Abstract
Targeted mutagenesis is an essential tool of reverse genetics that could be used experimentally to investigate basic plant biology or modify crop plants for improvement of important agricultural traits. Although targeted mutagenesis is routine in several model organisms including yeast and mouse, efficient and widely usable methods to generate targeted modifications in plant genes are not currently available. In this study we investigated the efficacy of a targeted-mutagenesis approach based on zinc-finger nucleases (ZFNs). In this procedure, ZFNs are used to generate double-strand breaks at specific genomic sites, and subsequent repair produces mutations at the break site. To determine whether ZFNs can cleave and induce mutations at specific sites within higher plant genomes, we introduced a construct carrying both a ZFN gene, driven by a heat-shock promoter, and its target into the Arabidopsis genome. Induction of ZFN expression by heat shock during seedling development resulted in mutations at the ZFN recognition sequence at frequencies as high as 0.2 mutations per target. Of 106 ZFN-induced mutations characterized, 83 (78%) were simple deletions of 1–52 bp (median of 4 bp), 14 (13%) were simple insertions of 1–4 bp, and 9 (8%) were deletions accompanied by insertions. In 10% of induced individuals, mutants were present in the subsequent generation, thus demonstrating efficient transmission of the ZFN-induced mutations. These data indicate that ZFNs can form the basis of a highly efficient method for targeted mutagenesis of plant genes.
Keywords: gene targeting, nonhomologous end joining
A major focus of plant biotechnology is genetic modification and improvement of crop plants. With this aim, large-scale genome and/or EST sequencing projects are underway for many important plant species including rice, maize, wheat, soybean, and tomato (www.ncbi.nlm.nih.gov/genomes/PLANTS/PlantList.html). The enormous amount of genome-sequence information becoming available has intensified the need for methods that can use this sequence information to generate targeted modifications in plant genes. Targeted-mutagenesis methods could be used experimentally to investigate plant gene function or for genetic modification of important crop plants. Such methods are especially important for species, including most crops, that lack readily available mutant collections (1, 2). Furthermore, targeted mutagenesis could facilitate development of genetically modified crops lacking transgenic DNA, including genes conferring resistance to antibiotics. To date, efficient and widely usable methods for targeted modification of higher plant genomes are not available.
The most widely used targeted-mutagenesis strategy is gene targeting (GT) by homologous recombination (3–6). Efficient GT procedures have been available for >20 years in yeast (7) and mouse (8). In these systems, DNA fragments are introduced into cultured cells, and repair by homologous recombination causes the introduced DNA to be incorporated into the homologous locus. Typically, GT events occur in a small proportion of treated cells (10-6 to 10-5 GT events per cell in yeast and 10-7 to 10-5 GT events per cell in mouse ES cells), and positive selection is required to identify targeting events. In yeast, most integrations occur at the homologous locus (7). By contrast, in mouse ES cells, most integrations occur at random nonhomologous sites (10-5 to 10-3 GT events per integration), and negative selection is required to enrich for targeting events (9).
Over the last 17 years, numerous attempts to achieve GT in higher plants have been reported. In most of these studies, DNA was introduced into cultured cells by direct gene transfer (10, 11) or Agrobacterium-mediated transformation (12–15). In general, GT efficiencies were low, on both a per-cell basis (<10-7 GT events per cell) and per-integration basis (10-6 to 10-4 GT events per integration), and negative selection to select against random integrations was not successful (16–19). Two exceptional studies reported relatively high GT ratios (≈10-3 GT events per integration), but these successes have not been repeated (20, 21). More recently, enhanced GT efficiencies were reported by using two alternative strategies. In the first study, DNA was introduced into Arabidopsis plants by using the vacuum-infiltration method of Agrobacterium-mediated transformation (22), resulting in targeting ratios of 7.2 × 10-4 GT events per integration (23). In the second strategy, the homologous sequence was flanked on both sides with negative-selectable marker genes and introduced into cultured rice cells by using Agrobacterium-mediated transformation; although the targeting ratio was low (6.5 × 10-4 GT events per integration), the stringent negative-selection scheme eliminated most random integrations, and ≈1% of survivors were the result of targeting events (24). Although promising, these strategies have not yet been tested at multiple loci, and thus, their general utility still needs to be assessed.
The low GT frequencies reported in higher plants may result from competition between homologous recombination and nonhomologous end joining (NHEJ) for repair of double-strand breaks (DSBs) (5, 6). The main pathway of DSB repair in higher plants seems to be NHEJ (5, 6, 25–28). As a consequence, the ends of a donor molecule are likely to be joined by NHEJ rather than participating in homologous recombination, thus reducing GT frequency. A large body of data indicates that DSB repair by NHEJ is extremely error-prone in plants. In plants, DSB repair by simple ligation of the two ends, with no sequence alteration, is rare. More often, DSBs are repaired by end-joining processes that generate insertions and/or deletions (26, 27). Taken together, these observations suggest that NHEJ-based strategies might be more effective than homologous recombination-based strategies for targeted mutagenesis in higher plants.
