Abstract
Hepcidin is a peptide that regulates iron homeostasis by inhibiting iron absorption by the small intestine and release of iron from macrophages. Its production is stimulated by iron overload and by inflammation. It has been suggested that IL-6 is the only cytokine that stimulates hepcidin transcription. However, mice with targeted disruption of the gene encoding IL-6 (IL-6–/–) respond to endotoxin by increasing the expression of hepcidin transcripts in the liver. We show that incubating murine hepatocytes with IL-6, IL-1α, and IL-1β strongly stimulates hepcidin transcription. IL-10 has little or no stimulatory effect, and IFN-β inhibits transcription of hepcidin. All of the hepcidin stimulatory activity of macrophages from IL-6–/– mice can be accounted for by IL-1 that they secrete. Hepatocytes from IL-6–/– mice, hfe–/– mice, and mice with a hypomorphic transferrin receptor 2 mutation responded to IL-6 and IL-1 by up-regulating hepcidin transcription. Nitric oxide does not seem to be involved in the stimulation of hepcidin transcription by cytokines: aminoguanidine does not inhibit the stimulation of hepcidin transcription by cytokines. IL-1 may play a significant role in the anemia of inflammation by up-regulating hepcidin.
Keywords: HFE, iron, liver, nitric oxide
Hepcidin has emerged as a central regulator of iron homeostasis. First described as a 25-amino acid antimicrobial peptide (1, 2), it was subsequently found to be a powerful negative regulator of iron absorption (3, 4). Befitting its role as an antimicrobial peptide, hepcidin is up-regulated in intact animals by the injection of endotoxin or turpentine.
Moreover, culture media conditioned by treatment of macrophages with LPS stimulate hepcidin transcription in cultures of primary hepatocytes (5). Because this stimulation was entirely blocked by anti-IL-6 antibody, it was concluded that the stimulation was due to IL-6 and that stimulation did not occur with IL-1 and TNF-α (6). Our data suggested, however, that some stimulation of hepcidin production occurred in IL-6–/– mice treated with LPS (7). These data have recently been confirmed by Rivera et al.† Accordingly, there must be substances other than IL-6 that stimulate hepcidin production. We now show that hepatocytes can be stimulated directly to produce hepcidin message by the cytokines IL-6, IL-1α, and IL-1β and that the stimulation of hepcidin production by macrophage-conditioned media can be accounted for entirely by these three cytokines.
Materials and Methods
All cytokines and cytokine-specific antibodies were obtained from R & D Systems. Aminoguanidine hemisulfate was purchased from Calbiochem. IL-6–/– mice were on a background of C57BL/6J (002650) and were obtained from The Jackson Laboratory. hfe–/– mice and transferrin receptor 2 (tfr2) mutant mice were kind gifts from Dr. William Sly (Saint Louis University, St. Louis) and Dr. Robert Fleming (Saint Louis University), respectively. The hfe–/– mice had been backcrossed into the 129 strain for 10 generations; the tfr2 mutant mice, transgenic mice with a hypomorphic mutation found in humans, were on an AKR background. Primary hepatocytes were isolated from perfused mouse livers as described in ref. 7. Peritoneal macrophages were isolated from IL-6–/– mice 4 days after i.p. injection of 1 ml of 3.85% thioglycollate Brewer's medium (Becton Dickinson). These macrophages were harvested by rinsing the peritoneal cavity twice with 5 ml of PBS. The peritoneal macrophages were placed in a 15-ml tube containing 5 ml of tissue culture medium (98% Williams medium, 2% human serum, penicillin, and streptomycin). The cells were pelleted by centrifugation at 300 × g and resuspended in Williams medium containing 2% human serum and plated at a density of 1 × 106 cells per ml. After 2 h, nonadherent cells were removed. The adherent macrophages were washed once, and fresh Williams medium containing 2% human serum was added. After 24 h in culture, conditioned media from the IL 6–/– macrophages were collected, passed through a 0.2-μm filter, and used to treat hepatocytes.
Incubation of primary mouse hepatocytes was carried out in 2.0% human serum with 0.5 μg/ml anti-human IL-6 antibody, unless otherwise indicated. Human serum was used instead of bovine serum because serum contains small amounts of IL-6, and anti-human IL-6 antibodies were available to neutralize this potentially confounding source of stimulatory activity. Twenty-four hours after isolation, the hepatocytes were treated with cytokines or conditioned media for 20–24 h.
Total RNA was extracted from hepatocytes by using the Versagene RNA Purification Kit (Gentra Systems). cDNA first-strand synthesis was performed with Moloney murine leukemia virus reverse transcriptase (Invitrogen) and oligo(dT) primer.
