Abstract
Decomposing transcriptional regulatory networks into functional modules and determining logical relations between them is the first step toward understanding transcriptional regulation at the system level. Modules based on analysis of genome-scale data can serve as the basis for inferring combinatorial regulation and for building mathematical models to quantitatively describe the behavior of the networks. We present here an algorithm called modem to identify target genes of a transcription factor (TF) from a single expression experiment, based on a joint probabilistic model for promoter sequence and gene expression data. We show how this method can facilitate the discovery of specific instances of combinatorial regulation and illustrate this for a specific case of transcriptional networks that regulate sporulation in the yeast Saccharomyces cerevisiae. Applying this method to analyze two crucial TFs in sporulation, Ndt80p and Sum1p, we were able to delineate their overlapping binding sites. We proposed a mechanistic model for the competitive regulation by the two TFs on a defined subset of sporulation genes. We show that this model accounts for the temporal control of the “middle” sporulation genes and suggest a similar regulatory arrangement can be found in developmental programs in higher organisms.
Keywords: gene expression, microarray, modem
Deciphering regulatory networks is a key step toward understanding gene regulation at a genomic scale. Both top-down and bottom-up approaches have been introduced. The top-down approach focuses on characterizing topologies of the networks from genome-wide measurements, such as large-scale surveys of network arrangements (1, 2), and is very useful in studying the organization of networks. The complementary bottom-up approach builds mechanistic models for each individual case, e.g., identifying the binding sites and target genes of a transcription factor (TF) (reviewed in ref. 3 and references therein), then specifies the roles of each TF in the networks, e.g., predicting under which cellular conditions a TF is activated (3–8). This approach next seeks to determine higher-order regulatory logic, e.g., how TFs cooperate with each other, and finally organizes all these pieces into functional networks. The bottom-up approach aims to explain the molecular basis of regulatory mechanisms. After a number of solid network structures are revealed, common regulatory rules are expected to emerge and, guided by the top-down approach, eventually general regulatory principles may be discovered.
We present here a bottom-up approach to decipher transcriptional regulatory networks (9), in which a key step is to accurately identify the binding sites and regulatory targets of transcription factors systematically. For the convenience of discussion, we use the term transcription module as an abbreviation for a TF, its binding sites, and target genes (9) throughout this article. To accurately reconstruct transcription modules, we have developed a computational algorithm called modem (Module construction using gene Expression and sequence Motif) based on a probabilistic model that integrates information from DNA sequences and large-scale gene expression data. We use “gene expression” broadly to refer to genome-wide measurements that detect transcriptional programs of a cell. These include, for example, standard mRNA microarray experiments, which measure the genomic change of transcript levels as a result of the activation or deactivation of specific transcription factors; microarray measurements of the effect of perturbation to a transcription factor by deletion or overexpression [referred to as TF perturbation experiments (TFPEs) (9), see below]; or TF genomic location measurements by chromatin immunoprecipitation followed by DNA microarray analysis (ChIP-chip) (10, 11). modem takes a consensus core motif, promoter sequences, and gene expression data as inputs. It outputs the probability of being a target for each gene and a position specific frequency matrix describing the binding site of the TF involved in the corresponding experiment. The ability of modem to construct transcription modules from a single expression measurement, which is usually a combined result of replicates, makes it distinct from other methods that require a large set of expression experiments (4–8). It allows the monitoring of the change of targets depending on the context, and the inference of network structure by integrating information from multiple sources (such as ChIP-chip and TFPE).
Combinatorial regulation and higher-order regulatory logic can be revealed by examining overlaps of target genes and binding sites between transcription modules. Once regulatory relationships are inferred with high confidence, mathematical models can be built to quantitatively study the specific regulatory logic. Such quantitative modeling may provide insight into underlying mechanisms and design principles, which can be tested by experiment.
We show here an example by inferring a network that regulates the yeast sporulation. The inferred network structure is consistent with the temporal observations during sporulation, and several hypotheses about meiosis are generated. The mechanistic model, built from transcription modules of two crucial sporulation TFs, Ndt80p and Sum1p, allows us to delineate the target genes and binding sites of Ndt80p and Sum1p, which partially overlap, and reveals the importance of the competitive regulation conveyed by Ndt80p and Sum1p for the sharp and precise temporal control of middle sporulation genes. This network structure laid the ground for further computational modeling and experimental manipulation of the sporulation program.
Methods
The modem algorithm constructs a transcription module for a TF based on a joint probability model describing the promoter sequence and gene expression data. The inputs to modem are as follows: (i) a single genome-wide microarray measurement related to a TF, such as ChIP-chip or TFPE; (ii) the core DNA motif recognized by the TF, typically six to eight bases long, either identified by applying the reduce algorithm (12) to the expression data or taken from databases; and (iii) the promoter sequences for all genes in the genome. The output of the algorithm is a refined description of the binding sites of the TF beyond the core motif, which is in the form of a position-specific frequency matrix (PSFM), and the probability of being a true target for each gene whose promoter contains the core motif (with a specified number of mismatches). This probability is then used to classify genes into target and nontarget categories. The main goal of modem is to accurately identify target genes given a core motif of a TF binding site; it also refines the input consensus core motif by outputting a PSFM that describes the motif beyond the consensus core.
