Abstract
Alcohol is likely to affect neurons nonselectively, and the understanding of its action in the CNS requires elucidation of underlying neuronal circuits and associated cellular processes. We have identified a Drosophila signaling system, comprising neurons expressing neuropeptide F (NPF, a homolog of mammalian neuropeptide Y) and its receptor, NPFR1, that acutely mediates sensitivity to ethanol sedation. Flies deficient in NPF/NPFR1 signaling showed decreased alcohol sensitivity, whereas those overexpressing NPF exhibited the opposite phenotype. Furthermore, controlled functional disruption of NPF or NPFR1 neurons in adults rapidly confers resistance to ethanol sedation. Finally, the NPF/NPFR1 system selectively mediates sedation by ethanol vapor but not diethyl ether, indicating that the observed NPF/NPFR1 activity reflects a specialized response to alcohol sedation rather than a general response to intoxication by sedative agents. Together, our results provide the molecular and neural basis for the strikingly similar alcohol-responsive behaviors between flies and mammals.
Keywords: acute ethanol response, neuropeptide Y, neuropeptide F receptor
Alcohol, a widely abused drug, impacts the functioning of the CNS in diverse animals. The behavioral responses to acute alcohol exposure are remarkably similar among humans, rodents, and even fruit flies. Alcohol induces an excitatory state at lower concentrations but exerts a sedative effect at higher doses (1, 2). The current knowledge about how this drug impacts the functioning of the CNS is rather limited. Ethanol is highly soluble in both water and lipids, and is likely to act on neurons in a nonselective manner. However, the sensitivity of neurons in different neural circuits to this drug may vary greatly (3). Thus, elucidation of neuronal circuits and underlying molecular mechanisms essential for alcohol sensitivity is a prerequisite for understanding how alcohol interferes with the functioning of the CNS.
Neuropeptides are a group of chemically diverse signal molecules implicated in modulating a broad spectrum of physiological processes and behaviors (4). Mammalian neuropeptide Y (NPY) is a 36 aa neuromodulator present abundantly in many regions of the CNS, and acts thorough a number of Y receptor subtypes (5). Mice lacking NPY or Y1 displayed increased ethanol consumption and resistance to alcohol sedation, whereas animals overexpressing NPY showed opposite behavioral phenotypes (6, 7). These results provide genetic evidence of a critical role of the NPY signaling system in acute ethanol response. However, the elucidation of the physiological role of the NPY system and the action sites has been difficult largely because of the complexity of mammalian models.
NPY family molecules have been found in diverse organisms (8-10). Neuropeptide F (NPF) is the sole member of the NPY family in the Drosophila genome, and its action is mediated by G protein-coupled seven-transmembrane receptors related to mammalian NPY receptors (11). Both NPY and NPF are present prominently, although not exclusively, in the CNS (5, 12). In response to sweet substances, the NPF neuronal circuit of Drosophila larvae underwent stable modifications in a quantitative fashion (12). These observations point to the role of the NPF system as a neural interface that integrates sensory inputs and modulates motor outputs accordingly. We postulate that the NPF system might play a broad role in modulating fly behavioral responses to diverse chemical cues.
In this report, we provide neuroanatomical and functional evidence that the NPF neuronal circuit plays a physiological role in acute modulation of alcohol sensitivity in adult flies. We found that targeted disruption of NPF/NPFR1 signaling or knockdown of npfr1 activity decreased fly sensitivity to ethanol sedation, whereas overexpression of NPF increased alcohol sensitivity. Moreover, controlled functional disruption of NPF or NPFR1 neurons rapidly triggered acute resistance to ethanol sedation, suggesting that the NPF pathway tonically controls acute alcohol response. We also show that the NPF system selectively mediates sedation by ethanol vapor but not diethyl ether. Previous studies (13-15) have shown that both mammalian NPY and fly NPY-like systems promote feeding response. Here, we demonstrate another functional parallel between the two neural systems in the modulation of alcohol sensitivity. The remarkable similarities in alcohol-related behavior and the control mechanism between flies and mammals validate the use of this invertebrate model for alcohol research.
Materials and Methods
Flies and Media. Synchronized fly eggs were collected onto apple juice agar plates containing yeast paste on the surface. Flies were reared on the same food at room temperature with exposure to natural lighting. Adult females, synchronized by collecting flies enclosed within a 12-h period, were used for all assays. The npfr1-gal4, npf-gal4, UAS-npf, and H1-lacZ flies were generated through P element-mediated transformation of y1 w76c23 flies, and the UAS-npfr1dsRNA flies were generated through transformation of w1118 flies. Other fly strains and transgenic lines included wild-type Canton S, y1 w76c23, w1118, UAS-DTI, UAS-GFP, 386Y-gal4, UAS-ANF-GFP (also named UAS-ANF-EMD), kindly provided by D. L. Deitcher (Cornell University, Ithaca, NY).
Immunostaining. The adult brains were dissected and fixed in freshly made 4% paraformaldehyde in PBS for 40 min at room temperature. The antisera and immunostaining procedure were described elsewhere (12). At least 12 tissues were examined for each group of flies, and the consensus patterns reported are derived from at least 80% of the tissues examined.
