Abstract
The development of breast cancer is a complex process that involves multiple genes at many stages, from initial cell cycle dysregulation to disease progression. To identify genetic variations that influence this process, we conducted a large-scale association study using a collection of German cases and controls and >25,000 SNPs located within 16,000 genes. One of the loci identified was located on chromosome 11q13 [odds ratio (OR) = 1.85, P = 0.017]. The initial association was subsequently tested in two independent breast cancer collections. In both sample sets, the frequency of the susceptibility allele was increased in the cases (OR = 1.6, P = 0.01). The susceptibility allele was also associated with an increase in cancer family history (P = 0.1). Fine mapping showed that the region of association extends ≈300 kb and spans several genes, including the gene encoding the nuclear mitotic apparatus protein (NuMA). A nonsynonymous SNP (A794G) in NuMA was identified that showed a stronger association with breast cancer risk than the initial marker SNP (OR = 2.8, P = 0.005 initial sample; OR = 2.1, P = 0.002 combined). NuMA is a cell cycle-related protein essential for normal mitosis that is degraded in early apoptosis. NuMA-retinoic acid receptor α fusion proteins have been described in acute promyelocytic leukemia. Although the potential functional relevance of the A794G variation requires further biological validation, we conclude that variations in the NuMA gene are likely responsible for the observed increased breast cancer risk.
Keywords: genome-wide association, single-nucleotide polymorphisms, coiled-coil domain, early apoptosis
The role of genetic factors in the epidemiology and pathogenesis of both sporadic and familial breast cancer is now well established (1, 2). Less than 10% of breast cancer patients develop the disease due to highly penetrant germ-line mutations in susceptibility genes such as BRCA1, BRCA2, and P53 (3–5). However, more common genetic variations with modest effects likely contribute to susceptibility in sporadic breast cancer patients (6–8). One strategy to identify such low penetrance genes and their predisposing variants is through association studies, in which the prevalence of alleles and genotypes for common polymorphisms is compared between cancer cases and matched controls (9). Ideally, all variants that may be directly involved in the disease etiology would be tested; however, these variants are largely unknown, and exhaustive typing of all common variations is currently not feasible. Therefore, the effect of causal variations on disease risk is measured indirectly by genetic markers assumed to be in linkage disequilibrium with those that are functionally relevant. Many breast cancer association studies using variations in candidate genes have been conducted with mixed success and low concordance between studies. Variations in certain metabolizing enzymes, such as cytochrome P450 enzymes (10), N-acetyltransferases (11), and glutathione-S-transferases (12) are among the most consistently reported ones to confer increased risk for breast cancer (13). More recently, variations in the G2 checkpoint kinase CHEK2 have been substantiated as risk factors for breast and other cancers (14, 15).
Lately, there has been increasing interest in the use of whole-genome association methods to identify genes involved in complex trait variation (16, 17). To date, however, few large-scale studies have been reported (18, 19). To identify genes and variants that influence breast cancer susceptibility, we conducted a large-scale case-control study using 25,000 genome-wide SNPs. To increase the likelihood of finding functionally relevant variations, we selected SNPs located within 10 kb of ≈16,000 known or predicted genes. In this report, we describe the identification of a 300-kb breast cancer susceptibility locus on chromosome 11q13 that harbors several potential candidate genes, including the gene encoding the nuclear mitotic apparatus protein NuMA [approved Human Genome Organization (HUGO) symbol, NUMA1].
Materials and Methods
Study Subjects. The sample used for the large-scale association study (referred to as the discovery sample) comprised 254 breast cancer patients attending the Frauenklinik Innenstadt, University of Munich (Munich). Lymph node status was positive at time of assessment in 94 cases (37%), and 18 cases (7%) had known distant metastases. Twenty-seven cases (11%) reported a family history of breast cancer (one or more first or second degree relatives), and 73 cases (29%) reported a family history of any cancer. The median age at diagnosis was 56 yr (range = 23–87 yr). A total of 268 controls with a median age of 57 yr (range = 17–88 yr) were recruited from patients with benign disease attending the clinic during the same period. Controls with a family history of breast or ovarian cancer were excluded. Both parents of each study participant were reported to be of German descent.
The German replication sample consisted of 188 cases and 150 controls recruited at the Department of Obstetrics and Gynecology, Technical University of Munich (Munich). The majority of breast cancer cases were recruited at preoperative visits, and female controls were recruited from healthy individuals or patients with nonmalignant diagnoses. Median age of diagnosis for cases was 59 yr (range = 22–87 yr), and median age of controls was 50 yr (range = 19–91 yr). All but two participants reported both parents to be of German descent. The two exceptions each reported one parent of non-German, Eastern European origin.
The Australian replication sample comprised 180 breast cancer cases recruited by the Pathology Department of Gold Coast Hospital or by the Genomics Research Center, Southport. Median age of diagnosis was 50 yr (range = 24–74 yr). Controls consisted of 180 healthy volunteers recruited through the Genomics Research Center. Only controls with no family history of cancer or precancerous conditions were included. Controls were individually age matched to cases (±5 yr). Median age of controls was 60 yr (range = 28–94 yr).