It is well known that a core motif alone does not contain enough information for identifying target genes of a TF with high sensitivity and specificity. Many genes in the genome with the core motif in their promoter regions are not targets, whereas many true targets have variant forms of the core motif. To distinguish true targets from false positives, modem uses two sources of information: (i) true targets tend to have additional sequence information in the flanking region of the core motif, and (ii) true targets have high ratio changes in TFPEs or ChIP-chip experiments. To obtain additional information from the flanking sequence, modem uses the core motif to extract matches (with a specified number of mismatches) and their flanking sequences (called extended motifs). These extended motifs (typically 20 bp long) and gene expression ratios are then used as input.
modem uses a mixture model to describe the joint probability for extended motifs and the associated expression ratios. The mixture model is specified by the following parameters: a prior percentage of true targets among all extended motifs (the mixing parameter), a PSFM for the extended motifs belonging to the true targets, a PSFM for the extended motifs belonging to the background, and two normal distributions for the expression ratios (logarithm transformed) of the true targets and the nontarget genes, respectively. The model is similar in spirit to the mixture model used by the meme algorithm for motif finding (13) with two important generalizations: (i) gene expression data are modeled together with the sequence and (ii) the background sequences are also modeled by a PSFM because all sequences under consideration contain the core motif, thus the background sequences are not random.
The parameters in the model are estimated through an iterative process by using expectation maximization. Probability for each gene being a true target is calculated thereafter. All genes containing the core motif are then classified as target or nontarget. The algorithm works in the following way. At the beginning, all extended motifs have the same sequence score because there is no additional information from the flanking sequences, and the algorithm assigns higher probability of being targets to genes with higher expression ratio (or lower expression ratio if the motif represses expression). These genes with higher probability contribute more to the PSFM of the motif in the next update. The updated PSFM is used in conjunction with expression ratios to reevaluate the probability of being a target for each gene. These steps are iterated until convergence. The mathematical details are presented in Supporting Text, which is published as supporting information on the PNAS web site.
Results
Construction of Transcription Modules from TFPEs. We first constructed transcription modules from TFPEs. TFPE experiments compare gene expression between a wild-type cell in which the TF of interest functions normally and a perturbed cell in which the TF is deactivated (or inappropriately activated) by a mutation, typically a constructed deletion (9). In a TFPE, the target genes of the perturbed TF should show significant expression changes. Thus, it is not surprising that the most significant motif identified by reduce (12) in a TFPE is typically the binding site of the corresponding TF. As a result, even if the binding site of a TF is unknown, modem still can construct the module based on the core motif suggested by reduce in a TFPE (9). In a typical TFPE, many nontarget genes will also change their expression because of indirect effects. These indirect targets will not be included in the module because of their lack of motif-matching sequences. About 30 TFPEs (9) are available in the public domain from which we have constructed the corresponding transcription modules.
Example: The Ndt80p module constructed from a TFPE (ectopic expression of Ndt80p in vegetative cells) (14) is shown as an example (Table 1). Ndt80p binds to the MSE site (CRCAAAW) and up-regulates its target genes in the middle stage of sporulation (Gene Ontology, www.geneontology.org). reduce (12) correctly identified CACAAAA as the most significant motif in the Ndt80p TFPE. Ninety-eight genes were predicted to be target genes of Ndt80p by modem with CACAAAA as the core motif (one mismatch allowed). Using Saccharomyces Genome Database Gene Ontology Term Finder (http://db.yeastgenome.org/cgi-bin/SGD/GO/goTermFinder) (15), we found that 24 of these genes are annotated to function in sporulation, whereas only 87 genes of the 7,271 annotated genes in the yeast genome are so annotated. This enrichment of biological process is very significant (chance probability is 10–24) and consistent with the function of Ndt80p. It is reasonable to suggest that those member genes with no known functions are involved in the middle stage of the sporulation based on the function of the module.
Table 1. Target genes (partial list) of Ndt80p identified from its TFPE by using a core motif CACAAAA with one mismatch allowed.
| Gene/ORF | Probability | Extended motif* | Expression ratio, log2 |
|---|---|---|---|
| SPS3 | 1.000 | TTAGCGACACAAAAGAGACCT | -5.644 |
| SPS4 | 1.000 | CGCGCGCCACAAAAACGTATC | -5.644 |
| SSP1 | 1.000 | CAGGCGACACAAAATCATGAA | -4.644 |
| CWP1 | 0.993 | AAGGTGCCACAAAAGAAAACA | -3.184 |
| SPO74 | 0.987 | CTTGTGACACAAAAGAGAACA | -3.059 |
| HSP12 | 0.921 | GGGGCGGCACAAAATAACATA | -2.644 |
| YFR032C | 0.918 | GAAGCGTCACAAATTAATAAC | -2.556 |
| IME2 | 0.837 | CTTTACCCAAAAAATAAAACT | -2.737 |
Informative positions are highlighted in bold.
