Abstract
Objective
To identify, purify, and characterize the proteins responsible for glutenase activity in the feces of healthy subjects and patients with celiac disease (CD).
Methods
Sixteen subjects were included in this study; 8 were healthy with no known food intolerances, and 8 were treated CD patients on a gluten-free diet. Fecal samples were homogenized, and precipitated proteins were purified by chromatography. Glutenase activity was evaluated by bioassays, zymography, and high-performance liquid chromatography with immunogenic 33-mer, 19-mer, and 13-mer gliadin peptides.
Results
The gastrointestinal elastase 3B (CEL3B), elastase 2A (CEL2A), and carboxypeptidase A1 (CBPA1) enzymes degraded human gluten. These proteins fully hydrolyzed 13-mer and 19-mer gliadin peptides that trigger immune-mediated enteropathy in individuals genetically predisposed to CD and partially digested a 33-mer. Feces from patients with CD showed more glutenase activity than feces from individuals without CD (171–466% higher). Peptidase activity against the gliadin peptides also increased in patients with CD.
Conclusion
The digestive tracts of patients with CD and healthy subjects have enzymatic machinery needed for gluten degradation. Patients with CD showed more gluten hydrolysis than did healthy individuals, although, in both cases, a fraction of 33-mer peptide remained intact. Gliadin peptides derived from gastrointestinal digestion, especially the 33-mer, can potentially be used by commensal microbiota from both CD-positive and CD-negative individuals, and differences in bacterial hydrolysis can modify its immunogenic capacity.
Keywords: Gluten, Gliadin, Celiac disease, Glutenase fecal activity, Gliadinase activity
Abbreviations: CD, celiac disease; HLA-DQ, human leukocyte antigen; LPLC, low-performance liquid chromatography; NCD, non-celiac-disease; CEL3B, gastrointestinal elastase 3B; CEL2A, gastrointestinal elastase 2A; CBPA1, human carboxypeptidase A1; ATIs, α-Amylase/trypsin inhibitors
Highlights
-
•
Protein from human faeces degrade gluten and gliadin-derived peptides.
-
•
Specific gastrointestinal proteases are involved in gliadin degradation.
-
•
Celiac patients can completely to degrade the 19- and 13-mer gliadin-derived peptides.
1. Introduction
Celiac disease (CD) is a chronic, small intestinal, immune-mediated enteropathy triggered by the exposure to gluten proteins in genetically predisposed individuals [1], [2], [3]. The pathogenesis of CD involves genetic and environmental factors. Susceptibility to CD is strongly associated with the human leukocyte antigen (HLA) genes of the major histocompatibility complex, and approximately 90% of patients express alleles coding for the HLA-DQ2 (HLA-DQ2.5 and HLA-DQ2.2) and HLA-DQ8 haplotypes [1], [4], [5]. The diet is the fundamental factor contributing to the development of CD. The gliadin prolamin (from gluten) and related prolamins (from wheat, barley, and rye) are resistant to complete digestion by human digestive enzymes due to their high glutamine and proline contents. Their digestion results in the production of large peptides (10 to ≥30 amino acids) that cross the small intestinal barrier, some of which (such as 13-, 19-, or 33-mer), are capable of triggering inflammatory processes associated with CD [6], [7], [8].
Nevertheless, although 30% of the general population carries a genetic predisposition for CD, only approximately 3% will develop this disease. Moreover, additional environmental cofactors may be required [9], including intestinal pathogens that can enhance gluten immunogenicity and toxicity, e.g., rotavirus infections [10]; an altered gut-microbiota composition [11], [12]; and some immune-modulatory drugs, e.g., IFN-alpha [13]. As such, intestinal dysbiosis has been associated with patients with CD [9], [14], and it has been purposed that gliadin-metabolizing bacteria may represent one of the missing environmental links in the development of CD [15], [16].
Previous findings have revealed that human feces show glutenase activity in both healthy individuals [17] and CD patients [9] and that significant differences in fecal glutenase activity may occur between these two groups [9]. These results suggest that human feces are potentially a good sample source for evaluating and identifying the proteases involved in gluten metabolism.
Accordingly, in this work, we identified and characterized the proteins responsible for fecal glutenase activity, and we compared the proteolytic profiles involved in degrading gluten peptides between healthy and individuals with CD.
2. Materials and methods
2.1. Fecal sampling
Sixteen subjects were included in this study; 8 were healthy with no known food intolerances (mean age 41; range 25–67), and 8 were CD patients on a gluten-free diet (mean age 42; range 27–57). The healthy subjects were symptom-free volunteers for whom CD was ruled out based on normal serum tissue Transglutaminase Antibody (tTGA) levels and an HLA-DQ phenotype that was not DQ2 or DQ8. Treated CD patients were diagnosed on the basis of positive tTGA and duodenal mucosa biopsy with villous atrophy (Marsh III in all of the cases). They complied with a strict gluten-free diet for at least 2 years, showed negative serology markers, and displayed complete recovery during the initial villous atrophy (Marsh 0 or Marsh I in the biopsy control). No participants in this study were treated with antibiotics in the month before the provided samples. The study complied with the Declaration of Helsinki guidelines, and all procedures involving human subjects were approved by the local ethics committee of our hospital. Written informed consent was obtained from all subjects. Fresh stools from both subject groups were collected and immediately stored at −80 °C. Previously, we showed that glutenase activity (evaluated by bioassay) in stool samples was not modified by freezing at −80 °C. Fecal samples were homogenized and processed immediately.
2.2. Purification of different fecal glutenase activities
All procedures were performed at 4 °C unless otherwise indicated.
Step 1: Sample preparation – Fecal samples were homogenized by mechanical stirring (90 min) in 10 mM Tris–HCl, pH 7.5 (1:5 w/v) and centrifuged (3,100 ×g, 30 min). The supernatant was re-centrifuged at 30,000 ×g for 15 min and filtered through a 0.44-μm nitrocellulose filter.
Step 2: Ammonium sulfate fractionation – Each filtered supernatant obtained from step 1 was precipitated with ammonium sulfate. The fraction precipitating between 35 and 65% ammonium sulfate (containing glutenase activity) was collected by centrifugation at 30,000 ×g, 15 min. The pellet was dissolved in 10 mM Tris–HCl (pH 7.5). Ammonium sulfate was eliminated by passing through a Sephadex G-25 (PD-10) column (GE Healthcare Live Sources, San Diego, CA) equilibrated with Tris-ClH (pH 7.5) according with manufacture indications.