Transgenic Constructs. To silence the npfr1 gene by using RNA interference, two 665-bp DNA fragments derived from the 5′ portion of the coding sequence were cloned into the downstream of the yeast upstream activating sequence (UAS) promoter in the pUAST vector as inverted repeats separated by a 619-bp DNA spacer. The resulting construct, pUAST-npfr1dsRNA, when integrated into the fly genome, can be used to stably express npfr1 hairpin-loop-structured RNA in the presence of a gal4 driver. The UAS-npf construct was made by inserting a full-length npf cDNA. The H1-lacZ transgene was cloned into a pCaSpeR-based vector that also contains a miniwhite gene (16).
Behavioral Assays. Synchronized 7-day-old yw females were used for the determination of the sedative effects of different ethanol doses. Active, well fed females were selected under CO2 and allowed to recover for at least 1 h before use. Typically, 20 females were used for each trial. Ethanol solutions of 10%, 31%, 42%, 54%, and 86% were made by mixing ethanol and water at the ratio (vol/vol)of1:9, 3:7, 4:6, 5:5, and 8:2, respectively. Ethanol solution (1 ml) at a desired concentration was added to a piece of folded Kimwipes tissue paper (11.4 × 21.5 cm) with edges sealed by using transparent tape and laid at the bottom of a 180-ml plastic fly bottle. Flies were transferred immediately into the bottle, which was then sealed with a paper lid and parafilm. The active flies remained on the top, and sedated flies that dropped to the bottom were counted at 5-min intervals. The assay conditions for the recovery of anesthetized flies were as follows. Twenty active females were placed into a 35-ml plastic vial closed with a cotton plug, to which 1 ml of 100% ethanol was added slowly to allow ethanol to soak in. In this case, sealing of the vials is optional because ethanol vapor rapidly reaches the intoxicating level in the vial. After exposure to ethanol vapor for 12 min, all flies tested laid motionlessly at the bottom of the vial. Subsequently, the old cotton plug was replaced with a fresh ethanol-free plug. The number of flies recovered from the ethanol sedation, as evidenced by their climbing and flying activities, was counted at a 2-min interval. The conditions for assaying acute response to ether sedation were the same except for the following alterations. Ether (33 μl) was added to a square cotton pad placed on the bottle top and sealed in with the plastic wrap.
Ethanol Content Assay. Ten active flies were first exposed to ethanol vapor in a 180-ml bottle covered with a cotton pad containing 1 ml of 100% ethanol and sealed with plastic wrap. The flies were subsequently allowed to recover for either 0 or 50 min in an alcohol-free 35-ml plastic vial. Afterward, flies were quickly frozen on dry ice, homogenized in 300 μl of 50 mM Tris buffer (pH 7.5), and spun at 13,500 rpm for 15 min in an Eppendorf microfuge. The cleared supernatant was recovered for further use. The assay for the alcohol concentration in the extract was performed by using an alcohol reagent kit (333-A, Sigma). The protein concentration was determined by using a protein assay kit (500-0113, Bio-Rad).
Results
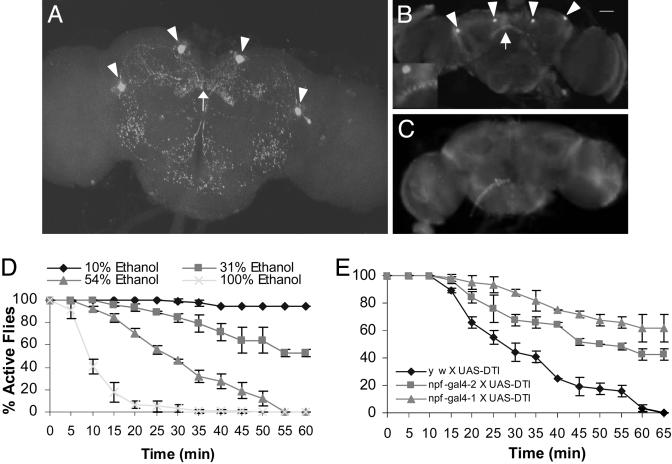
The NPF Neuronal Circuit in the Adult Brain. We analyzed the NPF distribution in the whole adult brain by using immunofluorescence staining. To obtain high specificity, the anti-NPF antiserum was preabsorbed against an amidated oligopeptide (C8) corresponding to the C-terminal structure of NPF, as described (12). The specificity of the preabsorbed antiserum was also verified genetically (see below), and by in situ RNA hybridization (12). The immunofluorescence staining revealed that NPF is prominently expressed in two pairs of neurons in the posterior side of the central brain, and NPF-positive projections displayed a largely symmetrical pattern that is stereotypic in both male and female brains (Fig. 1A). Extensive arborizations were observed at several regions in the posterior side of the central brain, whereas no NPF immunoactivity was detected in the optical lobes. One of the sites that exhibited intense NPF immunostaining was the fan-shaped body of the central complex, previously implicated in coordinating motor activities (17, 18). The NPF neuronal projections also innervate contralaterally the subesophageal ganglion, which might be important for regulating feeding and walking (19). NPF-containing arbors were also found in two lateral areas of the lower part of the central brain that appear to harbor the giant commissural interneurons of the giant fiber pathway (20). These data suggest that NPF neurons might coordinately modulate diverse sensory and motor neurons important for feeding, flight, and locomotion (21).