Real-time PCR was carried out on a Bio-Rad iCycler. Primers for analysis of murine hepcidin 1 (hepc1; GenBank accession no. AF503444) and murine ribosomal protein S18 (GenBank accession no. AK050626) as a housekeeping gene control were designed with the help of beacon designer (Bio-Rad). The hepcidin primers were designed to specifically amplify only hepcidin 1 and not hepcidin 2 (hepc2). All amplification primers and probes are listed in Table 1. The iCycler amplification system contained 20 mM Tris·HCl (pH 8.4), 50 mM KCl, 4.0 mM MgCl2, 200 μM dNTP, 0.625 units of iTaq DNA polymerase (Bio-Rad), 300 nM sense primer, 300 nM antisense primer, and 300 nM 6-carboxy fluorescein-labeled probe. Two microliters of synthesized cDNA or standards were used in a 25-μl amplification system, and all samples were analyzed in duplicate. After 40 amplification cycles, threshold cycle values were automatically calculated, and attomoles of starting cDNA were calculated from a standard curve ranging over 4 orders of magnitude. The hepcidin 1 standard curve ranged from 0.0136 to 136 attomoles per 25-μl reaction, and the ribosomal protein S18 standard curve ranged from 0.013 to 130 attomoles per reaction. The results were expressed as the ratio of hepcidin 1 to ribosomal protein S18 cDNA.
Table 1. Primers and fluorescent-labeled probes used in real-time PCR experiments.
| Gene | Sense primer 5′-3′ | Antisense primer 5′-3′ | 5′-6-FAM/3BHQ 1-3′ probe |
|---|---|---|---|
| Hepcidin 1 | TTGCGATACCAATGCAGAAGA | GATGTGGCTCTAGGCTATGTT | AGAGACACCAACTTCCCCATCTGC |
| Ribosomal protein S18 | ACTTTTGGGGCCTTCGTGTC | GCCCAGAGACTCATTTCTTCTTG | ACACCAAGACCACTGGCCGCAG |
6-FAM, 6-carboxy fluorescein; 3BHQ 1, 3-black hole quencher-1.
Results
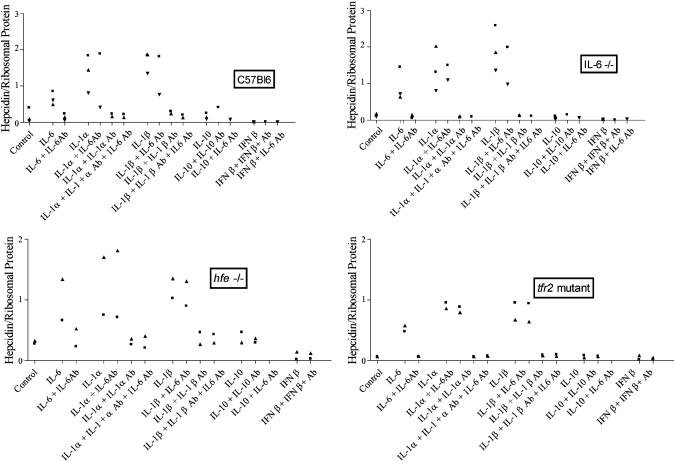
IL-6 and IL-1 Stimulate, and IFN-β Inhibits, Hepcidin Transcription by Murine Primary Hepatocytes. As shown in Fig. 1, IL-6 at a concentration of 20 ng/ml consistently stimulated expression of hepcidin in primary hepatocytes obtained from C57Bl6, IL-6–/–, hfe–/–, and tfr2 mutant mice. Maximum stimulation was attained in 24 h (data not shown). Anti-mouse IL-6 antibodies at a concentration of 2 μg/ml completely blocked this stimulation of hepcidin expression. Similarly, IL-1α (20 ng/ml) and IL-1β (20 ng/ml) each increased hepcidin mRNA expression, an increase that was completely blocked by the addition of anti-mouse IL-1α and anti-IL-1β-specific antibodies (2 μg/ml), respectively. Antibodies against mouse IL-6 did not block the induction of hepcidin expression by IL-1α and IL-1β, demonstrating that the IL-1α and IL-1β effects were not due to downstream activation of IL-6 synthesis. IL-10 (20 ng/μl) had little or no effect on hepcidin expression. IFN-β (250 units/ml), however, inhibited hepcidin expression by ≈3-fold.
Fig. 1.