Step3: Ion-exchange low-performance liquid chromatography (LPLC) – Samples from step 2 were injected into an LPLC system (BioLogic LP Systems, BioRad) equipped with a cation-exchange column (BioRad Macro-Prep High Q, 1,000 Å, 50 μm), equilibrated with 10 mM Tris–HCl buffer, pH 7.5. Next, the column was washed with the same buffer and eluted with a linear 0-to-0.3-M KCl gradient. Fractions (1 mL) were collected at a flow rate of 1 mL/min and assayed in MCG-1 plates to detect glutenase activity (see below). The non-retained proteins from a cation-exchange column were applied to an anion exchange column (BioRad Macro-Prep High S, 1,000 Å, 50 μm) equilibrated with 10 mM Tris–HCl (pH 7.5) or 10 mM MOPS (pH 6.5). The column was washed with either buffer and proteins were eluted using a linear KCl gradient (0–0.3 M). One-milliliter fractions were collected.
2.3. Evaluation of fecal glutenase activity in bioassays
Fecal glutenase activities were measured as described [17]. Briefly, fecal samples were spread on agar plates (MCG-1) containing gluten (1.5%), 20 g/L glucose, 0.05 g/L CaCl2, 0.07 g/L ZnSO4, 0.05 g/L l-cysteine, 0.1% Tween 80, 60 mM phosphate buffer (pH 6.5), and 1.5 g/L agar, and incubated at 37 °C for 24 h. The plates were evaluated by measuring the diameter of the halo formed. Trypsin, at different concentrations, was used to generate a standard curve. Fecal glutenase activity (FGA) was expressed as trypsin-activity equivalents/g feces [17].
2.4. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE)
SDS-PAGE (12%) was run as described by Laemmli [18], with modifications described by Helmerhorst and Wei [19]. Protein molecular weights were estimated using standard protein markers (97–14.4 kDa; GE Healthcare, Amersham LMW, UK). Gels were stained with Coomassie Brilliant Blue R-250.
2.5. Evaluation of gliadin activity by zymography
Gliadin degradation was assessed with 12% SDS-PAGE zymogram gels containing wheat gliadin (0.6 mg/mL; Sigma, St. Louis, MO), without β-mercaptoethanol. The protein samples were diluted (1/20) in 10 mM Tris–HCl (pH 7.5), and electrophoresis was performed at 100 V at 4 °C. Protein renaturing in the gels was achieved by washing twice for 30 min at room temperature in renaturing buffer containing 2% (v/v) Triton-X-100, 0.1 M NaCl, and 0.05 M Tris–HCl (pH 7.8). Subsequently, the Triton-X-100 was removed by washing (3 × 20 min) in developing buffer (0.05 M Tris–HCl, pH 7.8). After overnight in developing buffer at 37 °C, gels were stained for 30 min with 0.1% (w/v) Coomassie Brilliant Blue R-250.
2.6. Densitometric evaluation of the electrophoretic results
SDS-PAGE and zymogram gels were digitalized using a densitometer (Bio-Rad GS800) and Quantity One 1-D analysis software (Bio-Rad Laboratories Inc., Hercules, CA). Densitometry analyses were performed using lane-based background subtraction, followed by measuring the peak areas, and optical density (OD) values were used for statistical analysis. Each sample was analyzed on duplicate gels. Proteins were represented by comparing their relative mobilities with those of molecular weight standards.
2.7. Determining 33-mer, 19-mer, and 13-mer hydrolysis
Peptides were synthetized by Proteogenix SAS (Schiltigheim, France). The 33-mer (LQLQPFPQPQLPYPQPQLPYPQPQLPYPQPQPF), 19-mer (LGQQQPFPPQQPYPQPQPF), and 13-mer (LGQQQPFPPQQPY) peptides had purities of 95%, 97%, and 96%, respectively. Since the HPLC chromatograms of the commercial 19-mer and 13-mer peptides showed 2 peaks, purity was verified by the Laboratorio de Técnicas Instrumentales (Universidad de León, Spain), confirming that the appearance 2 peaks were caused by the assay pH and not by impurities (Figure 2, Figure 5, Figure 7).
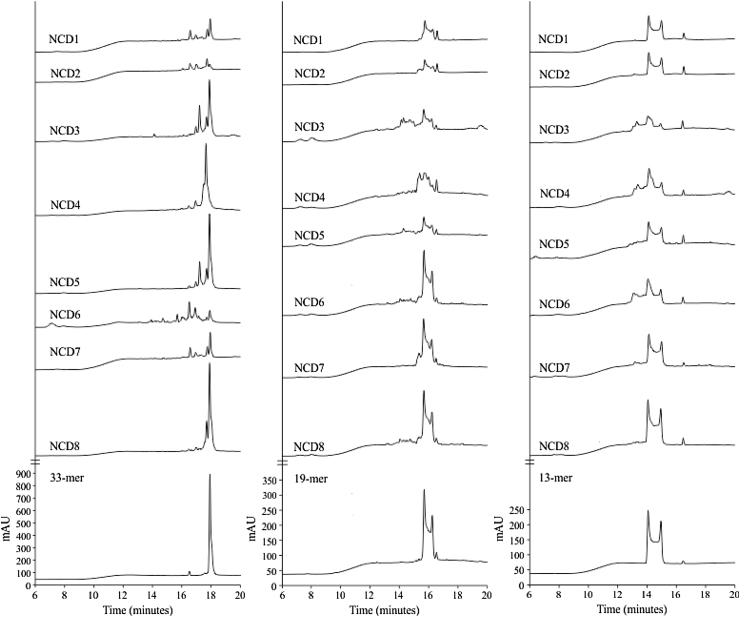
Figure 2.
Fecal hydrolytic activity against gliadin derived peptides of samples from non-celiac disease (NCD) volunteers. HPLC chromatograms generated after incubating the 33-, 19-, and 13-mer peptides for 60-min at 37 °C with desalted ecal protein extracts from healthy volunteers (NCD1–NCD8). The bottom of the figure shows the chromatographic migration of the peptides.
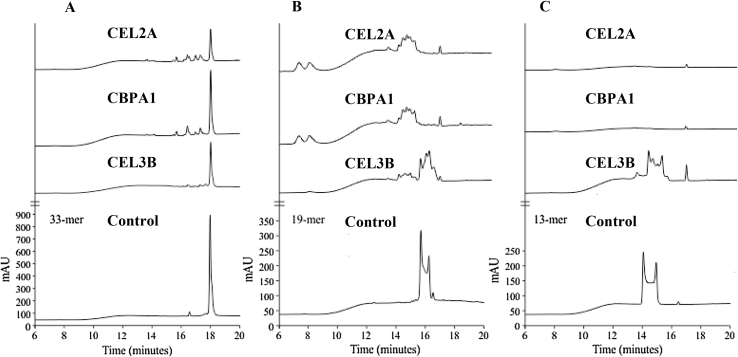
Figure 5.
