Abstract
DNA nonhomologous end-joining in vivo requires the DNA-dependent protein kinase (DNA-PK) and DNA ligase IV/XRCC4 (LX) complexes. Here, we have examined the impact of histone octamers and linker histone H1 on DNA end-joining in vitro. Packing of the DNA substrate into dinucleosomes does not significantly inhibit ligation by LX. However, LX ligation activity is substantially reduced by the incorporation of linker histones. This inhibition is independent of the presence of core histone octamers and cannot be restored by addition of Ku alone but can be partially rescued by DNA-PK. The kinase activity of DNA-PK is essential for the recovery of end-joining. DNA-PK efficiently phosphorylates histone H1. Phosphorylated histone H1 has a reduced affinity for DNA and a decreased capacity to inhibit end-joining. Our findings raise the possibility that DNA-PK may act as a linker histone kinase by phosphorylating linker histones in the vicinity of a DNA break and coupling localized histone H1 release from DNA ends, with the recruitment of LX to carry out double-stranded ligation. Thus, by using histone H1-bound DNA as a template, we have reconstituted the end-joining step of DNA nonhomologous end-joining in vitro with a requirement for DNA-PK.
Keywords: chromatin, DNA double-stranded break repair, nonhomologous end-joining
DNA nonhomologous end-joining (NHEJ) is the major mechanism for the repair of DNA double-stranded (ds) breaks (DSBs) in mammalian cells and functions to effect DNA rearrangement during V(D)J recombination. Five proteins forming two complexes are required for NHEJ in mammalian cells, namely DNA ligase IV/XRCC4 (LX) complex and the DNA-dependent protein kinase (DNA-PK) complex, which encompasses the two subunits of the Ku heterodimer and a large catalytic subunit (DNA-PKcs) (1, 2). Recently, a sixth protein, Artemis, has been shown to cleave the hairpin intermediate generated during V(D)J recombination (3). Although DNA-PK phosphorylation is required for the hairpin cleavage activity of Artemis and the biochemical activities of the DNA-PK and LX complexes have been established in vitro, little is known about the role of DNA-PK in DSB rejoining or its in vivo phosphorylation targets. Current models suggest that DNA-PK may regulate NHEJ (4–8).
Recently, we have shown that LX is most efficient on substrates ≈400 bp long and inefficient on short (≈50-bp) oligomers (9). Ku can stimulate LX ligation in a manner that requires freedom for Ku to translocate inwardly on the DNA (9–11). However, the presence of the intact DNA-PK complex does not provide any further stimulation of end-joining, and, indeed, its presence can be inhibitory (see Results). Our finding that efficient LX ligation in the presence of Ku requires inward translocation of Ku raises the question of how the presence of nucleosomes and the complex structural packaging of DNA into chromatin might impact the efficiency of NHEJ. The fundamental structural unit of chromatin is the nucleosome, an octameric complex of two copies of each core histone (H2A, H2B, H3, and H4) that associates with 146 bp of DNA (12). In addition, eukaryotic chromosomal DNA contains linker histones, such as histone H1. The structural unit encompassing histone H1 and the core histones associates with ≈166 base pairs and has been termed the chromatosome. Whereas nucleosomes exhibit “sliding” mobility on the DNA, linker histones are thought to stabilize the higher-order folding of reconstituted nucleosomes and restrict nucleosome movement (13, 14). It has been suggested that depletion of histone H1 may be a prerequisite for certain DNA processing events, such as transcriptional activation (15). Now that many repair processes can be reconstituted in vitro, studies are beginning to examine the impact of chromatin structure on repair. Nucleotide excision repair and base excision repair were shown to be impaired on nucleosome substrates relative to naked DNA (16–20) and repair efficiencies are recovered by the presence of ATP-dependent chromatin remodeling factors (21–23). In contrast, human DNA ligase I and Flap I endonuclease function efficiently on model nucleosome substrates (24, 25).
To address the impact of chromatin assembly on NHEJ, we used model reconstituted dinucleosome and chromatosome substrates. We show that incorporation of linker histones onto a DNA substrate dramatically inhibits LX activity. End-joining of histone H1-bound substrates is substantially recovered by the addition of DNA-PK, providing in vitro reconstitution of the end-joining step of NHEJ dependent on all of the known components.
Materials and Methods
Expression, Purification, and Immunological Analysis of LX, Ku70/80, and DNA-PK Complexes. LX and Ku70/80 were overexpressed in the Bac-To-Bac Baculovirus expression system (GIBCO/BRL) and purified to near-homogeneity as described in ref. 9. The details of LX complex purification can be found in Supporting Materials and Methods, which is published as supporting information on the PNAS web site. Aliquots of the purified complexes were stored at –80°C in the presence of 10% glycerol. DNA-PK was purified to near homogeneity from HeLa nuclear extracts (Computer Cell Culture Center, Seneffe, Belgium) as described in ref. 26.