Fig. 1.
Flies ablated of NPF neurons are resistant to ethanol sedation. Confocal imaging shows the NPF neuronal network in the posterior side of an adult brain. Two pairs of neurons (arrowheads) and nerve terminals in the fan-shaped body (arrow) show NPF immunoreactivity. NPF-positive axons innervate several regions in the central brain. (A) Immunostaining of the adult brain from yw X UAS-DTI flies reveal NPF immunoreactivity in four neurons (arrowheads) and the fan-shaped body (B, arrow) that is absent in the brain of experimental flies (C, npf-gal4 X UAS-DTI). (B Inset) Magnified view of the fan-shaped body. Twenty y w females were sealed in a 180-ml bottle for each assay. The effective doses of ethanol for fly sedation were determined through three separate trials, in the presence of 1 ml of 10%, 31%, 54%, and 100% ethanol solutions, respectively (D). The susceptibility to alcohol sedation of experimental and control flies was determined under the same conditions as above, except that 1 ml of 42% ethanol solution was used to minimize the toxic effect of ethanol (E). The males of yw and npf-gal4 (in the yw background) were crossed with homozygous UAS-DTI females (in the w background). Thus, the progeny from these crosses are isogenic, except for the npf-gal4 transgene. The Student-Newman-Keuls test was used for analyzing data of all behavioral assays. The error bars indicate SEM in all figures. Numbers of remaining active flies over the period of 20-65 min were statistically analyzed. The npf-gal4 X UAS-DTI lines were significantly more resistant than the control (P < 10-4).
Targeted Disruption of NPF Signaling. In an effort to determine whether the NPF signaling system is capable of responding to diverse external cues, we tested how flies deficient in NPF signaling respond to ethanol vapor. Two independent transgenic fly lines were generated that contain the gal4 coding sequence driven by an npf upstream regulatory sequence (npf-gal4). Using the Drosophila GAL4-UAS binary expression system (22), we found that npf-gal4 drives the expression of a GFP reporter in two pairs of neurons in the brain and subsets of endocrine cells in the midgut (see Fig. 6, which is published as supporting information on the PNAS web site, and data not shown). Targeted ablation of NPF cells was achieved by crossing the npf-gal4 lines with flies carrying the UAS-DTI construct encoding an attenuated diphtheria toxin (23). In the normal adult brain, intense NPF immunostaining was detected in four neuronal bodies in the central brain and in the neuronal processes in different areas, including the central complex (Fig. 1B). However, the NPF immunoreactivity was completely eliminated in the brain of the npf-gal4 X UAS-DTI progeny (Fig. 1C). In addition, NPF immunostaining in the midgut was also largely absent (data not shown).
We developed a simple assay to measure fly resistance to the sedative effect of ethanol vapor (also see Materials and Methods). Synchronized 7- to 10-day-old flies were used in all assays. For determination of the effective ethanol doses, 20 y w female flies were sealed into a 180-ml bottle together with 1 ml of ethanol solution (10%, 31%, 54%, or 100%) soaked in a piece of tissue paper laid at the bottom. All flies were initially active and flew to the bottle top. The 10% ethanol solution had little sedative effect on the flies even after 1 h, whereas ethanol solutions of 31% or higher caused an increasingly rapid accumulation of sedated flies at the bottom of the container in a dose-dependent manner (Fig. 1D). To evaluate the influence of the genetic background on fly ethanol response, we compared the ethanol resistance of Canton S wild-type and y w flies. Canton S females were slightly more resistant to alcohol sedation than were y w females in the presence of the 42% or 54% solution; for example, the time required for sedating 50% of the Canton S flies (T50%) was 40 min versus 28 min for y w flies with the 54% solution (data not shown). We also tested transgenic flies ablated of NPF neurons by using the same assay. For this experiment and subsequent experiments, unless stated otherwise, a 42% ethanol solution was chosen to provide effective sedation while minimizing the toxic effect of ethanol. Because npf-gal4 lines are in the y w background, we also included the y w X UAS-DTI flies as the control. At 65 min, all y w X UAS-DTI control flies were sedated, whereas 60% npf-gal4-1 X UAS-DTI flies and 42% npf-gal4-2 X UAS-DTI flies were still clinging to the bottle top (Fig. 1E). Thus, the targeted ablation of the NPF neurons caused at least a 30-min delay in T50% under the same conditions, indicating that NPF-neuron-deficient flies are significantly more resistant to ethanol sedation than are control flies.