Fecal hydrolytic activity from human fecal purified proteins against gliadin derived peptides. Chromatograms generated after incubating the 33-mer (A), 19-mer (B), and 13-mer (C) peptides for 60-min at 37 °C with the human fecal purified Elastase 2A (CEL2A), Carboxypeptidase 1A (CBPA1) and Elastase 3B (CEL3B) proteins. The bottom of the figure shows the chromatographic migration of the peptides.
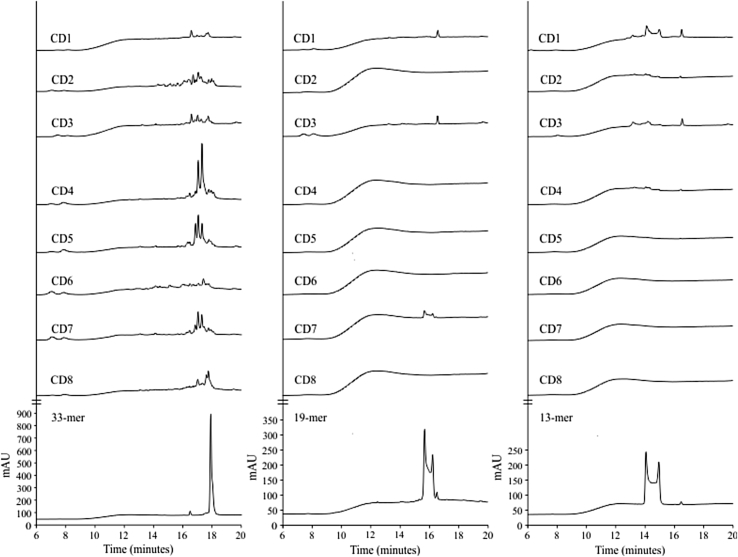
Figure 7.
Fecal hydrolytic activity against gliadin derived peptides of samples from celiac disease (CD) volunteers. HPLC chromatograms generated after incubating the 33-, 19-, and 13-mer peptides for 60-min at 37 °C with human fecal samples from CD patients (CD1–CD8). The bottom of the figure shows the chromatographic migration of peptides.
We assayed 33-mer, 19-mer, and 13-mer hydrolysis as described previously [20], with modifications. Reaction mixtures (40-μL) contained 3.4 μL (0.8 mg/mL) of desalted protein extract (purification step 2, described above) or purified proteins (above); 4.7 μL of 33-, 19-, or 13-mer peptide (60 μM); and 31.9 μL of phosphate-buffered saline (pH 7.3). Reactions were incubated at 37 °C for 60 min and stopped by boiling at 100 °C for 10 min. Each reaction was filtered using a 0.22-μm Cellulose Acetate Spin-X Centrifuge Tube Filter (Thermofisher), and 10-μL aliquots were subjected to reverse-phase HPLC using a C-18 column (LiChrospher 100 RP18 column, 5 μm, 4 × 250 mm, Teknokroma SL). The elution phases consisted of (A) MilliQ H2O containing 0.1% trifluoroacetic acid (TFA) (v/v) and (B) acetonitrile and 0.1% TFA (v/v). The gradient program started with 100% of solution A for 2 min and changed linearly to reach 100% of B solution at a flow rate of 1 mL/min over 20 min [20]. The eluate was monitored by measuring UV absorbance at 215 nm. Negative controls included runs with proteins denatured by boiling for 15 min, or 33-, 19-, and 13-mer peptides assayed without protein extract or purified enzymes.
2.8. Protein identification
Proteins with glutenase/gliadinase activity were identified as described previously [21]. Briefly, protein spots of interest were manually excised from stained SDS/PAGE gels by biopsy punches, placed in an Eppendorf tube, and washed twice with double-distilled water. Proteins were digested [22] and processed for further analysis, as described [23]. Samples were analyzed with a 4800 Proteomics Analyzer matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF/TOF) mass spectrometer (AB Sciex). A 4700 Proteomics Analyzer Calibration Mixture (Cal Mix 5; AB Sciex) was used for external calibration. All mass spectrometry (MS) spectra were internally calibrated using peptides from trypsin auto-digestion. MALDI-TOF/TOF MS analysis produced peptide mass fingerprints, and the peptides observed (up to 65/spot) were collected and represented as a list of monoisotopic molecular weights with a signal-to-noise (S/N) ratio >20, using 4000 Series Explorer v3.5.3 software (AB Sciex). All known contaminant ions (trypsin- and keratin-derived peptides) were excluded from later MS/MS analyses. Hence, from each MS spectra, the 10 most intensive precursors with an S/N greater >20 were selected for MS/MS analyses with CID (atmospheric gas was used) in 2-kV ion-reflector mode and precursor mass windows of ±7 Da. Default calibration was optimized for the MS/MS spectra. For protein identification, generic Mascot files combining MS and MS/MS spectra were created automatically and used to interrogate a non-redundant protein database with Mascot version 2.2.04 (Matrix Science) through the Global Protein Server version 3.6 (AB Sciex). The search parameters for peptide-mass fingerprints and tandem-MS spectra obtained were set as follows: i) NCBInr (2009.11.03) sequence databases were used, ii) the taxonomy of all entries (9,993,394 sequences, 3,409,286,210 residues) was included in the analysis, iii) fixed and variable modifications were considered (Cys as S-carbamidomethyl derivative and Met as oxidized methionine), iv) 1 missed cleavage site was allowed, v) precursor tolerance was 50 ppm and MS/MS-fragment tolerance was 0.3 Da, vi) peptide charge: 1+, and vii) trypsin was set as the enzyme. Protein candidates produced by this combined peptide mass fingerprinting/tandem MS search were considered valid when the global Mascot score was >83, with p < 0.05.
2.9. Statistical analysis
Statistical analysis was performed with Graph-Pad Prism software. When 2 groups were compared, an unpaired t-test was used. p < 0.05 was considered significant. Data are displayed as the mean ± SE.