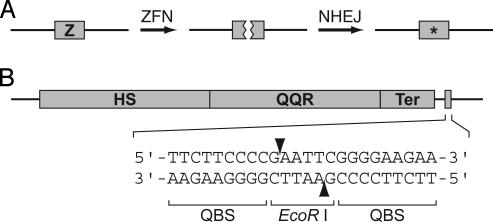
An NHEJ-based targeted-mutagenesis strategy was developed recently in Drosophila (29). This strategy, depicted in Fig. 1A, utilizes synthetic zinc-finger nucleases (ZFNs) to generate DSBs at specific genomic sites. Subsequent repair of the DSBs by NHEJ frequently produces deletions and/or insertions at the joining site. ZFNs are comprised of a nonspecific DNA-cleavage domain from the endonuclease Fok I and a DNA-binding domain composed of three Cys2His2 zinc fingers (30). The zinc-finger domains are engineered to bind to specific DNA sequences, and the nuclease domain generates DSBs at that site (29). The Fok I domain must dimerize to cut DNA (31), and the ZFN pairs function most efficiently in vivo when their binding sites are separated by precisely 6 bp (32).
Fig. 1.
Targeted mutagenesis using ZFNs. (A) Strategy for induction of mutations using ZFNs. ZFNs bind to their recognition sequence (Z) and generate DSBs. Repair of the DSBs by NHEJ frequently produces mutations (*) at the break site. (B) The HS::QQR-QEQ construct, which consists of the HS::QQR gene and its recognition sequence, QEQ. The HS::QQR gene consists of the QQR coding sequence (QQR) fused to a heat-shock promoter (HS) and a 3′ terminator sequence (Ter). (Upper) The narrow box downstream of HS::QQR is the QEQ sequence. (Lower) The QEQ sequence, which contains binding sites for the QQR ZFN (labeled QBS) and an intervening EcoRI site. Arrowheads indicate the sites of QQR cleavage, which are within the EcoRI site.
ZFNs have two demonstrated utilities. First, as depicted in Fig. 1 A, they can be used to generate mutations at specific genomic sites. In the Drosophila yellow gene, this procedure generated transmissible mutations at a frequency of ≈4 × 10-3 mutations per gamete (29). Recent experiments achieved frequencies of >10-1 mutations per gamete at another Drosophila locus (K. Beumer and D.C., unpublished data). Second, ZFNs can be used to enhance the frequency of GT by homologous recombination: in both Drosophila and cultured human cells, ZFN-induced DSBs stimulated GT frequency by 50- to 2,000-fold (refs. 33 and 34 and G. Bhattacharyya, K. Beumer, M. Bibikova, J. K. Trautman, and D.C., unpublished data). This latter application of ZFNs may facilitate development of GT methods in organisms, such as plants, in which GT occurs at low frequency.
In this report we test whether ZFNs can cleave and stimulate mutations at specific genomic sites in plants. We demonstrate targeted mutagenesis at frequencies as high as 0.2 mutations per target in Arabidopsis plants and transmission of the induced mutations to subsequent generations at high frequency. These data indicate that ZFNs can form the basis of a highly efficient method for targeted mutagenesis of plant genes.
Materials and Methods
Plant Growth. Seeds were germinated on plates containing Murashige and Skoog salts, 0.5% sucrose, and 25 μg/ml kanamycin. Ten-day-old seedlings were transferred to Scott's Redi-Earth and grown under 24-h illumination. Plants were watered three times per week.
Construction of HS::QQR-QEQ. HS::QQR-QEQ was generated in five steps. First, the nopaline synthase (nos) 3′ terminator was PCR-amplified by using pCAMBIA1304 (www.cambia.org) as a template and using the primers NOS5NH (5′-CCGCTAGCATCGT TCA A ACAT T TGGC-3′) and NOS3NH (5′-CCGCTAGCGATCTAGTAACATAGATG-3′). The PCR product was blunt-end-cloned into the EcoRV site of pBluescript II KS(-), generating clone pNos. Second, the promoter from the Arabidopsis HSP18.2 heat-shock gene (35) was PCR-amplified by using ecotype Columbia DNA as a template and the primers HSP914SpeI (5′-ACTAGACTCCACTAGTAAGCTTGCTGCAGCTTTGAC-3′) and HSP182NdeBam (5′-GTCAGTAGCGGGATCCAGCTGCCATATGTCGT TGCT T T TCGGGAGAC-3′). The PCR product was cloned into pCRII-TOPO. This clone then was digested with SpeI and BamHI, and the SpeI/BamHI fragment containing the HSP18.2 promoter was ligated in front of the nos 3′ terminator in pNos, generating clone pNos+HS. Third, an NdeI/BamHI fragment containing the QQR-L0 ZFN (32) was ligated between the HSP18.2 promoter and nos 3′ terminator in pNos+HS, generating clone pHS::QQR. Fourth, pHS::QQR was digested with KpnI and SacI, and the KpnI/SacI fragment containing the HS::QQR cassette was ligated into the KpnI and SacI sites of pCAMBIA2300 (www.cambia.org), generating clone HS::QQR/2300. Finally, the oligo QQR4 (5′-CTTCTTCCCCGAATTCGGGGAAGAAGGTAC-3′) was self-annealed to generate a double-stranded QQR-binding site with KpnI overhangs and ligated into the KpnI site of HS::QQR/2300, generating clone HS::QQR-QEQ/2300.
Plant Transformation. The HS::QQR-QEQ/2300 plasmid was transformed into Agrobacterium tumefaciens strain LBA4404 by electroporation and introduced into Arabidopsis ecotype Landsberg erecta by floral dipping (22). T1 transformants were selected on medium containing 25 μg/ml kanamycin and 15 μg/ml cefotaxime. T1 transformants were transplanted to soil and allowed to self-pollinate. Insert number was determined by scoring the ratio of resistant to sensitive T2 seedlings.