All subjects involved in our studies signed a written informed consent, and the institutional ethics committees of participating institutions approved the experimental protocols.
SNP Markers and Genotyping. A set of 25,494 SNP markers was selected from a collection of 125,799 experimentally validated polymorphic variations (20). This set was limited to SNPs located within gene coding regions, minor allele frequencies >0.02 (95% have frequencies >0.1), and a target intermarker spacing of 40 kb. SNP annotation is based on the National Center for Biotechnology Information (NCBI) dbSNP database, refSNP, build 118. Genomic annotation is based on NCBI Genome Build 34. Gene annotation is based on LocusLink genes for which NCBI provided positions on the Mapview FTP site.
DNA pools were formed by combining equimolar amounts of each sample as described (21, 22). For pooled DNA assays, 25 ng of case and control DNA pools was used for amplification at each site. All PCR and homogeneous MassEXTEND reactions were conducted by using standard conditions (22). Relative allele frequency estimates were derived from area under the peak calculations of mass spectrometry measurements from four analyte aliquots as described (22). For individual genotyping, the same procedure was applied except only 2.5 ng DNA was used and only one mass spectrometry measurement was taken. The following gene-specific primer sequences were used to genotype the SNPs discussed: for rs673478, PCR1, 5′-TAATACAAAGGTGGCAGCAG-3′; PCR2, 5′-TTGACAAGGATAAGGACAAG-3′; and Extend, 5′-AAGGGGAGGTCGACTGGG-3′; for rs3750913, PCR1, 5′-CACACTCACTCTCAGCTGTG-3′; PCR2, 5′-CCATCAGGCTGAGACTGAAG-3′; and Extend, 5′-CACTCTCAGCTGTGTGCTGGGCA-3′; for rs3018301, PCR1, 5′-ACTAAGAACCTTCCTGCTCG-3′; PCR2, 5′-TCTGTCCCATGTGAGTGTTG-3′; and Extend, 5′-ACCCTCATCACCTTTCAC-3′.
Statistical Analysis. Tests of association between disease status and each SNP by using pooled DNA were carried out in a similar fashion as explained in ref. 23. Sources of measurement variation included pool formation, PCR/mass extension, and chip measurement. When three or more replicate measurements of a SNP were available within a model level, the corresponding variance component was estimated from the data. Otherwise, the following historical laboratory averages were used: pool formation = 5.0 × 10–5, PCR/mass extension = 1.7 × 10–4, and chip measurement = 1.0 × 10–4. Tests of association using individual genotypes were carried out by using a χ2 test of heterogeneity based on allele and genotype frequencies. Selected tests of association involving contingency tables with rare or missing cells were carried out by using Fisher's exact test. The DerSimonian–Laird random effects metaanalysis method (24) was used for the analysis of replication samples to test for the consistency of association while permitting allele frequencies to differ among samples. All tests of allele frequencies involving only replication samples are one-sided, confirming the effect observed in the discovery sample. P values were derived by using the log odds of each contrast and their standard errors. Multiple approaches were explored in an effort to identify haplotypes demonstrating a stronger association with disease status than single sites. These approaches included analyses of five-SNP haplotypes and subsets thereof using the coalescent theory-based phase 2.0 (25) and the score method that relies on the expectation-maximization (EM) algorithm (26). No attempt was made to correct P values for multiple testing. Rather, P values are provided to compare the relative strength of association from multiple dependent (e.g., SNPs within samples) and independent (e.g., SNPs between samples) sources of information. P values <0.05 are referred to as statistically significant.
Results
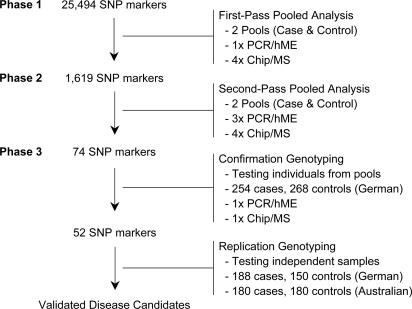
Identification of Breast Cancer Susceptibility Locus on 11q13. Using DNA samples from 254 clinically diagnosed breast cancer cases and 268 matched controls, we conducted a genome-wide study consisting of 25,494 SNPs to discover variants associated with increased breast cancer risk (27). The screening strategy involved DNA pooling and a three-phase filtering procedure (Fig. 1). In the first phase, a single PCR and primer extension reaction was conducted for each SNP on one DNA pool each for cases and controls, respectively. Four mass spectrometric measurements of each extension product were taken, and relative allele frequencies were calculated and compared between the pools. In the second phase, the 1,619 SNPs (≈5%) with the most statistically significant associations were tested again, this time in triplicate for each DNA pool, and the results were compared. In the third phase, the 74 most significant SNPs (≈5%) from step two were individually genotyped. Fifty-two SNPs were confirmed to have statistically significant differences between cases and controls (P < 0.05, Fig. 2). One of the loci identified was located on chromosome 11q13. A C-to-T transition in intron 1 of the gene LOC220074 (rs673478) was associated with breast cancer susceptibility [odds ratio (OR) = 1.8, P = 0.01]. The susceptibility allele (C) was increased in the case group from 4.7% to 7.7% (Table 1).