3. Results
3.1. Specific degradation of gluten in human feces
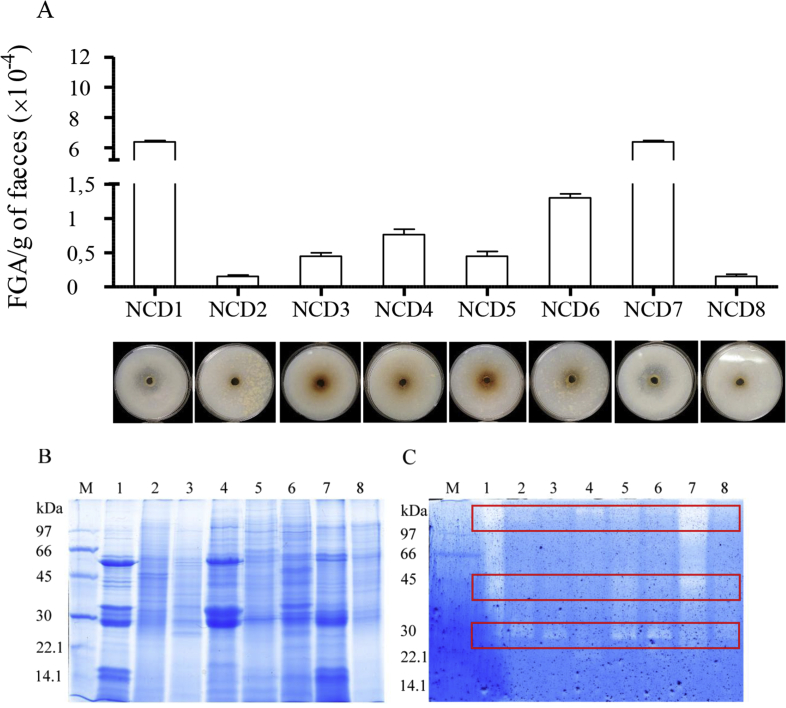
Previous data have demonstrated fecal hydrolytic activity against gluten and the gliadin-derived 33-mer peptide in healthy volunteers, CD patients, and first-degree relatives [9]. Here, we identified and characterized proteins responsible for these glutenase activities, first using feces from 8 healthy non-celiac-disease (NCD) volunteers. Glutenase activity was confirmed in all samples assayed (between 1500 and 63,000 FGA/g of faeces) by: i) running a bioassay (Figure 1A), ii) SDS-PAGE (Figure 1B), and iii) zymography, using gliadin the substrate and (Figure 1C). Despite the high fecal protein diversity and variability observed by SDS-PAGE (Figure 1B), densitometric analysis of zymograms revealed the presence of 3 principal zones with gliadinase activity at >97, 45–35, and 30–25 kDa (Figure 1C). By HPLC, we also observed that the fecal samples from NCD individuals showed hydrolytic activity against the 33-, 19- and 13-mer gliadin-derived peptides (Figure 2). However, complete peptide degradation was not observed in any case, even after a 60-min incubation (Figure 2).
Figure 1.
Fecal glutenase activity (FGA) of non-celiac disease (NCD) volunteers. (A) Bioassay results showed the gluten was degraded in solid gluten media. Data represented as the mean ± SEM, in terms of the number of units of trypsin/g feces. Representative SDS-PAGE (B) and gliadin zymogram (C) results from whole-protein fecal samples from non-CD volunteers (1–8). M: electrophoretic molecular weight marker. The boxed-in areas correspond to the 3 zones with gliadinase activity, as determined by densitometric analysis at >97, 45–35, and 30–25 kDa.
3.2. Digestive tract proteins are involved in gluten degradation
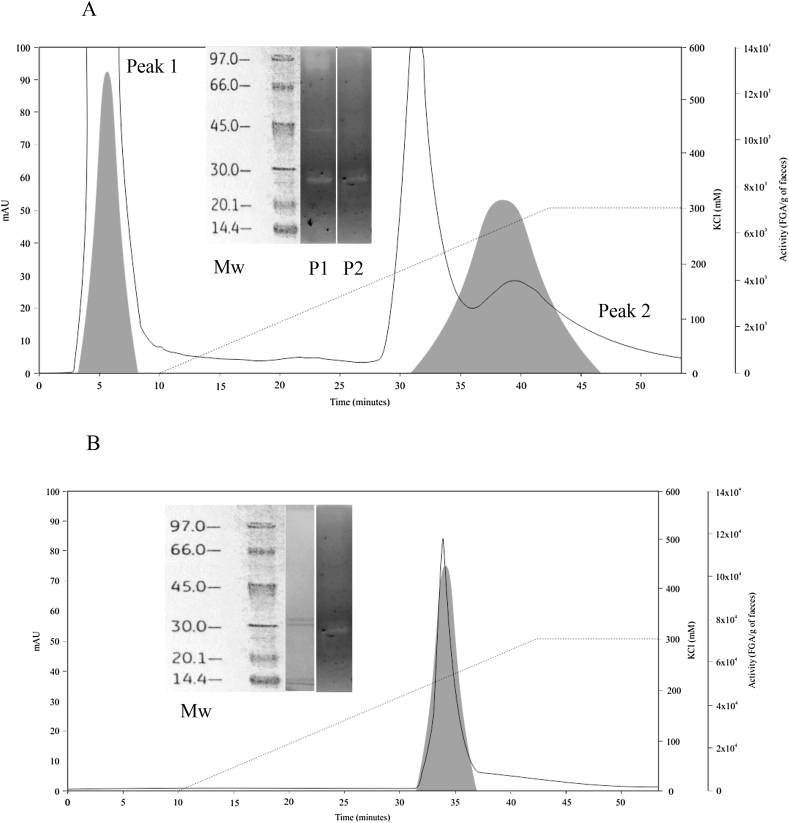
Protein purification was performed to identify the protein(s) responsible for fecal glutenase activity. Because all volunteer samples tested showed similar gliadinase activities (Figure 1C), we used the fecal sample from NCD volunteer number 1 (NCD 1) because of its high glutenase activity (Figure 1A) and accessibility (NCD 1 was a volunteer from our laboratory). Different ion-exchange chromatography columns were used for purification. First, 2.5 mg protein from feces (40 g), which was homogenized, ammonium sulfate-fractionated, and desalted, was applied to a cation-exchange chromatography (High Q column) and 2 areas with glutenase activity (evaluated in the bioassay) were obtained (Figure 3A): a non-retained fraction (peak 1) and a retained fraction (peak 2). Zymographic and densitometric evaluation of pooled fractions 4–8 from peak 1 (Figure 3A inset) showed gliadinase activity at >97, 45–35, and 30–25 kDa, while peak 2 fractions 37–45 showed activity between 30 and 25 kDa (Figure 3A inset). By re-chromatography of 3 pooled fractions from peak 2 (1.67 mg protein) using the same column (High Q) and conditions (10 mM Tris–HCl, pH 7.5), we obtained a single protein peak showing glutenase activity (Figure 3B). By electrophoresis and zymography of pooled fractions 32–37, we observed 2 proteins of 29 and 28 kDa with gliadinase activity (Figure 3B inset). Sequencing these purified molecules by MALDI-TOF/TOF and MS/MS analyses revealed that had molecular masses (Mr) of 29.8 and 29.7 kD, respectively. Amino acid sequence comparison showed high homology (Mowse score from Mascot database search: 475) with human elastase 3B (CEL3B and ELA3B, respectively), using the UniProt and BRENDA databases.