Heat Induction of QQR. T2 seeds from single-insert HS::QQR-QEQ lines (lines Q1–Q7) were germinated on selective medium. Seedlings were grown on plates at 20°C for 10 days. The plates then were wrapped in plastic wrap and immersed in water at 40°C for 2 h. Seedlings were grown for an additional 24 h at 20°C before DNA extraction.
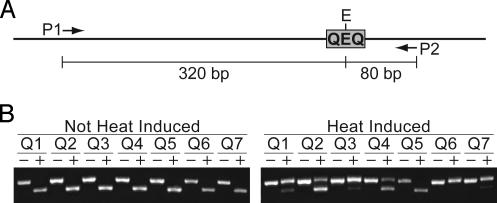
Detection of Mutations in QEQ. After heat induction, DNA was extracted from whole seedlings (36) and resuspended in 20 μl of TE buffer (10 mM Tris/1 mM EDTA, pH 8.0). DNA was also extracted from non-heat-shocked seedlings. The T2 seedlings analyzed were kanamycin-resistant and, thus, should segregate 1:2 for plants homozygous/hemizygous for the HS::QQR-QEQ insertion. To average out potential variation among individuals and between hemizygous and homozygous seedlings, 2-μl aliquots from each of 10 seedlings from a given line were pooled. Five microliters of pooled DNA was digested with EcoRI in a 10-μl volume. Two microliters of the digested DNA (representing 5% of the DNA yield from a single seedling) was used as template in a 20-μl PCR by using primers NOS5NH (5′-CCGCTAGCATCGTTCAAACATTTGGC-3′) and M13(-20) (5′-GTAAAACGACGGCCAGT-3′) that flank QEQ. PCR products could arise from DNA molecules containing QQR-induced mutations or from DNA molecules that were digested incompletely. To subtract those that arose from incomplete digestion, we carried out a second digest. Five microliters of the PCR product was digested with EcoRI in a volume of 10 μl, and 5 μl of each digest was analyzed on a 2% agarose gel. Longer exposures of the experimental (heat-induced) gel shown in Fig. 2B revealed faint 320-bp bands (Lower) in lines Q1, Q3, and Q7. The lower proportion of the 320-bp band in experimental seedlings most likely resulted from competition during the PCR: as the mutation frequency increases, the proportion of PCR products arising from DNA molecules containing mutations is increasingly favored; thus, at high mutation frequencies (e.g., lines Q1, Q3, Q6, and Q7), essentially all PCR products arise from DNA molecules containing mutations. To isolate molecules containing mutations in QEQ, the PCR product was ligated to pCRII-TOPO (Invitrogen) and transformed into Escherichia coli by electroporation. Plasmid DNA then was extracted from random clones and digested with EcoRI. The DNA sequence was determined for clones lacking EcoRI sites.
Fig. 2.
The QQR ZFN generates mutations at its recognition sequence in Arabidopsis cells. (A) Depiction of the QEQ sequence relative to the primers (P1 and P2) used to amplify the region. Primers P1 and P2 refer to primers NOS5NH and M13(-20), respectively (see Materials and Methods). PCR amplification with primers P1 and P2 produces a 400-bp product. Digestion of this PCR product with EcoRI produces fragments of 320 and 80 bp. (B) Gel assay to detect QQR-induced mutations. PCR with primers P1 and P2 was carried out, and the 400-bp PCR products were subjected to gel electrophoresis either with (+ lanes) or without (- lanes) prior EcoRI digestion. DNA fragments lacking EcoRI sites (400-bp, upper band) were detected in experimental seedlings (heat induced) but not in control seedlings (not heat induced).
Determination of Mutation Frequency. T2 seed was germinated on selective medium, heat-shocked as described above, and allowed to recover for 24 h. For each line, DNA was extracted (36) from 10 T2 seedlings and resuspended in 20 μl of TE buffer. To average out potential variation among individuals and between hemizygous and homozygous seedlings, 2-μl aliquots from each of the 10 seedling DNAs from a given line were pooled. Next, 2 μl of the pooled DNA was used as template in a 20-μl PCR by using primers NOS5NH and M13(-20) as described before. The PCR product then was ligated to pCRII-TOPO (Invitrogen) and transformed into E. coli by electroporation. Random clones were spotted to a replica grid, analyzed by colony PCR by using primers NOS5NH and M13(-20), and analyzed for the presence of the EcoRI site as described before. All EcoRI-minus clones were verified by growing 2-ml overnight cultures from the replica grid, extracting plasmid DNA, and sequencing.