Standard immunoblot analysis was performed by using anti-ATR [ataxia-telangiectasia-mutated kinase (ATM) and Rad3-related kinase] antibody (catalog no. PC538T, Calbiochem), anti-ATM (from M. A. Taylor, University of Birmingham, Birmingham, U.K.) and anti-DNA-PKcs (raised against recombinant DNA-PKcs peptide). Immunodepletion experiments were performed by incubating the indicated amounts of proteins with 2–5 μl of preimmune serum or anti-Ku/XRCC4 antibodies (raised against full-length recombinant proteins), coupled to 5 μl of Protein G beads (Amersham Biosciences) in buffer containing 20 mM Tris·HCl (pH 8.0), 10 mM 2-mercaptoethanol, and 150 mM NaCl for 2 h. Immunoprecipitated material and the beads were removed by centrifugation and the supernatant was assayed as described below.
Reconstitution of Chromatin from Purified Components. Dinucleosomes and chromatosomes were assembled by the salt dialysis method (27) by using purified chicken erythrocyte histone octamers and linker histones (28) or purified calf thymus linker histones (Roche) on radioactively labeled 445-bp dsDNA fragments produced from the Bluescript plasmid (Stratagene) as described in ref. 9. For the histone octamer preparation, an aliquot of purified chicken erythrocyte nuclei (≈6 mg of DNA) was resuspended in 10 ml of 50 mM Na-phosphate buffer (pH 6.8) containing 700 mM NaCl and protease inhibitors (complete protease inhibitor mixture, EDTA-free, Roche) and lysed by gentle stirring on ice for 30 min. Five grams of hydroxyapatite was added per tube, and the volume was increased to 40 ml with the same buffer. Linker histones and nonhistone proteins were removed by six repeated washes by using the same buffer. The pellet was resuspended in 40 ml of high-salt extraction buffer (50 mM Na-Phosphate buffer, pH 6.8/2.5 M NaCl) and gently stirred on ice for 15 min to elute core histones. Two eluates were collected, pooled, and concentrated on 20-ml Vivaspin columns (10-kDa cutoff size; Sartorius). Concentrated histone octamers were further purified by using gel filtration on a Superdex-200 HR 10/30 column (Amersham Biosciences). Linker histones were prepared by washing the suspension of purified nuclei (6 mg) with 2 × 10 ml of a buffer containing 10 mM Tris·HCl (pH 8.0), 350 mM NaCl, and 1 mM Na2EDTA to remove nonhistone proteins. Washed nuclei were resuspended in 10 ml of 50 mM Na-Phosphate buffer (pH 6.8) and 0.7 M NaCl and lysed on ice for 30 min with gentle stirring in the presence of protease inhibitors (Roche). Five grams of hydroxyapatite was added, and the volume was increased to 40 ml. Linker histones were extracted by stirring on ice for 15 min and reextracted with 40 ml of the same buffer. Combined eluates were concentrated on 20-ml Vivaspin columns (10-kDa cutoff size, Sartorius) and stored frozen at –80°C. The purity of histone octamers and linker histone preparations was assessed by using 12% SDS/PAGE. For large-scale dinucleosome reconstitution, 5.25 pmol of radioactively labeled 445-bp DNA fragments were mixed with purified histone octamers (3–5 μg) in a buffer containing 40 mM Tris·HCl (pH 8.0), 0.4 mM EDTA, 2 mM 2-mercaptoethanol, and 2.5 M NaCl. The reaction mixture was incubated for 1.5 h and dialyzed with decreasing NaCl concentrations by using a microdialyzer (GIBCO/BRL). The amount of histone octamers required for efficient packing was optimized empirically for each preparation and monitored by nucleoprotein agarose gel electrophoresis on 0.7% agarose gels. Samples were loaded in 10% glycerol loading buffer with no dyes and run at 30 mA. For chromatosome reconstitution, aliquots of reconstituted dinucleosomes containing ≈20 ng of DNA were mixed with increasing amounts of histone H1 (up to 20 ng) in 40 mM Tris·HCl, pH 8.0/0.4 mM EDTA/2 mM 2-mercaptoethanol and incubated for 30 min at room temperature. The efficiency of histone H1 binding was optimized empirically for each linker histone preparation and monitored as described above.
ds Ligation Assay. ds ligation assays on chromatinized and naked DNA were performed as described in ref. 9. Reactions with Ku heterodimer or DNA-PK were preincubated for 30 min at 30°C with the indicated amounts of proteins in the kinase reaction buffer (25 mM Hepes, pH 7.5/10 mM MgCl2/1mMDTT/0.1% Nonidet P-40/20% glycerol/4 mM ATP). Ligation was started by diluting the kinase reaction mixture 1:5 with water, adding an appropriate volume of 5× ligation buffer and the indicated amounts of LX, and transferring it to 37°C. Further details of the ds ligation assay can be found in Supporting Materials and Methods.