Fig. 1.
Schematic presentation of genome-wide association study from pool-based initial screen to replicated candidate genes. Phases 1 and 2 are conducted by using DNA pools yielding allele frequencies, and all subsequent steps involve genotyping of individual samples. hME, homogeneous Mass-EXTEND. See Results for more details.
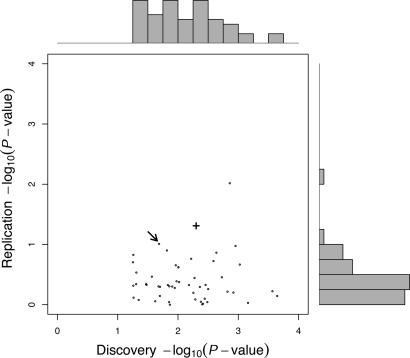
Fig. 2.
Distribution of discovery and replication P values for the 52 SNPs selected from the third phase (genotype confirmation) of the large-scale association study. The inner plot shows the P values (–log10 transformed) comparing allele frequencies between cases and controls in the discovery sample (x-axis, two-sided test) versus the comparison in the combined replication samples (y-axis, one-sided test based on effect observed in the discovery sample). The top and right outer margins show the univariate distributions of the discovery and replication P values, respectively. An arrow points to the result for marker rs673478. The result for the NuMA nonsynonymous variation rs3750913 identified during subsequent fine mapping is indicated as a bold +.
Table 1. Analysis of association of SNPs in LOC220074 and NuMA with breast cancer status.
| Sample | N* | MAF† | OR‡ | P value§ | Genotype frequencies¶ | P value∥ | |||
|---|---|---|---|---|---|---|---|---|---|
| rs673478 | |||||||||
| LOC220074 Intron 1 (T→C) | C | TT | TC | CC | |||||
| German | Controls | 265 | 0.047 | 1.85 | 0.017 | 0.91 | 0.09 | 0.00 | 0.04 |
| (Discovery) | Cases | 244 | 0.077 | 0.84 | 0.14 | 0.01 | |||
| German | Controls | 144 | 0.045 | 1.24 | 0.272 | 0.91 | 0.09 | 0.00 | 0.53 |
| (Replication) | Cases | 189 | 0.056 | 0.89 | 0.11 | 0.00 | |||
| Australian | Controls | 178 | 0.042 | 1.52 | 0.111 | 0.92 | 0.07 | 0.01 | 0.51 |
| (Replication) | Cases | 176 | 0.063 | 0.89 | 0.10 | 0.01 | |||
| Replication all | 1.38 | 0.098 | |||||||
| Total | 1.59 | 0.011 | |||||||
| rs3750913 | |||||||||
| NuMA Exon 16 (C→G, A794G) | G | CC | CG | GG | |||||
| German | Controls | 267 | 0.019 | 2.84 | 0.005 | 0.96 | 0.04 | 0.00 | 0.02 |
| (Discovery) | Cases | 246 | 0.051 | 0.90 | 0.09 | 0.00 | |||
| German | Controls | 143 | 0.017 | 2.31 | 0.055 | 0.97 | 0.03 | 0.00 | 0.09 |
| (Replication) | Cases | 190 | 0.040 | 0.92 | 0.08 | 0.00 | |||
| Australian | Controls | 180 | 0.028 | 1.43 | 0.198 | 0.94 | 0.06 | 0.00 | 0.54 |
| (Replication) | Cases | 179 | 0.039 | 0.93 | 0.07 | 0.01 | |||
| Replication all | 1.72 | 0.049 | |||||||
| Total | 2.13 | 0.002 | |||||||
| rs3018301 | |||||||||
| NuMA 5′ upstream (-510 G→A) | A | GG | GA | AA | |||||
| German | Controls | 260 | 0.052 | 1.68 | 0.042 | 0.90 | 0.09 | 0.01 | 0.09 |
| (Discovery) | Cases | 244 | 0.084 | 0.84 | 0.15 | 0.01 | |||
| German | Controls | 144 | 0.045 | 1.49 | 0.127 | 0.91 | 0.09 | 0.00 | 0.45 |
| (Replication) | Cases | 190 | 0.066 | 0.87 | 0.12 | 0.01 | |||
| Australian | Controls | 178 | 0.045 | 1.55 | 0.095 | 0.92 | 0.08 | 0.01 | 0.45 |
| (Replication) | Cases | 179 | 0.067 | 0.88 | 0.11 | 0.01 | |||
| Replication all | 1.52 | 0.042 | |||||||
| Total | 1.59 | 0.008 | |||||||
Number of subjects with genotypes.
Minor relative allele frequency.
OR with reference to allele with increased frequency in cases.
P value for test comparing allele frequencies between cases and controls; tests for German discovery and all samples (Total) are two-sided; tests in replication collections are one-sided, based on the effect observed in the discovery collection.