Transmission of QQR-Induced Mutations. T2 seed from lines Q1 and Q3 was germinated on selective medium and heat-shocked as described above. Heat-shocked T2 seedlings (103 T2 seedlings for line Q1 and 113 T2 seedlings for line Q3) were transplanted to soil, grown, and allowed to self-pollinate. For a mutation to transmit to the next generation, it must be induced in the L2 cells of the shoot apical meristem (37). Mutations induced in the shoot apical meristem of the seedling should form large sectors in the primary inflorescence. To prescreen T2 plants for the presence of these mutant sectors, we dissected the terminal flower cluster from the primary inflorescence of each plant, extracted DNA, and analyzed the DNA for the presence of mutations in QEQ as described above. Mutations were detected in 20 T2 plants in line Q1 and in 17 T2 plants in line Q3. Because these mutant sectors could have arisen from mutations induced in any of the shoot apical meristem layers (L1, L2, or L3), only some of these 37 plants were expected to give rise to mutant plants in the subsequent generation. From each of the 37 sublines containing mutant sectors, T3 seed was collected from the primary inflorescence. T3 seed was germinated on selective medium, 30 5-day seedlings from each subline were pooled, DNA was extracted from the pooled seedlings, and the DNA was analyzed for the presence of mutations in QEQ as described above. Mutations were detected in 14 of 20 sublines in line Q1 and 13 of 17 sublines in line Q3. These seedling pools were occasionally contaminated with seed-coat material, which could result in false positives. To identify the true positives, DNA was extracted from leaves of 10–15 individual T3 plants for each of the 27 sublines, and the DNA was analyzed for the presence of mutations in QEQ as described above. Mutations were detected in individual T3 seedlings in 10 of 14 sublines in line Q1 and 11 of 13 sublines in line Q3. Of those 21 lines that gave rise to transmitted mutations, an average of 32% of the T3 progeny in line Q1 and 58% of the T3 progeny in line Q3 harbored QQR-induced mutations.
Results
Experimental System. Our experimental system, summarized in legend for Fig. 1B, consisted of two components: HS::QQR and QEQ. HS::QQR is a chimeric gene construct consisting of a heat-shock promoter fused to a coding sequence encoding the QQR ZFN (31). QQR has a three-finger DNA-binding domain that recognizes the sequence 5′-GGGGAAGAA-3′ (31, 32). QEQ is a synthetic 24-bp oligonucleotide that contains the binding site for the QQR homodimer and an intervening EcoRI site (Fig. 1B). The EcoRI site lies within the QQR cleavage site and is lost if mutations are generated at the QQR break site (Fig. 1B). Molecules lacking EcoRI sites and containing QQR-induced mutations were detected by using a variety of strategies (discussed below).
We introduced the HS::QQR-QEQ construct into Arabidopsis plants, selected transgenic T1 seedlings, and identified seven single-locus lines. We refer to these lines as Q1–Q7. To address the questions outlined below, we subjected seedlings from lines Q1–Q7 to an inductive heat pulse and extracted DNA from these seedlings. We also extracted DNA from control seedlings that were not given an inductive heat pulse.
QQR Stimulates Mutations at Its Recognition Sequence in Arabidopsis Cells. To determine whether induction of QQR activity could induce mutations at its recognition sequence, we followed a procedure that enriched for DNA molecules lacking EcoRI sites. We digested the seedling DNAs with EcoRI, carried out PCR with the digested DNAs by using primers flanking the QEQ sequence (Fig. 2 A), digested the PCR products with EcoRI, and subjected the digested PCR products to gel electrophoresis. By using this procedure, DNA molecules containing QQR-induced mutations result in PCR products lacking EcoRI sites and, thus, undigested fragments after EcoRI digestion. As shown in Fig. 2B, undigested fragments were present in experimental (heat-induced) seedlings in six of the seven lines but were absent in control (not heat-induced) seedlings. The absence of undigested fragments in line Q5 suggested that this line did not contain QQR-induced mutations; additional experiments were not carried out with this line.
To verify that the heat-induced seedlings contained QQR-induced mutations, we cloned the PCR products, identified clones lacking EcoRI sites within the QEQ sequence, and determined the DNA sequence of 30 randomly picked EcoRI-minus clones. All 30 clones contained mutations within the QEQ sequence; sequences of these clones are discussed below. Taken together, these data indicate that the QQR ZFN can stimulate mutations at its recognition sequences in Arabidopsis cells.
Frequency of QQR-Induced Mutations. To determine the frequency of QQR-induced mutations in Arabidopsis cells, we determined the frequency of DNA molecules lacking EcoRI sites in the absence of enrichment. We carried out PCR with the (undigested) seedling DNAs by using primers flanking the QEQ sequence (Fig. 2 A), cloned the PCR products, and carried out EcoRI digestions of randomly picked clones. The DNA sequence of all EcoRI-minus clones was determined to verify that these clones contained mutations within the QEQ sequence. By using this procedure, the proportion of EcoRI-minus clones within the clone population should represent mutation frequency on a per-target basis. As summarized in Table 1, in heat-induced seedlings, 1.7–19.6% (n = 51–179) of clones contained QQR-induced mutations. Among the heat-induced seedlings, the average mutation frequency was 7.9%. By contrast, control seedlings, in which QQR expression was not induced by a heat pulse, gave rise to no (n = 93–139) EcoRI-minus clones. Taken together, these data suggest that QQR activity driven by a heat-shock promoter stimulated mutations at a frequency of ≈0.08 mutations per target in Arabidopsis seedling cells.