The effect of IL-6, IL-1α, IL-1β, IL-10 (20 ng/ml), IFN-β (250 units/ml), and cognate antibodies against these cytokines on the expression of hepcidin by primary murine hepatocytes. The target hepatocytes were derived from CB57Bl6, IL-6–/–, hfe–/–, and tfr2-mutant mice, as indicated. Each symbol represents one experiment in which two to four samples of primary murine hepatocytes were incubated with culture medium (control) or with the cytokine and antibody combination indicated.
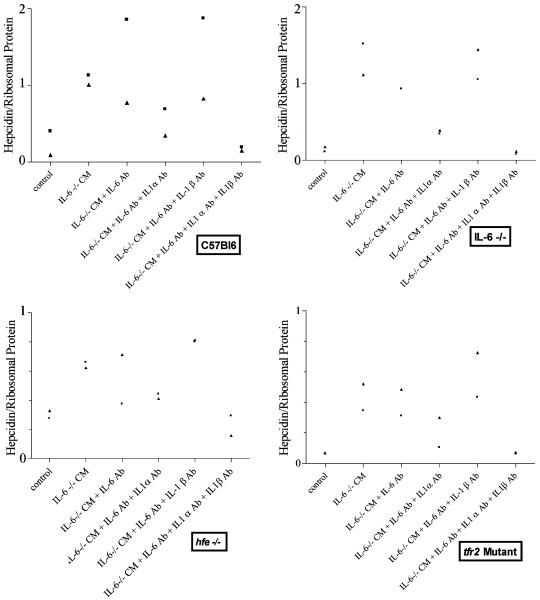
IL-1α and IL-1β Account for Stimulation of Hepcidin Expression by Primary Hepatocytes by IL-6–/– Macrophage-Conditioned Media. Hepatocytes from C57Bl6 wild-type mice were incubated for 20 h with 50% conditioned media from IL-6–/– peritoneal macrophages. As shown in Fig. 2, the IL-6–/– conditioned medium in the presence of anti-mouse IL-6 antibody retained the capacity to stimulate hepcidin expression, indicating that there were substances other than IL-6 that stimulated hepcidin transcription. Further addition of antibodies against mouse IL-1α partially inhibited the hepcidin-stimulatory effect of macrophage-conditioned media. In contrast, antibodies against IL-1β had little or no effect on the capacity of macrophage-conditioned media to induce hepcidin expression. The addition of all three anti-cytokine antibodies, those against IL-6, IL-1α, and IL-1β, abolished the stimulatory effect of macrophage-conditioned media on hepcidin expression.
Fig. 2.
The effect of adding 50% conditioned medium (CM) from IL-6–/– macrophages with and without antibodies against mouse IL-6, IL-1α, and/or IL-1β to primary murine hepatocytes derived from the strains of mice indicated in the boxes. Each symbol represents one experiment in which two to four samples of primary murine hepatocytes were incubated with culture medium (control) or with the antibody combination indicated for 24 h.
hfe–/– and tfr2 mutant hepatocytes were also induced to express more hepcidin mRNA in the presence of conditioned media from IL-6–/– peritoneal macrophages. The stimulation by IL-6–/– macrophage-conditioned media was only partially inhibited by IL-1α antibodies but was completely inhibited by the addition of all three antibodies.
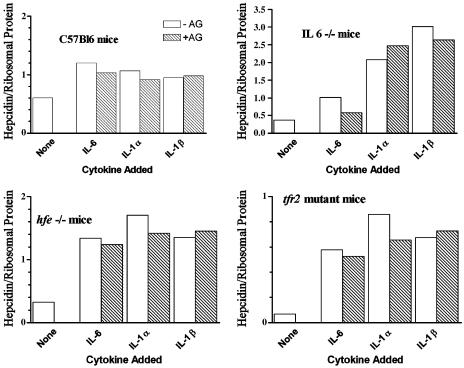
Inhibition of Nitric Oxide Synthase (NOS) Does Not Abrogate the Cytokine-Induced Stimulation of Hepcidin Transcription. To determine whether the induction of hepcidin expression by cytokines was mediated through nitric oxide, we examined the effect of aminoguanidine, a nonspecific inhibitor of NOSs. At a concentration of 1 mM, this NOS inhibitor had no effect on the stimulation of hepcidin expression by IL-6, IL-1α, and IL-1β in C57Bl6, IL-6–/–, hfe–/–, or tfr2-mutant mouse hepatocytes (Fig. 3).
Fig. 3.
The effect of aminoguanidine (AG), a nonspecific inhibitor of NOS, on hepcidin transcription in C57 BL, IL-6–/–, hfe–/–, and tfr2-mutant mice. Each bar represents the average results obtained from two to four samples of primary murine hepatocytes derived from the indicated strains of mice. Hatched bars represent hepcidin transcription in the presence of the inhibitor.