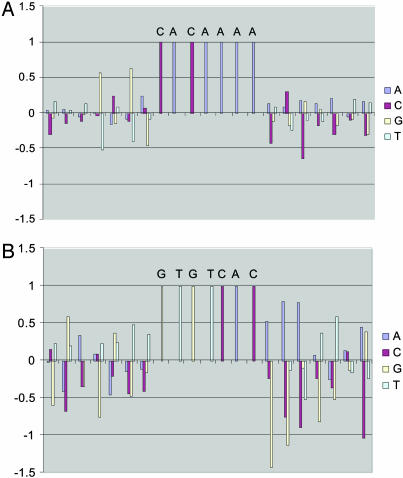
The Ndt80 example also illustrates the power of the modem algorithm in extracting flanking sequence information through the iterative process (see Methods). Although the initial input core motif is CACAAAA, modem found additional informative bases in the flanking region, e.g., G is predominant in positions 2 and 4 upstream of the core motif (Table 1 and Fig. 1). These two Gs are not critical to Ndt80p binding based on in vitro binding assay (E.J., C. Chin, I. Herskowitz, and H.L., unpublished data) but nevertheless play an important role in combinatorial regulation of Ndt80 and Sum1 that is critical in precisely regulating middle sporulation genes (see below).
Fig. 1.
Extended binding site profiles for Ndt80p and Sum1p. The position specific scoring functions of binding sites, i.e., the log ratio between the PSFM for the true targets of Ndt80p (A) and Sum1p (B) and that of the corresponding backgrounds. A positive score reflects that a base is favorable at the position. A negative score reflects that a base is unfavorable at the position. The initial input core motifs are shown in capital letters.
Construction of Transcription Modules from ChIP-Chip Data. ChIPchip technology has been used to determine localization of a particular TF's binding site and possible target genes of a TF (10, 11, 16, 17). For the latter purpose, a statistical algorithm was used to calculate a P value for each gene representing the significance of deviation from the background based on the observed two channel intensities (10, 17). Genes with P values less than a predefined threshold are considered as target genes (10, 17). This approach depends entirely on the fluorescence intensities and does not provide or use any information of binding motifs. We applied modem to ChIP-chip data, treating ratios of two channel intensities the same as mRNA expression ratios in TFPE.
ChIP-chip experiments for 106 TFs in yeast have been published by Lee et al. (17). We first used reduce to identify significant motifs in each experiment. If reduce finds, usually several, significant motifs, we then construct transcription modules for each of the motifs. We consider a module valid if the identified motif matches the known binding site of the TF or, if the binding site of the TF is unknown, the enriched function of member genes is consistent with that of the TF. Using this criterion, we were able to validate transcription modules (binding site and target genes) for 31 TFs, among which the binding sites of 15 TFs were previously known and 16 are previously uncharacterized (Table 3, which is published as supporting information on the PNAS web site). For the remaining factors, reduce mostly failed to find any significant motif. There are also a number of cases where the functions of genes in a module are apparently unrelated to the function of the TF (e.g., we found several modules contain many Y′ helicase genes).
For some of the cases, the failure to construct a valid module can be attributed to the fact that the ChIP-chip experiment was done under a condition where the TF was not activated. For example, no significant motif was found by reduce in the Pho4p ChIP-chip experiment. Consistently, the list of 99 Pho4p target genes suggested by Lee et al. (ref. 17 and http://web.wi.mit.edu/young/regulatory_network; 0.005 as a P value cutoff) only includes one known target, PHM6, of ≈15 previously identified by microarray experiments (18, 19). This example illustrates that accurately defining the binding site and targets of a TF requires a ChIP-chip experiment done under conditions that the TF is activated, which is a challenge for high-throughput methods.
Example: The Sum1p module constructed from ChIP-chip data (17) is shown as an example in Table 2. Sum1p is a transcriptional repressor that regulates sporulation and chromatin silencing (Gene Ontology). reduce found GTGTCAC as the most significant (P = 10–9) among motifs longer than 2 bases. Using GTGTCAC as the core motif (1 mismatch allowed), modem predicted 78 genes regulated by Sum1p, many of which were also in the set of 68 targets predicted by Lee et al. (17) with a threshold P = 0.001. Similar to the Ndt80p case, modem discovered additional flanking sequence information (Table 2 and Fig. 1). Several genes, NDT80, SSP1, CDA2, and MAM1, known to be regulated in sporulation, were not found by the analysis of Lee et al. (17). It is known that transcriptional regulation on Ndt80p by Sum1p is critical for the sporulation program (20, 21); missing the NDT80 gene as a target of Sum1p would make it difficult to infer the correct network topology. Genes with small ratios such as NDT80 (Table 2) were missed by the statistical analysis based on fluorescence data alone, but they were identified by modem because a strong sequence motif can compensate for the weak ratio. In general, modem is more sensitive in picking up known targets than the method based on ChIP-chip data alone. Among the 106 TFs studied by Lee et al. (17), 23 TFs have at least one target gene in the combined dataset from the Yeast Proteome Database (22), Saccharomyces cerevisiae Promoter Database (23), and TRANSFAC (24). We found that modem works better in six cases, equally good in 16 cases, and worse in one case (see Supporting Text for details).