Table 1. Frequency of QQR-induced mutations.
| Treatment | Line | Frequency, % | Identified/tested |
|---|---|---|---|
| Heat-induced | Q1 | 5.6 | 5/90 |
| Q2 | 3.8 | 3/80 | |
| Q3 | 19.6 | 18/92 | |
| Q4 | 1.7 | 3/179 | |
| Q6 | 7.7 | 4/52 | |
| Q7 | 19.6 | 10/51 | |
| Not induced | Q1 | 0 | 0/93 |
| Q3 | 0 | 0/95 | |
| Q7 | 0 | 0/139 |
Spectrum of QQR-Induced Mutations. To characterize the types of mutations induced by QQR expression, we determined the DNA sequence of 106 independent EcoRI-minus clones. In this analysis, identical sequences from the same seedling were not included to ensure that all mutations scored were derived independently. The results of this analysis are summarized in Table 2. Of 106 EcoRI-minus clones sequenced, 83 (78%) were simple deletions of 1–52 bp (median of 4 bp), 14 (13%) were simple insertions of 1–4 bp, and 9 (8%) were deletions accompanied by insertions.
Table 2. QQR-induced mutations.
| Lesion | Sequence | No. |
|---|---|---|
| Wild type | 5′-TTCTTCCCCGAATTCGGGGAAGAA-3′ | |
| -1 bp | TTCTTCCCC.AATTCGGGGAAGAA | 4 |
| TTCTTCCCCG.ATTCGGGGAAGAA | 4 | |
| TTCTTCCCCGAAT.CGGGGAAGAA | 6 | |
| TTCTTCCCCGAATT.GGGGAAGAA | 2 | |
| -2 bp | TTCTTCCCCG..TTCGGGGAAGAA | 1 |
| TTCTTCCCCGA..TCGGGGAAGAA | 4 | |
| TTCTTCCCCGAA..CGGGGAAGAA | 1 | |
| -3 bp | TTCTTCCCC...TTCGGGGAAGAA | 2 |
| TTCTTCCCCG...TCGGGGAAGAA | 1 | |
| TTCTTCCCCGA...CGGGGAAGAA | 6 | |
| TTCTTCCCCGAA...GGGGAAGAA | 1 | |
| -4 bp | TTCTTCCCC....TCGGGGAAGAA | 3 |
| TTCTTCCCCG....CGGGGAAGAA | 7 | |
| TTCTTCCCCGA....GGGGAAGAA | 1 | |
| -5 bp | TTCTTCCCC.....CGGGGAAGAA | 6 |
| TTCTTCCCCG.....GGGGAAGAA | 5 | |
| -6 bp | TTCTTCCCCG......GGGAAGAA | 5 |
| -7 bp | TTCTTCCC.......GGGGAAGAA | 1 |
| -8 bp | TTCTTCC........GGGGAAGAA | 1 |
| TTCTTCCC........GGGAAGAA | 1 | |
| -13 bp | T.............CGGGGAAGAA | 1 |
| -22 to -52 bp | 20 | |
| +1 bp | TTCTTCCCCGAAATTCGGGGAAGAA | 5 |
| TTCTTCCCCGAATTTCGGGGAAGAA | 6 | |
| +4 bp | TTCTTCCCCGAATTAATTCGGGGAA | 3 |
| -2 bp, +1 bp | TTCTTCCC.TAATTCGGGGAAGAA | 1 |
| -3 bp, +1 bp | TTCTTCCCC.T.TTCGGGGAAGAA | 1 |
| -5 bp, +1 bp | TTCTTCCCC..C..CGGGGAAGAA | 1 |
| -5 bp, +2 bp | TTCTTCCCCGA..TC.GGGAAGAA | 1 |
| -7 bp, +47 bp | TTCTTCCC.+47bp.GGGGAAGAA | 1 |
| -11 bp, +2 bp | T....CC.....TTCGGGGAAGAA | 1 |
| -12 bp, +97 bp | ...+97bp..AATTCGGGGAAGAA | 1 |
| -124 bp, +6 bp | ........GGAATT.......... | 1 |
| -287 bp, +235 bp | ..+235bp..AATTCGGGGAAGAA | 1 |
Transmission of QQR-Induced Mutations. We determined transmission frequency in two lines: Q1 and Q3. We subjected seedlings (103 from line Q1 and 113 from line Q3) to an inductive heat pulse and grew the induced plants. To transmit to a subsequent generation, a mutation must be induced in the L2 cells of the shoot apical meristem and form a sector in the primary inflorescence that incorporates the germ cells (37). To identify induced plants containing mutant sectors, we extracted DNA from the terminal flower cluster of each plant and analyzed these DNAs for the presence of mutations within QEQ. Finally, we collected progeny seed from the positive plants and scored progeny seedlings for the presence of QQR-induced mutations. Progeny containing QQR-induced mutations were identified in 10 of 103 induced plants in line Q1 and 11 of 113 induced plants in line Q3. Thus, 10% of the induced plants gave rise to transmitted mutations in these two lines.
Discussion
The QQR ZFN Generates Mutations at High Frequency in Arabidopsis Cells. We have shown that the QQR ZFN can generate mutations at specific sites within the Arabidopsis genome at frequencies as high as 0.2 mutations per target (Table 1). These data suggest that targeted mutagenesis of native loci could occur at frequencies as high as 0.2 mutations per gene or 0.4 mutations per cell (0.2 mutations per gene, two genes per cell). This mutation frequency clearly could enable efficient targeted mutagenesis in plants, in contrast to homologous recombination-based procedures, which have much lower mutation frequencies: typically <10-7 GT events per cell or 10-6 to 10-4 GT events per integration (10–15). The dramatically higher mutation frequencies we observed with a ZFN-based procedure compared to those obtained by using homologous recombination-based procedures supports the view that the main pathway for DSB repair in plants is NHEJ (5, 6, 25–27).
