inhibition of RIG-I-mediated signaling by Kaposi’s sarcoma-associated herpesvirus-encoded deubiquitinase ORF64. J Virol. 2011;85:10899–904. doi: 10.1128/JVI.00690-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gredmark-Russ S, Isaacson MK, Kattenhorn L, et al. A gammaherpesvirus ubiquitin-specific protease is involved in the establishment of murine gammaherpesvirus 68 infection. J Virol. 2009;83:10644–52. doi: 10.1128/JVI.01017-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li SW, Lai CC, Ping JF, et al. Severe acute respiratory syndrome coronavirus papain-like protease suppressed alpha interferon-induced responses through downregulation of extracellular signal-regulated kinase 1-mediated signalling pathways. J Gen Virol. 2011;92:1127–40. doi: 10.1099/vir.0.028936-0. [DOI] [PubMed] [Google Scholar]
- 67.Zheng D, Chen G, Guo B, Cheng G, Tang H. PLP2, a potent deubiquitinase from murine hepatitis virus, strongly inhibits cellular type I interferon production. Cell Res. 2008;18:1105–13. doi: 10.1038/cr.2008.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Clementz MA, Chen Z, Banach BS, et al. Deubiquitinating and interferon antagonism activities of coronavirus papain-like proteases. J Virol. 2010;84:4619–29. doi: 10.1128/JVI.02406-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sun L, Xing Y, Chen X, et al. Coronavirus papain-like proteases negatively regulate antiviral innate immune response through disruption of STING-mediated signaling. PLoS One. 2012;7:e30802. doi: 10.1371/journal.pone.0030802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang D, Fang L, Li P, et al. The leader proteinase of foot-and-mouth disease virus negatively regulates the type I interferon pathway by acting as a viral deubiquitinase. J Virol. 2011;85:3758–66. doi: 10.1128/JVI.02589-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.van Kasteren PB, Beugeling C, Ninaber DK, et al. Arterivirus and nairovirus ovarian tumor domain-containing Deubiquitinases target activated RIG-I to control innate immune signaling. J Virol. 2012;86:773–85. doi: 10.1128/JVI.06277-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.James TW, Frias-Staheli N, Bacik JP, et al. Structural basis for the removal of ubiquitin and interferon-stimulated gene 15 by a viral ovarian tumor domain-containing protease. Proc Natl Acad Sci U S A. 2011;108:2222–7. doi: 10.1073/pnas.1013388108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Karpe YA, Lole KS. Deubiquitination activity associated with hepatitis E virus putative papain-like cysteine protease. J Gen Virol. 2011;92:2088–92. doi: 10.1099/vir.0.033738-0. [DOI] [PubMed] [Google Scholar]
- 74.Li H, Zheng Z, Zhou P, et al. The cysteine protease domain of porcine reproductive and respiratory syndrome virus non-structural protein 2 antagonizes interferon regulatory factor 3 activation. J Gen Virol. 2010;91:2947–58. doi: 10.1099/vir.0.025205-0. [DOI] [PubMed] [Google Scholar]
- 75.Sun Z, Li Y, Ransburgh R, Snijder EJ, Fang Y. Nonstructural protein 2 of porcine reproductive and respiratory syndrome virus inhibits the antiviral function of interferon-stimulated gene 15. J Virol. 2012;86:3839–50. doi: 10.1128/JVI.06466-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Holzer B, Bakshi S, Bridgen A, Baron MD. Inhibition of interferon induction and action by the nairovirus Nairobi sheep disease virus/Ganjam virus. PLoS One. 2011;6:e28594. doi: 10.1371/journal.pone.0028594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Edelmann MJ, Kramer HB, Altun M, Kessler BM. Post-translational modification of the deubiquitinating enzyme otubain 1 modulates active RhoA levels and susceptibility to Yersinia invasion. FEBS J. 2010;277:2515–30. doi: 10.1111/j.1742-4658.2010.07665.x. [DOI] [PubMed] [Google Scholar]