Table 2. Target genes (partial list) of Sum1p identified from a ChIP-chip experiment by using a core motif GTGTCAC with one mismatch allowed.
| Gene/ORF | Probability | Extended motif* | Fluorescence ratio, log2 | Sum1P target† |
|---|---|---|---|---|
| SPS1 | 1.000 | TTTTTATGTGTCATTTTTTTT | 1.379 | Yes |
| SUM1 | 0.999 | ATCAAAAGTGTCAGCAAACAG | 1.220 | Yes |
| SPO74 | 0.996 | ATTTCTTGTGACACAAAAGAG | 1.000 | Yes |
| SPR3 | 0.980 | CTCTTTTGTGTCGCTAACAAA | 0.934 | Yes |
| CDA2 | 0.985 | TTGCGTTGCGTCACAAAATCA | 0.807 | No |
| SSP1 | 0.943 | TGATTTTGTGTCGCCTGTTTG | 0.824 | No |
| MAM1 | 0.930 | AAAATTAGTGACACAAAATAG | 0.696 | No |
| NDT80 | 0.902 | TAAATAGGTGACACAAAATGG | 0.705 | No |
Informative positions are highlighted in bold.
Denotes whether genes are identified as targets of Sum1p by Lee et al. (17).
Inference of Combinatorial Regulation and Regulatory Networks in Yeast Sporulation. Cooperation among several TFs (often called “combinatorial regulation”) is generally believed to be the source of complexity and sophistication in transcriptional regulatory programs. The modem outputs a PSFM for the TF binding site and a set of target genes, which lays the groundwork for building mechanistic model of combinatorial regulation.
Identification of active transcription modules. reduce was used previously to identify motifs in the microarray experiments that monitor temporal gene expression during sporulation (12, 14). We used those motifs and the modem algorithm to construct modules. The following picture emerged from comparing these motifs with known ones, such as URS1, MSE, and MCB sites, or motifs identified from TFPE or ChIP-chip data, such as Rap1 site (16), as well as examining the functions of genes in the modules (Fig. 6, which is published as supporting information on the PNAS web site). At the beginning (0.5 and 2 h), biosynthesis and metabolism are slowed down. The three repressed modules (represented by AAATTTT, GAGATGA, and TGAAAAA consensus motifs) include genes whose go process annotations are greatly enriched in particular functions. Namely, “rRNA processing,” “ribosome biogenesis,” and “ribosome assembly” annotations are enriched for the AAATTTT and GAGATGA modules, whereas “metabolism” and “cell growth” annotations are enriched for the TGAAAAA module (SGD Gene Ontology Term Finder) (15). TFs that recognize these motifs have not yet been identified. Another repressed module (from 0.5 to 5 h) is Rap1p module (motif CCCATAC) that includes metabolism and ribosomal biosynthesis genes. This observation suggests that Rap1p and other TFs reduce the rate of metabolism and biosynthesis in response to starvation that triggers the sporulation program. The URS1 module consists of the known early sporulation transcription factor complex Ime1p/Ume6p and its binding site DSGGCGGCND (14). This module contains many known early meiosis genes, such as HOP1, MEK1, RIM4, ZIP1, and IME2, as well as genes involved in metabolism, such as CAR1, CAT2, CRC1, ACS1, and PUT3. The observed induction of the URS1 module in the entire sporulation time course (12) suggests that the Ime1p/Ume6p complex has unknown regulatory functions in sporulation in addition to turning on the early genes.
The MCB module (consensus motif ACGCGT), which contains many cell-cycle and cell-proliferation genes like RAD17, RAD27, CDC21, CDC46, and SPO26, is induced between the early and the middle-late stages (0.5–7 h) (12). The MCB complex is critical in regulating cell cycle, but its roles in sporulation are not clear. Given RAD17 is a member of this module and regulates the checkpoint in sporulation, our hypothesis is that the MCB complex may regulate the switch between mitosis and meiosis by turning on or off RAD17 and other related genes.