From a targeted-mutagenesis point of view, a significant observation is that the vast majority of the QQR-induced mutations we characterized would produce functional gene knockouts if generated within coding sequence: of the 106 mutations, 82 (77%) would produce frame shifts, 15 (14%) would delete 1–3 amino acids, 7 (7%) would delete ≥8 amino acids, and 2 (2%) would change amino acids. Taken together, these data suggest that ZFNs targeted to coding sequences should generate functional gene knockouts at high frequency in plants.
The mutation frequencies we observed were somewhat variable: in six lines, mutation frequency varied ≈10-fold (0.017–0.196 mutations per target; Table 1), and in one line (line Q5), mutations were not detected. With our experimental system, both the ZFN gene and its target were introduced into the Arabidopsis genome. Thus, the variable mutation frequency most likely was caused by position effects resulting in variable expression of the HS::QQR transgene and/or variable accessibility of the QEQ target. The lack of mutations in line Q5 could possibly have resulted from rearrangement of the T-DNA insertion, which commonly occurs (38–41).
We do not know what limits the mutation frequency in these experiments. For example, it is feasible that use of a more active promoter to drive ZFN expression or a longer duration of heat shock would increase the efficiency of cleavage. Any products restored to the original sequence, by simple ligation or homologous recombination, would be susceptible to recutting by the enzyme. The mutant products we recovered should all be resistant to additional cleavage, although many of them retain the zinc-finger recognition sites, because the spacing between them has changed (32). However, the danger in overexpressing the ZFN is that noncanonical sites may be cut, producing a burden of DSBs that the cells cannot repair completely (29, 33).
The mutation frequencies we detected are likely to represent a minimum, because our experimental setup did not allow detection of all mutations. Deletions removing the primer-binding sites (Fig. 2 A), including deletions as small as 62 bp on one side, would eliminate PCR amplification and, thus, detection of the mutation. In other studies of NHEJ-repair products in plants, deletions of 0.2–2.0 kb were observed frequently (42–44). For example, Gorbunova and Levy (42) reported that ≈50% (n = 31) of NHEJ-generated deletions were >100 bp. These observations suggest that the actual mutation frequencies are significantly higher than those that we detected.
DSB Repair in Plants. The high mutation frequency we observed supports the view that DSB repair in plants is highly error-prone (26, 27). The mutations we recovered suggest mechanisms by which the mutations were generated. For example, the deletions most likely arose from exonuclease activity that enlarged breaks to gaps before end joining. In the cases of deletions <10 bp, joining may have proceeded by blunt-end ligation, because sequence matches at the joints (microhomologies of even a single base pair) were rare. The simple insertions (Table 2) were very small and seem to have arisen from partial filling in of the 5′-protruding single strands generated by QQR. Some (9 of 92) of the deletion products were associated with filler DNA of 1–235 bp in length. Two of the insertions (+97 and +235 bp) consisted of sequences from elsewhere in the genome (data not shown), suggesting involvement of a copy–release–join mechanism (45).
The mechanism of formation of the larger deletions may be different from that of the small deletions. In essentially all of these cases, short (1- to 6-bp), sometimes imperfect homologies are found at the junctions (data not shown). With the two long insertions for which the source of the extra DNA was identified, similar homologies are found at both new junctions (data not shown). These microhomologies imply a mechanism in which the alignment between the ends during repair is set by local base pairing. The joining reaction may proceed by ligation or by DNA synthesis using the short paired segment as a primer-template complex (46).
The profile of DSB-repair products we recovered in this study is similar to that observed in other plant studies (26, 27, 42, 43) with one exception. Kirik et al. (44) reported that Arabidopsis differs fundamentally from tobacco in DSB repair because only simple deletions were found in the former, whereas insertions accompanied deletions frequently in the latter. We found insertions, ranging from 1 to 235 bp, in 22% of the repair products that we characterized in Arabidopsis. The earlier study was constrained by a requirement for a relatively large deletion (>200 bp) to pass the initial selection. In contrast, our analysis is biased against deletions large enough to remove the binding sites for our PCR primers. Nonetheless, we found insertions associated with both small and substantial deletions.
Targeted Mutagenesis Using ZFNs in Plants. The crucial steps in a ZFN-based targeted-mutagenesis experiment are identifying a ZFN-binding site in a gene of interest and designing ZFNs that recognize that target site. The nuclease domain of the ZFN must dimerize to cut DNA (31), the DNA-binding domain of the ZFN is composed of three zinc fingers, and each zinc finger recognizes a triplet of nucleotides; thus, ZFN-binding sites are comprised of 18 bp. Because the ZFN pairs function most efficiently when their binding sites are separated by precisely 6 bp (32), a ZFN recognition sequence using only 5′-GNN-3′ triplets would consist of 5′-NNCNNCNNC(N6)GNNGNNGNN-3′, where N can be any nucleotide. Once a target site is chosen, the corresponding ZFNs are designed by combining the zinc-finger domains that bind to the specific set of triplets present in the target site. Zinc-finger domains have been identified that bind to most of the 5′-GNN-3′ and 5′-ANN-3′ triplets, and the selection of domains recognizing 5′-CNN-3′ and 5′-TNN-3′ triplets is in progress (47, 48). With the published 5′-(G/A)NN-3′ domains, it should be possible to identify an 18-bp target every 0.5–1.0 kb (47) and, thus, to target most plant genes.