Kinase Assay with Core Histones and Linker Histones As Substrates. Purified calf thymus linker histone H1 or individual core histones (Roche) was incubated with 5 μg of purified DNA-PK in a reaction mixture containing 25 mM Hepes (pH 7.5), 10 mM MgCl2, 1 mM DTT, 0.1% Nonidet P-40, 20% glycerol, 4 mM ATP, and 0.3 nCi (1 Ci = 37 GBq) of [γ-32P]ATP in the presence of 5 mg of dsDNA cellulose (Amersham Biosciences) and incubated for 1 h at 30°C. After incubation, 2 ml of buffer containing 10 mM Tris·HCl, pH 8.0, and 1.5 M NaCl was added to detach linker histones from the DNA and the reactions were spun on Vectaspin columns (Whatman) with 0.2-μm cutoff sizes to separate linker histones from dsDNA cellulose. The flowthrough was spun on a Vivaspin 500 concentrator (Sartorius) with a cutoff size of 50 kDa to separate linker histones from DNA-PK, and aliquots were analyzed by standard protein 12% SDS/PAGE.
Bulk Phosphorylation and Purification of Linker Histones. We incubated 25 μg of purified calf thymus linker histone H1 (Roche) with 25 μg of purified DNA-PK in scaled-up standard kinase reactions that were processed and concentrated as described above. Vivaspin columns were pretreated with nonionic detergent according to the manufacturer's instructions to minimize nonspecific protein adsorption. Control reactions were set up under identical conditions in the presence of [γ-32P]ATP and analyzed by SDS/PAGE as described above.
EMSA of Reconstituted Chromatin and Ku–DNA–H1 Complexes. To examine binding of H1 histones to Ku–DNA complexes, 70 fmol of the radioactively labeled, 445-bp AflIII-PstI Bluescript fragment was preincubated with 150 ng of recombinant purified Ku70/80 complex in 40 mM Tris·HCl, pH 8.0/0.4 mM EDTA/2 mM 2-mercaptoethanol and incubated for 30 min at room temperature. The indicated amounts of histone H1 were added to the reaction mixture and incubated for a further 30 min in the same buffer. The formation of Ku–DNA–H1 complexes was analyzed by nucleoprotein agarose gel electrophoresis as described above.
Results
Linker Histone H1 Inhibits Ligation by DNA LX. To examine the impact of chromatin structure on NHEJ, a dinucleosome substrate was generated by assembling purified chicken erythrocyte histone octamers on a radioactively labeled 445-bp dsDNA fragment. Chromatosomes were prepared by addition of histone H1 purified from chicken erythrocytes or calf thymus to the dinucleosome substrate. The efficiency of DNA-nucleoprotein assembly was monitored by nucleoprotein agarose gel electrophoresis (Fig. 1A Left). The assembled dinucleosomes were used as a substrate for ligation by baculovirus-expressed DNA LX complexes. Dinucleosomes were almost as efficient a substrate for LX ligation as naked DNA (Fig. 1 A, lanes 5 and 6). In marked contrast, the addition of histone H1 to the assembled nucleosome resulted in nearly complete inhibition of ligation (Fig. 1 A, lane 7). Similar levels of inhibition were obtained by using histone H1 derived from chicken erythrocytes or calf thymus (data not shown). No ligation was detected after immunodepletion of LX by using α-XRCC4, verifying that the activity can be attributed to LX (Fig. 1 A). Because linker histones have been reported to restrict the mobility of nucleosomes on DNA (14), we examined the impact of histone H1 on ligation in the absence of core histones. The addition of histone H1 to naked DNA also inhibited ligation by LX (Fig. 1B). Histone H1 inhibited LX ligation on naked DNA and dinucleosome substrates to similar extents (Fig. 1B). We conclude that the binding of histone H1 to the DNA is responsible for LX inhibition rather than its ability to restrict nucleosome mobility.
Fig. 1.
LX mediated ds ligation on reconstituted dinucleosomes and chromatosomes. (A) (Left) The efficiency of reconstitution of dinucleosomes and chromatosomes was analyzed by nucleoprotein agarose gel electrophoresis (lanes 2–4). Under empirically optimized conditions, nearly 100% efficiency of dinucleosome packing is achieved (compare lanes 1 and 2). (Right) The efficiency of ligation with naked DNA (lane 5), dinucleosome (lane 6), and chromatosome (lane 7) substrates. Immunodepletion of LX abolishes ligation (lanes 8–10). PI, Preimmune serum; α-LX, immunodepletion with α-XRCC4 antibodies. (B) Histone H1 inhibits LX ligation in the presence or absence of histone octamers. Reconstituted dinucleosomes (lanes 1–4) or corresponding equimolar concentrations of naked DNA (lanes 5–8) were preincubated with histone H1, and the resulting histone–DNA complexes were used as substrates for ligation by the LX complex. The presence of histone H1 causes a similar level of LX inhibition on both substrates. (All ligation reactions shown in all figures were repeated three to six times.)