- 78.Juris SJ, Shah K, Shokat K, Dixon JE, Vacratsis PO. Identification of otubain 1 as a novel substrate for the Yersinia protein kinase using chemical genetics and mass spectrometry. FEBS Lett. 2006;580:179–83. doi: 10.1016/j.febslet.2005.11.071. [DOI] [PubMed] [Google Scholar]
- 79.Barz C, Abahji TN, Trulzsch K, Heesemann J. The Yersinia Ser/Thr protein kinase YpkA/YopO directly interacts with the small GTPases RhoA and Rac-1. FEBS Lett. 2000;482:139–43. doi: 10.1016/s0014-5793(00)02045-7. [DOI] [PubMed] [Google Scholar]
- 80.Basseres E, Coppotelli G, Pfirrmann T, et al. The ubiquitin C-terminal hydrolase UCH-L1 promotes bacterial invasion by altering the dynamics of the actin cytoskeleton. Cell Microbiol. 2010;12:1622–33. doi: 10.1111/j.1462-5822.2010.01495.x. [DOI] [PubMed] [Google Scholar]
- 81.Coombs N, Sompallae R, Olbermann P, et al. Helicobacter pylori affects the cellular deubiquitinase USP7 and ubiquitin-regulated components TRAF6 and the tumour suppressor p53. Int J Med Microbiol. 2010 doi: 10.1016/j.ijmm.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 82.Akimana C, Al-Khodor S, Abu Kwaik Y. Host factors required for modulation of phagosome biogenesis and proliferation of Francisella tularensis within the cytosol. Plos One. 2010;5:e11025. doi: 10.1371/journal.pone.0011025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Brummelkamp TR, Nijman SMB, Dirac AMG, Bernards R. Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-kappa B. Nature. 2003;424:797–801. doi: 10.1038/nature01811. [DOI] [PubMed] [Google Scholar]
- 84.Lim JH, Jono H, Koga T, et al. Tumor suppressor CYLD acts as a negative regulator for non-typeable Haemophilus influenza-induced inflammation in the middle ear and lung of mice. PLoS One. 2007;2:e1032. doi: 10.1371/journal.pone.0001032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sakai A, Koga T, Lim JH, et al. The bacterium, nontypeable Haemophilus influenzae, enhances host antiviral response by inducing Toll-like receptor 7 expression: evidence for negative regulation of host anti-viral response by CYLD. FEBS J. 2007;274:3655–68. doi: 10.1111/j.1742-4658.2007.05899.x. [DOI] [PubMed] [Google Scholar]
- 86.Lim JH, Ha UH, Woo CH, Xu H, Li JD. CYLD is a crucial negative regulator of innate immune response in Escherichia coli pneumonia. Cell Microbiol. 2008;10:2247–56. doi: 10.1111/j.1462-5822.2008.01204.x. [DOI] [PubMed] [Google Scholar]
- 87.Lim JH, Stirling B, Derry J, et al. Tumor suppressor CYLD regulates acute lung injury in lethal Streptococcus pneumoniae infections. Immunity. 2007;27:349–60. doi: 10.1016/j.immuni.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 88.Bomberger JM, Ye S, Maceachran DP, et al. A Pseudomonas aeruginosa toxin that hijacks the host ubiquitin proteolytic system. PLoS Pathog. 2011;7:e1001325. doi: 10.1371/journal.ppat.1001325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bomberger JM, Barnaby RL, Stanton BA. The deubiquitinating enzyme USP10 regulates the post-endocytic sorting of cystic fibrosis transmembrane conductance regulator in airway epithelial cells. J Biol Chem. 2009;284:18778–89. doi: 10.1074/jbc.M109.001685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rytkonen A, Poh J, Garmendia J, et al. SseL, a Salmonella deubiquitinase required for macrophage killing and virulence. Proc Natl Acad Sci U S A. 2007;104:3502–7. doi: 10.1073/pnas.0610095104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Guiney DG, Fierer J. The Role of the spv Genes in Salmonella Pathogenesis. Front Microbiol. 2011;2:129. doi: 10.3389/fmicb.2011.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mesquita FS, Thomas M, Sachse M, et al. The Salmonella Deubiquitinase SseL Inhibits Selective Autophagy of Cytosolic Aggregates. PLoS Pathog. 2012;8:e1002743. doi: 10.1371/journal.ppat.1002743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ye Z, Petrof EO, Boone D, Claud EC, Sun J. Salmonella effector AvrA regulation of colonic epithelial cell inflammation by deubiquitination. Am J Pathol. 