It also should be possible to use a ZFN-based targeted-mutagenesis approach in essentially all plant species. Each pair of three-finger ZFNs has a recognition sequence of 18 bp, which should occur every 6.9 × 1010 (418) bp. Thus, three-finger ZFNs should provide ample specificity for plants with genome sizes of ≤1010 (e.g., rice, maize, and tomato). Where necessary, increased specificity can be achieved by using four-finger ZFNs that recognize sequences of 24 bp. A number of different synthetic transcription factors with zinc-finger DNA-binding domains have been shown to function in plant cells (49–52), supporting the idea that zinc fingers can provide access to many genomic targets.
The ZFN-based targeted-mutagenesis strategy described here is efficient enough that positive- and negative-selection schemes are not necessary to identify mutant individuals. In the lines we tested, 10% of induced individuals gave rise to mutants in the subsequent generation. Based on this result, direct analysis of just 100–200 individuals should be sufficient to identify several mutants. Because DSB repair has been found to be mutagenic in other plants (26, 27), a ZFN-based strategy is likely to be an efficient method for targeted mutagenesis in most plant species.
Acknowledgments
We thank Ramin Yadegari, Leslie Sieburth, and Anne Britt for critical review of the manuscript. This work was supported by National Institutes of Health Grant GM65173 (to D.C.) and National Science Foundation Grant IBN-0110035 (to G.N.D.).
Author contributions: A.L., C.L.P., D.C., and G.N.D. designed research; A.L. and C.L.P. performed research; A.L., C.L.P., D.C., and G.N.D. analyzed data; D.C. contributed new reagents/analytic tools; and A.L., D.C., and G.N.D. wrote the paper.
Abbreviations: GT, gene targeting; NHEJ, nonhomologous end joining; DSB, double-strand break; ZFN, zinc-finger nuclease; nos, nopaline synthase.
References
- 1.Alonso, J. M., Stepanova, A. N., Leisse, T. J., Kim, C. J., Chen, H., Shinn, P., Stevenson, D. K., Zimmerman, J., Barajas, P., Cheuk, R., et al. (2003) Science 301, 653-657. [DOI] [PubMed] [Google Scholar]
- 2.Till, B. J., Reynolds, S. H., Greene, E. A., Codomo, C. A., Enns, L. C., Johnson, J. E., Burtner, C., Odden, A. R., Young, K., Taylor, N. E., et al. (2003) Genome Res. 13, 524-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reiss, B. (2003) Int. Rev. Cytol. 228, 85-139. [DOI] [PubMed] [Google Scholar]
- 4.Puchta, H. (2002) Plant Mol. Biol. 48, 173-182. [PubMed] [Google Scholar]
- 5.Ray, A. & Langer, M. (2002) Trends Plant Sci. 7, 435-440. [DOI] [PubMed] [Google Scholar]
- 6.Britt, A. B. & May, G. D. (2003) Trends Plant Sci. 8, 90-95. [DOI] [PubMed] [Google Scholar]
- 7.Rothstein, R. (1991) Methods Enzymol. 194, 281-301. [DOI] [PubMed] [Google Scholar]
- 8.Capecchi, M. R. (1989) Science 244, 1288-1292. [DOI] [PubMed] [Google Scholar]
- 9.Vasquez, K. M., Marburger, K., Intody, Z. & Wilson, J. H. (2001) Proc. Natl. Acad. Sci. USA 98, 8403-8410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paszkowski, J., Baur, M., Bogucki, A. & Potrykus, I. (1988) EMBO J. 7, 4021-4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Halfter, U., Morris, P. C. & Willmitzer, L. (1992) Mol. Gen. Genet. 231, 186-193. [DOI] [PubMed] [Google Scholar]
- 12.Offringa, R., de Groot, M. J., Haagsman, H. J., Does, M. P., van den Elzen, P. J. & Hooykaas, P. J. (1990) EMBO J. 9, 3077-3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee, K. Y., Lund, P., Lowe, K. & Dunsmuir, P. (1990) Plant Cell 2, 415-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Offringa, R., Franke-van Dijk, M. E., De Groot, M. J., van den Elzen, P. J. & Hooykaas, P. J. (1993) Proc. Natl. Acad. Sci. USA 90, 7346-7350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Risseeuw, E., Offringa, R., Franke-van Dijk, M. E. & Hooykaas, P. J. (1995) Plant J. 7, 109-119. [DOI] [PubMed] [Google Scholar]
- 16.Risseeuw, E., Franke-van Dijk, M. E. & Hooykaas, P. J. (1997) Plant J. 11, 717-728. [DOI] [PubMed] [Google Scholar]
- 17.Gallego, M. E., Sirand-Pugnet, P. & White, C. I. (1999) Plant Mol. Biol. 39, 83-93. [DOI] [PubMed] [Google Scholar]
- 18.Xiaohui Wang, H., Viret, J. F., Eldridge, A., Perera, R., Signer, E. R. & Chiurazzi, M. (2001) Gene 272, 249-255. [DOI] [PubMed] [Google Scholar]