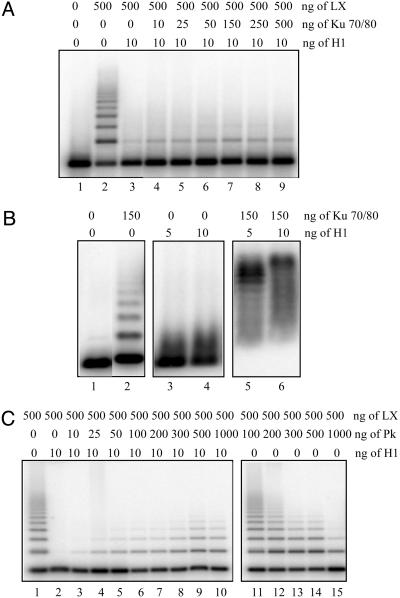
DNA-PK Rescues Ligation in the Presence of Histone H1. Previous studies have shown that Ku helps to recruit LX to the DNA ends (29, 30). We previously showed that Ku can stimulate ligation by LX over a narrow range of Ku/DNA ratios, with inhibition being observed at high Ku concentrations due to inhibition of inward translocation of Ku when multiple Ku molecules are bound to DNA substrates (9, 11). Therefore, we examined whether Ku could overcome the inhibition of ligation by histone H1. At all concentrations of Ku examined, including concentrations that stimulated ligation on naked DNA (50–150 ng; see ref. 9), the efficiency of ligation remained low (Fig. 2A). Analysis of DNA binding by EMSA, however, showed that Ku could bind to a histone H1-bound DNA substrate (Fig. 2B). We next examined whether the presence of the DNA-PK complex that included the DNA-PKcs might facilitate ligation in the presence of histone H1. After preincubation of histone H1-bound DNA with increasing concentrations of purified DNA-PK before addition of LX, a significant recovery of ligation was observed (Fig. 2C, lanes 2–10). A similar level of recovery of ligation was also observed in experiments by using a chromatosome substrate (dinuclesomes plus histone H1; data not shown). Thus, inhibition can be attributed to the presence of histone H1 alone, and all subsequent studies have been carried out by using DNA with bound histone H1 as a substrate. In contrast, the addition of DNA-PK to the naked DNA substrate showed no stimulation of ligation (Fig. 2C, lanes 11–15). Indeed, higher concentrations of purified DNA-PK were inhibitory. It is noteworthy that 500 ng of DNA-PK complex contains 150 ng of Ku, an amount that is able to stimulate the reaction on naked DNA (9). Taken together, these results show that the presence of the DNA-PK complex inhibits the stimulation of LX by Ku on naked DNA, yet reconstitutes ligation in the presence of histone H1. We next examined whether the stimulatory ability of DNA-PK requires its kinase activity. The recovery of ligation depends on the presence of ATP and is inhibited by LY294002, a specific DNA-PK inhibitor (Fig. 3 A and B) (31, 32). The inhibition of DNA-PK activity by LY294002 under these same conditions has been demonstrated in vivo (33). To verify that DNA-PK is the factor stimulating ligation and being inhibited by LY294002, we demonstrated that we were unable to detect either ATM or ATR in our purified DNA-PK preparation by Western blotting (Fig. 3C) and that immunodepletion with α-Ku80 antibodies abolished ligation in the presence of histone H1 but did not affect ligation on naked DNA (Fig. 3D). Taken together, these results demonstrate that the presence of DNA-PK in an inactive state is insufficient to rescue ligation and strongly supports the conclusion that the kinase activity of DNA-PK is essential for LX ligation in the presence of linker histones.
Fig. 2.
Rescue of LX ligation reaction by DNA-PK. (A) Addition of Ku does not overcome the histone H1-dependent inhibition of LX ligation, even at concentrations that stimulate ds ligation on naked DNA (see ref. 9). (B) Ku can bind to a histone H1-bound DNA substrate. EMSA analysis was carried out in the presence of Ku alone (lane 2), histone H1 alone (lanes 3 and 4), and both proteins (lanes 5 and 6). The DNA substrate was preincubated with the indicated amount of histone H1, and 150 ng of Ku was subsequently added, followed by a further 30-min incubation before EMSA. (C) Increasing amounts of purified DNA-PK overcome the histone H1-dependent inhibition of LX ligation. DNA–H1 complexes were preincubated with DNA-PK under conditions permitting efficient phosphorylation before initiation of LX ligation (lanes 1–10, with the peak at lane 9). DNA-PK inhibits ligation on naked DNA substrates under identical conditions (lanes 11–15).
Fig. 3.
The phosphorylation activity of DNA-PK is essential to rescue LX ligation in the presence of histone H1. (A) The recovery of LX ligation by DNA-PK is ATP-dependent. (B) The recovery of LX ligation is inhibited by the presence of the DNA-PK-specific inhibitor, LY294002. LY294002 does not inhibit LX ligation in the absence of DNA-PK (compare lanes 2 and 3). Note that the LX complex is preadenylated; therefore, ATP is not required for ligation in vitro. (C) ATM and ATR are not detected in purified DNA-PK. (D) Immunodepletion of Ku inhibits ligation. PI, Preimmune serum; α-Ku, α-Ku80 antibody.