2007;171:882–92. doi: 10.2353/ajpath.2007.070220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liao AP, Petrof EO, Kuppireddi S, et al. Salmonella type III effector AvrA stabilizes cell tight junctions to inhibit inflammation in intestinal epithelial cells. Plos One. 2008;3:e2369. doi: 10.1371/journal.pone.0002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mukherjee S, Keitany G, Li Y, et al. Yersinia YopJ acetylates and inhibits kinase activation by blocking phosphorylation. Science. 2006;312:1211–4. doi: 10.1126/science.1126867. [DOI] [PubMed] [Google Scholar]
- 96.Jones RM, Wu H, Wentworth C, et al. Salmonella AvrA Coordinates Suppression of Host Immune and Apoptotic Defenses via JNK Pathway Blockade. Cell Host Microbe. 2008;3:233–44. doi: 10.1016/j.chom.2008.02.016. [DOI] [PubMed] [Google Scholar]
- 97.Sweet CR, Conlon J, Golenbock DT, Goguen J, Silverman N. YopJ targets TRAF proteins to inhibit TLR-mediated NF-kappaB, MAPK and IRF3 signal transduction. Cell Microbiol. 2007;9:2700–15. doi: 10.1111/j.1462-5822.2007.00990.x. [DOI] [PubMed] [Google Scholar]
- 98.Zhou H, Monack DM, Kayagaki N, et al. Yersinia virulence factor YopJ acts as a deubiquitinase to inhibit NF-kappa B activation. J Exp Med. 2005;202:1327–32. doi: 10.1084/jem.20051194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Orth K, Xu Z, Mudgett MB, et al. Disruption of signaling by Yersinia effector YopJ, a ubiquitin-like protein protease. Science. 2000;290:1594–7. doi: 10.1126/science.290.5496.1594. [DOI] [PubMed] [Google Scholar]
- 100.Paquette N, Conlon J, Sweet C, et al. Serine/threonine acetylation of TGFbeta-activated kinase (TAK1) by Yersinia pestis YopJ inhibits innate immune signaling. Proc Natl Acad Sci U S A. 2012;109:12710–5. doi: 10.1073/pnas.1008203109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Catic A, Misaghi S, Korbel GA, Ploegh HL. ElaD, a Deubiquitinating Protease Expressed by E. coli. Plos One. 2007;2 doi: 10.1371/journal.pone.0000381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Misaghi S, Balsara ZR, Catic A, et al. Chlamydia trachomatis-derived deubiquitinating enzymes in mammalian cells during infection. Mol Microbiol. 2006;61:142–50. doi: 10.1111/j.1365-2958.2006.05199.x. [DOI] [PubMed] [Google Scholar]
- 103.Le Negrate G, Krieg A, Faustin B, et al. ChlaDub1 of Chlamydia trachomatis suppresses NF-kappaB activation and inhibits IkappaBalpha ubiquitination and degradation. Cell Microbiol. 2008;10:1879–92. doi: 10.1111/j.1462-5822.2008.01178.x. [DOI] [PubMed] [Google Scholar]
- 104.Tan KS, Chen Y, Lim YC, et al. Suppression of host innate immune response by Burkholderia pseudomallei through the virulence factor TssM. J Immunol. 2010;184:5160–71. doi: 10.4049/jimmunol.0902663. [DOI] [PubMed] [Google Scholar]
- 105.Shanks J, Burtnick MN, Brett PJ, et al. Burkholderia mallei tssM encodes a putative deubiquitinase that is secreted and expressed inside infected RAW 264.7 murine macrophages. Infect Immun. 2009;77:1636–48. doi: 10.1128/IAI.01339-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pearce MJ, Mintseris J, Ferreyra J, Gygi SP, Darwin KH. Ubiquitin-like protein involved in the proteasome pathway of Mycobacterium tuberculosis. Science. 2008;322:1104–7. doi: 10.1126/science.1163885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Burns KE, Darwin KH. Pupylation vs. Ubiquitylation: Tagging for Proteasome-Dependent Degradation. Cell Microbiol. 2010 doi: 10.1111/j.1462-5822.2010.01447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Imkamp F, Striebel F, Sutter M, et al. Dop functions as a depupylase in the prokaryotic ubiquitin-like modification pathway. EMBO Rep. 2010;11:791–7. doi: 10.1038/embor.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.`Imkamp F, Rosenberger T, Striebel F, et al. Deletion of dop in Mycobacterium smegmatis abolishes pupylation of protein substrates in vivo. Mol Microbiol. 2010;75:744–54. doi: 10.1111/j.1365-2958.2009.07013.x. [DOI] [PubMed] [Google Scholar]