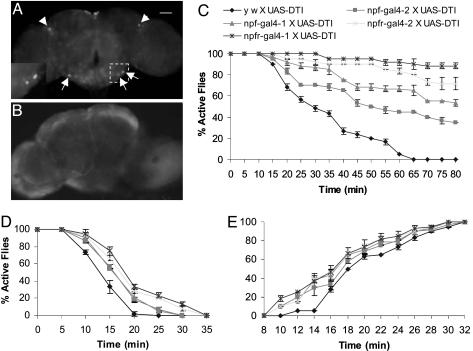
Targeted Disruption of NPFR1 Signaling. A previous pharmacological study (11) showed that one of the Drosophila G protein-coupled receptors, NPFR1, appears to be a receptor for NPF (11). The npfr1 transcripts were detected in fly heads by using RT-PCR (see Table 1, which is published as supporting information on the PNAS web site). We generated two independent transgenic fly lines containing gal4 driven by a 6.6-kb npfr1 upstream regulatory sequence (npfr1-gal4-1 and npfr1-gal4-2). Both npfr1-gal4 transgenic flies are derived from y w flies. The fly progeny from the npfr1-gal4 X UAS-GFP crosses consistently expressed GFP in a pair of dorsolateral neurons in the central brain and a small number of neurons in the subesophageal ganglion (Fig. 2A), suggesting NPFR1 neurons in one or both of the regions might be involved in mediating ethanol sensitivity.
Fig. 2.
NPFR1 neurons mediate ethanol response. GFP expression was detected in the brain of flies from the npfr1-gal4 X UAS-GFP crosses. (A) Arrowheads indicate two dorsolateral neurons in the central complex and three representative neurons in the subesophageal ganglia. (A Inset) Magnified view of the boxed area. (B) However, this GFP expression pattern was abolished in the brain of npfr1-gal4 X UAS-GFP/UAS-DTI flies. Control flies (UAS-GFP without a gal4 driver) showed no such GFP expression pattern in the brain (data not shown). (C) The alcohol sensitivity of experimental and control flies was determined under the same conditions as above. In three separate trials (n = 20 per trial), the significantly higher percentages of active flies were consistently observed for npfr1-gal4-1 and -2 X UAS-DTI flies than for npf-gal4-1 and -2 X UAS-DTI flies. The y w X UAS-DTI controls showed least ethanol resistance (P < 10-4, starting from 30 min). The flies were also tested for ethanol resistance under the same conditions, except a higher dose (1 ml of 86% ethanol) was used (D, P < 0.001, starting from 15 min). (E) To measure the rate of recovery from ethanol anesthetization, flies were exposed to concentrated ethanol vapor for 12 min, allowed to recover under the ethanol-free conditions, and the number of flies capable of climbing after the ethanol treatment was counted. The data of fly recovery over the time interval of 8-14 min after ethanol treatment were analyzed statistically. The control fly (y w X UAS-DTI) showed a delay in restoring normal locomotion compared with the four experimental flies (P < 10-4).
We have not been able to establish unequivocally a consistent immunostaining pattern of NPFR1 neurons in the adult brain by using an anti-NPFR1 peptide antiserum. As an alternative means to demonstrate that the DTI transgene is functional in npfr-gal4 X UAS-DTI flies, we generated fly progeny that harbored three transgenic constructs (npfr-gal4, UAS-GFP, and UAS-DTI). These flies, which presumably coexpress GFP and DTI in the same cells, indeed showed no detectable GFP expression in the corresponding neurons of the brain (Fig. 2 A and B). These data are also consistent with our observation that both npf-gal4 and npfr1-gal4 X UAS-DTI larvae display deficits in foraging activities (15).
We further tested the npfr1-gal4 X UAS-DTI flies for their resistance to ethanol vapor by using the same assay described above. Synchronized females were used in three separate trials (n = 20 per trial). Flies ablated of NPFR1 neurons consistently showed higher ethanol resistance than the control flies (y w X UAS-DTI;Fig. 2C). The NPFR1-neuron-deficient flies were significantly more resistant than the npf-gal4 X UAS-DTI flies, raising the possibility that NPFR1 neurons might receive convergent signals from another yet-unidentified ethanol-responsive neural circuit(s) in addition to the NPF pathway. The same five groups of flies were also tested in the presence of a higher ethanol dose. The assays were the same except that 1 ml of an 86% ethanol solution was used instead. In agreement with the previous results, the NPF- and NPFR1-neuron-deficient flies were significantly more resistant to ethanol sedation than were the control flies (Fig. 2D). Again, the two independent NPFR1-neuron-deficient transgenic lines showed the highest levels of ethanol resistance. Furthermore, functional knockdown of npfr1 by using RNA interference also confirmed ethanol resistance, suggesting that NPF and NPFR1 molecules directly mediate acute alcohol sensitivity (see Fig. 7, which is published as supporting information on the PNAS web site).
We also tested the rate of recovery of flies sedated by a brief exposure to ethanol vapor of high concentration (see Materials and Methods). Twenty sedated flies were kept in a 35-ml plastic vial, sedated after 12 min of exposure, and monitored over a 40-min period for recovery. The results show that NPF- and NPFR1-neuron-deficient flies both recovered significantly faster than the controls, suggesting they were sedated to a lesser degree than controls under the same conditions (Fig. 2E). NPFR1-neuron-deficient flies appeared to recover slightly faster than NPF-neuron-deficient flies. The recovery curves show that, although the control flies had a significant delay in recovery, the kinetics of all five recovery curves were similar. These observations suggest that the recovery process might be independent of NPF signaling.