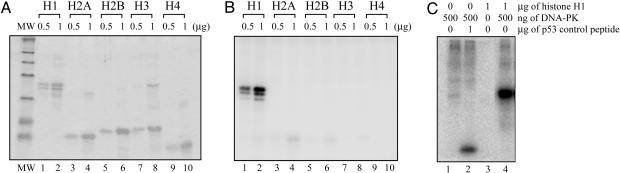
DNA-PK Efficiently Phosphorylates Histone H1 and Alters Its DNA-Binding Properties. Our findings raised the possibility that histone H1 might be a DNA-PK substrate and that phosphorylated histone H1 might less efficiently inhibit LX ligation. Whereas none of the core histones were phosphorylated by DNA-PK, DNA-PK efficiently phosphorylated histone H1 under the conditions used to examine ligation (Fig. 4 A and B). To estimate the efficiency of histone H1 as a DNA-PK substrate, we compared the ability of DNA-PK to phosphorylate equal amounts of histone H1 and a standard p53-derived peptide (Promega) routinely used to monitor DNA-PK activity. A stronger signal was detected for histone H1 compared to the p53 peptide, indicating that histone H1 is an efficient DNA-PK substrate (Fig. 4C).
Fig. 4.
Histone H1 is an efficient DNA-PK substrate. (A) SDS/PAGE analysis of histone H1 and core histones after phosphorylation by DNA-PK. After kinase assays, the histone substrates were purified by using Vivaspin concentrators with 50-kDa cutoff sizes, loaded onto 12% SDS/PAGE, and visualized by Coomassie blue staining. (B) PhosphorImager detection of [γ-32P]ATP by using the gel shown in A.(C) Comparison of DNA-PK-mediated phosphorylation of histone H1 to that of a standard p53-derived control substrate (Promega).
To investigate the impact of histone H1 phosphorylation on ligation by LX, we carried out bulk phosphorylation reactions, repurified phosphorylated histone H1, and compared the ability of nonphosphorylated (mock-treated) and phosphorylated histone H1 to bind DNA in an EMSA assay by using our standard ligation substrate. Binding of phosphorylated histone H1 resulted in a reduced band shift evident by “smearing” detected for all of the concentrations tested (Fig. 5A). This result suggests that phosphorylated histone H1 has impaired DNA-binding capacity. Consistent with the results shown in Fig. 1B, the addition of mock-treated (nonphosphorylated) histone H1 dramatically reduced DNA ligase IV activity (Fig. 5B, lanes 8–10), which could be overcome by the addition of DNA-PK to the ligation reaction (Fig. 5B, lane 7). The addition of prephosphorylated histone H1 in the presence of DNA-PK resulted in ligation comparable with that obtained by using nonphosphorylated histone H1 (Fig. 5B, lanes 2 and 7). Importantly, however, the addition of prephosphorylated histone H1 in the absence of DNA-PK resulted in increased ligation compared with that observed by using nonphosphorylated histone H1 (Fig. 5B, compare lanes 3–5 with lanes 8–10).
Fig. 5.
Phosphorylation of histone H1 affects DNA-binding and the ability of histone H1 to inhibit LX ligation. (A) EMSA analysis of the DNA-binding efficiency of histone H1 after phosphorylation by DNA-PK. Bulk phosphorylation reactions were carried out, and histone H1 was repurified from the reaction mixture as described in Materials and Methods. Nonphosphorylated (mock-treated) histone H1 was repurified from an identical reaction carried out in the absence of ATP. (B) LX ligation carried out after incubation of the DNA substrate with histone H1 prephosphorylated by DNA-PK or mock-treated nonphosphorylated histone H1. After in vitro phosphorylation, histone H1 was repurified to remove DNA-PK. Lanes 2 and 7 represent controls for which DNA-PK was added to the ligation reaction.
Taken together, these findings suggest that the mechanism underlying the rescue of ligation by DNA-PK is its capacity to phosphorylate histone H1, which alters DNA-binding capacity of histone H1 and results in a significant reduction in its ability to inhibit ligation catalyzed by LX.