Figure 3.
High Q chromatographic elution profiles of protein and glutenase/gliadinase activity from a non-CD volunteer fecal sample (NCD1). A: Elution profile of a fecal sample following homogenization, ammonium sulfate-based fractionation, and cation-exchange chromatography. Inset: electrophoretic weight markers (Mw) and zymogram profiles obtained from pooled fractions from peak 1 (P1, pooled fractions 4–8) and peak 2 (P2, pooled fractions 37–45), both of which had glutenase/gliadinase activity. B: Elution profile of proteins obtained from peak 2 (from Figure 3A) after re-chromatography using a High Q column. Inset: electrophoretic weight markers (Mw), electrophoretic profile, and zymogram profile obtained from pooled, retained fractions 32–37.
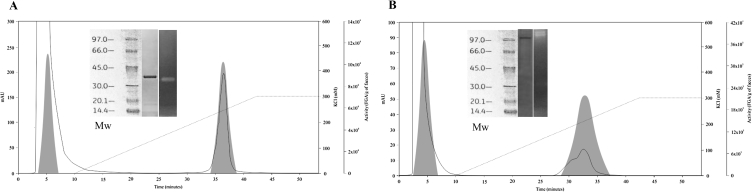
Three pooled peak-1 fractions (2.57 mg protein) were separated by anion-exchange chromatography (High S column; mobile phase: 10 mM Tris–HCl, pH 7.5), and we saw only 1 protein area with glutenase activity (Figure 4A). By electrophoresis and zymography of fraction 36, we observed only 1 band of ∼35 kDa (Figure 4A inset). Sequencing this purified protein revealed that it had a Mr of 29.4 kDa and showed a high homology grade (score: 601) with the human elastase 2A proteins CEL2A and ELA2A, using the UniProt and BRENDA databases, respectively. As human elastases are glycoproteins [24], the different Mr values found by electrophoresis and sequencing may have resulted from variable glycosylation.
Figure 4.
High S chromatographic elution profile of protein and glutenase/liadinase activity of peak-1 from high Q column. Elution profile of proteins from peak 1 after High Q chromatography (see Figure 1), using an anion-exchange column (High S) and 10 mM Tris–HCl, pH 7.5 (A) or 10 mM MOPS, pH 6.5 (B) as the mobile phase. Inset A: electrophoretic weight markers (Mw), electrophoretic profile, and zymogram profile obtained from the retained fraction 36. Inset B: Mw, electrophoretic profile, and zymogram profile of the retained fraction 34.
When the 3 pooled peak-1 fractions (2.57 mg protein) were separated with the High S column using a different mobile phase (10 mM MOPS, pH 6.5), a different peak with glutenase activity was identified (Figure 4B). By electrophoresis, we observed only 1 protein band (>97 kDa) in fraction 34 that, by zymography, also showed gliadinase activity (Figure 4B inset). Similarly, sequencing this purified protein from fractions 30–36 revealed it had an Mr of 104 kDa and that its amino acid sequence had a high homology grade (score: 193) with human carboxypeptidase A1 (CBPA1), using the UniProt database.
Next, we tested the glutenase and gliadinase activities of similar commercial proteases. By performing the bioassay and zymography, we found that recombinant mouse chymotrypsin-like elastase family member 3B (3.4.21.70), recombinant bovine chymotrypsin-like elastase family member 2A (3.4.21.71), recombinant rat carboxypeptidase A1 (3.4.17.1), all from MyBioSource (USA) and porcine elastase (3.4.21.36) from Sigma, (USA) showed hydrolytic activity against gluten and gliadin. Moreover, all these commercial proteases showed peptidase activity against the tree peptides tested (not show).
3.3. Specific digestive proteins hydrolyze toxic, gliadin-derived peptides
To establish the glutenase specificity of the isolated human proteins (CEL2A, CBPA1, and CEL3B), we studied their hydrolytic activity against the 33-, 19-, and 13-mer peptides. All 3 purified proteins showed hydrolytic activity against all 3 gliadin-derived peptides (Figure 5). The 33-mer gliadin fragment, which generates immune-adaptive response in CD, was not completely degraded under standard time assay (60 min) and, even after a 24-h incubation (not shown), although cleavage products were generated (Figure 5A). The CEL2A and CEL3B elastases were most effective in degrading the 33-mer (66% and 56% respectively). When CEL2A, CBPA1, and CEL3B were all 3 added (simultaneously or sequentially) to the peptide-hydrolysis reaction, 33-mer degradation did not increase after 60 min (not shown). CEL2A and CBPA1 (but not CEL3B) fully degraded the 19-mer and 13-mer peptides after a 60-min incubation (Figure 5, Figure 7C, respectively), although small peptides were observed after digestion with the 19-mer (but not the 13-mer).
3.4. Feces from patients with CD more efficiently degrade gluten and toxic gliadin-derived peptides
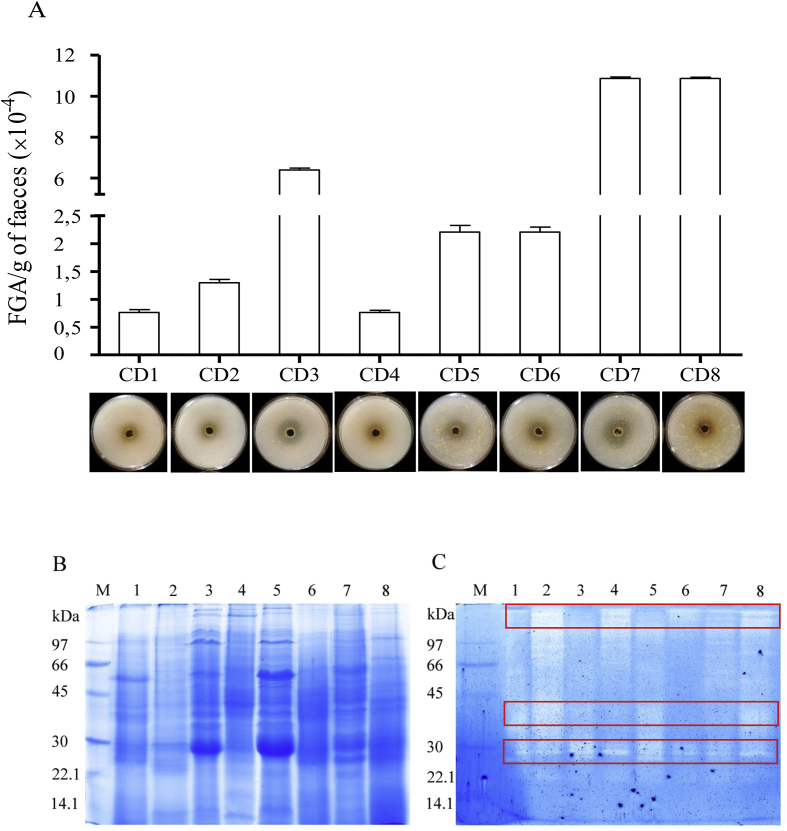
To uncover differences between NCD individuals and CD patients, we next evaluated fecal glutenase, gliadinase, and peptidase activities in samples from 8 CD patients on a gluten-free diet. Similar to samples from NCD volunteers, our bioassay results showed that glutenase activity was present in fecal samples from all CD patients tested (Figure 6A). Moreover, the fecal activity (7000–108,000 FGA/g feces) was 171–466% higher than observed in NCD samples (1500–63,000 FGA/g feces; Figure 1A).