Relative genotype frequencies.
P value for tests comparing genotype frequencies between cases and controls.
A conservative Bonferroni adjustment to yield an experiment-wide type I error rate of 0.05 would demand a test-wise P value on the order of 10–6. Given the modest sample size and low minor allele frequency, only variations with relatively large effects (OR > 4) would have adequate power to be detected. Instead, we chose to be more mindful of the role of type II error rates and apply a more liberal set of criteria in the initial phases of the study and verify true genetic effects by independent replication. In this study, we used two independent collections of breast cancer cases and controls from Germany and Australia, respectively, for replication. The distribution of P values obtained for each of the 52 SNPs selected for replication is presented in Fig. 2, comparing the results in the discovery sample with those in the combined replication samples. In both replication samples, the susceptibility allele was increased in the cases with ORs of 1.24 and 1.52, respectively (Table 1). The allele frequencies were very similar among the controls of all three samples. A combined analysis of all three samples resulted in an OR of 1.6 (P = 0.01). This finding was the second most significant result in the combined replication sample (see Fig. 2) and was therefore chosen to follow up. The most significant association identified was a variation in the ICAM region on chromosome 19q13.2, which was published recently (27).
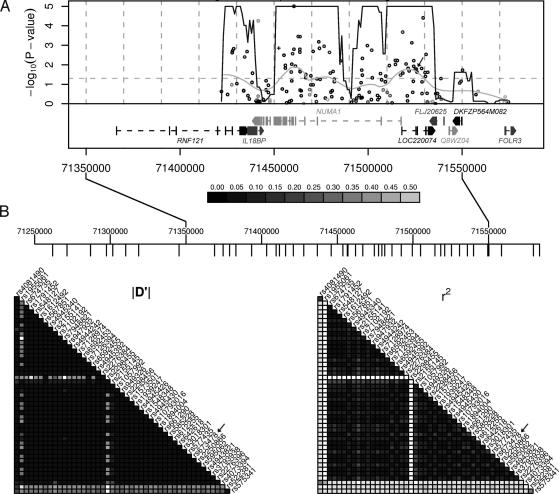
High-Linkage Disequilibrium in 300-kb Region. To fine map the region of association, we tested an additional 160 SNPs located within 100 kb of the initial marker using the discovery pools (Fig. 3A). Sixty-four of the 160 SNPs tested were significantly associated with breast cancer risk (P < 0.05). The region of highest significance extended >100 kb, suggesting relatively low haplotype diversity and a linkage disequilibrium (LD) block extending beyond the 5′ end of the region analyzed. To determine the complete extent of LD in this region, we analyzed the pair-wise relationships between the SNP genotypes available in the CEPH30 sample from the HapMap project (28) within a 500-kb window including our marker (Fig. 3B). According to this analysis, the LD block including the marker SNP rs673478 spans ≈300 kb. The fine mapping project included SNPs in all genes annotated in this window.
Fig. 3.
Association fine mapping of breast cancer susceptibility region on chromosome 11q13. (A) One hundred sixty public domain SNPs in a 100-kb window around the initial marker SNP (arrow) were compared between pools of cases and controls. Sixty-four of 160 SNPs were significant at P = 0.05 (horizontal dashed line). The nonsynonymous A794G SNP in exon 16 of NuMA is indicated (+). The x-axis corresponds to their chromosomal position, the y-axis to the test P values (–log10 scale). The continuous dark line presents the results of a goodness-of-fit test for an excess of significance (compared with 0.05) in a 10-kb sliding window assessed at 1-kb increments. The continuous light gray line is the result of a nonlinear smoothing function showing a weighted average of the P values across the region. The darkness of each point corresponds to the minor allele frequency of each SNP in the control sample (see legend below graph). The LocusLink gene annotations for NCBI genome build 34 are included. (B) Estimates of LD from HapMap CEPH30 data in a 300-kb region encompassing NuMA. Estimates of LD(|D′| and r2) are represented as gray-scale ranging from white (LD = 0) to black (LD = 1) at increments of 0.1. SNP locations are indicated as downward tick marks in the ruler above. The marker SNP rs673478 location is marked (arrow).
This 300-kb block contains seven annotated genes (Fig. 3A). The LOC220074 gene, in which the marker SNP is located, is a gene of unknown function. Downstream of this gene are another three genes encoding hypothetical proteins or proteins of unknown function: FLJ20625, Q8WZ04, and DKFZP564M082, all located around the 3′ end of the LD block. Upstream of LOC220074 are three genes that contain the majority of significant SNPs. RNF121 is a member of the RING finger 5 family, which is not well characterized to date. IL18BP codes for the Il-18-binding protein, which prevents the binding of IL-18 to its receptor, and thus inhibits IL-18-induced IFN-γ production (29). It is constitutively expressed in mononuclear cells, and its expression can be enhanced by IFN-γ. An elevated level of this protein is detected in the intestinal tissues of patients with Crohn's disease (30). Finally, the gene encoding the nuclear mitotic apparatus protein (NuMA) is located in the center of the LD block and covers ≈75 kb of genomic sequence.