Discussion
We have shown that the presence of two nucleosomal cores reconstituted on a 445-bp DNA fragment does not inhibit ligation by LX when compared with naked DNA, suggesting that nucleosomal cores do not restrict the access to or positioning of LX at the DNA end. In contrast, the addition of stoichiometric amounts of histone H1 completely inhibits ligation on substrates containing core nucleosomes as well as naked DNA substrates. Significantly, we show that this inhibitory effect can be at least partially overcome by the addition of DNA-PK in a reaction that requires DNA-PK kinase activity. Moreover, we show that phosphorylation of histone H1 by DNA-PK decreases its ability to bind DNA and inhibit LX ligation. Our findings are consistent with a model whereby DNA-PK phosphorylates histone H1 in the vicinity of the break, thereby decreasing its association with DNA. Decreased DNA binding of phosphorylated histone H1 has been demonstrated recently by using fluorescence recovery after photobleaching (34). We have shown that LX is most efficient ds ligase on DNA substrates of ≈400 bp and that the ability of Ku to stimulate LX ligation requires freedom for Ku to translocate along the DNA molecule (9). Histone H1 does not inhibit the ability of Ku to bind to DNA but does inhibit its ability to stimulate LX ligation. Histone H1 may, therefore, limit the ability of Ku to recruit LX and/or translocate inwards. Thus, we propose that, at a DNA end containing histone H1 molecules, Ku is able to bind but does not have freedom to move internally, a prerequisite for its ability to stimulate LX ligation. Recruitment and activation of DNA-PKcs results in localized histone H1 phosphorylation, its decreased DNA-binding potential, and, hence, restoration of the ability of Ku to recruit LX and stimulate ligation. Recently, we have shown that DNA-PK can contribute (together with ATM) to the phosphorylation of the histone variant, H2AX, in the presence of DNA DSBs (33). Thus, the ability of DNA-PK to phosphorylate components functioning in the higher-order structure of DNA is not without precedence. The recovery of LX ligation by the presence of DNA-PKcs, although marked, does not return to the level seen with naked DNA. One possible explanation is that we have observed that ATP slightly inhibits LX ligation in our in vitro assay. Thus, the ATP added to promote DNA-PK activity may also inhibit ligation. Secondly, our histone preparation is a mixture of multiple isoforms of H1. Although all isoforms contain multiple serine sites, the consensus DNA-PK motif (SQE) is found only in one isoform (National Center for Biotechnology Information Protein Database accession no. AAN06701). In vivo, there may be additional processes to remove histone H1 from the DNA as well as additional kinases acting on other isoforms. Interestingly, it has also been reported that histone H1 is dephosphorylated after ionizing radiation through an ATM-dependent mechanism (35). Although the effect is maximal after 1 h (when the majority of DSB rejoining has occurred), it raises the possibility that additional factors may impact histone H1 phosphorylation/dephosphorylation in response to DNA damage. Contrary to a previous report (36), we show that DNA-PK can efficiently phosphorylate histone H1.
Previous attempts to reconstitute NHEJ in vitro from fully purified components have failed to show a dependence on DNA-PK. Examination of a substrate containing histone H1 has provided the first demonstration of DNA-PK-dependent endjoining reconstituted by using purified or in-vitro-expressed proteins. In contrast, NHEJ activity by partially purified cell-free extracts has been shown to require DNA-PK activity in a reaction that uses naked DNA as a substrate (8, 37), and DNA-PK has been shown to be required for LX-mediated ligation in the presence of a partially purified factor of ≈200 kDa (8). Interestingly, we have observed that addition of naked DNA to cell-free extracts prepared under conditions that support NHEJ results in the binding of histone H1 to DNA (data not shown). Thus, it is possible that the requirement of DNA-PK for NHEJ in cell-free extracts can also be explained by a role for DNA-PK in phosphorylating linker histones. In vivo, differing levels of requirement for DNA-PK for NHEJ have been observed. The rejoining of radiation-induced chromosomal breaks is clearly DNA-PK-dependent (38). The rejoining of the coding junctions generated at V(D)J junctions requires DNA-PK, most likely because of its ability to phosphorylate and activate Artemis, which cleaves the hairpin junctions at the coding ends (3). In contrast, signal junctions are rejoined in a manner that is largely DNA-PK-independent (39, 40). The assay to monitor signal joint formation involves the introduction of naked DNA substrates into cells, and it is possible that rejoining occurs before the DNA interacts with histones.
Other examples of inhibition of DNA repair or DNA metabolic events by the core histones and the linker histones has been described. Histone H1 inhibits the activities of DNA topoisomerase I and DNA primase in a dose-dependent manner, reminiscent of that shown here for LX inhibition of ligation (41, 42). In contrast, DNA ligase I activity is not inhibited by the presence of nucleosomes and linker histone H1 (25). In this case, nick ligation was monitored, which occurs efficiently on short DNA oligonucleotides. It is possible that nick ligation does not require extensive contact or movement of the ligase along the DNA molecule. Nucleotide excision repair is another example for which the presence of nucleosomes is also inhibitory (16–20). Furthermore, chromatin remodeling factors, which serve to recover nucleotide excision repair on synthetic dinucleosomes, have been shown to be inhibited by the presence of histone H1 within the nucleosomal arrays (21, 22, 43). Interestingly, this inhibition can be overcome by linker histone phosphorylation with a known linker histone kinase, Xenopus Cdc2/Cyclin B. The linker histone phosphorylation was originally proposed to play a regulatory role in chromatin condensation by lowering the protein's affinity for DNA (44). The phosphorylation of histone H1 has also been shown to reduce the inhibitory activity of histone H1 on DNA primase activity (41) and linker histone phosphorylation by Cyclin-dependent kinase 2 was found to have a vital role in transcription activation of the mouse mammary tumor virus promoter by the glucocorticoid receptor (45). Finally, it has recently been shown that HHO1, a candidate linker histone in yeast, is inhibitory to DNA DSB repair by homologous recombination (46). These data, taken together with our results, suggest the possibility that linker histones inhibit many enzymes involved in DNA repair/metabolism. Thus, mechanisms must have evolved to release this inhibition and phosphorylation of histone H1 could represent one such mechanism. Phosphorylation of histone H1 by DNA-PK would allow release of histone H1 in a highly localized fashion within the vicinity of the break. Our results suggest that DNA-PK, with its extremely high affinity for DNA ends, and the ability to recruit the LX complex (47) would make an ideal candidate for a DSB-specific linker histone kinase.