We also examined the growth and reproduction of npf- and npfr1-gal4 X UAS-DTI flies. NPFR1-signaling-deficient and control flies showed no obvious difference in larval development and the rate of egg laying, although NPF-signaling-deficient flies showed only a slight decrease in larval development and the rate of egg laying (see Table 2, which is published as supporting information on the PNAS web site). Furthermore, additional control experiments were performed to rule out the potential effect of UAS-DTI on fly acute alcohol response (see Fig. 8, which is published as supporting information on the PNAS web site).
Controlled Disruption of NPF/NPFR1 Neuronal Activities. In the previous experiments (15), the NPF and NPFR1 neurons of the transgenic flies were likely ablated at an early larva stage. Therefore, it remains to be determined whether the phenotype of increased alcohol resistance reflects the physiological action of the NPF system or the secondary effect of some unknown developmental defects, resulting from a deficiency in NPF signaling. To address this issue, we attempted to conditionally disrupt NPF and NPFR1 neuronal function by using a temperature-sensitive allele of shibire, shits1. The shits1 allele encodes a semi-dominant-negative form of dynamin that can block neurotransmitter release at a restrictive temperature (>29°C), and has been widely used in fly behavioral studies (24). At the permissive temperature (23°C), the experimental flies (npf-gal4 or npfr1-gal4 X UAS-shits1) and controls (w X UAS-shits1) displayed similar response profiles to alcohol vapor (Fig. 3A). We then tested alcohol response at 30°C. The flies were preincubated at 30°C for 60 min before the assay. The experimental flies were consistently more resistant than were the controls. For example, the T50% of w X UAS-shits1 flies was ≈55 min, although the two experimental flies had a T50% of ≈85 min (Fig. 3B). These findings suggest that the NPF/NPFR1 neuronal pathway tonically mediates the acute alcohol response. The data also indicate that the increased alcohol resistance of flies ablated of NPF or NPFR1 neurons is unlikely due to some unknown genetic variations. These results also provide functional evidence that the neuropeptide release by NPF neurons may be coupled to a dynamin-mediated process. Consistent with this notion, it has been reported that pharmacological inhibition of norepinephrine uptake by neurons that corelease NPY and norepinephrine markedly reduces NPY release (25).
Fig. 3.
NPF/NPFR1 neurons acutely mediate ethanol response. At the permissive temperature of 23°C, both experimental flies (npf-gal4 and npfr1-gal4 X UAS-shits1) and controls (w X UAS-shits1) displayed comparable sensitivities to alcohol sedation (P > 0.2). The assay was performed in the presence of 1 ml of 42% ethanol solution. (A) In this experiment, the npf-gal4 and npfr1-gal4 flies are in the w background. At the restrictive temperature of 30°C, the experimental flies showed significant increases in ethanol resistance (P = 0.01). The flies were preincubated at 30°C for 60 min before the assay. (B) In this assay, 1 ml of 36% ethanol solution was used instead. The results suggest that the activities of NPF and NPFR1 neurons are tonically required for mediating acute alcohol response.
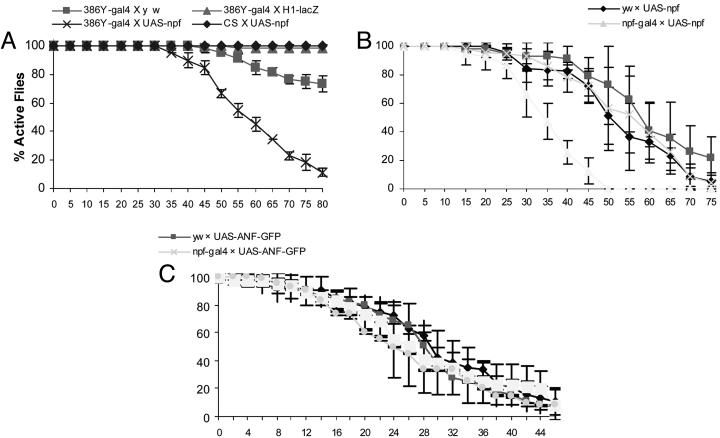
Effect of NPF Overexpression on Acute Alcohol Sensitivity. We wondered whether overexpression of NPF could cause increased alcohol sensitivity. An enhancer-trap gal4 line (386Y-gal4) was chosen because it was previously shown to drive reporter expression in brain cells, including peptidergic neurons (26). Transgenic flies containing UAS-npf driven by 386Y-gal4 showed strong ectopic expression of NPF in the adult brain (data not shown) and were tested for alcohol resistance. In this case, a 31% ethanol solution was used, because it conferred a weak sedative effect on control flies in earlier experiments. Because the influence of the genetic background on fly ethanol response was more pronounced at the reduced ethanol concentration, we decided to add more control crosses to exclude other trivial reasons that might cause increased sensitivity to ethanol. We included three different types of control flies, 386Y-gal4 X y w or H1-lacZ, and Canton S X UAS-npf, each of which contains either 386Y-gal4 or UAS-npf alone, and also provides an assessment of the effects due to variations in genetic background. We found that flies overexpressing NPF were highly sensitive to alcohol, even at the reduced concentration, whereas all of the control flies showed high resistance to ethanol vapor under the same condition (Fig. 4A). We further examined the ethanol sensitivity of npf-gal4 X UAS-npf flies that overexpress NPF in the NPF neurons. In this experiment, all transgenic flies are in the y w background. Again, the experimental flies (npf-gal4 X UAS-npf) were much more sensitive to ethanol than were the control flies (e.g., yw X UAS-npf or npf-gal4 X UAS-ANF-GFP;Fig. 4B). Taken together, these results provide direct evidence that NPF is a critical component for acute modulation of ethanol response.