Nonsynonymous Variation in NuMA Strongly Associated with Disease. NuMA is the best-described and most likely candidate in this region. The gene encodes a 236-kDa protein with a very large coiled-coil central domain (31, 32). NuMA is a cell cycle-related protein that is present throughout the nucleus in the interphase, and localizes to the spindle apparatus at mitosis (31, 33). Several studies have shown that it is essential for normal mitosis as an organizer of the mitotic spindle and for nuclear reassembly in late mitosis (34–37).
Because we had individually genotyped only one SNP in LOC220074, we selected four additional SNPs throughout the NuMA gene from the 64 SNPs that showed significant differences between cases and controls in the pool-based fine-mapping experiment and genotyped them in all three sample collections. The SNPs were located in the 5′ upstream region (rs3018301), intron 3 (rs4945430), exon 16 (rs3750913), and intron 25 (rs2852365). Except for the intron 25 marker, all SNPs confirmed to be statistically significant. The results for two of these SNPs are included in Table 1. The third significant SNP, rs4945430, was in almost complete LD with rs3018301 (r2 = 0.99) and was therefore not presented. All SNPs genotyped had low minor allele frequencies, and LD analysis confirmed low haplotype diversity in the region in agreement with the results obtained from the HapMap project (Fig. 3B). Interestingly, the most significant association was observed for rs3750913, a C-to-G polymorphism in exon 16 encoding an alanine-to-glycine variation at residue 794 (A794G). In the discovery sample, the susceptibility allele (glycine) was increased from 1.9% in the controls to 5.1% in the case group (OR = 2.8, P = 0.005), and the combined significance of all samples was P = 0.002 (OR = 2.1; Table 1). It is noteworthy that the effect was stronger in the German replication sample than in the Australian sample. All other publicly annotated coding variations in the 300-kb window (including ten nonsynonymous SNPs in NuMA, three nonsynonymous SNPs in IL18BP, and one nonsynonymous SNP in FLJ20625) were tested as part of the fine mapping and were rare in our sample. None of these SNPs showed significantly different frequencies between case and control pools. Analyses of complete haplotypes as well as those consisting of subsets of the five genotyped SNPs did not reveal any haplotype with stronger association than that observed for A794G (data not shown).
Correlation of Association with Family History of Cancer. The discovery collection of German breast cancer patients included information related to family history, age of onset, severity of disease, and type of treatment. The genotyped SNPs were tested for association with these variables (Table 2). The A794G SNP showed association with a positive family history of cancer (P = 0.1), which was reported by 73 cases. Of those homozygous for the protective allele (C), only 27% had a positive family history of cancer, compared with 39% of the heterozygotes. The one homozygote for the susceptible allele also reported family history of cancer. Comparing A794G allele frequencies between cases with a cancer family history and all controls resulted in a larger estimated genetic effect (OR = 4.45, P = 0.001). There was no significant correlation with family history of breast cancer per se, but the sample number in this group was very low (n = 25). There was also no correlation with age of diagnosis or predisposition to the development of metastases. It is noteworthy, however, that the individual homozygous for the glycine allele was younger than the lower quartile of the alanine homozygotes and received radiation therapy, which only 50% of the remaining group underwent.
Table 2. Association of rs3750913 with clinical indicators in the discovery sample.
| Genotype | N | CC (N = 222) | GC (N = 23) | GG (N = 1) | P value |
|---|---|---|---|---|---|
| Age of diagnosis (yr) | 235 | 49/56/63* | 51/57/61 | 43/43/43 | 0.441 |
| Familial history cancer | 254 | 27% (60) | 39% (9) | 100% (1) | 0.101 |
| Familial history breast cancer | 254 | 10% (23) | 9% (2) | 0% (0) | 1 |
| Lymph node metastases | 254 | 31% (68) | 22% (5) | 100% (0) | 0.8 |
| Organ metastasis | 254 | 7% (16) | 4% (1) | 0% (0) | 1 |
| Radiation therapy | 254 | 49% (109) | 52% (12) | 100% (1) | 0.22 |
Quantitative variables are expressed as first quartile/median/third quartile. Categorical variables are expressed as column percentage (count).
Discussion
In an association study by using SNPs in nearly 16,000 genes we obtained evidence that a 300-kb region on chromosome 11q13 encompassing the NuMA gene influences breast cancer risk. The effect proved to be relatively consistent in two independent samples. Of the seven genes in the defined LD block, five encoded either hypothetical or poorly described proteins, and one, IL18BP, is mainly expressed in mononuclear cells and was previously implicated in Crohn's disease. We will not attempt to speculate on a potential involvement of these genes in breast cancer predisposition but elaborate on NuMA as a likely candidate for the observed association. The NuMA gene codes for a component of the nuclear matrix and relocalizes to the spindle poles during mitosis (33), where it forms a complex with cytoplasmic dynein and dynactin (37). Evidence that NuMA is interacting with splicing factors (38) and with the putative transcription factor GAS41, glioma-amplified sequence 41 (39), suggests that NuMA might also be involved in gene regulation. Furthermore, NuMA seems to be a key protein in apoptotic events. Several studies have shown that NuMA is degraded in early apoptotic cell death (40, 41), induced through cleavage by caspase-3 and/or caspase-6 (42, 43). Silencing of the NuMA gene results in an apoptotic phenotype in HeLa cells (44), suggesting an antiapoptotic function of the protein. Also interesting in this context is that NuMA is preferentially expressed in proliferating cells (45).