- 19.Thykjaer, T., Finnemann, J., Schauser, L., Christensen, L., Poulsen, C. & Stougaard, J. (1997) Plant Mol. Biol. 35, 523-530. [DOI] [PubMed] [Google Scholar]
- 20.Miao, Z. H. & Lam, E. (1995) Plant J. 7, 359-365. [DOI] [PubMed] [Google Scholar]
- 21.Kempin, S. A., Liljegren, S. J., Block, L. M., Rounsley, S. D., Yanofsky, M. F. & Lam, E. (1997) Nature 389, 802-803. [DOI] [PubMed] [Google Scholar]
- 22.Bechthold, N. & Ellis, J. P. G. (1993) C. R. Acad. Sci. 316, 1194-1199. [Google Scholar]
- 23.Hanin, M., Volrath, S., Bogucki, A., Briker, M., Ward, E. & Paszkowski, J. (2001) Plant J. 28, 671-677. [DOI] [PubMed] [Google Scholar]
- 24.Terada, R., Urawa, H., Inagaki, Y., Tsugane, K. & Iida, S. (2002) Nat. Biotechnol. 20, 1030-1034. [DOI] [PubMed] [Google Scholar]
- 25.Vergunst, A. C. & Hooykaas, P. J. J. (1999) Crit. Rev. Plant Sci. 18, 1-31. [Google Scholar]
- 26.Gorbunova, V. V. & Levy, A. A. (1999) Trends Plant Sci. 4, 263-269. [DOI] [PubMed] [Google Scholar]
- 27.Britt, A. B. (1999) Trends Plant Sci. 4, 20-25. [DOI] [PubMed] [Google Scholar]
- 28.Siebert, R. & Puchta, H. (2002) Plant Cell 14, 1121-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bibikova, M., Golic, M., Golic, K. G. & Carroll, D. (2002) Genetics 161, 1169-1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim, Y. G., Cha, J. & Chandrasegaran, S. (1996) Proc. Natl. Acad. Sci. USA 93, 1156-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith, J., Bibikova, M., Whitby, F. G., Reddy, A. R., Chandrasegaran, S. & Carroll, D. (2000) Nucleic Acids Res. 28, 3361-3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bibikova, M., Carroll, D., Segal, D. J., Trautman, J. K., Smith, J., Kim, Y. G. & Chandrasegaran, S. (2001) Mol. Cell. Biol. 21, 289-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Porteus, M. H. & Baltimore, D. (2003) Science 300, 763. [DOI] [PubMed] [Google Scholar]
- 34.Bibikova, M., Beumer, K., Trautman, J. K. & Carroll, D. (2003) Science 300, 764. [DOI] [PubMed] [Google Scholar]
- 35.Takahashi, T. & Komeda, Y. (1989) Mol. Gen. Genet. 219, 365-372. [DOI] [PubMed] [Google Scholar]
- 36.Murray, M. G. & Thompson, W. F. (1980) Nucleic Acids Res. 8, 4321-4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Irish, V. F. & Jenik, P. D. (2001) Curr. Opin. Genet. Dev. 11, 424-430. [DOI] [PubMed] [Google Scholar]
- 38.Laufs, P., Autran, D. & Traas, J. (1999) Plant J. 18, 131-139. [DOI] [PubMed] [Google Scholar]
- 39.Nacry, P., Camilleri, C., Courtial, B., Caboche, M. & Bouchez, D. (1998) Genetics 149, 641-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Castle, L. A., Errampalli, D., Atherton, T. L., Franzmann, L. H., Yoon, E. S. & Meinke, D. W. (1993) Mol. Gen. Genet. 241, 504-514. [DOI] [PubMed] [Google Scholar]
- 41.Tax, F. E. & Vernon, D. M. (2001) Plant Physiol. 126, 1527-1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gorbunova, V. & Levy, A. A. (1997) Nucleic Acids Res. 25, 4650-4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Salomon, S. & Puchta, H. (1998) EMBO J. 17, 6086-6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kirik, A., Salomon, S. & Puchta, H. (2000) EMBO J. 19, 5562-5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Merrihew, R. V., Marburger, K., Pennington, S. L., Roth, D. B. & Wilson, J. H. (1996) Mol. Cell. Biol. 16, 10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lehman, C. W., Trautman, J. K. & Carroll, D. (1994) Nucleic Acids Res. 22, 434-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Segal, D. J. (2002) Methods 26, 76-83. [DOI] [PubMed] [Google Scholar]
- 48.Liu, Q., Xia, Z., Zhong, X. & Case, C. C. (2002) J. Biol. Chem. 277, 3850-3856. [DOI] [PubMed] [Google Scholar]
- 49.Guan, X., Stege, J., Kim, M., Dahmani, Z., Fan, N., Heifetz, P., Barbas, C. F., III, & Briggs, S. P. (2002) Proc. Natl. Acad. Sci. USA 99, 13296-13301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Segal, D. J., Stege, J. T. & Barbas, C. F., III (2003) Curr. Opin. Plant Biol. 6, 163-168. [DOI] [PubMed] [Google Scholar]
- 51.Stege, J. T., Guan, X., Ho, T., Beachy, R. N. & Barbas, C. F., III (2002) Plant J. 32, 1077-1086. [DOI] [PubMed] [Google Scholar]
- 52.Ordiz, M. I., Barbas, C. F., III, & Beachy, R. N. (2002) Proc. Natl. Acad. Sci. USA 99, 13290-13295. [DOI] [PMC free article] [PubMed] [Google Scholar]