Discussion
Derived from an antimicrobial peptide, hepcidin appears to assume twin roles in the regulation of iron homeostasis (9). On one hand, it regulates the total body iron content. Animals that are loaded with iron produce increased amounts of hepcidin, which serves to inhibit absorption of iron from the bowel and its release into the circulation from macrophages. The fact that defects in hfe, tfr2, and hemojuvelin all result in suboptimal hepcidin levels has led to the suggestion that the various heritable forms of hemochromatosis (iron storage disease) are due, at least in part, to the inability of iron to stimulate the production of hepcidin (10–12). How iron overload stimulates hepcidin production remains unknown; treatment of isolated hepatocytes with iron (3, 6) or with iron-laden macrophages (unpublished data) decreases hepcidin production.
On the other hand, hepcidin plays a role in the innate immune system. For example, its production in experimental animals is stimulated by inflammation caused by the injection of endotoxin (3). It is likely that in this instance, hepcidin serves to deprive invading microorganisms of iron, again by inhibiting its absorption from the bowel and its release from macrophages. More is known about the regulation of hepcidin by inflammation than about its regulation by iron overload, but many of the data are contradictory. It has been suggested that the only cytokine that stimulates hepcidin production by human hepatocytes (6) and in HepB3 cells (5) is IL-6 and that IL-1 has no stimulatory effect, but we have shown that IL-6-deficient mice retain the ability to respond to endotoxin injections (7). It was suggested in one study that hfe–/– mice are unable to respond to endotoxin (13), but in other studies, a normal response to endotoxin was documented in such animals (7, 14).
The present study clarifies the spectrum of cytokines that have the capacity to stimulate hepcidin transcription. We show that hepcidin transcription was stimulated in murine hepatocytes not only by IL-6 but also by IL-1α and IL-1β. Indeed, hepcidin stimulation by both forms of IL-1 was consistently greater than stimulation by IL-6. When antibodies to these three cytokines were added to macrophage-conditioned media made in IL-6–/– mice, all stimulation of hepcidin transcription was abolished, indicating that these cytokines are responsible for virtually all of the stimulating effect of macrophage-conditioned media. IFN-β inhibited hepcidin transcription, confirming results previously reported by Nemeth et al. (6). hfe and tfr2, each of which seems to be required for the stimulation of hepcidin production by iron overload, are not required for cytokine-stimulated signaling. The response to cytokines in the hfe–/– and tfr2 mutant hepatocytes was unmistakable, but the degree of stimulation in these mutant mice seemed to be modestly less than in the C57Bl6 control mice. Although one cannot rule out with certainty that the response may be slightly blunted in hepatocytes bearing these mutations, as has been suggested (13), the difference observed was small and could easily have been due to strain differences or experimental variation.
How cytokines simulate hepcidin transcription is unknown. The cytokine that is most potent in up-regulating hepcidin, IL-1, has the capacity to increase synthesis of nitric oxide (15, 16). Nitric oxide can modulate iron homeostasis by increasing binding of iron regulatory protein (IRP) 1 to its iron-responsive element (IRE) targets and by accelerating the degradation of IRP2, preventing its binding to its IRE targets. It therefore seemed possible that nitric oxide might serve as a mediator in increasing hepcidin transcription, common to both the iron-overload and inflammatory pathways. However, we found that the addition of aminoguanidine, a nonspecific NOS inhibitor, had no effect on the stimulation of hepcidin transcription in hepatocytes.
The previous reports that found IL-1 does not stimulate hepcidin transcription were based on studies carried out in human primary hepatocytes or in a human hepatocyte cell line, but we regard it unlikely that a major species difference exists. We have observed that HepG2 cells show a response to IL-1; it is a smaller response than that observed in primary murine hepatocytes, but immortalized hepatocytes cell lines, in general, respond less robustly to cytokine stimulation than do primary cultures (unpublished data). Northern blots from human hepatocytes published previously do show stimulation of hepcidin production by IL-1α at 24 h (6). It was suggested that this IL-1 effect was an indirect one, IL-1 having stimulated hepatocytes to produce IL-6 (6). Our studies show clearly that indirect stimulation of IL-6 by IL-1 is not the case. A robust IL-1 stimulating effect was demonstrated by using IL-6–/– cells; such cells could not have produced IL-6. Moreover, the addition of anti-IL-6 antibody did not ablate the IL-1 effect. IL-1 induces hypoferremia (17), up-regulates ferritin (18), and has been thought to play a primary role in the anemia of chronic inflammation (8). It may well be that it is stimulation of hepcidin by IL-1 that plays a more important role in the anemia of inflammation than does IL-6.