Another line of evidence that makes NuMA a plausible candidate for cancer predisposition is the existence of NuMA-retinoic acid receptor α (RARα) fusion proteins in acute promyelocytic leukemia (46). Even though the contribution of the NuMA gene disruption to the phenotype is unclear, this observation was the first to implicate a mitotic apparatus protein in the molecular pathogenesis of human malignancy. The comparison of NuMA-RARα mice with mice carrying other RAR fusion proteins strongly suggests that disruption of the NuMA function plays a role in the development of myeloid leukemia (47).
The only verified polymorphic nonsynonymous residue in NuMA, A794G, proved to be the most strongly associated with breast cancer risk, and individuals carrying the glycine allele have a >2-fold higher risk compared with individuals not carrying this allele. The residue is located in coil 3 of the large central coiled-coil domain of NuMA (32), an α-helical structure that is important for oligomer formation and potentially for protein–protein interactions during mitosis. A conserved domain search at the National Center for Biotechnology Information (NCBI) web site revealed that A794G is located in a domain that has homology to the SMC (structural maintenance of chromosome) family of chromosome segregation ATPases. This family includes proteins that are involved in cell division, chromosome partitioning, and DNA repair, such as RAD18 and SMC1, a protein that has been shown to interact with BRCA1 (48). An alignment of NuMA protein sequences from all species available at NCBI's GenBank (human, mouse, rat, and frog) showed that residue A794 is conserved in all species (data not shown).
A functional implication of the variant in the etiology of breast cancer, however, can only be hypothesized at this point. Biological validation of this variant is necessary to determine whether the variation leads to changes in the structural or functional properties of the protein. The G794 allele is also significantly more common in cases with a family history of any cancer. Therefore, it might be worthwhile to also analyze sample collections of other cancer types for a correlation of this variant with disease status. As an essential protein for chromosomal segregation and cell proliferation, NuMA is a conceivable candidate for predisposition to different forms of cancer.
In summary, a large-scale association study discovered variations in NuMA that might be involved in the predisposition to breast cancer, a finding that supports the promise of more comprehensive genome-wide studies. Disruption or constitutive activation of NuMA could lead to perturbation of cellular functions. Loss-of-function might render the cell less efficient in the execution of mitotic events. On the other hand, a gain-of-function variation might disrupt proper cleavage of NuMA and prohibit apoptosis. Additional analysis of the A794G and other potentially functional variants in the gene, including promoter sites, will be necessary to gain further knowledge about the biological basis of this disease association. As a consequence of the relatively low frequency of the variant, the statistical significance of some of the observations made in this study is limited. As shown for a variant in the G2 checkpoint kinase 2, CHEK2*1100delC, which has a similarly low frequency in the population, the observed effect of the variation is relatively modest but consistent among most studies (14). In our study, the genetic effect of NuMA*G794 has been demonstrated in three independent breast cancer samples. Additional independent and ideally larger samples are needed to support and extend these findings. Despite its relatively low frequency and modest effect, this region may be an important component of breast cancer risk in the population. Based on the combined data from this study, the A794G variation has an estimated population-attributable fraction (attributable risk) of 3.5%, comparable with BRCA1 and BRCA2 mutations combined, which are substantially rarer (49). In combination with other predisposing factors, such as CHEK2*1100delC and the ICAM variations [which we have previously reported (27)], variations in NuMA may prove to be useful to assess breast cancer risk and potentially help decide on the appropriate timing and selection of treatment.
Acknowledgments
We thank all patients participating in this study and the members of Sequenom's genotyping team for their excellent support in producing the genetic data for this research.
Author contributions: S.K., R.R., M.K., U.S.-B., L.R.G., F.E., J.R., C.R.C., M.R.N., and A.B. designed research; S.K., R.B.R., C.R.H., M.R.N., and A.B. analyzed data; S.K. wrote the paper; R.B.R. performed research; G.M. contributed new reagents/analytic tools; R.R. clinical database management; G.M. executed SNP selection; M.K., U.S.-B., L.R.G., F.E., and J.R. sample collection and management.
Abbreviations: LD, linkage disequilibrium; NuMA, nuclear mitotic apparatus protein; OR, odds ratio.