Figure 6.
Fecal glutenase activity (FGA) of CD patients. (A) Bioassay results showing that the gluten was degraded in solid gluten media. The data shown represent the mean ± SEM (units of trypsin/g faeces). Representative SDS-PAGE (B) and gliadin zymogram (C) results of several whole-protein fecal samples from patients with CD. The lanes were loaded with samples from different patients with CD (1–8). The boxed-in areas correspond to the 3 zones with gliadinase activity, as determined by densitometric analysis at >97, 45–35, and 30–25 kDa. M: electrophoretic molecular weight marker. The boxed-in areas correspond to the 3 zones with gliadinase activity, as determined by densitometric analysis at >97, 45–35, and 30–25 kDa.
Electrophoresis and zymography (using gliadin as a substrate) results also revealed gliadinase activity in all CD fecal samples tested (Figure 6C). As observed with the NCD volunteer samples (Figure 1B), high fecal protein diversity and variability was present in the CD fecal samples (Figure 6B), and 3 principal zones with gliadinase activity were detected by zymography (>97, 45–35, and 30–25 kDa; Figure 6C). Moreover, densitometric evaluation of proteolytic cleavage products in both the NCD and CD zymographs (Figure 1, Figure 6C, respectively) revealed that the gliadinase activity was >3-fold higher in samples from patients with CD (not shown). Thus, feces from patients with CD had increased glutenase activity even when the patients were on a gluten-free diet. Moreover, the zymography results (Figure 6C) revealed similar gluten-degradation patterns, compared with the NCD samples (Figure 1C). Figure 7 shows that fecal samples from patients with CD displayed hydrolytic activity against the 33-, 19-, and 13-mer gliadin-derived peptides. Moreover, as previously reported using 33-mer peptides [9], our results revealed elevated fecal peptidase activity in CD patient samples against the 3 peptides assayed (Figure 2, Figure 7). Degradation of the 33-mer was 37% higher in CD patients. Nevertheless, and similar to the NCD samples, degradation of the 33-mer was incomplete (Figure 7A) even after a 24-h incubation (not shown). All fecal samples had sufficient activity to completely degrade the gliadin-derived 19-mer and 13-mer peptides derived during a 60-min incubation, except for samples from patient CD7 (with the 19-mer) and CD1 and CD3 (with the 13-mer) (Figure 7B,C). Moreover, after a 24-h incubation, all CD samples tested completely degraded 19- and 13-mer (not shown).
4. Discussion
We confirm that human feces contain proteins specifically capable of gluten degradation (Figure 1, Figure 2), which can hydrolyze the potentially toxic 33-, 19-, and 13-mer gliadin-derived peptides (Figure 2). Protein-purification (Figure 3, Figure 4) and amino acid-sequencing results indicated that CEL3B, CEL2A, and CBPA1 were responsible for fecal glutenase activity. These proteases are commonly present in the human small intestine, the digestive tract that is affected by CD and where the main protein degradation process takes place [25], [26]. Thus, our results allow us to conclude that the human digestive tract has the necessary tools for gluten-protein hydrolysis. Nevertheless, different authors have indicated that gluten is not completely degraded by proteolytic gastrointestinal activity and consequently, gliadin-derived peptides such as 33-, 19- and 13-mer can accumulate and trigger inflammatory process associated with CD [8], [27]. Using partially purified extracts (ammonium sulfate-fractioned), we verified this (Figure 2). However, as revealed in Figure 5, purified CEL2A and CBPA1 hydrolyzed the 19- and 13-mer (Figure 5), and only a minor portion of the 33-mer peptide remained intact (33%). These results confirmed previous data [27] indicating that >30% of the gliadin-reactive peptides remain intact after in vitro gastrointestinal digestion. The fact that ammonium sulfate-extracted proteins did not completely degrade the 19- and 13-mer (Figure 2), even after a 24-h incubation (not shown), suggests that human feces contains molecules that can inhibit CEL2A and CBPA1 activity. α-Amylase/trypsin inhibitors as ATIs from wheat cereal or other protease inhibitors (serpins such as elafin which has been related in celiac disease) as well as other factors (enzymes or microbes or blocking proteases) from bacterial, host or diet origin could be present in the crude extract and reduce the activity of proteases involved in gliadin digestion [28]. This inhibitory effect could promote the in vivo accumulation of different gliadin-derived peptides, which could be hydrolyzed by other proteases present in the gastrointestinal tract or excreted in the feces. In this sense, the presence of gluten (or gluten-derived peptides) and bacteria from the intestinal microbiota with glutenase activity have already been identified in human feces [14], [16], [17], [27], [29], [30].
In this work, surprisingly, we did not observe the presence of fecal bacterial proteins with glutenase activity. Because our method of fecal-protein extraction involved isolating soluble proteins without cellular rupture, it is probable that, under these conditions, microbial proteins were not present in our protein extracts. In previous studies, gluten-enrichment culture media were necessary to ensure efficient isolation and identification of fecal bacteria communities involved in gluten metabolism [9]. Moreover, comparison of the peptide-degrading capacity of the ammonium sulfate protein extract from NCD volunteer 1 (Figure 2) with the corresponding purified proteins (Figure 5), revealed that this ammonium sulfate extract had more 33-mer-degrading capacity (from 66–56 to 78%). These differences suggest that additional proteins are present in human feces that can participate in degrading the 33-mer. Thus, our results do not rule out the presence of other proteases in human feces that participate in gliadin metabolism or toxic peptide hydrolysis, as Caminero et al. [16] recently published (discussed below). It is possible that other proteases (in minor concentration, less activity or non-stable) could be masked and not being characterized in this study. Moreover, peptidases could be acting in gluten metabolism and not being detected by bioassay and zymography.