Fig. 4.
Flies overexpressing NPF are more sensitive to ethanol vapor. (A) The ethanol sensitivity was tested for female flies overexpressing NPF (386Y-gal4 X UAS-npf) and three controls, 386Y-gal4 X y w, H1-lacZ, and CS X UAS-npf. The control fly H1-lacZ harbors the transgenic construct containing a lacZ reporter driven by an embryonic enhancer from the hairy gene and a miniwhite gene (see Materials and Methods) that provides a control for the effect of the white gene product on host ethanol response. The UAS-npf and H1-lacZ flies are in the y w background. The assay was performed by using a 31% ethanol solution. Flies were tested in three separate trials (n = 20 per trial). The 386Y-gal4 X UAS-npf flies are more sensitive to ethanol than the controls starting from 50 min (P < 0.0001). (B) The NPF-overexpressing files (npf-gal4 X UAS-npf) showed more sensitivity to ethanol vapor than the controls (y w X UAS-npf, y w X UAS-ANF-GFP, and npf-gal4 X UAS-ANF-GFP) when exposed to a 31% ethanol solution. All of the fly lines used are in the y w background. P = 0.026 starting from 30 min. (C) The ether sensitivity of y w and npf-gal4 X UAS-npf flies was also measured by using an assay where 33 μl of ether was added to a cotton pad sealed in a 180-ml bottle. Each assay typically uses 20 females, and at least three separate trials were performed. The T50% values of the flies were similar (P > 0.79).
Acute Ether Response of NPF-Overexpressing Flies. We also tested NPF-overexpressing flies for their responses to diethyl ether (Fig. 4C). In this case, 33 μl of ether was added to a cotton pad sealed in a 180-ml plastic bottle that contained 20 flies. We found that experimental flies (npf-gal4 X UAS-npf) and controls (e.g., y w X UAS-npf) displayed no significant difference in ether sensitivity (P > 0.7). Therefore, these results indicate that NPF selectively modulates the sedative effect of ethanol vapor, but not diethyl ether, and the observed NPF/NPFR1 activity is not a general response to intoxication by sedative agents.
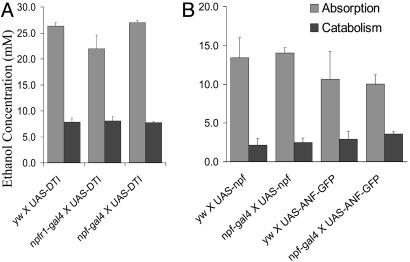
Ethanol Absorption and Metabolism. It is possible that the reduced ethanol sensitivity of NPF-signaling-deficient flies might be caused by decreased ethanol absorption and/or increased ethanol catabolism. To test this hypothesis, we measured the alcohol content in the bodies of the control and experimental flies that displayed either increased or decreased alcohol resistance. The comparable ethanol levels were detected in the extracts of y w and npf-gal4 X UAS-DTI flies (Fig. 5A). Furthermore, the alcohol levels in the extracts of NPF-overexpressing flies and controls were also similar (Fig. 5B). Based on these observations, we conclude that the altered ethanol responses by NPF-signaling-deficient or NPF-overexpressing flies are not due to changes in their pharmacokinetic properties.
Fig. 5.
NPF signaling does not affect alcohol absorption and catabolism. The ethanol content in both control and experimental flies was measured using an alcohol-dehydrogenase based assay, as described earlier. (A) The body alcohol content of experimental (npf-gal4 X UAS-DTI) and control (yw, npfr1-gal4 X UAS-DTI) flies; the flies were exposed to ethanol vapor for 20 min and subsequently allowed to recover for 0 or 50 min. (B) The flies were exposed to ethanol vapor for 12 min before the recovery. No significant difference in body alcohol contents was detected among the flies tested (ANOVA, P > 0.05).
Discussion
We have identified the NPF neural signaling system that acutely mediates fly sensitivity to alcohol sedation. Flies deficient in NPF/NPFR1 signaling displayed decreased ethanol sensitivity, whereas flies overexpressing NPF showed hypersensitive response to alcohol sedation. Moreover, controlled disruption of NPF/NPFR1 neuronal activities in adult flies rapidly conferred increased ethanol resistance. These findings suggest that this Drosophila NPY-like system is a physiological regulator of acute alcohol response.
The NPF neuronal circuit has also been shown to respond to chemosensory stimuli by sugar, and promote feeding response (12, 15). In the present work, we demonstrate that it has a role in acute response to ethanol. Therefore, the NPF system appears to be capable of regulating fly responses to diverse external cues. We propose that the NPF neuronal circuit should be a useful platform for unraveling the physiological roles of genes in behavioral regulation in the context of functionally characterized neurons.