- 110.Srivastav S, Kar S, Chande AG, Mukhopadhyaya R, Das PK. Leishmania donovani Exploits Host Deubiquitinating Enzyme A20, a Negative Regulator of TLR Signaling, To Subvert Host Immune Response. J Immunol. 2012;189:924–34. doi: 10.4049/jimmunol.1102845. [DOI] [PubMed] [Google Scholar]
- 111.White RR, Miyata S, Papa E, et al. Characterisation of the Trichinella spiralis deubiquitinating enzyme, TsUCH37, an evolutionarily conserved proteasome interaction partner. PLoS Negl Trop Dis. 2011;5:e1340. doi: 10.1371/journal.pntd.0001340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Frickel EM, Quesada V, Muething L, et al. Apicomplexan UCHL3 retains dual specificity for ubiquitin and Nedd8 throughout evolution. Cell Microbiol. 2007;9:1601–10. doi: 10.1111/j.1462-5822.2007.00896.x. [DOI] [PubMed] [Google Scholar]
- 113.Ponts N, Yang J, Chung DW, et al. Deciphering the ubiquitin-mediated pathway in apicomplexan parasites: a potential strategy to interfere with parasite virulence. Plos One. 2008;3:e2386. doi: 10.1371/journal.pone.0002386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Artavanis-Tsakonas K, Weihofen WA, Antos JM, et al. Characterization and structural studies of the Plasmodium falciparum ubiquitin and Nedd8 hydrolase UCHL3. J Biol Chem. 2010;285:6857–66. doi: 10.1074/jbc.M109.072405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Artavanis-Tsakonas K, Misaghi S, Comeaux CA, et al. Identification by functional proteomics of a deubiquitinating/deNeddylating enzyme in Plasmodium falciparum. Mol Microbiol. 2006;61:1187–95. doi: 10.1111/j.1365-2958.2006.05307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hunt P, Afonso A, Creasey A, et al. Gene encoding a deubiquitinating enzyme is mutated in artesunate- and chloroquine-resistant rodent malaria parasites. Mol Microbiol. 2007;65:27–40. doi: 10.1111/j.1365-2958.2007.05753.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ponder EL, Albrow VE, Leader BA, et al. Functional characterization of a SUMO deconjugating protease of Plasmodium falciparum using newly identified small molecule inhibitors. Chem Biol. 2011;18:711–21. doi: 10.1016/j.chembiol.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Fang W, Price MS, Toffaletti DL, et al. Pleiotropic Effects of Deubiquitinating Enzyme Ubp5 on Growth and Pathogenesis of Cryptococcus neoformans. Plos One. 2012;7:e38326. doi: 10.1371/journal.pone.0038326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Blade J, Cibeira MT, Rosinol L. Bortezomib: a valuable new antineoplastic strategy in multiple myeloma. Acta Oncol. 2005;44:440–8. doi: 10.1080/02841860510030002. [DOI] [PubMed] [Google Scholar]
- 120.Chen D, Frezza M, Schmitt S, Kanwar J, Dou QP. Bortezomib as the First Proteasome Inhibitor Anticancer Drug: Current Status and Future Perspectives. Curr Cancer Drug Targets. 2011 doi: 10.2174/156800911794519752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Curran MP, McKeage K. Bortezomib: a review of its use in patients with multiple myeloma. Drugs. 2009;69:859–88. doi: 10.2165/00003495-200969070-00006. [DOI] [PubMed] [Google Scholar]
- 122.Ooi MG, Hayden PJ, Kotoula V, et al. Interactions of the Hdm2/p53 and proteasome pathways may enhance the antitumor activity of bortezomib. Clin Cancer Res. 2009;15:7153–60. doi: 10.1158/1078-0432.CCR-09-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Pham LV, Tamayo AT, Li C, et al. Degrasyn potentiates the antitumor effects of bortezomib in mantle cell lymphoma cells in vitro and in vivo: therapeutic implications. Mol Cancer Ther. 2010;9:2026–36. doi: 10.1158/1535-7163.MCT-10-0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Perry JW, Ahmed M, Chang KO, et al. Antiviral Activity of a Small Molecule Deubiquitinase Inhibitor Occurs via Induction of the Unfolded Protein Response. PLoS Pathog. 