Our results (Figure 6B,C) suggested that gluten degradation in feces from patients with CD is caused by the same proteases that we purified and identified in NCD individuals, namely human elastase 3B, elastase 2A, and carboxypeptidase A1. Moreover, our data confirmed previous findings reported by Caminero et al. [9] and revealed that fecal CD samples have up to 2–5-fold more glutenase activity than NCD individuals (Figure 1, Figure 6A). We also observed that, unlike NCD samples (Figure 2) ammonium sulfate protein extracts from CD patients were capable of completely degrading the 19- and 13-mer gliadin-derived peptides (Figure 7). Importantly, the patients with CD that provided fecal samples were on a gluten-free diet; thus, ATIs from wheat cereal were not present. Accordingly, the total degradation capacity shown by feces from patients with CD against the 19- and 13-mer support a potential inhibitory effect proposed to ATIs against digestive enzymes with gliadinase activity [28].
Our data revealed that CD patients can more efficiently degrade gliadin than can NCD individuals, even with a gluten-free diet. It is possible that patients with CD have developed greater ability to degrade gluten, gliadin, and toxic peptides than NCD individuals as a protective mechanism. As Bernardo et al. [15] proposed, increased gluten proteolysis in the human gut might not be the solution for CD, but rather the origin of CD. Increased gluten proteolysis can generate different patterns of gluten-protein degradation between CD and NCD individuals and promote differences in the intestinal microbiota involved in gluten metabolism. Differences in intestinal flora occur between NCD and CD individuals [14], and it has been proposed that dysbiosis arises in patients with CD [5]. Olivares et al. [31] and De Angelis et al. [32], reported that increased pro-inflammatory bacteria correlated with low levels of probiotic microorganisms in patients with CD. This microbiota displacement involves differences in bacterial glutenase activity that can be fundamental in gluten toxicity. After gastrointestinal gluten degradation by elastase 3B, elastase 2A, and carboxypeptidase A1, the gluten-degraded products may be used by intestinal microbiota in CD and NCD individuals to generate peptides with different immunogenic capacities. Recently, differences in 33-mer-degradation patterns were observed between microorganisms isolated from NCD and CD individuals, which correlated with different immunogenic capacities [16]. With this in mind, we hypothesize that patients with CD have increased gluten-digestion activity, which modifies the pattern of gluten-derived peptides and promotes the selective proliferation of microorganisms that generate more immunogenic peptides. As gluten products are found in human feces [9], it may be essential to determine the immunogenic capacities of gliadin products present in the feces from non-treated CD and NCD individuals to test this hypothesis. In addition, it will be essential to establish whether gliadin metabolism occurs in individuals with a genetic predisposition to CD who do not develop the disease, which is a current focus in our lab.
Funding
This research was supported by grants from the Dirección General de Investigación (SAF2015-64306-R) and the Junta de Castilla y León (LE283U14). Jenifer Pérez-Andrés received a grant from the Ministerio de Educación Cultura y Deporte del Gobierno de España (SAF2015-64306R).
Acknowledgments
We thank Eduardo Fernández and Antonina Martínez for providing technical assistance.
Conflict of interest
None declared.
References
- 1.Sanz Y., De Pama G., Laparra M. Unraveling the ties between celiac disease and intestinal microbiota. International Reviews of Immunology. 2011;30:207–218. doi: 10.3109/08830185.2011.599084. [DOI] [PubMed] [Google Scholar]
- 2.Ludvigsson J.F., Leffler D.A., Bai J.C., Biagi F., Fasano A., Green P.H. The Oslo definitions for coeliac disease and related terms. Gut. 2013;62:43–52. doi: 10.1136/gutjnl-2011-301346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kurien M., Trott N., Sanders D.S. Long-term care for patients with coeliac disease in the UK: a review of the literature and future directions. Journal of Human Nutrition and Dietetic. 2016;29:617–623. doi: 10.1111/jhn.12379. [DOI] [PubMed] [Google Scholar]
- 4.Sanz Y. Microbiome and gluten. Annals of Nutrition & Metabolism. 2015;67:28–41. doi: 10.1159/000440991. [DOI] [PubMed] [Google Scholar]
- 5.Vivas S., Vaquero L., Rodríguez-Martín L., Caminero A. Age-related differences in celiac disease: Specific characteristics of adult presentation. World Journal of Gastrointestinal Pharmacology and Therapeutics. 2015;6:207–212. doi: 10.4292/wjgpt.v6.i4.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clayburgh D.R., Shen L., Turner J.R. A porous defense: the leaky epithelial barrier in intestinal disease. Laboratory Investigation. 2004;84:282–291. doi: 10.1038/labinvest.3700050. [DOI] [PubMed] [Google Scholar]
- 7.Thomas K.E., Sapone A., Fasano A., Vogel S.N. Gliadin stimulation of murine macrophage inflammatory gene expression and intestinal permeability are MyD88-dependent: role of the innate immune response in Celiac disease. Journal of Immunology (Baltimore Md.: 1950) 2006;176:2512–2521. doi: 10.4049/jimmunol.176.4.2512. [DOI] [PubMed] [Google Scholar]
- 8.Araya R.E., Castro M.F.G., Carasi P., McCarville J.L., Jury J., Mowat A.M. Mechanisms of innate immune activation by gluten peptide p31-43 in mice. American Journal of Physiology, Gastrointestinal and Liver Physiology. 2016;311:G40–G49. doi: 10.1152/ajpgi.00435.2015. [DOI] [PubMed] [Google Scholar]
- 9.Caminero A., Nistal E., Herrán A.R., Pérez-Andrés J., Ferrero M.A., Vaquero Ayala L. Differences in gluten metabolism among healthy volunteers, coeliac disease patients and first-degree relatives. British Journal of Nutrition. 2015;114:1157–1167. doi: 10.1017/S0007114515002767. [DOI] [PubMed] [Google Scholar]
- 10.Stene L.C., Honeyman M.C., Hoffenberg E.J., Haas J.E., Sokol R.J., Emery L. Rotavirus infection frequency and risk of celiac disease autoimmunity in early childhood: a longitudinal study. American Journal of Gastroenterology. 2006;101:2333–2340. doi: 10.1111/j.1572-0241.2006.00741.x. [DOI] [PubMed] [Google Scholar]