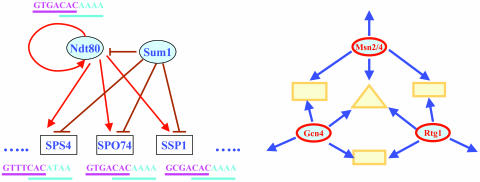
Building more complete network elements by combining modules. The MSE module (motif CRCAAAW; ref. 14) remains highly induced through the middle (5 h) and late stages (11.5 h). Recently, researchers found that Sum1p as well as Ndt80p can recognize the MSE site and, thus, provide negative control on the transcription of NDT80 and other middle sporulation genes (21). Xie et al. (25) showed that Sum1p binds to different MSE sites with significantly different affinities. To understand how the specificity is achieved, we analyzed the Sum1p module constructed from the ChIP-chip experiment (17) and the Ndt80p module constructed from the TFPE (14) (see above). The PSFMs for binding motifs show that Sum1p and Ndt80p prefer the DNA segments GTGTCACAAA and GNGNCACAAAA, respectively (Fig. 1). The obvious overlap between the two binding sites (underlined) may explain why the binding affinity of Sum1p to the MSE site depends on sequence context. During preparation of this article, we learned that Vershon and colleagues (26) have showed experimentally that Ndt80p and Sum1p do indeed bind to overlapping but distinguished motifs, completely consistent with our findings.
By constructing Ndt80p and Sum1p modules (Fig. 2 Left), we can see a distinctive regulatory arrangement: Ndt80p induces, whereas Sum1p represses, a set of middle genes, Ndt80p autoregulates itself, and Sum1p inhibits transcription of NDT80. This arrangement, combined with the overlap of binding sites of Ndt80p and Sum1p, provides a mechanism that has several attractive features as a regulatory system.
Fig. 2.
Network architectures inferred from transcriptional regulatory relationships. (Left) Controlled/autoregulation network architecture in sporulation. Overlapping binding site of Ndt80p and Sum1p are shown in magenta and cyan, respectively. (Right) Multiinput network architecture in amino acid starvation (9).
First, it becomes possible to selectively regulate Sum1p's targets. The general repressor Sum1p constitutively represses its target genes, including a set of genes that need to be transcribed by Ndt80p in the middle stage of sporulation. Because of their overlapping binding sites, Ndt80p and Sum1p would compete to regulate a subset of middle genes; the concentration of Ndt80p increases as sporulation proceeds and it takes over in the middle stage of sporulation. Those Sum1p-only targets (not bound by Ndt80p) remain repressed and presumably are not functioning in sporulation.
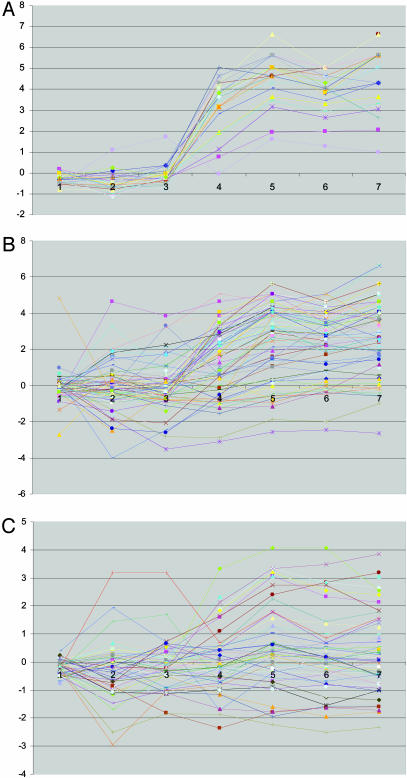
Second, this regulatory arrangement provides precise temporal control of the shared middle genes by the Ndt80p and Sum1p modules, making the notable quick and complete response of these genes possible. We examined the expression profiles of genes in the Ndt80p and Sum1p modules (Fig. 3). The apparent pattern is that all 25 genes except DAL4 shared by the two modules are precisely up-regulated in the middle stage (5 h, the fourth time point) of sporulation. It is not surprising to see NDT80 in this group and nine other genes (CDA2, SPS4, SPO77, DTR1, SPO21, SSP1, SPS1, SPR3, and SPO74) annotated to function in sporulation or meiosis. YSW1 is expressed specifically in spores. Another four genes, CRR1, HXT14, MIP6, and PES4, are involved in various biological processes. The remaining nine genes have no known functions, and it is reasonable to predict that they may play roles in sporulation. It is important to note that genes regulated only by Ndt80p or Sum1p do not have such precise temporal regulation (Fig. 3). Our hypothesis is that the genes regulated by both Ndt80 and Sum1 play critical roles that require synchronous appearance to ensure the proper proceeding of sporulation. The ability to make temporally well-defined changes is a general and important feature of development and, thus, we hypothesize that similar regulatory arrangements may be implemented in diverse developmental processes in different organisms. Similar mechanism may also be used to define sharp spatial boundaries in developing embryos.
Fig. 3.
Gene expression profiles for differentially regulated genes during sporulation. Expression profiles in sporulation for genes in both the Ndt80p and Sum1p modules (A), only in the Ndt80p module (B), and only in the Sum1p module (C). The x axis represents time points in (0, 0.5, 2, 5, 7, 9, and 11.5 h) sporulation experiments (14), and the y axis is the log ratio of gene expression level.