Supplementary Material
Acknowledgments
This work was supported by the Medical Research Council, the Human Frontiers Science Program, the Primary Immunodeficiency Association, the Leukaemia Research Fund, and European Union Grant FIGH-CT1999.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: NHEJ, nonhomologous end-joining; ds, double-stranded; DSB, ds breaks; DNA-PK, DNA-dependent protein kinase; DNA-PKScs, DNA-PK catalytic subunit; LX, ligase IV/XRCC4; ATM, ataxia-telangiectasia-mutated kinase.
References
- 1.Jeggo, P. A. (1998) Adv. Genet. 38, 185–211. [DOI] [PubMed] [Google Scholar]
- 2.Grawunder, U., Zimmer, D., Fugmann, S., Schwarz, K. & Lieber, M. R. (1998) Mol. Cell 2, 477–484. [DOI] [PubMed] [Google Scholar]
- 3.Ma, Y., Pannicke, U., Schwarz, K. & Lieber, M. R. (2002) Cell 108, 781–794. [DOI] [PubMed] [Google Scholar]
- 4.Leber, R., Wise, T. W., Mizuta, R. & Meek, K. (1998) J. Biol. Chem. 273, 1794–1801. [DOI] [PubMed] [Google Scholar]
- 5.Ding, Q., Reddy, Y. V., Wang, W., Woods, T., Douglas, P., Ramsden, D. A., Lees-Miller, S. P. & Meek, K. (2003) Mol. Cell. Biol. 23, 5836–5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan, D. W., Chen, B. P., Prithivirajsingh, S., Kurimasa, A., Story, M. D., Qin, J. & Chen, D. J. (2002) Genes Dev. 16, 2333–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang, J. & Dynan, W. S. (2002) Nucleic Acids Res. 30, 667–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Udayakumar, D., Bladen, C. L., Hudson, F. Z. & Dynan, W. S. (2003) J. Biol. Chem. 278, 41631–41635. [DOI] [PubMed] [Google Scholar]
- 9.Kysela, B., Doherty, A. J., Chovanec, M., Stiff, T., Ameer-Beg, S. M., Vojnovic, B., Girard, P. M. & Jeggo, P. A. (2003) J. Biol. Chem. 278, 22466–22474. [DOI] [PubMed] [Google Scholar]
- 10.Walker, J. R., Corpina, R. A. & Goldberg, J. (2001) Nature 412, 607–614. [DOI] [PubMed] [Google Scholar]
- 11.Ramsden, D. A. & Gellert, M. (1998) EMBO J. 17, 609–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luger, K., Mader, A. W., Richmond, R. K., Sargent, D. F. & Richmond, T. J. (1997) Nature 389, 251–260. [DOI] [PubMed] [Google Scholar]
- 13.Fletcher, T. M. & Hansen, J. C. (1996) Crit. Rev. Eukaryotic Gene Expression 6, 149–188. [DOI] [PubMed] [Google Scholar]
- 14.Pennings, S., Meersseman, G. & Bradbury, E. M. (1994) Proc. Natl. Acad. Sci. USA 91, 10275–10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zlatanova, J., Caiafa, P. & Van Holde, K. (2000) FASEB J. 14, 1697–1704. [DOI] [PubMed] [Google Scholar]
- 16.Cleaver, J. E. (1977) Nature 270, 451–453. [DOI] [PubMed] [Google Scholar]
- 17.Wang, Z. G., Wu, X. H. & Friedberg, E. C. (1991) J. Biol. Chem. 266, 22472–22478. [PubMed] [Google Scholar]
- 18.Hara, R., Mo, J. & Sancar, A. (2000) Mol. Cell. Biol. 20, 9173–9181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nilsen, H., Lindahl, T. & Verreault, A. (2002) EMBO J. 21, 5943–5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beard, B. C., Wilson, S. H. & Smerdon, M. J. (2003) Proc. Natl. Acad. Sci. USA 100, 7465–7470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ura, K., Araki, M., Saeki, H., Masutani, C., Ito, T., Iwai, S., Mizukoshi, T., Kaneda, Y. & Hanaoka, F. (2001) EMBO J. 20, 2004–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hara, R. & Sancar, A. (2003) Mol. Cell. Biol. 23, 4121–4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hara, R. & Sancar, A. (2002) Mol. Cell. Biol. 22, 6779–6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huggins, C. F., Chafin, D. R., Aoyagi, S., Henricksen, L. A., Bambara, R. A. & Hayes, J. J. (2002) Mol. Cell 10, 1201–1211. [DOI] [PubMed] [Google Scholar]