2012;8:e1002783. doi: 10.1371/journal.ppat.1002783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Burkholder KM, Perry JW, Wobus CE, et al. A small molecule deubiquitinase inhibitor increases localization of inducible nitric oxide synthase to the macrophage phagosome and enhances bacterial killing. Infect Immun. 2011;79:4850–7. doi: 10.1128/IAI.05456-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bartholomeusz GA, Talpaz M, Kapuria V, et al. Activation of a novel Bcr/Abl destruction pathway by WP1130 induces apoptosis of chronic myelogenous leukemia cells. Blood. 2007;109:3470–8. doi: 10.1182/blood-2006-02-005579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kapuria V, Peterson LF, Fang D, et al. Deubiquitinase inhibition by small-molecule WP1130 triggers aggresome formation and tumor cell apoptosis. Cancer Res. 2010;70:9265–76. doi: 10.1158/0008-5472.CAN-10-1530. [DOI] [PubMed] [Google Scholar]
- 128.Liu Y, Lashuel HA, Choi S, et al. Discovery of inhibitors that elucidate the role of UCH-L1 activity in the H1299 lung cancer cell line. Chem Biol. 2003;10:837–46. doi: 10.1016/j.chembiol.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 129.Ratia K, Pegan S, Takayama J, et al. A noncovalent class of papain-like protease/deubiquitinase inhibitors blocks SARS virus replication. Proc Natl Acad Sci U S A. 2008;105:16119–24. doi: 10.1073/pnas.0805240105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chen X, Chou CY, Chang GG. Thiopurine analogue inhibitors of severe acute respiratory syndrome-coronavirus papain-like protease, a deubiquitinating and deISGylating enzyme. Antivir Chem Chemother. 2009;19:151–6. doi: 10.1177/095632020901900402. [DOI] [PubMed] [Google Scholar]
- 131.Altun M, Kramer HB, Willems LI, et al. Activity-based chemical proteomics accelerates inhibitor development for deubiquitylating enzymes. Chem Biol. 2011;18:1401–12. doi: 10.1016/j.chembiol.2011.08.018. [DOI] [PubMed] [Google Scholar]
- 132.Reverdy C, Conrath S, Lopez R, et al. Discovery of specific inhibitors of human USP7/HAUSP deubiquitinating enzyme. Chem Biol. 2012;19:467–77. doi: 10.1016/j.chembiol.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 133.Menard R, Sulea T. Selective inhibition of USP7. Chem Biol. 2012;19:437–8. doi: 10.1016/j.chembiol.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Tian X, Sh Isamiddinova N, Peroutka RJ, et al. Characterization of Selective Ubiquitin and Ubiquitin-Like Protease Inhibitors Using a Fluorescence-Based Multiplex Assay Format. Assay Drug Dev Technol. 2010 Dec 6; doi: 10.1089/adt.2010.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Colland F, Formstecher E, Jacq X, et al. Small-molecule inhibitor of USP7/HAUSP ubiquitin protease stabilizes and activates p53 in cells. Mol Cancer Ther. 2009;8:2286–95. doi: 10.1158/1535-7163.MCT-09-0097. [DOI] [PubMed] [Google Scholar]
- 136.Koterski JF, Nahvi M, Venkatesan MM, Haimovich B. Virulent Shigella flexneri causes damage to mitochondria and triggers necrosis in infected human monocyte-derived macrophages. Infect Immun. 2005;73:504–13. doi: 10.1128/IAI.73.1.504-513.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wessel D, Flugge UI. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem. 1984;138:141–3. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- 138.Edelmann MJ, Iphofer A, Akutsu M, et al. Structural basis and specificity of human otubain 1-mediated deubiquitination. Biochem J. 2009;418:379–90. doi: 10.1042/BJ20081318. [DOI] [PubMed] [Google Scholar]
- 139.Peddinti D, Nanduri B, Kaya A, et al. Comprehensive proteomic analysis of bovine spermatozoa of varying fertility rates and identification of biomarkers associated with fertility. BMC Syst Biol. 2008;2:19. doi: 10.1186/1752-0509-2-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