Parallel activities have been reported between mammalian NPY and Drosophila NPF in feeding regulation (13-15). Here, we demonstrate another functional parallel between the two signaling molecules in ethanol response (6, 7). To many animals, including fruit flies, rodents, and humans, ethanol is a natural component in a variety of their food sources. At lower concentrations, ethanol is likely to serve as a volatile food cue. We previously showed that the Drosophila NPY-like signaling system enhances the motivation of food acquisition (15). Therefore, it is inviting to speculate that the NPY-like system might be evolved to function as an alcohol sensor to facilitate food searching. It would be interesting to test whether NPY/NPF signaling is activated by ethanol at a low concentration.
In Caenorhabditis elegans, an NPY-like receptor, NPR-1, has been recently reported to suppress the development of acute tolerance of ethanol intoxication (27). However, the ligand of NPR-1 is FMRFamide-like, rather than NPY-like (28), and its role in acute alcohol sensitivity has not been reported. Therefore, future work will be needed to clarify the functional relationship among mammalian NPY, Drosophila NPF, and nematode NPR-1, and to determine whether the NPY and NPY-like systems regulate different aspects of behavioral response to alcohol.
Several NPY receptor subtypes have been identified that mediate NPY action through coupling to heterotrimeric G proteins that down-regulate the intracellular level of cAMP (5). Consistent with this work, pharmacological analysis showed binding of NPF to the NPFR1 receptor inhibited forskolin-stimulated adenylate cyclase activity in cultured cells (11). We propose that Y1- and NPFR1-mediated alcohol sensitivity in mammals and fruit flies may require down-regulation of second-messenger cAMP and subsequent reduction of protein kinase A activity (29-31).
Immunofluorescence staining reveals that NPF-positive axon arbors are widely present in the central brain. Interestingly, a 6.6-kb npfr1 promoter sequence drives GFP expression in a small number of neurons that appear to be adjacent to, or overlap with, the NPF arbors in the subesophageal and dorsolateral regions. The subesophageal ganglia have been implicated in coordinated locomotion (19, 21), whereas the neurobiological role of neurons in the dorsolateral regions is not clear. It would also be interesting to determine whether ethanol-responsive NPFR1 neurons play a role in mediating the fly wake-sleep cycle.
It remains unclear how the NPF neuronal circuit responds to ethanol but not to ether. One possible explanation is that the cell membrane components of NPF neurons may selectively interact with volatile agents. For example, the binding of ethanol, but not ether, to NPF neurons may elicit an intracellular response(s) that subsequently leads to increased release of NPF and possibly other neurotransmitter(s) as well. In support of this notion, ethanol has been shown to selectively interact with the Drosophila potassium channel protein, Shaw2 (32, 33). It will be interesting to determine whether Shaw2 is expressed in NPF neurons, and if so, whether it might mediate the selective response to ethanol vapor. Furthermore, a number of cell membrane proteins have been biochemically identified as potential molecular targets of volatile anesthetics (3, 34). The NPF neuronal circuit offers a useful platform to test the physiological relevance of these targets in alcohol response.
Supplementary Material
Acknowledgments
We thank D. L. Deitcher, T. Kitamoto (University of Iowa, Iowa City), J. H. Park (University of Tennessee, Knoxville), K. Scott (University of California, Berkeley), and L. Stevens (University of Texas, Austin) for providing fly lines and J. S. Willis for critical reading of the manuscript. This work was supported by National Institutes of Health Grant DK-58348 (to P.S.).
Author contributions: P.S. designed research; T.W., C.A.P., D.X., and Q.W. performed research; T.W., C.A.P., D.X., Q.W., and P.S. analyzed data; and P.S. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: NPF, neuropeptide F; NPY, neuropeptide Y; UAS, upstream activating sequence; T50%, the time required for sedating 50% of the Canton S flies.
References
- 1.Singh, C. M. & Heberlein, U. (2000) Alcohol Clin. Exp. Res. 24, 1127-1136. [PubMed] [Google Scholar]
- 2.Parr, J., Large, A., Wang, X., Fowler, S. C., Ratzlaff, K. L. & Ruden, D. M. (2001) J. Neurosci. Methods 30, 93-99. [DOI] [PubMed] [Google Scholar]
- 3.Yaksh, T., Lynch, C. I., Zepol, W. M., Maze, M., Biebuyck, J. F. & Saidman, L. J. (1998) Anesthesia Biologic Foundations (Lippincott-Raven, Philadelphia).
- 4.Sandman, C. A., Strand, F. L., Beckwith, B., Chronwall, B. M., Flynn, F. W. & Nachman, R. J. (1999) Neuropeptides: Structure and Function in Biology and Behavior (New York Acad. Sci., New York).