References
- 1.Greene, M. H. (1997) Mayo Clin. Proc. 72, 54–65. [DOI] [PubMed] [Google Scholar]
- 2.Balmain, A., Gray, J. & Ponder, B. (2003) Nat. Genet. 33, 238–244. [DOI] [PubMed] [Google Scholar]
- 3.Miki, Y., Swensen, J., Shattuck-Eidens, D., Futreal, P. A., Harshman, K., Tavtigian, S., Liu, Q., Cochran, C., Bennett, L. M., Ding, W., et al. (1994) Science 266, 66–71. [DOI] [PubMed] [Google Scholar]
- 4.Wooster, R., Neuhausen, S. L., Mangion, J., Quirk, Y., Ford, D., Collins, N., Nguyen, K., Seal, S., Tran, T., Averill, D., et al. (1994) Science 265, 2088–2090. [DOI] [PubMed] [Google Scholar]
- 5.Malkin, D., Li, F. P., Strong, L. C., Fraumeni, J. F., Jr., Nelson, C. E., Kim, D. H., Kassel, J., Gryka, M. A., Bischoff, F. Z., Tainsky, M. A., et al. (1990) Science 250, 1233–1238. [DOI] [PubMed] [Google Scholar]
- 6.Dunning, A. M., Healey, C. S., Pharoah, P. D., Teare, M. D., Ponder, B. A. & Easton, D. F. (1999) Cancer Epidemiol. Biomarkers Prev. 8, 843–854. [PubMed] [Google Scholar]
- 7.Coughlin, S. S. & Piper, M. (1999) Cancer Epidemiol. Biomarkers Prev. 8, 1023–1032. [PubMed] [Google Scholar]
- 8.Lichtenstein, P., Holm, N. V., Verkasalo, P. K., Iliadou, A., Kaprio, J., Koskenvuo, M., Pukkala, E., Skytthe, A. & Hemminki, K. (2000) N. Engl. J. Med. 343, 78–85. [DOI] [PubMed] [Google Scholar]
- 9.Cardon, L. R. & Bell, J. I. (2001) Nat. Rev. Genet. 2, 91–99. [DOI] [PubMed] [Google Scholar]
- 10.Feigelson, H. S., Coetzee, G. A., Kolonel, L. N., Ross, R. K. & Henderson, B. E. (1997) Cancer Res. 57, 1063–1065. [PubMed] [Google Scholar]
- 11.Ambrosone, C. B., Freudenheim, J. L., Marshall, J. R., Graham, S., Vena, J. E., Brasure, J. R., Michalek, A. M., Laughlin, R., Nemoto, T. & Shields, P. G. (1995) Ann. N.Y. Acad. Sci. 768, 250–252. [DOI] [PubMed] [Google Scholar]
- 12.Helzlsouer, K. J., Selmin, O., Huang, H. Y., Strickland, P. T., Hoffman, S., Alberg, A. J., Watson, M., Comstock, G. W. & Bell, D. (1998) J. Natl. Cancer Inst. 90, 512–518. [DOI] [PubMed] [Google Scholar]
- 13.Smith, G., Stanley, L. A., Sim, E., Strange, R. C. & Wolf, C. R. (1995) Cancer Surv. 25, 27–65. [PubMed] [Google Scholar]
- 14.The CHEK2 Breast Cancer Case-Control Consortium (2004) Am. J. Hum. Genet. 74, 1175–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kilpivaara, O., Vahteristo, P., Falck, J., Syrjakoski, K., Eerola, H., Easton, D., Bartkova, J., Lukas, J., Heikkila, P., Aittomaki, K., et al. (2004) Int. J. Cancer 111, 543–547. [DOI] [PubMed] [Google Scholar]
- 16.Collins, F. S., Guyer, M. S. & Charkravarti, A. (1997) Science 278, 1580–1581. [DOI] [PubMed] [Google Scholar]
- 17.Goldstein, D. B., Ahmadi, K. R., Weale, M. E. & Wood, N. W. (2003) Trends Genet. 19, 615–622. [DOI] [PubMed] [Google Scholar]
- 18.Ozaki, K., Ohnishi, Y., Iida, A., Sekine, A., Yamada, R., Tsunoda, T., Sato, H., Hori, M., Nakamura, Y. & Tanaka, T. (2002) Nat. Genet. 32, 650–654. [DOI] [PubMed] [Google Scholar]
- 19.Kammerer, S., Burns-Hamuro, L. L., Ma, Y., Hamon, S. C., Canaves, J. M., Shi, M. M., Nelson, M. R., Sing, C. F., Cantor, C. R., Taylor, S. S. & Braun, A. (2003) Proc. Natl. Acad. Sci. USA 100, 4066–4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nelson, M. R., Marnellos, G., Kammerer, S., Hoyal, C. R., Shi, M. M., Cantor, C. R. & Braun, A. (2004) Genome Res. 14, 1664–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buetow, K. H., Edmonson, M., MacDonald, R., Clifford, R., Yip, P., Kelley, J., Little, D. P., Strausberg, R., Koester, H., Cantor, C. R. & Braun, A. (2001) Proc. Natl. Acad. Sci. USA 98, 581–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bansal, A., van den Boom, D., Kammerer, S., Honisch, C., Adam, G., Cantor, C. R., Kleyn, P. & Braun, A. (2002) Proc. Natl. Acad. Sci. USA 99, 16871–16874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barratt, B. J., Payne, F., Rance, H. E., Nutland, S., Todd, J. A. & Clayton, D. G. (2002) Ann. Hum. Genet. 66, 393–405. [DOI] [PubMed] [Google Scholar]