Given the topology of the regulatory arrangement (Fig. 2 Left) and the information of binding-site sequences, we can build a mechanistic model to analyze its behavior. Such analysis may provide a quantitative description of how the system works and reveal essential features of the system. The experimentally observed behavior needs to be reproduced by the model with parameters in a reasonable range.
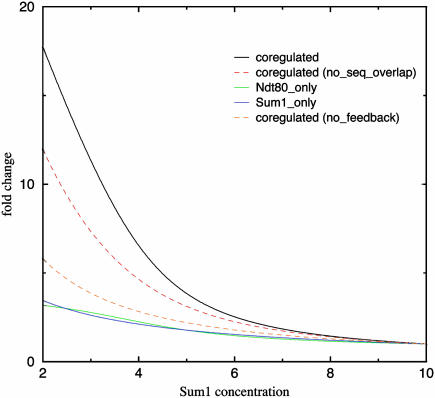
We adapted a physical chemistry model proposed by Shea and Ackers (27) and generalized by Buchler et al. (28) to study the regulatory arrangement in Fig. 2 Left (for details, see Supporting Text and Fig. 7, which is published as supporting information on the PNAS web site). According to the arrangement, the transcript level of NDT80 is controlled by the concentrations of the active Sum1p (Fig. 2 Left) and Ndt80p. With the simplifying assumption that the active protein level of Ndt80p is proportional to its transcript level (neglecting possible posttranscriptional regulation), Ndt80p level can be determined self-consistently as a function of Sum1p level. Thus, the transcript level of the regulatory targets of Ndt80p and/or Sum1p are all determined by the Sum1p level. It has been shown that Sum1p concentration decreases during sporulation (29). Therefore, we analyzed responses of genes to the change of active Sum1p. First, we found similar differential regulation as observed in experiments (Fig. 4). As Sum1p level decreases (but still higher than the concentration at which Sum1p site is half occupied in the absence of competition), genes belonging to both the Ndt80p and Sum1p modules (including Ndt80p itself) are sharply induced; genes regulated only by Ndt80p are induced slightly, but the expression changes are much smaller than those of the dually regulated genes, whereas genes regulated only by Sum1p remain repressed (only a small induction). Second, we found the positive autofeedback and sequence competition necessary for the sharp induction of dually regulated genes. Without the autofeedback, dually regulated genes are only weakly induced. If the binding of Ndt80p and Sum1p is not exclusive, the level of induction decreases significantly (Fig. 4). We speculate that autofeedback and competition between activator and repressor may be a general feature of network design, where sharp temporal or spatial profile is needed and the control is at the promoter level, such as in yeast sporulation and fly embryonic development.
Fig. 4.
Dependence of gene transcription levels on the active concentration of Sum1p, produced by the mechanistic model derived from the network arrangement in Fig. 2 Left. Green, blue, and black lines represent genes regulated by Ndt80p only, Sum1p only, and dually regulated genes, respectively. The red and orange dashed lines represent the hypothetical cases for the dually regulated genes with no sequence competition (between Ndt80p and Sum1p) and no autofeedback of Ndt80, respectively. All of the other parameters are kept the same. Sum1p level is measured relative to the concentration at which Sum1p site is half-occupied in the absence of competition. Transcript level is measured as fold changes by using the level at Sum1p concentration equals 10 as baseline. See Supporting Text for the detail of the model and the choice of parameters.
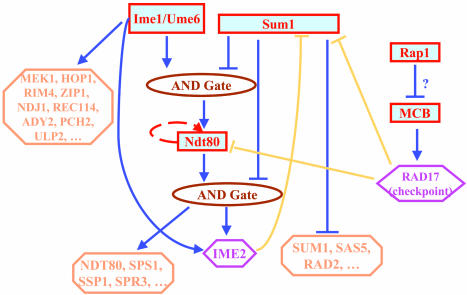
Inference of the network topology. Based on the transcription modules and the existing knowledge, we inferred the networks that regulate sporulation (Fig. 5). Because the network topology is quite complex, we mainly focused on transcriptional cascades. Various network regulatory strategies, such as AND gate, autoregulation (Ndt80p), and feed-forward loop (Ime1p/Ume6p and Ndt80p regulate IME2), have been exploited. Regulation provided by this network is consistent with temporal events occurring in sporulation (20, 29). Ime1p/Ume6p complex is activated first and transcribes a set of early genes, such as MEK1, HOP1, etc., as well as IME2. Ime2p is a kinase that inhibits the activity of Sum1p (29). As the activity of Sum1p decreases and Ime1p/Ume6p complex keeps active, the first AND gate is open to transcribe NDT80. Because of the autoregulation, once NDT80 is transcribed, its activity is increased dramatically. The second AND gate is then open to transcribe the middle genes and also keeps the transcript level of IME2 high to continuously inhibit the activity of Sum1p. This picture is likely to be an oversimplification of the reality because gene regulation during sporulation is very complex. If the appropriate experimental data such as TFPEs or ChIP-chip under the right conditions for all TFs in the yeast genome were available, we would be able to obtain a more complete network structure.