Acknowledgments
This work was supported by National Institutes of Health Grant DK53505 and the Stein Endowment Fund. This is manuscript no. 17090-MEM of The Scripps Research Institute.
Author contributions: P.L. and E.B. designed research; P.L., H.P., T.G., and L.W. performed research; P.L., H.P., T.G., L.W., and E.B. analyzed data; and P.L. and E.B. wrote the paper.
Abbreviations: NOS, NO synthase; tfr2, transferrin receptor 2.
Footnotes
Rivera, S., Gabayan, V. & Ganz, T. (2004) Blood 104, 875a (abstr.).
References
- 1.Krause, A., Neitz, S., Magert, H. J., Schulz, A., Forssmann, W. G., Schulz-Knappe, P. & Adermann, K. (2000) FEBS Lett. 480, 147–150. [DOI] [PubMed] [Google Scholar]
- 2.Park, C. H., Valore, E. V., Waring, A. J. & Ganz, T. (2001) J. Biol. Chem. 276, 7806–7810. [DOI] [PubMed] [Google Scholar]
- 3.Pigeon, C., Ilyin, G., Courselaud, B., Leroyer, P., Turlin, B., Brissot, P. & Loreal, O. (2001) J. Biol. Chem. 276, 7811–7819. [DOI] [PubMed] [Google Scholar]
- 4.Nicolas, G., Bennoun, M., Devaux, I., Beaumont, C., Grandchamp, B., Kahn, A. & Vaulont, S. (2001) Proc. Natl. Acad. Sci. USA 98, 8780–8785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nemeth, E., Rivera, S., Gabayan, V., Keller, C., Taudorf, S., Pedersen, B. K. & Ganz, T. (2004) J. Clin. Invest. 113, 1271–1276. [DOI] [PubMed] [Google Scholar]
- 6.Nemeth, E., Valore, E. V., Territo, M., Schiller, G., Lichtenstein, A. & Ganz, T. (2003) Blood 101, 2461–2463. [DOI] [PubMed] [Google Scholar]
- 7.Lee, P. L., Peng, H., Gelbart, T. & Beutler, E. (2004) Proc. Natl. Acad. Sci. USA 101, 9263–9265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maury, C. P., Andersson, L. C., Teppo, A. M., Partanen, S. & Juvonen, E. (1988) Ann. Rheum. Dis. 47, 972–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ganz, T. (2004) Curr. Opin. Hematol. 11, 251–254. [DOI] [PubMed] [Google Scholar]
- 10.Nemeth, E., Roetto, A., Garozzo, G., Ganz, T. & Camaschella, C. (October 14, 2004) Blood, 10.1182/blood-2004-08-3042. [DOI] [PubMed]
- 11.Ganz, T. (2003) Blood 102, 783–788. [DOI] [PubMed] [Google Scholar]
- 12.Papanikolaou, G., Samuels, M. E., Ludwig, E. H., MacDonald, M. L., Franchini, P. L., Dube, M. P., Andres, L., MacFarlane, J., Sakellaropoulos, N., Politou, M., et al. (2004) Nat. Genet. 36, 77–82. [DOI] [PubMed] [Google Scholar]
- 13.Roy, C. N., Custodio, A. O., De Graaf, J., Schneider, S., Akpan, I., Montross, L. K., Sanchez, M., Gaudino, A., Hentze, M. W., Andrews, N. C., et al. (2004) Nat. Genet. 36, 481–485. [DOI] [PubMed] [Google Scholar]
- 14.Frazer, D. M., Wilkins, S. J., Millard, K. N., McKie, A. T., Vulpe, C. D. & Anderson, G. J. (2004) Br. J. Haematol. 126, 434–436. [DOI] [PubMed] [Google Scholar]
- 15.Geller, D. A., de Vera, M. E., Russell, D. A., Shapiro, R. A., Nussler, A. K., Simmons, R. L. & Billiar, T. R. (1995) J. Immunol. 155, 4890–4898. [PubMed] [Google Scholar]
- 16.Guan, Z., Buckman, S. Y., Baier, L. D. & Morrison, A. R. (1998) Am. J. Physiol. 274, F673–F679. [DOI] [PubMed] [Google Scholar]
- 17.Alvarez-Hernandez, X., Liceaga, J., McKay, I. C. & Brock, J. H. (1989) Lab. Invest. 61, 319–322. [PubMed] [Google Scholar]
- 18.Torti, F. M. & Torti, S. V. (2002) Blood 99, 3505–3516. [DOI] [PubMed] [Google Scholar]