- 5.Michel, M. C., Beck-Sickinger, A., Cox, H., Doods, H., N., Herzog, H., Larhammar, D., Quirion, R., Schwartz, T. & Westfall, T. (1998) Pharmacol. Rev. 50, 143-150. [PubMed] [Google Scholar]
- 6.Thiele, T. E., Marsh, D. J., Ste. Marie, L., Bernstein, I. L. & Palmiter, R. D. (1998) Nature 396, 366-369. [DOI] [PubMed] [Google Scholar]
- 7.Thiele, T. E., Koh, M. T. & Pedrazzini, T. (2002) J. Neurosci. 22, RC208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Larhammar, D. (1996) Regul. Pept. 62, 1-11. [DOI] [PubMed] [Google Scholar]
- 9.Brown, M. R., Crim, J. W., Arata, R. C., Cai, H. N., Chun, C. & Shen, P. (1999) Peptides (Tarrytown, NY) 20, 1035-1042. [DOI] [PubMed] [Google Scholar]
- 10.Riehle, M. A., Garczynski, S. F., Crim, J. W., Hill, C. A. & Brown, M. R. (2002) Science 298, 172-175. [DOI] [PubMed] [Google Scholar]
- 11.Garczynski, S. F., Brown, M. R., Shen, P., Murray, T. F. & Crim, J. W. (2002) Peptides (Tarrytown, NY) 23, 773-780. [DOI] [PubMed] [Google Scholar]
- 12.Shen, P. & Cai, H. N. (2001) J. Neurobiol. 47, 16-25. [DOI] [PubMed] [Google Scholar]
- 13.Bannon, A. W., Seda, J., Carmouche, M., Francis, J. M., Norman, M. H., Karbon, B. & McCaleb, M. L. (2000) Brain Res. 868, 79-87. [DOI] [PubMed] [Google Scholar]
- 14.Segal-Lieberman, G., Trombly, D. J., Juthani, V., Wang, X. & Maratos-Flier, E. (2003) Am. J. Physiol. 284, E1131-E1139. [DOI] [PubMed] [Google Scholar]
- 15.Wu, Q., Wen, T., Lee, G., Park, J. H., Cai, H. N. & Shen, P. (2003) Neuron 39, 147-161. [DOI] [PubMed] [Google Scholar]
- 16.Cai, H. N. & Shen, P. (2001) Science 291, 493-495. [DOI] [PubMed] [Google Scholar]
- 17.Ilius, M., Wolf, R. & Heisenberg, M. (1994) J. Neurogenet. 9, 189-206. [DOI] [PubMed] [Google Scholar]
- 18.Varnam, C. J., Strauss, R., Belle, J. S. & Sokolowski, M. B. (1996) J. Neurogenet. 11, 99-115. [DOI] [PubMed] [Google Scholar]
- 19.Bausenwein, B., Muller, N. R. & Heisenberg, M. (1994) J. Comp. Neurol. 340, 255-268. [DOI] [PubMed] [Google Scholar]
- 20.King, D. G. & Wyman, R. J. (1980) J. Neurocytol. 9, 753-770. [DOI] [PubMed] [Google Scholar]
- 21.Burrows, M. (1996) The Neurobiology of an Insect Brain (Oxford Univ. Press, Oxford).
- 22.Brand, A. H. & Perrimon, N. (1993) Development (Cambridge, U.K.) 118, 401-415. [DOI] [PubMed] [Google Scholar]
- 23.Han, D. D., Stein, D. & Stevens, L. M. (2000) Development (Cambridge, U.K.) 127, 573-583. [DOI] [PubMed] [Google Scholar]
- 24.Kitamoto, T. (2002) J. Neurogenet. 16, 205-228. [DOI] [PubMed] [Google Scholar]
- 25.Haass, M., Cheng, B., Richardt, G., Lang, R. E. & Schomig, A. (1989) Naunyn-Schmiedeberg's Arch. Pharmacol. 339, 71-78. [DOI] [PubMed] [Google Scholar]
- 26.Taghert, P. H., Hewes, R. S., Park, J. H., O'Brien, M. A., Han, M. & Peck, M. E. (2001) J. Neurosci. 21, 6673-6686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davies, A. G., Bettinger, J. C., Thiele, T. R., Judy, M. E. & McIntire, S. L. (2003) Neuron 42, 731-743. [DOI] [PubMed] [Google Scholar]
- 28.Rogers, C., Reale, V., Kim, K., Chatwin, H., Li, C., Evans, P. & de Bono, M. (2003) Nat. Neurosci. 6, 1178-1185. [DOI] [PubMed] [Google Scholar]
- 29.Tabakoff, B. & Hoffman, P. L. (1998) Adv. Second Messenger Phosphoprotein Res. 32, 173-193. [DOI] [PubMed] [Google Scholar]
- 30.Moore, M. S., DeZazzo, J., Luk, A. Y., Tully, T., Singh, C. M. & Heberlein, U. (1998) Cell 93, 997-1007. [DOI] [PubMed] [Google Scholar]
- 31.Dohrman, D. P., Chen, H. M., Gordon, A. S. & Diamond, I. (2002) Alcohol Clin. Exp. Res. 26, 407-415. [PubMed] [Google Scholar]
- 32.Covarrubias, M. & Rubin, E. (1993) Proc. Natl. Acad. Sci. USA 90, 6957-6960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Covarrubias, M., Vyas, T. B., Escobar, L. & Wei, A. (1995) J. Biol. Chem. 270, 19408-19416. [DOI] [PubMed] [Google Scholar]
- 34.Antkowiak, B. (2001) Naturwissenschaften 88, 201-213. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.