Fig. 5.
Regulatory networks in the yeast sporulation. All blue links are based on the modules we constructed, and only the three yellow links are taken from literature.
Discussion
We presented the modem algorithm for identifying the binding site and target genes of a TF and demonstrated its utility for analyzing ChIP-chip and TFPE data. Compared with other approaches (ref. 3 and references therein) for identifying binding sites and target genes of TFs, the modem algorithm has a number of advantages. Although clustering-based module-inference methods depend on gene expression data under multiple conditions (6–8), modem can construct a module from a single microarray experiment such as ChIP-chip or TFPE. The feature is particularly useful in monitoring how regulatory targets of transcription factors change upon biological context (such as different time points during cell cycle or during environmental perturbations) and provides a way to integrate data from a variety of sources. In addition, because the modem algorithm is based on a joint probability model that combines sequence information and gene expression data, its sensitivity and specificity is higher than methods based on either binding site motif or express data alone. modem is also distinct from methods for motif finding by combining sequence and expression data (12, 30, 31) because it focuses on target identification.
We showed that we could implicate genes (and especially regulators) of previously unknown function in processes such as sporulation. Such inferences can, and no doubt will, be tested by experiment. We could also infer the logic of regulation, especially the ways in which actual modules are combined. Notable in this regard is the finding of overlapping DNA binding motifs. Specifically, it would be interesting to construct adjacent or overlapping binding sites of Ndt80p and Sum1p (or other pairs of regulators) in the promoter region of a reporter gene and test whether appropriately regulatory logic results. It would also be interesting to make specific single-base mutations at the binding site that affect the binding of one factor and see how it changes the temporal profile of the expression. Similarly, the properties of the network architectures proposed here and those likely to emerge from future studies can and should be tested experimentally.
Bioinformatics can identify components and links between components in the networks. To understand the underlying regulatory mechanism, we need mechanistic models. We believe the combination of bioinformatics and molecular modeling is necessary to understand the general design principles used by various regulatory systems. We showed that a physical chemistry model can capture the main features of the controlled/autoregulation architecture of Sum1p/Ndt80p. The predictions from this model are waiting for experimental test.
As more and more microarray and ChIP-chip experiments done under various conditions become available, we will be able to construct transcription modules and monitor activation for more and more TFs. We expect other regulatory architectures like the controlled/autoregulation architecture of Sum1p/Ndt80p in sporulation (Fig. 2 Left) to be discovered. It is interesting to compare the logical structures and dynamic features of different network architectures and correlate their characteristics to the biological processes in which they are implemented. For instance, the two network architectures shown in Fig. 2 are quite different: controlled/autoregulation motif provides precise temporal control and sharp response in sporulation (see Results), whereas multiinput architecture may enable both general and pathway specific response to the lack of amino acid in the environment (9). As more data are becoming available in yeast as well as in other organisms, it would be interesting to examine whether these network architectures are general, e.g., whether the controlled/autoregulation architecture is more generally exploited by different developmental processes.
Supplementary Material
Acknowledgments
This work was supported in part by National Institutes of Health Grants HG01315 and GM46406 (to D.B.) and GM70808 (to H.L.); the Departmental Faculty Startup Fund and Academic Senate Research Grant from the University of California, San Diego; supercomputer time at the National Center for Supercomputing Applications through a Small Allocation Grant; and a David and Lucile Packard fellowship (to H.L.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ChIP-chip, chromatin immunoprecipitation followed by DNA microarray analysis; PSFM, position-specific frequency matrix; TF, transcription factor; TFPE, TF perturbation experiment.