- 11.Fava F., Danese S. Intestinal microbiota in inflammatory bowel disease: friend of foe? World Journal of Gastroenterology. 2011;17:557–566. doi: 10.3748/wjg.v17.i5.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sellitto M., Bai G., Serena G., Fricke W.F., Sturgeon C., Gajer P. Proof of concept of microbiome-metabolome analysis and delayed gluten exposure on celiac disease autoimmunity in genetically at-risk infants. PLoS One. 2012;7:e33387. doi: 10.1371/journal.pone.0033387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cammarota G., Cuoco L., Cianci R., Pandolfi F., Gasbarrini G. Onset of coeliac disease during treatment with interferon for chronic hepatitis C. Lancet (London, England) 2000;356:1494–1495. doi: 10.1016/S0140-6736(00)02880-4. [DOI] [PubMed] [Google Scholar]
- 14.Nistal E., Caminero A., Vivas S., Ruiz de Morales J.M., Sáenz de Miera L.E., Rodríguez-Aparicio L.B. Differences in faecal bacteria populations and faecal bacteria metabolism in healthy adults and celiac disease patients. Biochimie. 2012;94:1724–1729. doi: 10.1016/j.biochi.2012.03.025. [DOI] [PubMed] [Google Scholar]
- 15.Bernardo D., Garrote J.A., Nadal I., León A.J., Calvo C., Fernández-Salazar L. Is it true that coeliacs do not digest gliadin? Degradation pattern of gliadin in coeliac disease small intestinal mucosa. Gut. 2009;58:886–887. doi: 10.1136/gut.2008.167296. [DOI] [PubMed] [Google Scholar]
- 16.Caminero A., Galipeau H.J., McCarville J.L., Johnston C.W., Bernier S.P., Russell A.K. Duodenal bacteria from patients with celiac disease and healthy subjects distinctly affect gluten breakdown and immunogenicity. Gastroenterology. 2016;151:670–683. doi: 10.1053/j.gastro.2016.06.041. [DOI] [PubMed] [Google Scholar]
- 17.Caminero A., Nistal E., Arias L., Vivas S., Comino I., Real A. A gluten metabolism study in healthy individuals shows the presence of faecal glutenasic activity. European Journal of Nutrition. 2012;51:293–299. doi: 10.1007/s00394-011-0214-3. [DOI] [PubMed] [Google Scholar]
- 18.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 19.Helmerhorst E.J., Wei G. Experimental strategy to discover microbes with gluten-degrading enzyme activities. Proceedings of SPIE – The International Society for Optical Engineering. 2014:9112. doi: 10.1117/12.2058730. Published online first: 5 May 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caminero A., Herrán A.R., Nistal E., Pérez-Andrés J., Vaquero l., Vivas S. Diversity of the cultivable human gut microbiome involved in gluten metabolism: isolation of microorganisms with potential interest for coeliac disease. FEMS Microbiology Ecology. 2014;88:309–319. doi: 10.1111/1574-6941.12295. [DOI] [PubMed] [Google Scholar]
- 21.Vasco-Cárdenas M.F., Baños S., Ramos A., Martín J.F., Barreiro C. Proteome response of Corynebacterium glutamicum to high concentration of industrially relevant C₄ and C₅ dicarboxylic acids. Journal of Proteomics. 2013;85:65–88. doi: 10.1016/j.jprot.2013.04.019. [DOI] [PubMed] [Google Scholar]
- 22.Havlis J., Thomas H., Sebela M., Shevchenko A. Fast-response proteomics by accelerated in-gel digestion of proteins. Analytical Chemistry. 2003;75:1300–1306. doi: 10.1021/ac026136s. [DOI] [PubMed] [Google Scholar]
- 23.Jami M.-S., Barreiro C., García-Estrada C., Martín J.F. Proteome analysis of the penicillin producer Penicillium chrysogenum: characterization of protein changes during the industrial strain improvement. Molecular & Cellular Proteomics. 2010;9:1182–1198. doi: 10.1074/mcp.M900327-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Magnelli P.E., Bielik A.M., Guthrie E.P. Identification and characterization of protein glycosylation using specific endo- and exoglycosidases. Journal of Visualized Experiments. 2011;58:e3749. doi: 10.3791/3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Erickson R.H., Kim Y.S. Digestion and absorption of dietary protein. Annual Review of Medicine. 1990;41:133–139. doi: 10.1146/annurev.me.41.020190.001025. [DOI] [PubMed] [Google Scholar]
- 26.Gray G.M. Comprehensive physiology. John Wiley & Sons, Inc; 2010. Dietary protein processing: intraluminal and enterocyte surface events; pp. 411–420. [Google Scholar]
- 27.Comino I., Real A., Vivas S., Síglez M.Á., Caminero A., Nistal E. Monitoring of gluten-free diet compliance in celiac patients by assessment of gliadin 33-mer equivalent epitopes in feces. American Journal of Clinical Nutrition. 2012;95:670–677. doi: 10.3945/ajcn.111.026708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cuccioloni M., Mozzicafreddo M., Ali I., Bonfili L., Cecarini V., Eleuteri A.M. Interaction between wheat alpha-amylase/trypsin bi-functional inhibitor and mammalian digestive enzymes: kinetic, equilibrium and structural characterization of binding. Food Chemistry. 2016;213:571–578. doi: 10.1016/j.foodchem.2016.07.020. [DOI] [PubMed] [Google Scholar]
- 29.Berger M., Sarantopoulos C., Ongchangco D., Sry J., Cesario T. Rapid isolation of gluten-digesting bacteria from human stool and saliva by using gliadin-containing plates. Experimental Biology and Medicine (Maywood, N.J.) 2015;240:917–924. doi: 10.1177/1535370214564748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei G., Tian N., Valery A.C., Zhong Y., Schuppan D., Helmerhorst E.J. Identification of pseudolysin (lasB) as an aciduric gluten-degrading enzyme with high therapeutic potential for celiac disease. American Journal of Gastroenterology. 2015;110:899–908. doi: 10.1038/ajg.2015.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olivares M., Neef A., Castillejo G., Palma G.D., Varea V., Capilla A. The HLA-DQ2 genotype selects for early intestinal microbiota composition in infants at high risk of developing coeliac disease. Gut. 2015;64:406–417. doi: 10.1136/gutjnl-2014-306931. [DOI] [PubMed] [Google Scholar]
- 32.De Angelis M., Vannini L., Di Cagno R., Cavallo N., Minervini F., Francavilla R. Salivary and fecal microbiota and metabolome of celiac children under gluten-free diet. International Journal of Food Microbiology. 2016;239:125–132. doi: 10.1016/j.ijfoodmicro.2016.07.025. [DOI] [PubMed] [Google Scholar]