- 24.DerSimonian, R. & Laird, N. (1986) Control Clin. Trials 7, 177–188. [DOI] [PubMed] [Google Scholar]
- 25.Stephens, M. & Donnelly, P. (2003) Am. J. Hum. Genet. 73, 1162–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schaid, D. J., Rowland, C. M., Tines, D. E., Jacobson, R. M. & Poland, G. A. (2002) Am. J. Hum. Genet. 70, 425–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kammerer, S., Roth, R. B., Reneland, R., Marnellos, G., Hoyal, C. R., Markward, N. J., Ebner, F., Kiechle, M., Schwarz-Boeger, U., Griffiths, L. R., et al. (2004) Cancer Res. 64, 8906–8910. [DOI] [PubMed] [Google Scholar]
- 28.Gibbs, R. A., Belmont, J. W., Hardenbol, P., Willis, T. D., Yu, F., Yang, H., Ch'ang, L. Y., Huang, W., Liu, B., Shen, Y., et al. (2003) Nature 426, 789–796.14685227 [Google Scholar]
- 29.Aizawa, Y., Akita, K., Taniai, M., Torigoe, K., Mori, T., Nishida, Y., Ushio, S., Nukada, Y., Tanimoto, T., Ikegami, H., et al. (1999) FEBS Lett. 445, 338–342. [DOI] [PubMed] [Google Scholar]
- 30.Corbaz, A., ten Hove, T., Herren, S., Graber, P., Schwartsburd, B., Belzer, I., Harrison, J., Plitz, T., Kosco-Vilbois, M. H., Kim, S. H., et al. (2002) J. Immunol. 168, 3608–3616. [DOI] [PubMed] [Google Scholar]
- 31.Compton, D. A., Szilak, I. & Cleveland, D. W. (1992) J. Cell Biol. 116, 1395–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang, C. H., Lambie, E. J. & Snyder, M. (1992) J. Cell Biol. 116, 1303–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lydersen, B. K. & Pettijohn, D. E. (1980) Cell 22, 489–499. [DOI] [PubMed] [Google Scholar]
- 34.Kallajoki, M., Weber, K. & Osborn, M. (1991) EMBO J. 10, 3351–3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang, C. H. & Snyder, M. (1992) Mol. Biol. Cell 3, 1259–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Compton, D. A. & Cleveland, D. W. (1993) J. Cell Biol. 120, 947–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Merdes, A., Ramyar, K., Vechio, J. D. & Cleveland, D. W. (1996) Cell 87, 447–458. [DOI] [PubMed] [Google Scholar]
- 38.Zeng, C., He, D., Berget, S. M. & Brinkley, B. R. (1994) Proc. Natl. Acad. Sci. USA 91, 1505–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harborth, J., Weber, K. & Osborn, M. (2000) J. Biol. Chem. 275, 31979–31985. [DOI] [PubMed] [Google Scholar]
- 40.Weaver, V. M., Carson, C. E., Walker, P. R., Chaly, N., Lach, B., Raymond, Y., Brown, D. L. & Sikorska, M. (1996) J. Cell Sci. 109, 45–56. [DOI] [PubMed] [Google Scholar]
- 41.Gueth-Hallonet, C., Weber, K. & Osborn, M. (1997) Exp. Cell Res. 233, 21–24. [DOI] [PubMed] [Google Scholar]
- 42.Hirata, H., Takahashi, A., Kobayashi, S., Yonehara, S., Sawai, H., Okazaki, T., Yamamoto, K. & Sasada, M. (1998) J. Exp. Med. 187, 587–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taimen, P. & Kallajoki, M. (2003) J. Cell Sci. 116, 571–583. [DOI] [PubMed] [Google Scholar]
- 44.Harborth, J., Elbashir, S. M., Bechert, K., Tuschl, T. & Weber, K. (2001) J. Cell Sci. 114, 4557–4565. [DOI] [PubMed] [Google Scholar]
- 45.Taimen, P., Viljamaa, M. & Kallajoki, M. (2000) Exp. Cell Res. 256, 140–149. [DOI] [PubMed] [Google Scholar]
- 46.Wells, R. A., Catzavelos, C. & Kamel-Reid, S. (1997) Nat. Genet. 17, 109–113. [DOI] [PubMed] [Google Scholar]
- 47.Sukhai, M. A., Wu, X., Xuan, Y., Zhang, T., Reis, P. P., Dube, K., Rego, E. M., Bhaumik, M., Bailey, D. J., Wells, R. A., Kamel-Reid, S. & Pandolfi, P. P. (2004) Oncogene 23, 665–678. [DOI] [PubMed] [Google Scholar]
- 48.Yazdi, P. T., Wang, Y., Zhao, S., Patel, N., Lee, E. Y. & Qin, J. (2002) Genes Dev. 16, 571–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.King, M. C., Marks, J. H. & Mandell, J. B. (2003) Science 302, 643–646. [DOI] [PubMed] [Google Scholar]