References
- 1.Milo, R., Shen-Orr, S., Itzkovitz, S., Kashtan, N., Chklovskii, D. & Alon, U. (2002) Science 298, 824–827. [DOI] [PubMed] [Google Scholar]
- 2.Shen-Orr, S. S., Milo, R., Mangan, S. & Alon, U. (2002) Nat. Genet. 31, 64–68. [DOI] [PubMed] [Google Scholar]
- 3.Li, H. & Wang, W. (2003) Curr. Opin. Genet. Dev. 13, 611–616. [DOI] [PubMed] [Google Scholar]
- 4.Beer, M. A. & Tavazoie, S. (2004) Cell 117, 185–198. [DOI] [PubMed] [Google Scholar]
- 5.Gao, F., Foat, B. C. & Bussemaker, H. J. (2004) BMC Bioinformatics 5, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Segal, E., Shapira, M., Regev, A., Pe'er, D., Botstein, D., Koller, D. & Friedman, N. (2003) Nat. Genet. 34, 166–176. [DOI] [PubMed] [Google Scholar]
- 7.Ihmels, J., Bergmann, S. & Barkai, N. (2004) Bioinformatics 20, 1993–2003. [DOI] [PubMed] [Google Scholar]
- 8.Bar-Joseph, Z., Gerber, G. K., Lee, T. I., Rinaldi, N. J., Yoo, J. Y., Robert, F., Gordon, D. B., Fraenkel, E., Jaakkola, T. S., Young, R. A. & Gifford, D. K. (2003) Nat. Biotechnol. 21, 1337–1342. [DOI] [PubMed] [Google Scholar]
- 9.Wang, W., Cherry, J. M., Botstein, D. & Li, H. (2002) Proc. Natl. Acad. Sci. USA 99, 16893–16898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ren, B., Robert, F., Wyrick, J. J., Aparicio, O., Jennings, E. G., Simon, I., Zeitlinger, J., Schreiber, J., Hannett, N., Kanin, E., et al. (2000) Science 290, 2306–2309. [DOI] [PubMed] [Google Scholar]
- 11.Iyer, V. R., Horak, C. E., Scafe, C. S., Botstein, D., Snyder, M. & Brown, P. O. (2001) Nature 409, 533–538. [DOI] [PubMed] [Google Scholar]
- 12.Bussemaker, H. J., Li, H. & Siggia, E. D. (2001) Nat. Genet. 27, 167–171. [DOI] [PubMed] [Google Scholar]
- 13.Bailey, T. L. & Elkar, C. (1994) in Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology (ISMB' 94) (Am. Assoc. Art. Intell., Menlo Park, CA), pp. 28–36.
- 14.Chu, S., DeRisi, J., Eisen, M., Mulholland, J., Botstein, D., Brown, P. O. & Herskowitz, I. (1998) Science 282, 699–705. [DOI] [PubMed] [Google Scholar]
- 15.Boyle, E. I., Weng, S., Gollub, J., Jin, H., Botstein, D., Cherry, J. M. & Sherlock, G. (2004) Bioinformatics 20, 3710–3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lieb, J. D., Liu, X., Botstein, D. & Brown, P. O. (2001) Nat. Genet. 28, 327–334. [DOI] [PubMed] [Google Scholar]
- 17.Lee, T. I., Rinaldi, N. J., Robert, F., Odom, D. T., Bar-Joseph, Z., Gerber, G. K., Hannett, N. M., Harbison, C. T., Thompson, C. M., Simon, I., et al. (2002) Science 298, 799–804. [DOI] [PubMed] [Google Scholar]
- 18.Carroll, A. S., Bishop, A. C., DeRisi, J. L., Shokat, K. M. & O'Shea, E. K. (2001) Proc. Natl. Acad. Sci. USA 98, 12578–12583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ogawa, N., DeRisi, J. & Brown, P. O. (2000) Mol. Biol. Cell 11, 4309–4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vershon, A. K. & Pierce, M. (2000) Curr. Opin. Cell Biol. 12, 334–339. [DOI] [PubMed] [Google Scholar]
- 21.Pak, J. & Segall, J. (2002) Mol. Cell. Biol. 22, 6417–6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Costanzo, M. C., Crawford, M. E., Hirschman, J. E., Kranz, J. E., Olsen, P., Robertson, L. S., Skrzypek, M. S., Braun, B. R., Hopkins, K. L., Kondu, P., et al. (2001) Nucleic Acids Res. 29, 75–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu, J. & Zhang, M. Q. (1999) Bioinformatics 15, 607–611. [DOI] [PubMed] [Google Scholar]
- 24.Matys, V., Fricke, E., Geffers, R., Gossling, E., Haubrock, M., Hehl, R., Hornischer, K., Karas, D., Kel, A. E., Kel-Margoulis, O. V., et al. (2003) Nucleic Acids Res. 31, 374–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie, J., Pierce, M., Gailus-Durner, V., Wagner, M., Winter, E. & Vershon, A. K. (1999) EMBO J. 18, 6448–6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pierce, M., Benjamin, K. R., Montano, S. P., Georgiadis, M. M., Winter, E. & Vershon, A. K. (2003) Mol. Cell. Biol. 23, 4814–4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shea, M. A. & Ackers, G. K. (1985) J. Mol. Biol. 181, 211–230. [DOI] [PubMed] [Google Scholar]
- 28.Buchler, N. E., Gerland, U. & Hwa, T. (2003) Proc. Natl. Acad. Sci. USA 100, 5136–5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kassir, Y., Adir, N., Boger-Nadjar, E., Raviv, N. G., Rubin-Bejerano, I., Sagee, S. & Shenhar, G. (2003) Int. Rev. Cytol. 224, 111–171. [DOI] [PubMed] [Google Scholar]
- 30.Liu, X. S., Brutlag, D. L. & Liu, J. S. (2002) Nat. Biotechnol. 20, 835–839. [DOI] [PubMed] [Google Scholar]
- 31.Conlon, E. M., Liu, X. S., Lieb, J. D. & Liu, J. S. (2003) Proc. Natl. Acad. Sci. USA 100, 3339–3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.