- 25.Chafin, D. R., Vitolo, J. M., Henricksen, L. A., Bambara, R. A. & Hayes, J. J. (2000) EMBO J. 19, 5492–5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dvir, A., Stein, L. Y., Calore, B. L. & Dynan, W. S. (1993) J. Biol. Chem. 268, 10440–10447. [PubMed] [Google Scholar]
- 27.Wolffe, A. P. & Ura, K. (1997) Methods 12, 10–19. [DOI] [PubMed] [Google Scholar]
- 28.Stein, A. (1989) Methods Enzymol. 170, 585–603. [DOI] [PubMed] [Google Scholar]
- 29.Nick McElhinny, S. A., Snowden, C. M., McCarville, J. & Ramsden, D. A. (2000) Mol. Cell. Biol. 20, 2996–3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cary, R. B., Peterson, S. R., Wang, J., Bear, D. G., Bradbury, E. M. & Chen, D. J. (1997) Proc. Natl. Acad. Sci. USA 94, 4267–4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Izzard, R. A., Jackson, S. P. & Smith, G. C. (1999) Cancer Res. 59, 2581–2586. [PubMed] [Google Scholar]
- 32.Rosenzweig, K. E., Youmell, M. B., Palayoor, S. T. & Price, B. D. (1997) Clin. Cancer Res. 3, 1149–1156. [PubMed] [Google Scholar]
- 33.Stiff, T., O'Driscoll, M., Rief, N., Iwabuchi, K., Lobrich, M. & Jeggo, P. A. (2004) Cancer Res. 64, 2390–2396. [DOI] [PubMed] [Google Scholar]
- 34.Dou, Y., Bowen, J., Liu, Y. & Gorovsky, M. A. (2002) J. Cell Biol. 158, 1161–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo, C. Y., Wang, Y., Brautigan, D. L. & Larner, J. M. (1999) J. Biol. Chem. 274, 18715–18720. [DOI] [PubMed] [Google Scholar]
- 36.Carter, T., Vancurova, I., Sun, I., Lou, W. & DeLeon, S. (1990) Mol. Cell. Biol. 10, 6460–6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baumann, P. & West, S. C. (1998) Proc. Natl. Acad. Sci. USA 95, 14066–14070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blunt, T., Finnie, N. J., Taccioli, G. E., Smith, G. C. M., Demengeot, J., Gottlieb, T. M., Mizuta, R., Varghese, A. J., Alt, F. W., Jeggo, P. A. & Jackson, S. P. (1995) Cell 80, 813–823. [DOI] [PubMed] [Google Scholar]
- 39.Lieber, M. R., Hesse, J. E., Lewis, S., Bosma, G. C., Rosenberg, N., Mizuuchi, K., Bosma, M. J. & Gellert, M. (1988) Cell 55, 7–16. [DOI] [PubMed] [Google Scholar]
- 40.Taccioli, G. E., Cheng, H.-L., Varghese, A. J., Whitmore, G. & Alt, F. W. (1994) J. Biol. Chem. 269, 7439–7442. [PubMed] [Google Scholar]
- 41.Takada, S., Magira, T. & Yamamura, M. (1989) Biochem. Biophys. Res. Commun. 160, 711–714. [DOI] [PubMed] [Google Scholar]
- 42.Richter, A. & Kapitza, M. (1991) FEBS Lett. 294, 125–128. [DOI] [PubMed] [Google Scholar]
- 43.Horn, P. J., Carruthers, L. M., Logie, C., Hill, D. A., Solomon, M. J., Wade, P. A., Imbalzano, A. N., Hansen, J. C. & Peterson, C. L. (2002) Nat. Struct. Biol. 9, 263–267. [DOI] [PubMed] [Google Scholar]
- 44.Roth, S. Y. & Allis, C. D. (1992) Trends Biochem. Sci. 17, 93–98. [DOI] [PubMed] [Google Scholar]
- 45.Bhattacharjee, R. N., Banks, G. C., Trotter, K. W., Lee, H. L. & Archer, T. K. (2001) Mol. Cell. Biol. 21, 5417–5425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Downs, J. A., Kosmidou, E., Morgan, A. & Jackson, S. P. (2003) Mol. Cell 11, 1685–1692. [DOI] [PubMed] [Google Scholar]
- 47.Dynan, W. S. & Hoo, S. (1998) Nucleic Acids Res. 26, 1551–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.